Abstract
Objectives: As the United States (U.S.) population rapidly ages, the incidence of Alzheimer's Disease and Related Dementias (ADRDs) is rising, with racial/ethnic minorities affected at disproportionate rates. Much research has been undertaken to test, sequence, and analyze genetic risk factors for ADRDs in Caucasian populations, but comparatively little has been done with racial/ethnic minority populations. We conducted a scoping review to examine the nature and extent of the research that has been published about the genetic factors of ADRDs among racial/ethnic minorities in the U.S.
Design: Using an established scoping review methodological framework, we searched electronic databases for articles describing peer-reviewed empirical studies or Genome-Wide Association Studies that had been published 2005–2018 and focused on ADRD-related genes or genetic factors among underrepresented racial/ethnic minority population in the U.S.
Results: Sixty-six articles met the inclusion criteria for full text review. Well-established ADRD genetic risk factors for Caucasian populations including APOE, APP, PSEN1, and PSEN2 have not been studied to the same degree in minority U.S. populations. Compared to the amount of research that has been conducted with Caucasian populations in the U.S., racial/ethnic minority communities are underrepresented.
Conclusion: Given the projected growth of the aging population and incidence of ADRDs, particularly among racial/ethnic minorities, increased focus on this important segment of the population is warranted. Our review can aid researchers in developing fundamental research questions to determine the role that ADRD risk genes play in the heavier burden of ADRDs in racial/ethnic minority populations.
Keywords: genetic risk factors, Alzheimer's disease, race, ethnicity, minority, review
Introduction
As the United States (U.S.) population rapidly ages, the incidence of Alzheimer's Disease and related dementias (ADRD) is on the rise (1, 2). Alzheimer's Disease (AD) is the sixth leading cause of death in the U.S. and the fifth leading cause of death for those age 65 years and older (1, 2). In the U.S., 5.7 million people are living with AD, which is projected to grow to 13.9 million adults (3.3% of the population) by 2060 (2). Although the primary risk factor for ADRD is age, race, and ethnicity are also associated with ADRD (2–4).
The U.S. population is becoming more racially and ethnically diverse, with Census projections showing that the country will be a “majority–minority” nation by 2050. That is, racial/ethnic minorities will comprise more than 50% of the population by this date (5). African Americans are twice as likely as Non-Hispanic Whites to have AD, while Hispanics are 1.5 times as likely to have AD compared to their Non-Hispanic White counterparts (1). Also by 2050 in the U.S., it is estimated that the proportion of racial/ethnic minorities who suffer from AD will double in size compared to current figures (6). Regarding rates of diagnoses, in particular, African Americans are diagnosed later in the course of ADRD than White patients. Quinones et al. (7) suggest that this is likely due to cultural factors and normalization of ADRD symptoms as part of the usual aging process. There are also noted disparities in cognitive decline and impairment with racial/ethnic minorities suffering greater cognitive decline after ADRD diagnosis compared to other groups (8–10), potentially related to socio-economic resources, such as education quality, development of cognitive reserve, financial means, and early midlife stressors (7). Racial/ethnic health disparities in the U.S. proliferate, are multilayered, and are rooted in a variety of structural and historical inequalities that continue to disproportionately burden racial/ethnic minorities. These disparities underscore the need to examine the factors underlying ADRD in racial/ethnic minority U.S. populations.
As our population ages and the size of our minority populations increase in the U.S., understanding the burden of ADRD on our aging populations can aid in providing insight into the most appropriate and effective public health actions. For example, to provide the best care and community support for aging minority populations, it is valuable to understand any patterns of genetic risk factors to address comorbid disease management, environmental, and socio-economic factors that may affect ADRD prevention, diagnosis, and progression. Similarly, more precise knowledge of differences in prevalence of ADRD in minority populations is useful for policy planning when allocating resources, ensuring access to care, and improving quality of care (11).
Much research has been undertaken to test, sequence, and analyze genetic risk factors for ADRD in White populations, but comparatively little has been done with racial/ethnic minority populations (12). In fact, examining genetic factors in heath disparities research has sometimes led to intense controversy (13, 14), oftentimes for concern of the racialization of medicine, misuse of pharmacogenomics, and racial biology (15–17). In studies that have explored ADRD genetic risk factors in minorities, the study sizes have been relatively small, making the conclusions about genetic associations less powerful. Some data appears to show differences in the genetic etiologies between Caucasians and African Americans, especially relating to the APOE gene, which needs to be explored further (11, 18, 19).
There are multiple types of AD classified by age at onset and method of inheritance. The two main categories from a genetic perspective are Early Onset Alzheimer's Disease (EOAD) and Late Onset Alzheimer's Disease (LOAD). According to the National Institute on Aging website, EOAD is also referred to as Familial Alzheimer's Disease and follows an autosomal dominant inheritance pattern, meaning that only one allele from either parent is required to cause disease. EOAD is caused by mutations in three genetic loci, APP, PSEN1, and PSEN2 (20–22). Late Onset Alzheimer's Disease, which is also referred to as Sporadic Alzheimer's Disease, is polygenic, meaning that multiple genes along with environmental factors contribute to the risk of AD, age of onset, and severity of disease (20, 21). APOE is one of the most well-established genetic risk factors for LOAD and has implications for risk of other types of AD (20, 21).
The APOE e4 allele is a strong risk factor for Sporadic or Late-Onset Familial AD, with the degree of risk increased with two copies of the allele (homozygous e4/e4), but possession of an e4 allele is not in itself necessary to produce AD or sufficient alone to cause the disease (23). Homozygosity in genetics refers to an individual with two copies of the same allele at a particular genetic loci or gene, while heterozygosity refers to the presence of two different alleles at a loci or gene (24). The effects of homozygosity and heterozygosity for the e4 allele has been studied extensively in European American populations, with homozygotes having a 12 times increased risk of LOAD, and heterozygotes having a 2–3 times increased risk of LOAD (18, 19). In African American populations and Hispanic populations, e4 heterozygosity or homozygosity does not correlate with increased risk of AD, indicating that other genetic and environmental factors are responsible for the increased incidence and prevalence of AD in these populations (25–27).
Examining genetic risk factors for ADRDs in minority populations can deepen our understanding of the interaction between biological or genetic factors and socio-ecological determinants of health. It also has the potential to aid in preventive care and early diagnosis for these populations with greater incidence of ADRDs (28). To better understand the risk profile of racial/ethnic minorities who are impacted by ADRD, research should be conducted to comprehend the disease mechanism in these populations, including influential genetic risk factors. If advances in genomic medicine continue to be valid, reliable, and promising, racial/ethnic minorities should be afforded the opportunities to participate in research at similar rates as their White counterparts (13). Other systematic reviews have been conducted in this general subject area. These reviews have had a more segmented focus, with some examining one specific gene and others focusing on a specific population (29, 30). Additional scoping or systematic reviews were focused on a single type of ADRD, such as Lewy Body Dementia or LOAD (31, 32). To explore this gap in the literature, we conducted a scoping review to examine the nature and extent of research that has been published about the genetic factors of ADRDs among racial/ethnic minorities in the U.S.
Methods
Search Strategy and Selection Criteria
Our study protocol was developed using the established and peer-reviewed scoping review methodological framework and updated based on prior ADRD-focused scoping reviews (33–38). Scoping reviews are a useful format used to explore fields of study not already well-explored or defined. A scoping review is a “preliminary assessment of the potential size and scope of available research literature. It aims to identify the nature and extent of research evidence” [(39), p. 31]. Scoping reviews can be utilized for a variety of research purposes including discovering the scope of existing research in a field of study, in order to identify gaps in the literature for future study. Scoping reviews can also be used to explore the need for a systematic review and the potential value of a systematic review (34, 38).
