Abstract
Background/purpose
Adolescents undergoing fixed orthodontic therapy have an increased risk of oral diseases due to additional plaque accumulation sites. However, the effect of fixed orthodontics appliances (FOAs) on the colonization of Candida albicans (Ca) and Streptococcus mutans (Sm), two synergistic oral pathogens, is largely unknown and was, therefore, the primary objective of this pilot investigation.
Material and methods
Sixteen children aged 10–15 years were enrolled, nine in the FOA and seven in the control groups. Saliva and occlusal plaque were collected, and the Ca and Sm levels were quantified with a quantitative real-time polymerase chain reaction (qPCR) assay.
Results
A trend of higher Ca levels was observed in the saliva and occlusal plaque of the FOA group, while the control group contained higher levels of Sm. Furthermore, for Sm levels, a positive correlation between saliva and occlusal plaque was shown in both the FOA and control groups; in contrast, Ca levels were negatively correlated between these samples only in the FOA group. Between Ca and Sm, a positive correlation was observed in saliva and occlusal plaque in the control group; however, this relationship was disrupted in the FOA group.
Conclusion
Our preliminary study demonstrated that the presence of FOAs disturbs the colonization of Ca and Sm within the oral cavity. This perturbation might increase orthodontic patients’ risk for Ca- and Sm-related diseases.
Keywords: Candida albicans, Streptococcus mutans, Orthodontic, qPCR
Introduction
A significant portion of the pediatric population at the adolescent stage suffers malocclusion and jaw problems as part of their growth and development. Malocclusions are one of the most common oral health problems.1 These conditions are often managed with orthodontic treatment, which includes fixed and removable orthodontic appliances.2 One of the most common fixed orthodontic treatments is the application of fixed orthodontic appliances (FOAs).3
The introduction of FOAs in the oral cavity greatly affects oral hygiene by increasing food retention, which leads to plaque accumulation.4 This complex oral biofilm is composed of a network of bacteria, fungi, and other microorganisms. Studies have demonstrated that FOAs can elevate the colonization of fungi such as Candida spp. and gram-positive bacteria, including Streptococcus spp. and Lactobacillus spp. in the oral cavity. This heightened level of pathogens might be responsible for complications associated with orthodontic treatment, such as caries and gingivitis.5, 6, 7, 8
Among these microorganisms, Candida albicans is a common fungal opportunistic pathogen present in the oral cavity of 62% of preschool children and 71% of school children.9 Previous studies demonstrated that C. albicans has the ability to colonize the dentin and enamel and functions as a reservoir for the spread of the microorganism.10,11 Of the bacterial species, Streptococcus mutans has a well-established role in the etiology of caries.12
The co-occurrence of C. albicans and S. mutans has been described in children with dental caries.13,14 Recently, more emphasis has been placed on the cross-kingdom interactions between C. albicans and S. mutans in the oral cavity. The cross-talk between the two species has been suggested to mutually enhance their colonization.15,16 In the presence of an orthodontic appliance, this could set a stage for children to be more prone to caries development. Previously, some studies have demonstrated the presence of S. mutans and C. albicans on removable orthodontic appliances,17 but little is known about their colonization in the presence of the FOAs. This pilot study aimed to investigate the impact of FOAs on C. albicans and S. mutans levels and their correlation in saliva and occlusal plaque in children.
Material and methods
Ethics
This study was conducted in accordance with the Declaration of Helsinki, reviewed and approved by the Institutional Review Board (#16–000282). Written informed consent was obtained from parents or legal guardians of all the subjects that participated in the study.
Subjects recruitment
Sixteen participants were recruited from the pediatric patient population of the University of California, Los Angeles (UCLA) Children's Dental Centre. The enrolled subjects included 9 subjects with fixed orthodontic appliances (FOAs) and 7 without FOAs (control). Participants were eligible based on the following inclusion criteria: healthy children (ASA physical status, class I) aged 10–15 years old, who were not taking any medication, and no antibiotic use within the last 6 months. The FOAs had an additional requirement of metal fixed orthodontic appliances (FOAs), while the control group must have no FOAs or removable appliances. Participants were excluded from the study if they had generalized rampant caries, periodontitis, halitosis, open sores or ulcerations, chronic systemic diseases, reduced saliva production, or any other medical conditions that could influence the oral microbiome. All participants were asked to avoid eating or drinking for at least 1 h before sample collection and received routine oral hygiene care such as tooth brushing instructions, professional teeth cleaning, and topical fluoride application.
