Abstract
Tumor cells actively release large quantities of exosomes, which pivotally participate in the regulation of cancer biology, including head and neck cancer (HNC). Exosome biogenesis and release are complex and elaborate processes that are considered to be similar to the process of exocyst-mediated vesicle delivery. By analyzing the expression of exocyst subunits and their role in patients with HNC, we aimed to identify exocyst and its functions in exosome biogenesis and investigate the molecular mechanisms underlying the regulation of exosome transport in HNC cells. We observed that exocysts were highly expressed in HNC cells and could promote exosome secretion in these cells. In addition, downregulation of exocyst expression inhibited HN4 cell proliferation by reducing exosome secretion. Interestingly, immunofluorescence and electron microscopy revealed the accumulation of multivesicular bodies (MVBs) after the knockdown of exocyst. Autophagy, the major pathway of exosome degradation, is not activated by this intracellular accumulation of MVBs, but these MVBs are consumed when autophagy is activated under the condition of cell starvation. Rab11a, a small GTPase that is involved in MVB fusion, also interacted with the exocyst. These findings suggest that the exocyst can regulate exosome biogenesis and participate in the malignant behavior of tumor cells.
Keywords: exocyst, exosome biogenesis, head and neck cancer, Rab11a, multivesicular bodies, intraluminal vesicles
Graphical abstract
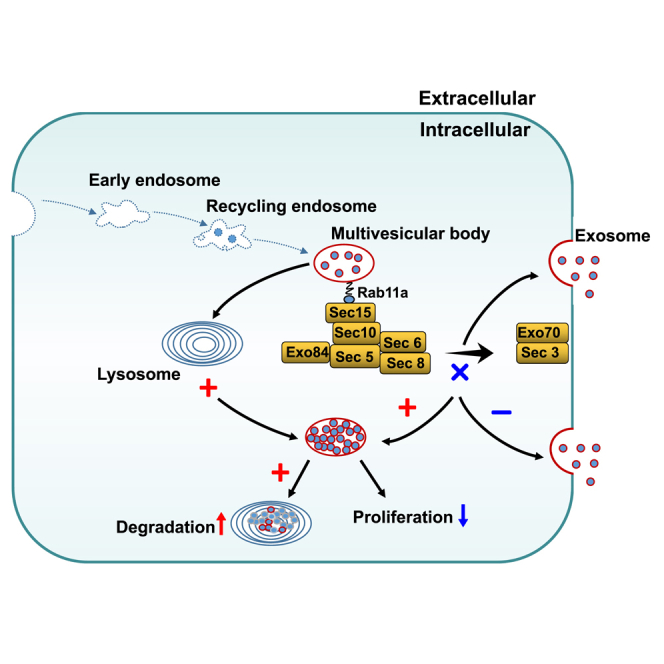
Exosomes play a role in the occurrence and development of head and neck cancer (HNC); however, the specific mechanism for their secretion remains unclear. We found that the multivesicular body gets transported to the plasma membrane for exosome secretion by exocyst via binding with Rab11a in HNC.
Introduction
Head and neck cancer (HNC) is a common malignancy and is the seventh most common cancer in the world, with approximately 890,000 new cases worldwide and approximately 450,000 HNC-related deaths in 2018.1 HNC is a group of tumors that originate from tissues or organs in the head and neck region, excluding the eye, ear, brain, and esophagus. Accordingly, HNC includes thyroid cancer, laryngeal cancer, nasopharyngeal carcinoma, and oral and maxillofacial tumors.2 More than 90% of HNC cases are of squamous cell carcinoma (SCC) arising from epithelial cells in the mucosa.3 The major therapeutic approaches for HNC are surgery, chemoradiotherapy, targeted therapy, and immunotherapy. Despite extensive advances in HNC treatment, the 5-year overall survival rate of advanced HNC remains low, at approximately 50%, thereby posing a major clinical challenge.1,4 Therefore, it is crucial to discover novel treatments for HNC.