The databases used to conduct the search were PubMed, CINAHL, and Science Direct. We chose to limit the search to those articles published from 2005 to 2019, as 2005 is when next generation DNA sequencing was available, allowing for more extensive genetic studies with larger sample sizes (40). We conducted a search within the databases using a combination of three concepts: (1) ADRD Genes, (2) Populations and Minority Groups, and (3) ADRDs. The search used a combination of terms from the three concepts to find articles relevant to our research questions. Specific ADRD candidate gene terms were chosen by recent data from Genome Wide Association Studies (GWAS) (41, 42). Some included terms were: APOE, beta Amyloid Protein Precursor, CD2AP, Genetic Predisposition to Disease, PSEN1, PSEN2, STM2, APP, TREM2, African American, Alaska Native, Arabs, Asian American, Ethnic Groups, Hispanic American, Native American, Jews, Minority Groups, Alzheimer's Disease, Dementia, Lewy Bodies, Lewy Body Disease. Inclusion criteria for the review were (1) articles published after January 1, 2005, (3) available in English, (3) peer reviewed empirical studies or Genome-Wide Association Studies (GWAS) (4) that focus on or include an underrepresented minority population in the U.S., (5) that focus on ADRDs, and (6) that focus on ADRD-related genes or genetic factors.
Data Extraction and Synthesis
The study selection process included three interrelated steps: Title/abstract reviews, full-article reviews, and reviewers' examination of reference lists from full articles to identify articles for possible inclusion (43). First, five out of nine of our team members were randomly assigned to review the 1,134 article titles and abstracts in Covidence systematic review online software, with each abstract randomly assigned to two reviewers. Two team members were designated as arbitrators for review discrepancies. When a discrepancy occurred between reviewers (e.g., one “Yes, include in the review” and one “No, do not include in the review”), the designated team members arbitrated the discrepancy. When both randomly assigned reviewers marked an abstract as “Yes” for inclusion, Covidence automatically moved it into the full article review list. Once all titles and abstracts were reviewed twice and all discrepancies arbitrated, the research team then performed a complete review of the resulting 115 articles. Seven team members were randomly assigned a set of articles for full review and the same inclusion and exclusion criteria were used. A data abstraction tool was developed to facilitate review of the full articles and to abstract relevant data. The tool included 21 questions to aid in summarizing the key characteristics of each article. Discrepancies on final article selection and data extraction were then arbitrated by two team members with consultation with the rest of the research team. Once all full articles had been determined, the abstracted data were converted to a Microsoft Excel file for management.
Results
Studies Identified
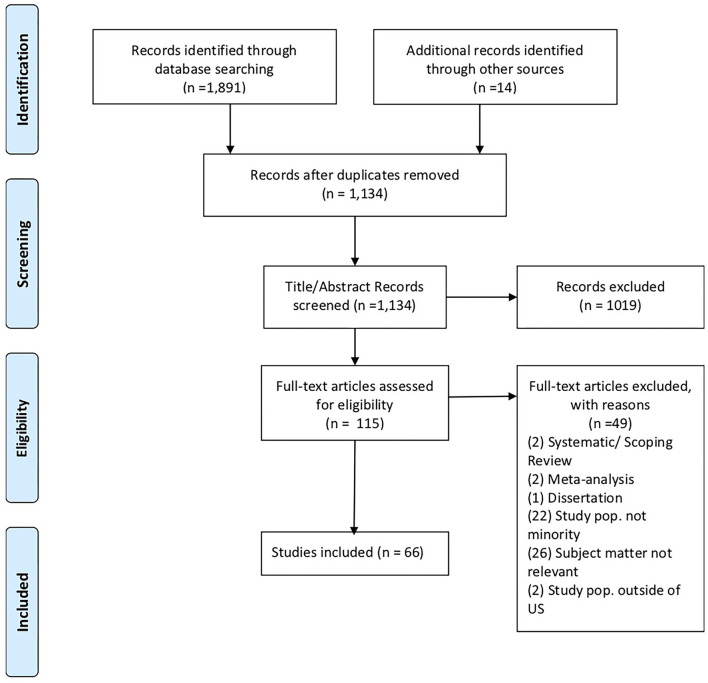
From the searches in all three databases there were a total of 1,891 articles and 14 additional articles identified from reference lists, for a total of 1,905. We removed 771 duplicates, for a total 1,134 articles for the abstract review stage. During the title abstract review we excluded 1,019 articles due to the following reasons: published outside of the date range, article not available in English, dissertation, metanalysis, systematic review, scoping review, not focused on ADRD, not focused on minority U.S. population, not focused on ADRD genetic factors. After title abstract review, 115 articles remained for full text review. An additional 49 articles were excluded during the full-text review stage if the criteria were not met through examination of the full article. The full text review resulted in 66 included articles (see Figure 1).
Figure 1.
PRISMA chart. Source: Moher et al. (43).
Populations and Genes Examined
Tables 1, 2 present the general characteristics of the studies included in the full-text review. Table 3 presents a detailed listing of the characteristics of the articles that were included in the full-text review. Among the resulting 66 studies, most of the studies (n = 41, 62%) were focused on African Americans as the population of interest followed by those focusing on the Hispanic population (n = 28, 42%). Asian American populations were examined in seven out of the 66 studies (11%), and Native American/Alaska Natives populations were included in only one study (1.5%) (Table 1).
Table 1.
Characteristics of studies included in the full-text review (N = 66).
| Characteristic | Number | Percentage (%) |
|---|---|---|
| Publication year | ||
| 2005–2006 | 7 | 10.6 |
| 2007–2008 | 6 | 9.1 |
| 2009–2010 | 5 | 7.6 |
| 2011–2012 | 9 | 13.6 |
| 2013–2014 | 15 | 22.7 |
| 2015–2016 | 9 | 13.6 |
| 2017–2018 | 15 | 22.7 |
| Race/Ethnicity a | ||
| African American | 41 | 62.1 |
| Hispanic American | 28 | 42.4 |
| Asian American | 7 | 10.6 |
| Native American/Alaska Native | 1 | 1.5 |
| Sample size | ||
| 0–100 | 3 | 4.5 |
| 101–500 | 11 | 16.7 |
| 501–1,000 | 10 | 15.2 |
| 1,001–1,500 | 11 | 16.7 |
| 1,501–2,000 | 7 | 10.6 |
| 2,001–2,500 | 4 | 6.1 |
| 2,501–3,000 | 2 | 3.0 |
| 3,001–3,500 | 2 | 3.0 |
| 3,501–4,000 | 0 | 0.0 |
| 4,001–4,500 | 1 | 1.5 |
| 4,501 or more | 15 | 22.7 |
| Type of study | ||
| Case-control | 18 | 27.3 |
| Cross-sectional | 15 | 22.7 |
| Cohort | 12 | 18.2 |
| Genome Wide Association Study (GWAS) | 9 | 13.6 |
| Longitudinal | 5 | 7.6 |
| Other | 5 | 7.6 |
| Case report/case study | 2 | 3.0 |
Some articles included multiple races/ethnicities in the study sample.
Table 2.
Type of ADRD and risk genes identified in full-text review articles (N = 66).
| Characteristic | Number | Percentage (%) |
|---|---|---|
| Type of ADRD a | ||
| Lewy Body Dementia | 1 | 1.5 |
| Mild Cognitive Impairment | 2 | 3.0 |
| Cognitive Decline | 2 | 3.0 |
| Vascular Dementia | 4 | 6.1 |
| Early onset AD (EOAD) | 7 | 10.6 |
| Alzheimer's Disease | 13 | 19.7 |
| Type of ADRD not specified | 20 | 30.3 |
| Late Onset AD (LOAD) | 26 | 39.4 |
| ADRD risk genes identified b | ||
| PSEN2 | 1 | 1.5 |
| AKAP9 | 2 | 3.0 |
| GRIN3B | 2 | 3.0 |
| SORL1 | 2 | 3.0 |
| CR1 | 3 | 4.5 |
| APP | 4 | 6.1 |
| PSEN1 | 4 | 6.1 |
| ABCA7 | 6 | 9.1 |
| CLU | 6 | 9.1 |
| PICALM | 7 | 10.6 |
| APOE | 43 | 65.2 |
| Other | 42 | 63.6 |
Some articles examined more than one type of ADRD.
Some articles included multiple risk genes.
Table 3.