Questionnaire and oral examination
Each participant completed a questionnaire, which provided demographic information, oral hygiene habits, and dental treatment history. Oral clinical evaluation and radiographic exams were performed by trained clinical examiners at the UCLA School of Dentistry. The participant's dental caries status was recorded using decayed, missing, and filled tooth (DMFT) criteria proposed by the World Health Organization.18
Oral sample collection
All participants were asked to withhold their teeth cleaning and avoid eating and drinking at least 1 h before sample collection. Two oral samples were collected from each individual: saliva and occlusal plaque. Participants were instructed to rinse with water for the saliva collection to remove all saliva from the mouth. Then, 2–3 mL of unstimulated saliva was collected by drooling/spitting directly into a 50 mL Falcon conical sterile tube (Fisher Scientific, Pittsburg PA, USA) kept on ice during the collection. The posterior occlusal plaque was collected using one Pikster™ (Erskine oral care, Marian del Rey, CA, USA) per each quadrant, and all 4 quadrants were pooled (four-quadrant sample) into a sterile 1.5 mL micro-centrifuge tube (Eppendorf, AG, Hamburg, Germany) containing glycerol to a final concentration of 25%. All the collected samples, either saliva or plaque, were frozen immediately and stored at −80 °C.
Extraction of genomic DNA
Total genomic DNA was extracted from our participant's oral samples (saliva and occlusal plaque) using the Epicenter MasterPure™ DNA purification kit (Lucigen Corporation, Middleton, WI, USA) following the manufacturer's instructions with modifications.19 The extracted DNA was stored at −20 °C until further use.
Real-time quantitative polymerase chain reaction (qPCR) and sensitivity test
The fungi and C. albicans levels in the saliva and occlusal plaque samples were assessed by real-time quantitative polymerase chain reaction (qPCR), using the fungi-specific (ITS1, ITS2)20,21 and the C. albicans-specific (SAP)22,23 primers (Table 1). In parallel, we also performed qPCR to assess the total bacteria and S. mutans levels, using the universal 16S RNA gene primers for bacteria24 and S. mutans specific glucosyltransferases (gtfB)25 gene primers (Table 1).
Table 1.
Oligonucleotide primer sequences used in this study.
| Target | Primer name | Sequence (5′-3′) | AT (ºC) | Amplicon size (bp) |
|---|---|---|---|---|
| Fungi20,21 | ITS1-F | CTTGGTCATTTAGAGGAAGTAA | 60 | Vary in different species |
| ITS2-R | GCTGCGTTCTTCATCGATGC | |||
| Candida albicans22,23 | SAP-F | CTGCTGATATTACTGTTGGTTC | 58 | 263 |
| SAP-R | CCACCAATACCAACG GTATC | |||
| Bacteria24 | Eub338 | ACTCCTACGGGAGGCAGCAG | 64 | 200 |
| Eub518 | ATTACCGCGGCTGCTGG | |||
| Streptococcus mutans25 | gtf B–F | CTACACTTTCGGGTGGCTTG | 64 | 250 |
| gtf B-R | GAAGCTTTTCACCATTAGAAGCTG |
AT- Annealing temperature; AL- Amplicon length.
ITS - Internal Transcribed Spacer Region; SAP - Secreted Aspartyl Proteinase.
gtf B- codes glucosyltransferase B.
The qPCR reactions were performed using Bio-Rad iCycler Thermal Cycler with iQ5 Multicolour Real-Time PCR Detection System (BioRad iQ 5 RTPCR QPCR, BioRad, Hercules, CA, USA). Amplification was carried out as followed: the cycling condition was 95 °C for 10 min, followed by 40 cycles of denaturation for 30 s at 95 °C, then primers annealing at 60 °C for fungi or 58 °C for C. albicans-specific primers for 30 s, and extension at 72 °C for 30 s. For bacterial universal and S. mutans specific primers, we use similar amplification conditions as described above, except for the annealing temperature, which was 64 °C for both primers. All amplifications and detections were carried out in a BioRad iQ5 qPCR 96-well reaction plate with optical caps (BioRad) and performed three times. In addition, the iQ5 Optical System Software generated quantification cycle values (Cq) which were analyzed along with melting point data. Additional information on the analysis is in Appendix 1.
Statistical analyses
Statistical analysis with one-way ANOVA followed by Kruskal–Wallis multiple comparisons test was performed using GraphPad (GraphPad Prism version 8.0.0, GraphPad Software, San Diego, California USA), and difference significance were defined as follows: ∗P ≤ 0.01, ∗∗P ≤ 0.001 and ∗∗∗P ≤ 0.0001.