Exosomes are extracellular vesicles with a diameter of 30–150 nm that contain proteins, lipids, and genetic materials, including DNA, RNA, and miRNA, perceived to be carriers of these cargoes between diverse locations in the body.5 Most cell types can release exosomes into the extracellular space under both physiological and pathological conditions.6,7 Exosomes perform an important range of extracellular functions, which include interactions with the cellular microenvironment through morphogen signaling, immunologic mediation, cell recruitment, and horizontal transfer of genetic material.8 Tumor cells actively release large quantities of exosomes, which are mobile and participate in crucial steps to regulate cancer biology, including tumor growth, tumorigenesis, immune escape, angiogenesis, metastasis, and resistance to therapies.9, 10, 11 Increasing evidence has demonstrated the role of exosomes in all stages of cancer by mediating intercellular communication and signal transduction, including HNC.12 For example, exosomes produced by hypoxic oral SCC cells can induce epithelial-mesenchymal transition in normoxic cells and promote the invasion of cancer cells by inducing macrophage activation.13,14 In addition, laryngeal cancer (LC)-derived exosomes can reduce LC radiosensitivity via the long non-coding RNA HOTAIR/microRNA-454-3p/E2F2 axis.15 Thus, the biogenesis mechanism of exosomes needs to be evaluated in detail.
Exosome biogenesis is a complex and elaborate process, which starts in the endosomal system. Early endosomes mature into late endosomes or multivesicular bodies (MVBs) and, during the process of MVB formation, the outer endosomal membrane invaginates to produce intraluminal vesicles (ILVs) inside the organelles.16 MVBs can then either merge with lysosomes to degrade their cargoes or fuse with the plasma membrane to release ILVs into the extracellular space as exosomes.17 As part of endosomal trafficking, the docking and fusion of MVBs with the plasma membrane and exosome release play crucial roles not only in the delivery of messengers to the extracellular environment but also in the functioning of membrane lysosome homeostasis and autophagy. Any defect in this release process may have a serious impact on numerous aspects of cell biology, including cell growth and metabolism. Current research shows that the docking and fusion of MVBs with the plasma membrane are regulated by small GTPases of the Rab family, including Rab27a, Rab27b, and Rab11a.18, 19, 20 However, the specific molecular mechanisms involved in the transport of Rab protein and exosome biogenesis remain unclear.
The exocyst is a conserved octameric complex that is composed of eight subunits: namely, Sec10, Sec6, Sec8, Sec5, Sec15, and Exo84, and two membrane-targeting subunits Sec3 and Exo70.21 The exocyst localizes to dynamic areas of the plasma membrane where it mediates the delivery of Golgi-derived secretory vesicles through its role in vesicular trafficking, tethering, and fusion.22, 23, 24 The exocyst complex is assembled hierarchically. Sec10 is a central component of the exocyst and is considered to serve as a bridge between the plasma membrane and the remaining exocyst complex.25 Sec3 and likely some Exo70 proteins are located at the plasma membrane and assist in the formation of the plasma membrane-associated target membrane-soluble NSF attachment receptor proteins (t-SNAREs) as well as interact with GTPases at the plasma membrane to organize the cytoskeleton in preparation for vesicle delivery.23,26, 27, 28 Besides its role in exosome biogenesis, the exocyst contributes to the regulation of a wide array of cellular processes, including cell growth, division, motility, and polarization, which links to human diseases.22
As the docking and fusion process of MVBs is similar to the process involved in exocyst-mediated vesicle delivery, we hypothesize that the exocyst plays a critical role in the guiding, docking, and fusing of MVBs with the plasma membrane and exosome release. This new finding provides potential therapeutic targets for the clinical treatment of cancer metastasis, particularly HNC metastasis.