Detailed listing of studies included in the full-text review.
| Author and year | Study design | URM group | Data source | Sample size* | Type of ADRD | Gene(s) included |
|---|---|---|---|---|---|---|
| Akomolafe et al. (44) | Case-control | African American | MIRAGE Study | 511 cases, 679 controls* |
EOAD, LOAD | NOS3, APOE |
| Arnold et al. (45) | Cohort | Puerto Rican | Original data | 283 | EOAD, LOAD | PSEN1 |
| Beeri et al. (46) | Longitudinal, cohort | African American | ACCORD-MIND Study | 466 | Cognitive Decline | HP |
| Borenstein et al. (47) | Prospective, cohort | Japanese American | The Kame Project | 1,859 | Alzheimer's disease | APOE |
| Borenstein et al. (48) | Prospective, cohort | Japanese American | The Kame Project | 1,859 | Vascular Dementia and Alzheimer's Disease | APOE |
| Bressler et al. (49) | GWAS | African Americans | The ARIC Study | 10,359* | LOAD | APOE, ABCA7, BIN1, CD2AP, CDS33, CELF1, EPHA1, MS4A4E, NME8, PICALM, PKT2B, ZCWPW1, |
| Campos et al. (50) | Case-control | Hispanic Americans, Amerindians | Original data | 56 cases, 56 controls* | Alzheimer's Disease | APOE |
| Carrion-Baralt et al. (51) | Cohort | Puerto Ricans | Original data | 87 | Alzheimer's Disease | APOE |
| Conway et al. (52) | Case-control, targeted sequencing | African Americans | Mayo Clinic | 5,924 cases, 5,173 controls* | EOAD, LOAD, Lewy Body Dementia | ABI3, APOE, PLCG2 |
| Cukier et al. (53) | Case-control | African Americans, Caribbean Hispanics | HIHG and ADGC data sets | 149 cases, 137 controls* | LOAD | ABC1, ABCA7 |
| Desai et al. (54) | Case-control | African Americans | ADRC data set | 1,059 cases, 716 controls* | LOAD | BDNF |
| Edwards-Lee et al. (55) | Family study | African Americans | Original data | 7 | EOAD (autosomal dominant) | APP, PS1, MAPT |
| Erlich et al. (56) | Case-control study | African Americans | MIRAGE Study | 520 cases, 677 controls* | Alzheimer's Disease | PON1, PON2, PON3 |
| Fitten et al. (57) | Cross-sectional study | Hispanic Americans | ADRC data set, OVMC data set | 290* | Alzheimer's Disease, Vascular Dementia | APOE |
| Ghani et al. (58) | Case-control, GWAS | Hispanic Americans | Washington Heights-Inwood Columbia Aging Project, Estudio Familiar de Influencia Genetica de Alzheimer Study | 547 cases, 542 controls* | LOAD | APOE, CLU, PICALM, BIN1 |
| Gonzalez et al. (59) | Cohort study | Hispanic Americans | The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) | 10,887* | Alzheimer's Disease | APOE |
| Harwood et al. (60) | Cross-sectional study | African Americans, Hispanic Americans | Original data | 685 | Alzheimer's Disease | APOE |
| He et al. (61) | Cross-sectional study | African Americans, Hispanic Americans | Original data | 439 | Mild Cognitive Impairment (MCI) | APOE |
| Hendrie et al. (62) | Case-control study | African Americans | Original data | 221 cases, 218 controls | MCI, Dementia, Alzheimer's Disease | APOE |
| Hohman et al. (63) | Case-control, GWAS | African Americans | ADGC | 1,840 cases, 3,804 controls | LOAD | APOE, STM2, ABCA7, CR1, PICALM, BIN1, EPHA1, CD33, SLC24A4, GRIN3B, FERMT2, MS4A6A |
| Janicki et al. (64) | Cohort study | African Americans, Hispanic Americans | Washington Heights Inwood Columbia Aging Project (WHICAP) | 1,686* | Alzheimer's Disease | APOE, CYP19 |
| Jin et al. (65) | Case-control | African Americans | Knight-ADRC + NIA-LOAD, Mayo Clinic, Indiana University, WHICAP, Emory University | 906 cases, 2,487 controls* | LOAD | TREM2 |
| Janicki et al. (66) | Prospective Cohort study | African Americans, Hispanic Americans | WHICAP | 1,686* | Alzheimer's Disease | ESR1 |
| Kim et al. (67) | Longitudinal prospective community-based study | African Americans | IIDP | 1,858* | AD, Dementia | CD2AP, CBS, DTWD2, DYNC111, JRKL-AS1, BIRC8, HCY |
| Kuller et al. (68) | Longitudinal cohort study | African Americans | Pittsburgh Cardiovascular Health Study | 532 | LOAD | APOE |
| Kunkle et al. (69) | Case-control study, GWAS | African Americans | HIHG/CWRU, NIMH Genetic Studies of Alzheimer's Disease Cohort, NCRAD/NIA-LOAD, African American Alzheimer's Disease Genomics Coalition (AAADGC) | 2,762 cases, 2,812 controls* | LOAD | ABCA7 |
| Kwon et al. (70) | Cohort study | African Americans, Hispanic Americans | Original data | 1,309* | LOAD | APOE |
| Lee et al. (71) | Nested Case-control study, prospective | African Americans, Hispanic Americans | Original data | 296 cases, 428 controls* | AD | SORL1 |
| Lee et al. (72) | Family-based cohort study, GWAS | Caribbean Hispanic | Original data | 1,161 individuals from 209 families | Familial LOAD | APOE, PSEN1, 5q15, 7q36.3, 14q32.12, 17q25.1, 17p13 |
| Lee (73) | Family-based case-control and unrelated case-control study, GWAS | Caribbean Hispanics | ADRC | 693 cases, 442 controls* | LOAD | APOE, 12p13 |
| Lee (74) | Nested case-control GWAS | Caribbean Hispanics | WHICAP and EFIGA datasets | 549 cases, 544 controls | LOAD | CLU, PICALM, BIN1, PSEN1, GHITM, C10orf99, PCDH21, LRT2, LRT1, RGR, DGKB, HPCAL1, ODC1 |
| Lee (75) | Family-based cohort study | Caribbean Hispanics | WHICAP and EFIGA datasets | 2,888 | EOAD, LOAD | PSEN1, SNX25, PDLIM3, SORBS2, SH3RF3, NPHP1 |
| Livney (76) | Cross-sectional study | African American, Hispanic Americans | Original data | 1,341 | AD | APOE |
| Logue (77) | Case-control study | African Americans | MIRAGE, GenerAAtions, ADNI, GenADA, NIA-LOAD, FHS | 3,568 cases, 6,205 controls* | APOE, PVRL2, CLU, PICALM, BIN1, EPHA1, MS4A, ABCA7, and CD33, TOMM40 | |
| Logue et al. (78) | Case-control | African Americans | MIRAGE Study, GenerAAtions Study | 422 cases, 394 controls | EOAD, LOAD | AKAP9, APOE, BIN1, CLU, CR1, PICALM, MS4A6E, CD2AP, CD33, ABCA7, EPHA1, SORL1, ACE, PSEN1, PSEN2, APP |
| Logue et al. (79) | Case-control | African Americans | MIRAGE Study, GenerAAtions Study, National Cell Repository for Alzheimer's Disease (NCRAD), Ibadan/Indianapolis (INDY) Study | 489 cases, 472 controls | LOAD | ABCA7, AKAP9, KIAA0196, KANSL1, CNN2, TRIM2 |
| Marden et al. (80) | Cohort | African Americans | Health and Retirement Study (HRS) | 7,690* | AD and Dementia | APOE, BIN1, CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, MS4A4E, CD2AP |
| Marden et al. (81) | Cohort | African Americans | HRS | 8,253* | AD | APOE, CLU, CR1, PICALM |
| McAninch et al. (82) | Cohort | African Americans | Original data | 12,348* | AD | DIO2 |
| Melville et al. (83) | Case-control | African Americans | MIRAGE Study, ADNI Study | 1,146 cases, 956 controls* | AD, MCI | APOE, PICALM, F5/SELP, LHFP, GCFC2, SYNPR, TTC27 |
| Mez et al. (84) | Case-control | African Americans | ADGC, GenerAAtions, MIRAGE, CHAP | 1,825 cases, 3,784 controls | LOAD | APOE, ABCA7, COBL, SLC10A2 |
| Mount et al. (85) | Cross-sectional, retrospective | African Americans | ADCR | 65 | LOAD | APOE |
| Murrell et al. (86) | Cohort | African Americans | Original data | 480 | LOAD | APOE |
| N'Songo et al. (87) | Cohort | African Americans | Original data | 198 cases, 350 controls | EOAD | APP, PSEN1, PSEN2 |
| O'Bryant et al. (88) | Cohort | Mexican Americans | Project FRONTIER, TARCC |
1,628 | MCI | APOE |
| O'Bryant et al. (89) | Cohort, CBPR | Mexican Americans | Project FRONTIER, TARCC |
1,069* | MCI, AD | APOE |
| Olarte et al. (90) | Population-based, case series | Hispanics | HCFA | 680 | Sporadic and familial AD | APOE |
| Pedraza et al. (91) | Case-control | African Americans | Mayo Clinic Alzheimer's Disease Research Center Data, Mayo Clinic Study of Aging, Mayo Clinic LOAD-GWAS | 476 cases, 2,443 controls* | LOAD | CLU, CR1, PICALM |
| Peila et al. (92) | Nested case-control | Japanese-Americans | Honolulu-Asia Aging Study (HAAS), Honolulu Heart Program (HHP) | 283 cases, 573 controls | AD, Vascular Dementia | APOE, TGF-β1 |
| Petrovich et al. (93) | Longitudinal, cohort | Japanese-Americans | The Honolulu-Asia Aging Study | 375 | AD | APOE |
| Qian et al. (94) | Prospective, cohort | Latinos | NACC, Rotterdam Study, Framingham Heart Study, and Sacramento Area Latino Study (SALSA) | 16,844* | AD | APOE |