Results
Study subjects
The 16 pediatric participants enrolled in this pilot study included 9 subjects with FOAs and 7 control individuals. The participants’ age, gender, ethnicity, DMFT index, oral hygiene parameters, and duration of orthodontic treatment are summarized in Table 2. In brief, the gender distribution was 12 (75%) females and 4 (25%) males, the age ranged from 10 to 15 years with an average of 12.4 (±1.5) and ethnicity included 81.3% Hispanic and 18.7% non-Hispanic, and the caries experience was 0.2 (±0.4) DMFT score. Overall, there were no significant differences between the FOA and control groups for all of the parameters mentioned above. In addition, subjects with FOAs had an average of 15.4 (±5.8) months in treatment duration, ranging from 10 to 27 months.
Table 2.
Demographic and oral hygiene information obtained from the study questionnaire.
| Group Characteristic | Control |
FOAs |
Total |
|||
|---|---|---|---|---|---|---|
| (n=7) | % | (n=9) | % | (n=16) | % | |
|
Gender | ||||||
| Female | 4 | 57.1 | 8 | 88.9 | 12 | 75.0 |
| Male |
3 |
42.9 |
1 |
11.1 |
4 |
25.0 |
|
Ethnicity | ||||||
| Hispanic | 5 | 71.4 | 8 | 88.9 | 13 | 81.3 |
| Non-Hispanic |
2 |
28.6 |
1 |
11.1 |
3 |
18.7 |
|
Age (years) | ||||||
| 11.9 (± 1.3) |
12.9 (± 1.6) |
12.4 (±1.5) |
||||
|
DMFT index | ||||||
| 0.3 (± 0.5) |
0.1 (± 0.3) |
0.2 (± 0.4) |
||||
|
Orthodontic appliances period (months) | ||||||
| 15.4 (±5.8) |
||||||
|
Professional dental cleaning frequency | ||||||
| None | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Once a year | 1 | 14.3 | 1 | 11.1 | 2 | 12.5 |
| More than once a year |
6 |
85.7 |
8 |
88.9 |
14 |
87.5 |
|
Tooth brushing frequency | ||||||
| Not brushing | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Once a day | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| More than once per day |
7 |
100.0 |
9 |
100.0 |
16 |
100.0 |
|
Flossing frequency | ||||||
| Not flossing | 1 | 14.3 | 2 | 22.2 | 3 | 18.8 |
| Less than once per day | 2 | 28.6 | 4 | 44.4 | 6 | 37.5 |
| Once per day | 3 | 42.9 | 3 | 33.3 | 6 | 37.5 |
| More than once per day |
1 |
14.3 |
0 |
0.0 |
1 |
6.3 |
|
Mouthrinse use frequency | ||||||
| Not using mouthrinse | 2 | 28.6 | 6 | 66.7 | 8 | 50.0 |
| Less than once per day | 2 | 28.6 | 0 | 0.0 | 2 | 12.5 |
| Once per day | 0 | 0.0 | 3 | 33.3 | 3 | 18.8 |
| More than once per day | 3 | 42.9 | 0 | 0.0 | 3 | 18.8 |
Control – without Fixed Orthodontic Appliances; FOA – with Fixed Orthodontic Appliances.
Levels of C. albicans and fungi in saliva and occlusal plaque
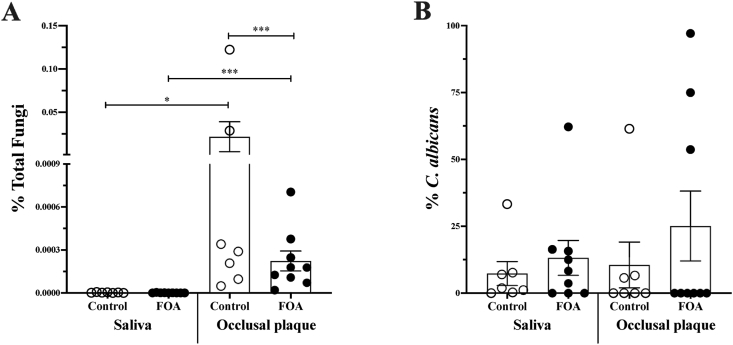
The introduction of FOAs in the oral cavity may impact the maintenance of oral hygiene, plaque accumulation, and microbial colonization. We quantified the levels of C. albicans and total fungi in saliva and occlusal plaque samples collected from subjects with or without FOAs via qPCR. Overall, the fungi levels in all samples were low. However, the control group had a significantly higher fungi load in the occlusal plaque samples (Fig. 1A). Higher levels of C. albicans in the saliva and occlusal plaque of the FOAs were apparent but not statistically significant (Fig. 1B).
Figure 1.