Results
Exocyst controls exosomes secretion
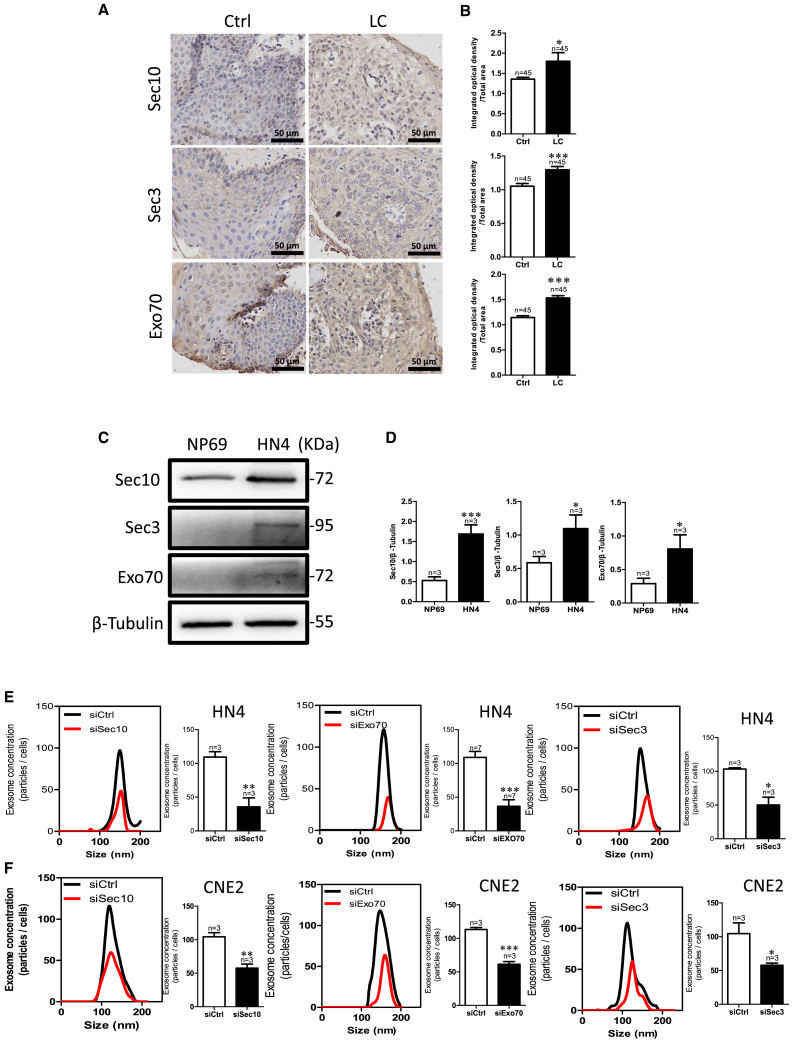
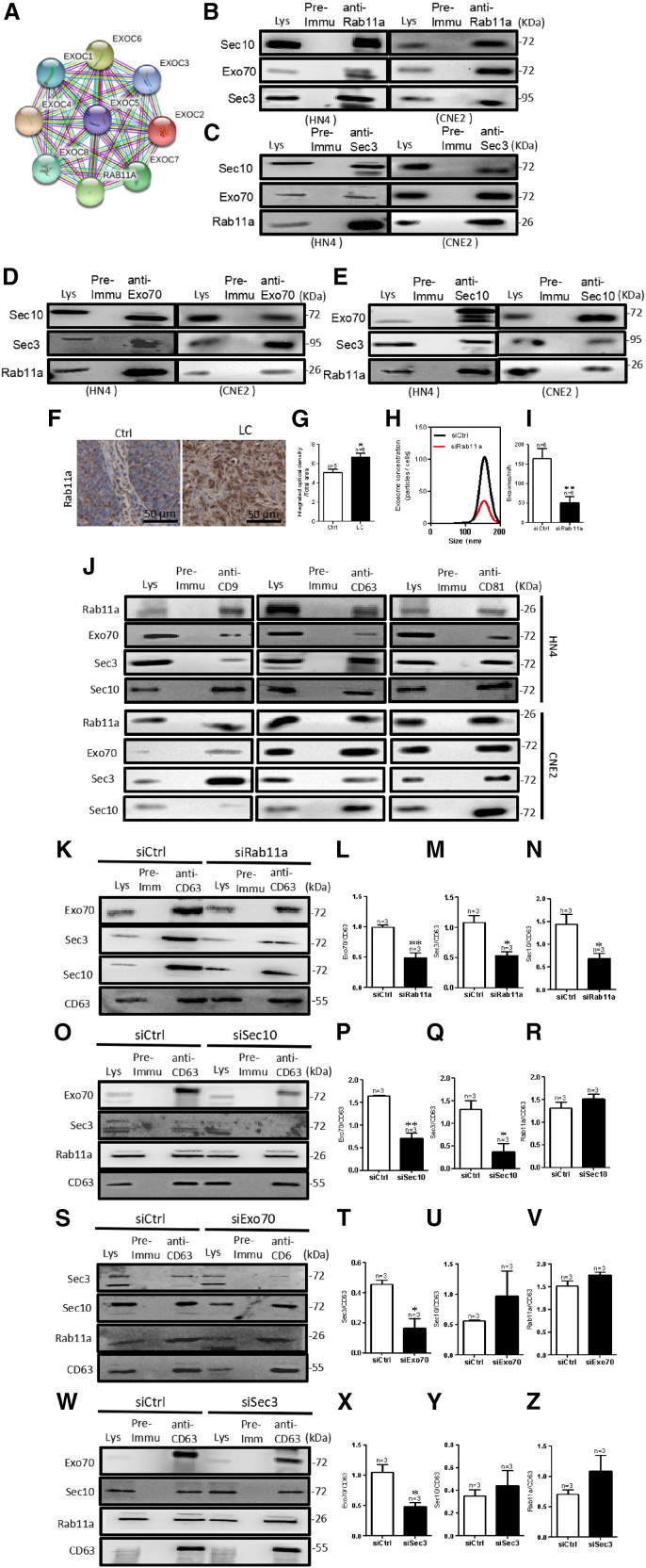
Consistent with a previous finding that tumor cells actively produce and release more exosomes,29 we found that the expression level of exosome-specific markers (CD9, CD63, and CD81) was higher in LC tissues than in adjacent healthy tissue (Figures S1A and S1B). Using the online database HNCDB, we found that a positive correlation between exocyst subunits and exosome markers (Figures S1C–S1J), with the expression level of exocyst subunits being higher in HNC tissues than in normal tissues, especially being highest in tumor grade 4 versus normal and grade 1–3 tissues (Figures S1K–S1R). We confirmed this result in LC tissue and HNC cell line, HN4 cells. The exocyst is composed of eight subunits, of which two units of Sec3 and Exo70 proteins are located at the plasma membrane, whereas Sec10 acts as a central bridge for these two modules. We selected Sec10, Sec3, and Exo70 for the exocyst complex. Immunohistochemistry results revealed that the expression of these three proteins was significantly higher in LC tissues than in the adjacent healthy tissue (Figures 1A and 1B). Consistent with the observations in LC tissues, such exocyst subunits were also more highly expressed in HN4 cells than in NP69 cells, which is a normal human nasopharyngeal epithelial cell line (Figures 1C and 1D).
Figure 1.
Regulation of exosome release by exocysts in HNC
(A and B) Sec10, Sec3, and Exo70 expression in LC tissue compared with that in Ctrl. (A) Representative images of immunohistochemistry staining. (B) Data summary of immunohistochemistry staining. LC, laryngocarcinoma tissue; Ctrl, adjacent normal tissues. ∗∗p < 0.01, ∗∗∗p < 0.001 by t test. Scale bar, 50 μm. (C and D) Sec10, Sec3, and Exo70 expression in HN4 and NP69 cells. (C) Representative images of western blotting. (D) The summary data of western blotting compared with NP69 cells. (E and F) Representative NTA traces and quantification of exosomes after knockdown of the expression of exocyst subunits by transfecting HN4 (E) and CNE2 (F) cells with specific small interfering (si)RNA for Sec10, Sec3, or Exo70 protein. ∗p < 0.05, ∗∗p < 0.05, ∗∗∗p < 0.001 by t test.