| Rajabli et al. (95) | Case-control | African Americans, Hispanic Americans | HGDP (Human Genome Diversity Project) | 1,986 cases, 3,899 controls* | LOAD | APOE |
| Reitz et al. (96) | Case-control | African Americans and Caribbean Hispanics | Toronto dataset, NIA-LOAD, MIRAGE Caucasian dataset, MIRAGE African American dataset, Miami Caucasian, Caribbean Hispanic dataset | 2,809 cases, 3,482 controls | AD | SORCS1, APP, Aβ, SORL1 |
| Reitz et al. (97) | Case-control | Caribbean Hispanics | DMS-IV, NINCDS-ADRDA | 160 cases, 294 controls | LOAD | IDE, KIF1, HHEX |
| Reitz et al. (98) | Case-control | African Americans | CHAP, MARS/CORE, UM/VU | 1,968 cases, 3,928 controls | LOAD | ABCA7, APOE |
| Rippon et al. (99) | Family-based cohort study | Latinos | NINDCS-ADRDA | 1,498 | Familial AD | APOE |
| Roses et al. (100) | Cohort | African Americans, Japanese Americans | Bryan ADRC Database/Repository, Coriell Cell Repositories | 447* | LOAD | TOMM40, APOE |
| Sacyzynsky et al. (101) | Cohort | Japanese-Americans | The Honolulu Heart Program, Cooperative Lipoprotein Study | 929 | Dementia | APOE |
| Sawyer et al. (102) | Prospective cohort | African Americans | Duke EPESE Study | 2,076* | Cognitive decline (CD) | APOE |
| Simino et al. (103) | Cohort | African Americans | CHARGE, the NHLBI Exome Sequencing Project | 1,414* | AD | Amyloid-β, KLKB1, F12, PLIN2, ITPRIP |
| Tosto et al. (104) | Cohort | Caribbean Hispanics | NIA-LOAD, EFIGA | 8,116* | LOAD | APOE ε4 |
| Vardarajan et al. (105) | Case-control | African Americans | ADGC | 8,309 cases, 7,366 controls* | AD | APP, KIAA1033, SNX1, SORL1, SNX3, RAB7A |
| Vardarajan et al. (106) | Family and cohort-based genetic association study | Caribbean Hispanics | Original data | 464 familial subjects—(350 affected, 114 unaffected), 498 unrelated controls | LOAD | SORL1 |
| Weiner et al. (107) | Case-control | Choctaw Indians | Original data (Choctaw Indians) and UT Southwestern Alzheimer's Disease Center (ADC) | 78 cases, 39 controls* | AD | APOE |
| Yu et al. (108) | Longitudinal, cohort | African Americans | Religious Orders Study (ROS), Rush Memory and Aging Project (MAP), Minority Aging Research Study (MARS) | 2,388* | AD | APOE, TOMM40 |
Article included multiple races/ethnicities in the study sample.
There were many different study designs represented in our results. The most common study design was a case control study design, with 18 included articles using this design. The next most frequently found study design was cross-sectional with 15 included studies in this category. There were nine GWAS which is expected because candidate risk genes for ADRD in minority populations have not been fully established. There were five longitudinal studies in the results and two case studies. Lastly, there were five studies that could not be classified into one of these categories (Table 1).
Many different types of ADRDs were represented in our search results. The most frequently examined type of AD in our results was LOAD (n = 26, 40%), followed by AD (n = 12, 18%) and EOAD (n = 7, 11%). Vascular Dementia was the focus of four articles out of the total 66 results (n = 4, 6%). Both Mild Cognitive Impairment (MCI) and Cognitive Decline were examined in two articles each (n = 2, 3%). Lewy Body Dementia was the subject of one article (n = 1, 1.5%). Lastly, there were 20 articles that did not specify a particular ADRD designation (n = 20, 30%) (Table 2).
In terms of specific ADRD risk genes, APOE was examined in most studies, with 44 out of 66 included studies examining this genetic risk factor. Other potential ADRD risk genes that were examined by multiple studies included ABCA7, CLU, CR1, PICALM, APP, PSEN1, SORL1 and AKAP9, APP, and PSEN1 are well-established genetic risk factors for EOAD, but in total, they were examined in only eight out of 66 included studies (Table 2).
Discussion
Our findings provide an overview of the published literature examining the association between genetic factors and ADRD risk among racial/ethnic minorities in the U.S. These findings help to illuminate knowledge gaps and suggest whether further study should be undertaken to assess more comprehensively the role that ADRD genes play in AD rates and disease outcomes for minority populations.
Regarding the extent of the genes examined in the studies that we found, APOE was examined in most studies, with 44 out of 66 included studies examining this genetic risk factor. This corresponds with extant ADRD genetic risk factor research findings in general, as APOE is the most well-established genetic risk factor for Sporadic or LOAD (23). We found that well-established ADRD genetic risk factors for Caucasian populations including APOE, APP, PSEN1, and PSEN2 have not been studied to the same degree in minority U.S. populations. The APOE genotype has been shown to be less predictive of ADRD risk in African American, Asian American, Hispanic American, and Native American populations (26, 27, 29, 98). Other genetic risk factors may play a larger role in ADRD genetic risk in these populations, with potential candidates including genes with various functions such as ABCA7, CLU, CR1, PICALM, SORL1, AKAP9, and TREM2 (26, 27, 29, 98, 109). These genes were noted in our review, however with far less frequency than APOE. Preliminary findings indicate that there may be a more complex polygenic profile of ADRD genetic risk in these populations, and this has potential implications for the possible polygenic nature of ADRD risk in all populations (27, 59, 87).
In comparison to the amount of research that has been conducted on Caucasians in the U.S., we found that some minority communities were vastly underrepresented in the research, namely Hispanics, Native Americans, and Asian Americans. Though the number of studies on ADRD genetic risk factors in minority populations has increased over time, especially for certain populations such as African Americans, more comprehensive studies with large sample sizes should be performed to establish key genetic risk factors for these populations as well (27, 109–112). Among the studies in our review, sample size for non-GWAS studies started as low as N = 19 for a case report design. As the sample size increases and more diverse persons are included, additional, more statistically sound conclusions can be made about the associations between genetic expression and disease outcome.
Additionally, comparative studies with both minority and majority population group samples would be useful in examining genetic risk factors, as well as the effects of environment and other factors. Studies exploring genetic risk factors in these populations is warranted to determine the role that both genes and environmental factors play in increased ADRD risks in these populations. A larger, systematic review of existing literature on genetic risk factors for minority U.S. populations would be an appropriate next step in better understanding the existing study landscape with intentions toward implementing GWAS and meta-analyses for diverse U.S. populations.
Knowledge gaps in the disease mechanism among racial/ethnic minority populations is a critical indicator of inequities in genetics and genomics research in these communities, as well as a lack of equity in the health care system for these groups (112). Advancements in genetic medicine and genomic research proliferate, unfortunately not at the same rate for all persons. The impact that disproportionate expansion, innovation, and progress in the field can have on health disparities is significant (12, 112). With that in mind, it is also important to acknowledge that while genetic inquiry is crucial to understanding the disparities present in ADRD, it is not the sole risk factor. Other factors such as environment and socio-environmental context, are implicated in the distribution of racial health disparities, and in fact, the complex interplay of all these factors contribute to many disease outcomes (12, 113, 114).