Candida albicans and fungi levels quantification in saliva and occlusal plaque. Fungi-specific (ITS1, ITS2) and the C. albicans-specific (SAP) primers were used to evaluate the levels of fungi and C. albicans using qPCR. Percentage of total fungi (A), and C. albicans (B) within the total fungi, in the saliva and occlusal plaque collected from subjects with (FOA, ) and without fixed orthodontic appliances (Control,
) and without fixed orthodontic appliances (Control, ). The groups significant differences were analyzed with one-way ANOVA followed by Kruskal–Wallis multiple comparisons test; ∗P <0.01, ∗∗P < 0.001, ∗∗∗P < 0.0001.
). The groups significant differences were analyzed with one-way ANOVA followed by Kruskal–Wallis multiple comparisons test; ∗P <0.01, ∗∗P < 0.001, ∗∗∗P < 0.0001.
Levels of S. mutans and bacteria in saliva and occlusal plaque
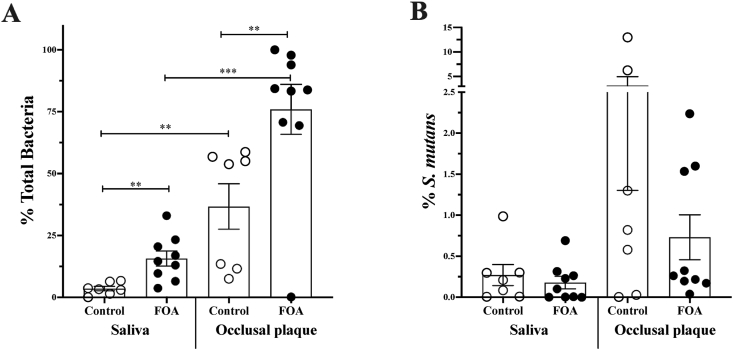
In parallel to fungi and C. albicans levels, we also quantified the S. mutans levels in the saliva and occlusal plaque. A significantly higher level of total bacteria was detected in the FOA group of both saliva and occlusal plaque samples (Fig. 2A). Moreover, independent of the study group, the occlusal plaque site displayed higher levels of total bacteria when compared to the saliva samples. While there were no statistically significant differences in either sample type, we observed a trend of higher S. mutans levels in the occlusal plaque in the control group (Fig. 2B).
Figure 2.
Streptococcus mutans and total bacteria levels quantification in saliva and occlusal plaque. Universal bacterial specific (Eub1, Eub2) and the S. mutans specific (gtfB, glucosyltransferase B) primers were used to evaluate the level of total bacteria and S. mutans in using qPCR. Percentage of total bacteria (A) and S. mutans(B), within the total bacteria, in the saliva and occlusal plaque collected from subjects with (FOA, ) and without fixed orthodontic appliances (Control,
) and without fixed orthodontic appliances (Control, ). The groups differences significance was analyzed with one-way ANOVA followed by Kruskal–Wallis multiple comparisons test; ∗P < 0.01, ∗∗P < 0.001, ∗∗∗P < 0.0001.
). The groups differences significance was analyzed with one-way ANOVA followed by Kruskal–Wallis multiple comparisons test; ∗P < 0.01, ∗∗P < 0.001, ∗∗∗P < 0.0001.
Correlation between levels of C. albicans and S. mutans
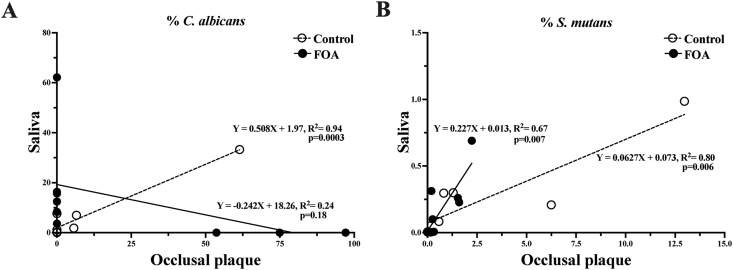
We investigated the possible correlations between the different microorganism's levels (Fungi, bacteria, C. albicans, and S. mutans) in saliva and occlusal plaque using linear regression analysis. In occlusal plaque and saliva, C. albicans levels were positively correlated in the control group (R2=0.94; p = 0.0003), while in the FOA group, the correlation was negative (R2=0.24; p = 018) (Fig. 3A). However, the linear correlation between the occlusal plaque and the saliva S. mutans levels was positive in both the control (R2 = 0.94; p = 0.0003) and the FOA groups (R2 = 0.67; p = 0.007) (Fig. 3B).
Figure 3.
Correlation between saliva and occlusal plaque C. albicans and S. mutans levels in FOA and control groups. The graphics represent the linear regression analysis: correlation between saliva and the occlusal plaque C. albicans(A) and S. mutans(B) levels on the subjects with (FOA, ) and without fixed orthodontic appliances (Control,
) and without fixed orthodontic appliances (Control, ).