Of additional consideration as an important implication of this research, particularly for minority populations, is the potential of stigma related to ADRD diagnosis. Some groups have been found to consider dementia as a normal part of aging (115), while others may find shame in an AD diagnosis or the need to keep such health information private (116–118). We highlight these studies as further evidence of the need to focus research in racially and ethnically diverse communities. Furthermore, we acknowledge that such research should consider both quantitative and qualitative approaches.
This study is not without limitations. First, while we conducted a systematic and structured process for the scoping review, we did not evaluate the quality of the evidence presented or the authors' research methods as part of this review. Second, some studies more clearly identified the characteristics of interest for our review than others, and as such, some of the data presented was left to the interpretation of the authorship team. Third, we acknowledge that there is limited generalizability of our findings to research that has been conducted in the U.S. among racial/ethnic minorities. That said, we find that an important strength of this review is in identifying the knowledge gaps in examining and understanding the genetic factors associated with ADRD among racial/ethnic minority populations, which is of growing disease and economic burden in the U.S.
Conclusion
Based our findings, we recommend that additional studies be undertaken to map out and more deeply explore ADRD genetic risk factors among racial/ethnic minority populations in the U.S. at levels comparable to non-minority populations. An increased number of larger scale studies of racially/ethnically diverse persons can aid researchers in making more powerful conclusions about genetic associations in ADRD among populations most affected. Examining genetic risk factors for ADRDs in minority populations can deepen our understanding of the interaction between biological or genetic factors and socio-ecological determinants of health. Furthermore, understanding the role of genetic predisposing factors has the potential to increase preventive health measures and screening, which could lead to reduced time to diagnosis and improved ADRD disease management. Lastly, ethical concerns about the impact that this knowledge of genetic risk factors may have on the health and well-being of individuals must be addressed as we continue to obtain more data on these genetic factors. As our population ages and the size of our minority populations increase in the U.S., understanding the burden of ADRD on our aging populations can aid in providing insight into the most appropriate and effective public health actions.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
LR conceptualized the study, was a scoping reviewer, and contributed to the manuscript narrative. LI and DF were scoping reviewers, contributed to the manuscript narrative, and helped to edit the manuscript. NR was a scoping reviewer and contributed to the manuscript narrative. BA-C, AR, KL, SU, and QM were scoping reviewers and helped to edit the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Alzheimer's Association Report: 2020 Alzheimer's Disease Facts and Figures . Alzheimers Dement. (2020). 16:391–460. 10.1002/alz.12068 [DOI] [PubMed] [Google Scholar]
- 2.Hebert LE, Weuve J, Scherr PA, Evans DA. Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology. (2013). 80:1778–83. 10.1212/WNL.0b013e31828726f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthews KA, Xu W, Gaglioti AH, Holt JB, Croft JB, Mack D, et al. Racial and Ethnic Estimates of Alzheimer's Disease and Related Dementias in the United States (2015-2060) in Adults Aged ≥ 65 years. Alzheimers Dement. (2019) 15:17–24. 10.1016/j.jalz.2018.06.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, Zissimopoulos JM. Racial and ethnic differences in trends in dementia prevalence and risk factors in the United States. Alzheimers Dement. (2018) 4:510–20. 10.1016/j.trci.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steenland K, Goldstein FC, Levey A, Wharton W. A meta-analysis of Alzheimer's disease incidence and prevalence comparing African-Americans and Caucasians. J Alzheimers Dis. (2016) 50:71–6. 10.3233/JAD-150778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colby SL, Ortman JM. Projections of the size and composition of the U.S. population: 2014 to 2060. Population estimates and projections: current population reports. Washington, DC: US Census Bureau; (2015). Available online at: http://www.census.gov/content/dam/Census/library/publications/2015/demo/p25-1143.pdf (accessed March 22, 2021). [Google Scholar]
- 7.Shin J, Doraiswamy PM. Underrepresentation of African Americans in Alzheimer's trials: a call for affirmative action. Front Aging Neurosci. (2016) 8:123. 10.3389/fnagi.2016.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quiñones AR, Kaye J, Allore HG, Botoseneanu A, Thielke SM. An agenda for addressing multimorbidity and racial and ethnic disparities in Alzheimer's disease and related dementia. Am J Alzheimers Dis Other Demen. (2020) 35:153331752096087. 10.1177/1533317520960874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson RL, Fain MJ, Butler AE, Ehiri JE, Carvajal SC. The role of social and behavioral risk factors in explaining racial disparities in age-related cognitive impairment: a structured narrative review. Aging, Neuropsychol Cogn. (2019) 27:173–96. 10.1080/13825585.2019.1598539 [DOI] [PubMed] [Google Scholar]
- 10.Weuve J, Barnes LL, Mendes de Leon CF, Rajan KB, Beck T, Aggarwal NT, et al. Cognitive aging in black and white Americans. Epidemiology. (2018) 29:151–9. 10.1097/EDE.0000000000000747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuelsdorff M, Okonkwo OC, Norton D, Barnes LL, Graham KL, Clark LR, et al. Stressful life events and racial disparities in cognition among middle-aged and older adults. J Alzheimers Dis. (2020) 73:671–82. 10.3233/JAD-190439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froehlich TE, Bogardus ST Jr, Inouye SK. Dementia and race: are there differences between African Americans and Caucasians?. J Am Geriatr Soc. (2001) 49:477–84. 10.1046/j.1532-5415.2001.49096.x [DOI] [PubMed] [Google Scholar]
- 13.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. (2019) 51:584–91. 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moonesinghe R, Jones W, Honoré PA, Truman BI, Graham G. Genomic medicine and racial/ethnic health disparities: promises, perils, and the challenges for health care and public health policy. Ethn Dis. (2009) 19:473–8. [PubMed] [Google Scholar]
- 15.Hunt LM, Megyesi MS. Genes, race and research ethics: who's minding the store?. J Med Ethics. (2008) 34:495–500. 10.1136/jme.2007.021295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SS. Pharmacogenomics and the challenge of health disparities. Publ Health Genomics. (2009) 12:170–9. 10.1159/000189630 [DOI] [PubMed] [Google Scholar]
- 17.Rotimi CN. Are medical and nonmedical uses of large-scale genomic markers conflating genetics and 'race'?. Nat Genet. (2004) 36(11 Suppl):S43–7. 10.1038/ng1439 [DOI] [PubMed] [Google Scholar]
- 18.Scharff DP, Mathews KJ, Jackson P, Hoffsuemmer J, Martin E, Edwards D. More than Tuskegee: understanding mistrust about research participation. J Health Care Poor Underserved. (2010) 21:879–97. 10.1353/hpu.0.0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg CN, Sinha N, Gluck MA. The effects of APOE and ABCA7 on cognitive function and Alzheimer's disease risk in African Americans: a focused mini review. Front Hum Neurosci. (2019) 13:387. 10.3389/fnhum.2019.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter H, Wisniewski T. Apolipoprotein E: essential catalyst of the Alzheimer amyloid cascade. Int J Alzheimers Dis. (2012) 2012:489428. 10.1155/2012/489428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. (2007) 39:17–23. 10.1038/ng1934 [DOI] [PubMed] [Google Scholar]
- 22.Bertram L, Tanzi RE. Alzheimer disease risk genes: 29 and counting. Nat Rev Neurol. (2019) 15:191–2. 10.1038/s41582-019-0158-4 [DOI] [PubMed] [Google Scholar]
- 23.Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer's disease revisited. Alzheimers Dement. (2016) 12:733–48. 10.1016/j.jalz.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 24.Roberts JS, Patterson AK, Uhlmann WR. Genetic testing for neurodegenerative diseases: Ethical and health communication challenges. Neurobiol Dis. (2020) 141:104871. 10.1016/j.nbd.2020.104871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elston RC, Satagopan JM, Sun S. Genetic terminology. Methods Mol Biol. (2012) 850:1–9. 10.1007/978-1-61779-555-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graff-Radford NR, Besser LM, Crook JE, Kukull WA, Dickson DW. Neuropathologic differences by race from the National Alzheimer's Coordinating Center. Alzheimers Dement. (2016) 12:669–77. 10.1016/j.jalz.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan AL, Chidgey C, Peng P, Masters CL, Roberts BR. The neurobiology and age-related prevalence of the ε4 allele of apolipoprotein E in Alzheimer's Disease Cohorts. J Mol Neurosci. (2016) 60:316–24. 10.1007/s12031-016-0804-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mez J, Marden JR, Mukherjee S, Brewster P, Hamilton JL, Gilsanz P, et al. P2-076: Alzheimer's disease genetic risk variants beyond Apoe ε4 predict mortality in the adult changes in thought (ACT) study. Alzheimers Dement. (2016) 12:P637–8. 10.1016/j.jalz.2016.06.1281 [DOI] [Google Scholar]
- 29.Berkowitz CL, Mosconi L, Rahman A, Scheyer O, Hristov H, Isaacson RS. Clinical application of APOE in Alzheimer's prevention: a precision medicine approach. J Prev Alzheimers Dis. (2018) 5:245–52. 10.14283/jpad.2018.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang M-X, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. (1998) 279:751–5. 10.1001/jama.279.10.751 [DOI] [PubMed] [Google Scholar]
- 31.Huang M, Wang D, Xu Z, Xu Y, Xu X, Ma Y, et al. Lack of genetic association between TREM2 and Alzheimer's disease in East Asian population: a systematic review and meta-analysis. Am J Alzheimers Dis Other Dement. (2015) 30:541–6. 10.1177/1533317515577128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huq AJ, Fransquet P, Laws SM, Ryan J, Sebra R, Masters CL, et al. Genetic resilience to Alzheimer's disease in APOE ε4 homozygotes: a systematic review. Alzheimers Dement. (2019) 15:1612–23. 10.1016/j.jalz.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 33.Sanghvi H, Singh R, Morrin H, Rajkumar AP. Systematic review of genetic association studies in people with Lewy Body Dementia. Int J Geriatr Psychiatry. (2020) 35:436–48. 10.1002/gps.5260 [DOI] [PubMed] [Google Scholar]
- 34.Andrews SJ, McFall GP, Booth A, Dixon RA, Anstey KJ. Association of Alzheimer's disease genetic risk loci with cognitive performance and decline: a systematic review. J Alzheimers Dis. (2019) 69:1109–36. 10.3233/JAD-190342 [DOI] [PubMed] [Google Scholar]
- 35.Arksey H, O'Malley L. Scoping studies: towards a methodological framework. Int J Soc Res Methodol. (2005) 8:19–32. 10.1080/1364557032000119616 [DOI] [Google Scholar]
- 36.Peters MD, Godfrey CM, Khalil H, McInerney P, Parker D, Soares CB. Guidance for conducting systematic scoping reviews. Int J Evid Based Healthc. (2015) 13:141–6. 10.1097/XEB.0000000000000050 [DOI] [PubMed] [Google Scholar]
- 37.Friedman DB, Becofsky K, Anderson LA, Bryant LL, Hunter RH, Ivey SL, et al. Public perceptions about risk and protective factors for cognitive health and impairment: a review of the literature. Int Psychogeriatr. (2015) 27:1263–75. 10.1017/S1041610214002877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resciniti NV, Tang W, Tabassum M, Pearson JL, Spencer SM, Lohman MC, et al. Knowledge evaluation instruments for dementia caregiver education programs: a scoping review. Geriatr Gerontol Int. (2020) 20:397–413. 10.1111/ggi.13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tricco AC, Lillie E, Zarin W, O'Brien KK, Colquhoun H, Levac D, et al. Prisma Extension for Scoping Reviews (PRISMA-SCR): checklist and explanation. Ann Intern Med. (2018) 169:467–73. 10.7326/M18-0850 [DOI] [PubMed] [Google Scholar]
- 40.Grant MJ, Booth A. A typology of reviews: an analysis of 14 review types and associated methodologies. Health Info Libr J. (2009) 26:91–108. 10.1111/j.1471-1842.2009.00848.x [DOI] [PubMed] [Google Scholar]
- 41.Ureña S, Ingram LA, Leith K, Lohman MC, Resciniti N, Rubin L, et al. Mentorship and training to increase diversity of researchers and practitioners in the field of aging and Alzheimer's disease: a scoping review of program characteristics. J Aging Health. (2021) 33:48–62. 10.1177/0898264320953345 [DOI] [PubMed] [Google Scholar]
- 42.Tanzi RE, Bertram L. New frontiers in Alzheimer's disease genetics. Neuron. (2001) 32:181–4. 10.1016/S0896-6273(01)00476-7 [DOI] [PubMed] [Google Scholar]
- 43.Kilpinen H, Barrett JC. How next-generation sequencing is transforming complex disease genetics. Trends Genet. (2013) 29:23–30. 10.1016/j.tig.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 44.Akomolafe A, Lunetta KL, Elrich PM, Cupples LA, Baldwin CT, Huyck M, et al. Genetic association between endothelial nitric oxide synthase and Alzheimer disease. Clin Genet. (2006) 70:49–56. 10.1111/j.1399-0004.2006.00638.x [DOI] [PubMed] [Google Scholar]
- 45.Arnold SE, Vega IE, Karlawish JH, Wolk DA, Nunez J, Negron M, et al. Frequency and clinicopathological characteristics of presenilin 1 Gly206Ala mutation in Puerto Rican Hispanics with dementia. J Alzheimers Dis. (2013) 33:1089–95. 10.3233/JAD-2012-121570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beeri MS, Lin HM, Sano M, Springer RR, Liu X, Bendlin BB, et al. Association of the haptoglobin gene polymorphism with cognitive function and decline in elderly african american adults with type 2 diabetes: findings from the action to control cardiovascular risk in diabetes-memory in diabetes (accord-mind) study. JAMA Netw Open. (2018) 7:e184458. 10.1001/jamanetworkopen.2018.4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borenstein AR, Wu Y, Mortimer JA, Schellenberg GD, McCormick WC, Bowen JD, et al. Developmental and vascular risk factors for Alzheimer's disease. Neurobiol Aging. (2005) 26:325–34. 10.1016/j.neurobiolaging.2004.04.010 [DOI] [PubMed] [Google Scholar]
- 48.Borenstein AR, Wu Y, Bowen JD, McCormick WC, Uomoto J, McCurry SM, et al. Incidence rates of dementia, Alzheimer disease, and vascular dementia in the Japanese American population in Seattle, WA: the Kame Project. Alzheimer Dis Assoc Disord. (2014) 28:23–9. 10.1097/WAD.0b013e3182a2e32f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bressler J, Mosley TH, Penman A, Gottesman RF, Windham BG, Knopman DS, et al. Genetic variants associated with risk of Alzheimer's disease contribute to cognitive change in midlife: The atherosclerosis risk in communities study. Am J Med Genet B Neuropsychiatr Genet. (2017) 174:269–82. 10.1002/ajmg.b.32509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campos M, Edlan S, Peavy G. An exploratory study of APOE-ε4 genotype and risk of Alzheimer's disease in Mexican Hispanics. J Am Geriatr Soc. (2013) 61:1038-y40. 10.1111/jgs.12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrión-Baralt J, Meléndez-Cabrero J, Rodríguez-Ubiñas H, Schmeidler J, Beei M, Angelo G, et al. Impact of APOE ε4 on the cognitive performance of a sample of non-demented Puerto Rican Nonagenarians. J Alzheimer Dis. (2009) 18:533–40. 10.3233/JAD-2009-1160 [DOI] [PubMed] [Google Scholar]
- 52.Conway O, Carrasquillo M, Wang X, Bredenberg J, Reedy J, Strickland S, et al. ABI3 and PLCG2 missense variants as risk factors for neurodegenerative diseases in Caucasians and African Americans. Mol. Neurodegener. (2018) 13. 10.1186/s13024-018-0289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cukier H, Kunkle B, Vardarajan B, Rolati S, Hamilton-Nelson K, Kohli M, et al. ABCA7 frameshift deletion associated with Alzheimer disease in African Americans. Neurol. Genet. (2016) 2:34–8. 10.1212/NXG.0000000000000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desai P, DeKosky S, Kamboh I. Genetic variation in the cholesterol 24-hydroxylase (CYP46) gene and the risk of Alzheimer's disease. Neurosci. Lett. (2002) 328:9–12. 10.1016/s0304-3940(02)00443-3 [DOI] [PubMed] [Google Scholar]