).
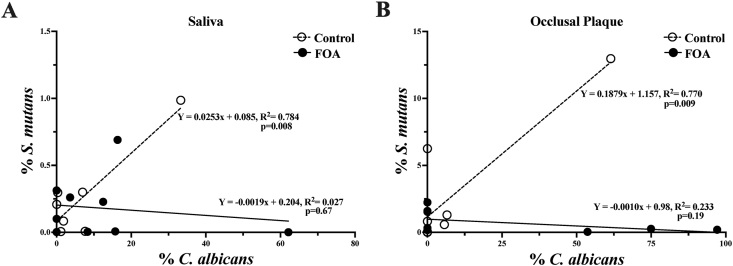
In addition, when comparing C. albicans and S. mutans levels, positive correlations were observed in saliva (R2 = 0.78; p = 0.008) and occlusal plaque (R2 = 0.77; p = 0.009) in the control group (Fig. 4A), but not in FOA subjects (Fig. 4B).
Figure 4.
Correlation between S. mutans and C. albicans levels in the FOA and control groups. The graphics represent the linear regression analysis between S. mutans and C. albicans levels in saliva (A) and occlusal plaque (B) of the subjects with (FOA, ) and without fixed orthodontic appliances (Control,
) and without fixed orthodontic appliances (Control, ).
).
Discussion
Malocclusion is a prevalent problem in the pediatric population, which most commonly has been managed with FOAs. The introduction of FOAs affects oral hygiene maintenance, stimulating microbial accumulation and promoting plaque formation. Thus, disturbance of the normal oral microflora environment by long-term use of fixed orthodontic appliances may increase the vulnerability of the patients to oral disease. For example, imbalances in bacterium–fungus interactions are known to influence the transition from a healthy to a diseased state.26 Particularly in dental caries, high levels of S. mutans and C. albicans have been detected and are known to function synergistically in the plaque biofilms of children.14,27,28 However, little is known about their levels and interactions in individuals with FOAs. To explore this important question, this pilot clinical study investigated the effect of FOAs on C. albicans and S. mutans levels and their correlation in saliva and occlusal plaque in children.
The oral cavity consists of complex microbial ecosystems, including teeth, tongue, and saliva. Saliva has immense potential as a diagnostic fluid for the identification and monitoring of diseases. The relative ease of collection of saliva further strengthens large-scale diagnostic purposes.29 Importantly, saliva reflects the composition of the oral microbiome, mirroring the local supragingival and subgingival microbiota, and the overall oral health.30
In the oral cavity, S. mutans is known to be most prevalent in the occlusal surfaces of the teeth, while it is unclear how this affects the colonization of its partner, C. albicans.31 Previously, it has been shown that fungi comprise a small portion of the salivary oral microbiome.32 They are often found in the oral cavity of healthy individuals, with C. albicans being the most predominant species (approximately 60%–70%).33 Several Candida spp. have been isolated from caries in both children and adults with a prevalence ranging from 66% to 97% in pediatric populations and 31%–56% in adult populations.10
The use of quantitative real-time PCR (qPCR) with species-specific primers provides an accurate and sensitive method for detecting and quantifying individual species. This method has been shown to have a high sensitivity for the quantification of S. mutans and C. albicans.13,34,35 Therefore, our study applied the qPCR detection method for C. albicans and S. mutans quantification in saliva and occlusal plaque samples. Consistent with previous studies,32,36 the overall levels of fungi detected in all samples were generally low (Fig. 1A). Both the control and FOA groups displayed high levels of total fungi in the occlusal plaque (Fig. 1A). However, the C. albicans levels were higher in occlusal plaque only in the FOA group compared to saliva (Fig. 1B). For total bacteria, significantly higher abundance was observed in the occlusal plaque of both groups (Fig. 2A). This pattern could be explained by tooth surface architecture. Pits and fissures within occlusal surfaces provide a special topography, making plaque removal very difficult and harboring higher microbial loads.37
Comparing between groups, the FOA subjects had significant lower total fungi levels in occlusal plaque than the control (Fig. 1A). Within the fungi, we detected higher levels of C. albicans, although not statistically significant, in saliva and occlusal plaque in the FOA group compare to the control (Fig. 1B). The increase in C. albicans level in the presence of the fixed orthodontic appliance is in accordance with previous studies both in vivo and in vitro systems.38,39 However, a previous investigation revealed a lack of significant changes in salivary C. albicans in the presence of the FOA even though the study used a different method of detection.40
In the presence of the FOA, lower S. mutans levels were observed in both sample types. This was more pronounced in the occlusal plaque than saliva; however, it was not statistically significant (Fig. 2B). Taken together, we observed the trend that the FOA group had higher C. albicans and low S. mutans levels in saliva and occlusal plaque. However, the levels of S. mutans and C. albicans may not always correspond to their synergistic relationship. A previous study demonstrated a positive correlation between C. albicans and S. mutans in saliva but a negative relationship in dental plaque.13 However, the correlation between C. albicans and S. mutans levels in the presence of the FOA is not well understood.
In our study, the C. albicans levels in saliva and occlusal plaque were positively correlated in the control group, while a negative correlation was observed in the FOA group (Fig. 3A). The presence of the FOAs may contribute to the negative correlation as it acts as a retentive surface and may serve as a reservoir for fungal accumulation. This may prevent the release of the fungi to saliva or localization on the occlusal surfaces.2
In terms of S. mutans, we observed a positive correlation between the occlusal plaque and the saliva levels in both the control and the FOA groups (Fig. 3B). These results are in agreement with previous studies that demonstrated the preferential localization of S. mutans for occlusal surfaces.31 Ultimately, we compared C. albicans and S. mutans relationship and found positive correlations for their levels in saliva and occlusal plaque in the control group, but not the FOA subjects (Fig. 4).
The ratio of S. mutans and C. albicans may contribute to the health and disease state. When the FOAs are inserted in the oral cavity, disruption of the balance occurs. This may favor the colonization of C. albicans and other partner bacteria, which can be explored in future studies. Although our pilot study was cross-sectional in nature, we demonstrated that the FOAs disrupt the existing relationship between C. albicans and S. mutans species that may impact the future oral health of orthodontic patients. Future studies should consider expanding the investigation into a longitudinal study with a larger sample size.
In conclusion, the presence of FOAs perturbed the correlation between C. albicans and S. mutans. Specifically, there was a significant increase in the overall bacterial load and a trend of higher C. albicans accumulation on the occlusal surfaces of the FOA group, which might increase the risk of orthodontic patients for fungal and bacterial related diseases. Therefore, it is essential to explore further the impact of FOAs treatment on the individual's oral microbiome and the association with oral health and diseases.
Declaration of competing interest
All authors declare no conflict of interests relevant to this article.
Acknowledgements
The authors would like to thank Drs. Joshua Lin, Alejandra Rivera, Loliya Stewart, William Traynor, and dental student volunteers for their support during subject recruitment. The authors would also thank Drs. Renate Lux and Bhumika Shokeen for critical review of this manuscript.
F.Y was supported by the following grants: 81670979 and 31300424 from Natural Science Foundation of China, 16-5-1-64-jch from Bureau of Science and Technology, Qingdao, grants from Qingdao Outstanding Health Professional Development Fund, and grant by the China Scholarship Council for one year visiting scholar at UCLA. This study was also supported in part by C3 Jian, Inc.
Appendix 1.
Supplementary material and methods
Extraction of genomic DNA
Total genomic DNA was extracted from our participant's oral samples (saliva and occlusal plaque) using the Epicenter MasterPure™ DNA purification kit (Lucigen, USA) following the manufacturer's instructions with modifications. Briefly, prior to the kit purification protocol, samples were subjected to mechanical grinding with glass beads followed by lysozyme treatment for 2 h at 37 °C. The quantity and quality of DNA were measured with NanoDrop 2000 (Thermo Fisher Scientific, USA). The extracted DNA was stored at −20 °C until further use.
Real -time quantitative polymerase chain reaction (qPCR) and sensitivity test
The fungi and C. albicans levels in the saliva and occlusal plaque samples were assessed by real-time quantitative polymerase chain reaction (qPCR), using the fungi-specific (ITS1, ITS2) and the C. albicans-specific (SAP) primers (Table 1). To assess the total bacteria and S. mutans levels, using the universal 16S RNA gene primers for bacteria and S. mutans specific glucosyltransferases (gtfB) gene primers (Table 1). Briefly, the reaction mix consisted of 0.5 μM of each primer with 1X SYBR Green Master Mix (BioRad) and template DNA. For template DNA, 100 ng of saliva and 15 ng of occlusal plaque DNA was used in a reaction volume of 20 uL, while 100 ng of DNA was used for fungal (C. albicans SN152) and bacterial (S. mutans UA140) genomic DNA.
To generate standard curves for real-time PCR, a tenfold serial dilution of reference species (C. albicans SN152, or S. mutans UA140) DNA, initial 10 ng, was used as templates and the threshold at the different dilution points averaged. Standard curves were generated with linear relationships with universal primers. The fungi-specific (ITS1, ITS2) and the C. albicans-specific (SAP) primers (Table 1) were verified using genomic DNA from different fungi and C. albicans (SN152) reference strains. Additionally, specificity of the universal 16S rRNA-specific and S. mutans-specific primers (glucosyltransferase B, gftB) (Table 1) was confirmed using genomic DNA from common oral bacterial species and S. mutans (U140) reference strains. Moreover, the respective melting peaks were used to assess the specificity of the amplicon.