- 55.Edwards-Lee T, Ringman JM, Chung J, Werner J, Morgan A, Hyslop G, et al. An African American family with early-onset Alzheimer disease and an APP (T714I) mutation. Neurology. (2005) 64:23. 10.1212/01.WNL.0000149761.70566.3E [DOI] [PubMed] [Google Scholar]
- 56.Elrich P, Lunetta K, Cupples A, Huyck M, Green R, Baldwin C, et al. Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum Mol Genet. (2006) 15:77–85. 10.1093/hmg/ddi428 [DOI] [PubMed] [Google Scholar]
- 57.Fitten J, Ortiz F, Fairbanks L, Bartzokis G, Lu P, Klein E, et al. Younger age of dementia diagnosis in a Hispanic population in southern California. Int J Geriatr. (2014) 29:586–93. 10.1002/gps.4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghani M, Sato C, Lee J, Reitz C, Moreno D, Mayeux R, et al. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: Genome-wide survey of runs of homozygosity. JAMA Neurol. (2013) 70:1261–7. 10.1001/jamaneurol.2013.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González HM, Tarraf W, Jian X, Vasquez PM, Kaplan R, Thyagarajan B, et al. Apolipoprotein E genotypes among diverse middle-aged and older latinos: study of latinos-investigation of neurocognitive aging results (HCHS/SOL). Sci Rep. (2018) 8:17578. 10.1038/s41598-018-35573-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harwood DG, Kalechstein A, Barker WW, et al. The effect of alcohol and tobacco consumption, and apolipoprotein E genotype, on the age of onset in Alzheimer's disease. Int J Geriatr Psychiatry. (2010) 25:511–8. 10.1002/gps.2372 [DOI] [PubMed] [Google Scholar]
- 61.He J, Farias S, Martinez O, Reed B, Mungas D, Decarli C. Differences in brain volume, hippocampal volume, cerebrovascular risk factors, and apolipoprotein E4 among mild cognitive impairment subtypes. Arch Neurol. (2009) 66:1393–9. 10.1001/archneurol.2009.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hendrie HC, Murrell J, Baiyewu O, Lane KA, Purnell C, Ogunniyi A, et al. APOE ε4 and the risk for Alzheimer disease and cognitive decline in African Americans and Yoruba. Int Psychogeriatr. (2014) 26:977–85. 10.1017/S1041610214000167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hohman TJ, Cooke–Bailey JN, Reitz C, Jun G, Naj A, Beecham GW, et al. Global and local ancestry in African-Americans: Implications for Alzheimer's disease risk. Alzheimers Dement. (2016) 12:233–43. 10.1016/j.jalz.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janicki SC, Park N, Cheng R, Schupf N, Clark LN, Lee JH. Aromatase variants modify risk for Alzheimer's disease in a multiethnic female cohort. Dement Geriatr Cogn Disord. (2013) 35:340–6. 10.1159/000343074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janicki SC, Park N, Cheng R, Clark LN, Lee JH, Schupf N. Estrogen receptor α variants affect age at onset of Alzheimer's disease in a multiethnic female cohort. Dement Geriatr Cogn Disord. (2014) 38:200–13. 10.1159/000355559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin SC, Carrasquillo MM, Benitez BA, Tara S, Carrell D, Patel D, et al. TREM2 is associated with increased risk for Alzheimer's disease in African Americans. Mol Neurodegener. (2015) 10:19. 10.1186/s13024-015-0016-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S, Nho K, Ramanan VK, Lai D, Foroud TM, Lane K, et al. Genetic Influences on Plasma Homocysteine Levels in African Americans and Yoruba Nigerians. J Alzheimers Dis. (2016) 49:991–1003. 10.3233/JAD-150651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuller LH, Lopez OL, Becker JT, Chang Y, Newman AB. Risk of dementia and death in the long-term follow-up of the Pittsburgh Cardiovascular Health Study-Cognition Study. Alzheimers Dement. (2016) 12:170–83. 10.1016/j.jalz.2015.08.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kunkle BW, Carney RM, Kohli MA, Naj AC, Nelson KLH, Whitehead PL, et al. Targeted sequencing of ABCA7 identifies splicing, stop-gain and intronic risk variants for Alzheimer disease. Neurosci Lett. (2017) 649:124–9. 10.1016/j.neulet.2017.04.014 [DOI] [PubMed] [Google Scholar]
- 70.Kwon OD, Khaleeq A, Chan W, Pavlik VN, Doody RS. Apolipoprotein E polymorphism and age at onset of Alzheimer's disease in a quadriethnic sample. Dement Geriatr Cogn Disord. (2010) 30:486–91. 10.1159/000322368 [DOI] [PubMed] [Google Scholar]
- 71.Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, et al. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. (2007) 64:501–6. 10.1001/archneur.64.4.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JH, Cheng R, Rogaeva E, Meng Y, Stern Y, Santana V, et al. Further examination of the candidate genes in chromosome 12p13 locus for late-onset Alzheimer disease. Neurogenetics. (2008) 9:2. 10.1007/s10048-008-0122-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JH, Barral S, Cheng R, Chacon I, Santana V, Williamson J, et al. Age-at-onset linkage analysis in Caribbean Hispanics with familial late-onset Alzheimer's disease. Neurogenetics. (2008) 9:127–38. 10.1007/s10048-007-0103-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. (2011) 68:320–8. 10.1001/archneurol.2010.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JH, Cheng R, Vardarajan B, Lantigua R, Dumeyeer DR, Ortmann W, et al. Genetic Modifiers of Age at Onset in Carriers of the G206A Mutation in PSEN1 With Familial Alzheimer Disease Among Caribbean Hispanics. JAMA Neurol. (2015) 72:1043–51. 10.1001/jamaneurol.2015.1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Livney MG, Clark CM, Karlawish JH, Cartmell S, Negron M, Nunez J, et al. Ethnoracial differences in the clinical characteristics of Alzheimer's disease at initial presentation at an urban Alzheimer's disease center. Am J Geriatr Psychiatry. (2011) 19:430–9. 10.1097/JGP.0b013e3181f7d881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RCP, et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. (2011) 68:1569–79. 10.1001/archneurol.2011.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Logue MW, Schu M, Vardarajan BN, Farell J, Bennett DA, Buxbaum JD, et al. Two rare AKAP9 variants are associated with Alzheimer's disease in African Americans. Alzheimers Dement. (2014) 10:609–18. 10.1016/j.jalz.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Logue MW, Lancour D, Farrell J, Simhina I, Fallin MD, Lunetta KL, et al. Targeted Sequencing of Alzheimer Disease Genes in African Americans Implicates Novel Risk Variants. Front Neurosci. (2018) 12:592. 10.3389/fnins.2018.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marden JR, Walter S, Tchetgen Tchetgen EJ, Kawachi I, Glymour MM. Validation of a polygenic risk score for dementia in black and white individuals. Brain Behav. (2014) 4:687–97. 10.1002/brb3.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marden JR, Mayeda ER, Walter S, Vivot A, Tchetgan EJT, Kawachi I, et al. Using an Alzheimer Disease Polygenic Risk Score to Predict Memory Decline in Black and White Americans Over 14 Years of Follow-up. Alzheimer Dis Assoc Disord. (2016) 30:195–202. 10.1097/WAD.0000000000000137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McAninch E, Rajan K, Evans D, Jo S, Chaker L, Peeters R, et al. A common DIO2 polymorphism and Alzheimer disease dementia in African and European Americans. J Clin Endocrinol Metab. (2018) 103:505–30. 10.1210/jc.2017-01196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Melville S, Buros J, Parrado A, Vardarajan B, Logue M, Shen L, et al. Multiple loci influencinghippocampal degeneration identified by genome scan. Ann Neurol. (2012) 72:108–20. 10.1002/ana.23644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mez J, Chung J, Jun G, Kriegel J, Bourias A, Sherva R, Logue M, et al. Two novel loci, COBL andSLC10A2, for Alzheimer's disease in African Americans. Alzheimer's Dement. (2017) 13:45-81. 10.1016/j.jalz.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mount D, Ashley A, Lah J, Levey A, Goldstein F. Is ApoE ε4 Associated with Cognitive Functioning in African Americans Diagnosed with Alzheimer Disease? An Exploratory Study. South Med J. (2009) 102:945–8. 10.1097/SMJ.0b013e3181b21b82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murrell JR, Price B, Lane KA, Baiyewu O, Gureje O, Ogunnieyi A, et al. Association of apolipoprotein E genotype and Alzheimer disease in African Americans. Arch Neurol. (2006) 63:431–4. 