References
- 1.Zou J., Meng M.M., Law C.S., Rao Y., Zhou X.D. Common dental diseases in children and malocclusion. Int J Oral Sci. 2018;10:7. doi: 10.1038/s41368-018-0012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brusca M.I., Chara O., Sterin-Borda L., Rosa A.C. Influence of different orthodontic brackets on adherence of microorganisms in vitro. Angle Orthod. 2007;77:331–336. doi: 10.2319/0003-3219(2007)077[0331:IODOBO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Cobourne M.T.F.P., Dibiase A.T., Ahmed S. Wiley; 2012. Clinical cases in orthodontics. [Google Scholar]
- 4.Klaus K., Eichenauer J., Sprenger R., Ruf S. Oral microbiota carriage in patients with multibracket appliance in relation to the quality of oral hygiene. Head Face Med. 2016;12:28. doi: 10.1186/s13005-016-0125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitas A.O., Marquezan M., Nojima Mda C., Alviano D.S., Maia L.C. The influence of orthodontic fixed appliances on the oral microbiota: a systematic review. Dent Press J Orthod. 2014;19:46–55. doi: 10.1590/2176-9451.19.2.046-055.oar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko-Adams C., Cioffi I., Dufour D., Nainar S.M.H., Levesque C.M., Gong S.G. Short-term effects of fixed orthodontic appliance on concentrations of mutans streptococci and persister cells in adolescents. Am J Orthod Dentofacial Orthop. 2020;157:385–391. doi: 10.1016/j.ajodo.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Naranjo A.A., Triviño M.L., Jaramillo A., Betancourth M., Botero J.E. Changes in the subgingival microbiota and periodontal parameters before and 3 months after bracket placement. Am J Orthod Dentofacial Orthop. 2006;130:275. doi: 10.1016/j.ajodo.2005.10.022. e17-22. [DOI] [PubMed] [Google Scholar]
- 8.Reichardt E., Geraci J., Sachse S., et al. Qualitative and quantitative changes in the oral bacterial flora occur shortly after implementation of fixed orthodontic appliances. Am J Orthod Dentofacial Orthop. 2019;156:735–744. doi: 10.1016/j.ajodo.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Ghasempour M., Sefidgar S.A.A., Eyzadian H., Gharakhani S. Prevalence of Candida albicans in dental plaque and caries lesion of early childhood caries (ECC) according to sampling site. Caspian J Int Med. 2011;2:304–308. [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira D., Seneviratne C.J., Koga-Ito C.Y., Samaranayake L.P. Is the oral fungal pathogen Candida albicans a cariogen? Oral Dis. 2018;24:518–526. doi: 10.1111/odi.12691. [DOI] [PubMed] [Google Scholar]
- 11.Rozkiewicz D., Daniluk T., Zaremba M.L., et al. Oral Candida albicans carriage in healthy preschool and school children. Adv Med Sci. 2006;51:187–190. [PubMed] [Google Scholar]
- 12.Forssten S.D., Bjorklund M., Ouwehand A.C. Streptococcus mutans, caries and simulation models. Nutrients. 2010;2:290–298. doi: 10.3390/nu2030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachtiar E.W., Bachtiar B.M. Relationship between Candida albicans and Streptococcus mutans in early childhood caries, evaluated by quantitative PCR. F1000Res. 2018;7:1645. doi: 10.12688/f1000research.16275.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Carvalho F.G., Silva D.S., Hebling J., Spolidorio L.C., Spolidorio D.M.P. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol. 2006;51:1024–1028. doi: 10.1016/j.archoralbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Ellepola K., Truong T., Liu Y., et al. Multi-omics analyses reveal synergistic carbohydrate metabolism in Streptococcus mutans-Candida albicans mixed-species biofilms. Infect Immun. 2019;87:600339–600419. doi: 10.1128/IAI.00339-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D., Sengupta A., Niepa T.H.R., et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. 2017;7:41332. doi: 10.1038/srep41332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rammohan S.N., Juvvadi S.R., Gandikota C.S., Challa P., Manne R., Mathur A. Adherence of Streptococcus mutans and Candida albicans to different bracket materials. J Pharm BioAllied Sci. 2012;4:S212–S216. doi: 10.4103/0975-7406.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Organization W.H. World Health Organization; Geneva: 1997. Oral health surveys : basic methods. [Google Scholar]
- 19.Agnello M., Cen L., Tran N.C., Shi W., McLean J.S., He X. Arginine improves pH homeostasis via metabolism and microbiome modulation. J Dent Res. 2017;96:924–930. doi: 10.1177/0022034517707512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardes M., Bruns T.D. Its pimers with enhanced specificity for Basidiomycetes - application to the identification of Mycorrhizae and Rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 21.White T.J.B.T., Lee S., Taylor J.W. Academic Press Inc; New York: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. [Google Scholar]
- 22.Flahaut M., Sanglard D., Monod M., Bille J., Rossier M. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J Clin Microbiol. 1998;36:395–401. doi: 10.1128/jcm.36.2.395-401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyzer G., de Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung W.S., Yang I.L.H., Lim W.H., Baek S.H., Kim T.W., Ahn S.J. Adhesion of mutans streptococci to self-ligating ceramic brackets: in vivo quantitative analysis with real-time polymerase chain reaction. Eur J Orthod. 2015;37:565–569. doi: 10.1093/ejo/cju090. [DOI] [PubMed] [Google Scholar]
- 26.Morales D.K., Hogan D.A. Candida albicans interactions with bacteria in thecontext of human health and disease. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raja M., Hannan A., Ali K. Association of oral Candidal carriage with dental caries in children. Caries Res. 2010;44:272–276. doi: 10.1159/000314675. [DOI] [PubMed] [Google Scholar]
- 28.Yang X.Q., Zhang Q., Lu L.Y., Yang R., Liu Y., Zou J. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch Oral Biol. 2012;57:1048–1053. doi: 10.1016/j.archoralbio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Dawes C., Wong D.T.W. Role of saliva and salivary diagnostics in the advancement of oral health. J Dent Res. 2019;98:133–141. doi: 10.1177/0022034518816961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belstrøm D. The salivary microbiota in health and disease. J Oral Microbiol. 2020;12:1723975. doi: 10.1080/20002297.2020.1723975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvalho J.C., Ekstrand K.R., Thylstrup A. Dental plaque and caries on occlusal surfaces of first permanent molars in relation to stage of eruption. J Dent Res. 1989;68:773–779. doi: 10.1177/00220345890680050401. [DOI] [PubMed] [Google Scholar]
- 32.Fábián T.K., Hermann P., Beck A., Fejérdy P., Fábián G. Salivary defense proteins: their network and role in innate and acquired oral immunity. Int J Mol Sci. 2012;13(4):4295–4320. doi: 10.3390/ijms13044295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salehi B., Kregiel D., Mahady G., Sharifi-Rad J., Martins N., Rodrigues C.F. Management of Streptococcus mutans-Candida spp. oral biofilms' infections: paving the way for effective clinical interventions. J Clin Med. 2020;9:517. doi: 10.3390/jcm9020517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi E.J., Lee S.H., Kim Y.J. Quantitative real-time polymerase chain reaction for Streptococcus mutans and Streptococcus sobrinus in dental plaque samples and its association with early childhood caries. Int Paed Dent. 2009;19:141–147. doi: 10.1111/j.1365-263X.2008.00942.x. [DOI] [PubMed] [Google Scholar]
- 35.Childers N.K., Osgood R.C., Hsu K.L., et al. Real-time quantitative polymerase chain reaction for enumeration of Streptococcus mutans from oral samples. Eur J Oral Sci. 2011;119:447–454. doi: 10.1111/j.1600-0722.2011.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seed P.C. The human mycobiome. Cold Spring Harb Perspect Med. 2014;5:a019810. doi: 10.1101/cshperspect.a019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumaran P. Clinical evaluation of the retention of different pit and fissure sealants: a 1-year study. Int J Clin Pediatr Dent. 2013;6:183–187. doi: 10.5005/jp-journals-10005-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arslan S.G., Akpolat N., Kama J.D., Ozer T., Hamamei O. One-year follow-up of the effect of fixed orthodontic treatment on colonization by oral Candida. J Oral Pathol Med. 2008;37:26–29. doi: 10.1111/j.1600-0714.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 39.Saloom H.F., Mohammed-Salih H.S., Rasheed S.F. The influence of different types of fixed orthodontic appliance on the growth and adherence of microorganisms (in vitro study) J Clin Exp Dent. 2013;5:e36–e41. doi: 10.4317/jced.50988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouvelis G., Papadimitriou A., Merakou K., Doulis I., Karapsias S., Kloukos D. A prospective cohort study assessing the impact of fixed orthodontic appliances on saliva properties and oral microbial flora. Oral Health Prev Dent. 2021;19:67. doi: 10.3290/j.ohpd.b898961. [DOI] [PubMed] [Google Scholar]