10.1001/archneur.63.3.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.N'Songo A, Carrasquillo MM, Wang X, Burgess JD, Nguyen T, Asmann YW, et al. African American exome sequencing identifies potential risk variants at Alzheimer disease loci. Neurol Genet. (2017) 3:e141. 10.1212/NXG.0000000000000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O'Bryant SE, Johnson L, Balldin V, Edwards M, Barbar R, Williams B, et al. Characterization of Mexican Americans with mild cognitive impairment and Alzheimer's disease. J Alzheimers Dis. (2013) 33:373–9. 10.3233/JAD-2012-121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Bryant SE, Johnson L, Reisch J, Edwards M, Hall J, Barbar R, et al. Risk factors for mild cognitive impairment among Mexican Americans. Alzheimers Dement. (2013) 9:622–31. 10.1016/j.jalz.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olarte L, Schupf N, Lee JH, Tang MX, Santana V, Williamson J, et al. Apolipoprotein E epsilon4 and age at onset of sporadic and familial Alzheimer disease in Caribbean Hispanics. Arch Neurol. (2006) 63:1586–90. 10.1001/archneur.63.11.1586 [DOI] [PubMed] [Google Scholar]
- 91.Pedraza O, Allen M, Jennette K, Carrasquilo M, Crook J, Serie D, et al. Evaluation of memory endophenotypes for association with CLU, CR1, and PICALM variants in black and white subjects. Alzheimers Dement. (2014) 10:205–13. 10.1016/j.jalz.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peila R, Yucesoy B, White LR, Johnson V, Kashon ML, Wu K, et al. A TGF-beta1 polymorphism association with dementia and neuropathologies: the HAAS. Neurobiol Aging. (2007) 28:1367–73. 10.1016/j.neurobiolaging.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 93.Petrovitch H, Ross GW, He Q, Lock JU, Markesbery W, Davis D, et al. Characterization of Japanese-American men with a single neocortical AD lesion type. Neurobiol Aging. (2008) 29:1448–55. 10.1016/j.neurobiolaging.2007.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qian J, Wolters FJ, Beiser A, Haan M, Ikram MA, Karlawish J, et al. APOE-related risk of mild cognitive impairment and dementia for prevention trials: An analysis of four cohorts. PLoS Med. (2017) 14:e1002254. 10.1371/journal.pmed.1002254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rajabli F, Feliciano BE, Celis K, Nelson KLH, Whitehead PL, Adams LD, et al. Ancestral origin of ApoE ε4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. (2018) 14:e1007791. 10.1371/journal.pgen.1007791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reitz C, Tokuhiro S, Clark LN, Conrad C, Vonsattel JP, Hazrati LN, et al. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer's disease risk. Ann Neurol. (2011) 69:47–64. 10.1002/ana.22308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reitz C, Cheng R, Schupf N, Lee JH, Mehta PD, Rogaeva E, et al. Association between variants in IDE-KIF11-HHEX and plasma amyloid β levels. Neurobiol Aging. (2012) 199:e13–7. 10.1016/j.neurobiolaging.2010.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moher D, Liberati A, Tetzlaff J, Altman DG, the PRISMA Group . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rippon GA, Tang MX, Lee JH, Lantigua R, Medrano M, Mayeux R. Familial Alzheimer disease in Latinos: interaction between APOE, stroke, and estrogen replacement. Neurology. (2006) 66:35–40. 10.1212/01.wnl.0000191300.38571.3e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roses AD, Lutz MW, Saunders AM, Goldgaber D, Saul R, et al. African-American TOMM40'523-APOE haplotypes are admixture of West African and Caucasian alleles. Alzheimers Dement. (2014) 10:592–601. 10.1016/j.jalz.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 101.Saczynski JS, White L, Peila RL, Rodriguez BL, Launer LJ. The relation between apolipoprotein A-I and dementia: the Honolulu-Asia aging study. Am J Epidemiol. (2007) 165:91–9. 10.1093/aje/kwm027 [DOI] [PubMed] [Google Scholar]
- 102.Sawyer K, Sachs-Ericsson N, Preacher KJ, Blazer DG. Racial differences in the influence of the APOE epsilon 4 allele on cognitive decline in a sample of community-dwelling older adults. Gerontology. (2009) 55:105–20. 10.1159/000137666 [DOI] [PubMed] [Google Scholar]
- 103.Simino J, Wang Z, Bressler J, Chouraki V, Yang Q, Younkin SG, et al. Whole exome sequence-based association analyses of plasma amyloid-β in African and European Americans; the Atherosclerosis Risk in Communities-Neurocognitive Study. PLoS One. (2017) 12:e0180046. 10.1371/journal.pone.0180046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tosto G, Bird TD, Tsuang D, Bennett DA, Boeve BF, Crushaga C, et al. Polygenic risk scores in familial Alzheimer disease. Neurology. (2017) 88:1180–86. 10.1212/WNL.0000000000003734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vardarajan BN, Bruesegem SY, Harbour ME, et al. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. (2012) 33:e15–e30. 10.1016/j.neurobiolaging.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vardarajan BN, Zhang Y, Lee JH, Cheng R, Bohm C, Ghani M, et al. Coding mutations in SORL1 and Alzheimer disease. Ann Neurol. (2015) 77:215–27. 10.1002/ana.24305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weiner MF, Hynan LS, Rossetti H, Womack KB, Rosenberg RN, Gong YH, et al. The relationship of cardiovascular risk factors to Alzheimer disease in Choctaw Indians. Am J Geriatr Psychiatry. (2011) 19:423–9. 10.1097/JGP.0b013e3181e89a46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu L, Lutz MW, Wilson RS, Burns DK, Roses AD, Saunders AM, et al. APOE ε4-TOMM40 '523 haplotypes and the risk of Alzheimer's disease in older Caucasian and African Americans. PLoS One. (2017) 12:e7–e9. 10.1371/journal.pone.0180356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang L-S, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ϵ4, and the risk of late-onset Alzheimer disease in African Americans. JAMA. (2013) 309:1483–92. 10.1001/jama.2013.2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barnes LL, Leurgans S, Aggarwal NT, Shah RC, Arvanitakis Z, James BD, et al. Mixed pathology is more likely in black than white decedents with Alzheimer dementia. Neurology. (2015) 85:528–34. 10.1212/WNL.0000000000001834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kunkle BW, Schmidt M, Klein H-U, Naj AC, Hamilton-Nelson KL, Larson EB, et al. Novel Alzheimer disease risk loci and pathways in African American individuals using the african genome resources panel: a meta-analysis. JAMA Neurol. (2021) 78:102–13. 10.1001/jamaneurol.2020.3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nussbaum RL. Genome-wide association studies, Alzheimer disease, and understudied populations. JAMA. (2013) 309:1527–8. 10.1001/jama.2013.3507 [DOI] [PubMed] [Google Scholar]
- 113.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. (2019) 570:514–8. 10.1038/s41586-019-1310-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McGinnis JM, Williams-Russo P, Knickman JR. The case for more active policy attention to health promotion. Health Aff (Millwood). (2002) 21:78–93. 10.1377/hlthaff.21.2.78 [DOI] [PubMed] [Google Scholar]
- 115.Adler NE, Newman K. Socioeconomic disparities in health: pathways and policies. Health Aff (Millwood). (2002) 21:60–76. 10.1377/hlthaff.21.2.60 [DOI] [PubMed] [Google Scholar]
- 116.Mahoney DF, Cloutterbuck J, Neary S, Zhan L. African American, Chinese, and Latino family caregivers' impressions of the onset and diagnosis of dementia: cross-cultural similarities and differences. Gerontologist. (2005) 45:783–92. 10.1093/geront/45.6.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jang Y, Kim G, Chiriboga D. Knowledge of Alzheimer's disease, feelings of shame, and awareness of services among Korean American elders. J Aging Health. (2010) 22:419–33. 10.1177/0898264309360672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu D, Hinton L, Tran C, Hinton D, Barker JC. Reexamining the relationships among dementia, stigma, and aging in immigrant Chinese and Vietnamese family caregivers. J Cross Cult Gerontol. (2008) 23:283–99. 10.1007/s10823-008-9075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.