To directly assess the effect of the exocyst on exosome secretion, we analyzed exosome concentration after knockdown of the exocyst by specific siRNA against each of Sec10, Sec3, and Exo70 in HN4 cells (Figures S2A–S2E and S2G). As shown in Figure 1E, HN4 cells with exocyst knockdown secreted significantly fewer exosomes than those expressing a control siRNA (Figures 1E, S2I, and S2J). Another HNC cell line, CNE2, got similar results (Figure 1F). These results suggested that the exocyst might contribute to controlling exosome secretion in HNC.
Exocyst depletion inhibits cell proliferation
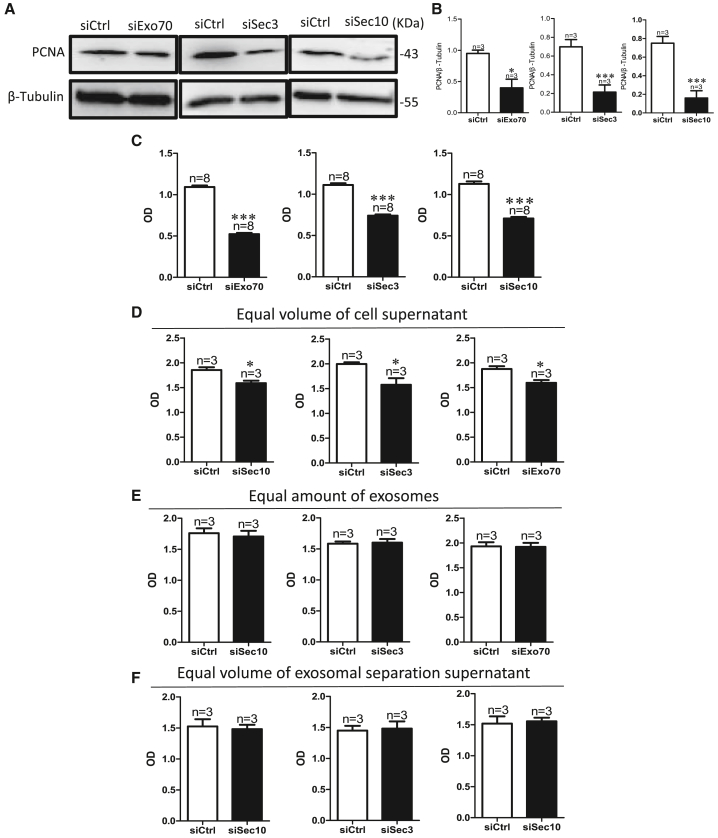
Interestingly, we found that exocyst knockdown inhibited the proliferation of HN4 cells (Figures 2A–2C), but did not affect apoptosis and cell migration (Figures S3A–S3H). To further explore whether exosome concentration or exosome content affects the inhibition of cell proliferation induced by exocyst depletion, we assessed proliferation using cell counting kit-8 (CCK-8) assays after treating HN4 cells with the same amount of exosomes or equal volumes of cell culture medium with or without exosomes from HN4 cells transfected with a scrambled, Sec3 siRNA, Sec10 siRNA, or Exo70 siRNA. The results showed that the proliferation of HN4 cells was inhibited by an equal volume of the whole supernatant from HN4 cells transfected with siRNAs of exocyst subunits, but not by equal amounts of exosomes (Figures 2D and 2E), This suggested that exocyst knockdown induced the abatement of exosome concentration but did not affect exosome content; furthermore, low exosome concentration induced a decrease in the proliferation to other cells through intercellular communication. To further determine whether only the amount of exosomes affects cell proliferation, we treated HN4 cells with equal volumes of cell culture medium without exosomes from HN4 cells transfected with a scrambled, Sec3 siRNA, Sec10 siRNA, or Exo70 siRNA; the results showed that cell proliferation did not differ in these four groups, which suggested that other components in the supernatant, except exosomes, do not affect cell proliferation (Figure 2F). These results showed that exocyst depletion inhibits cell proliferation by reducing the amount of exosomes secretion.
Figure 2.
Exocyst depletion inhibits cell proliferation by reducing exosome concentration
(A–C) Proliferation detection after knockdown of the expression of exocyst subunits by transfecting HN4 cells with specific small interfering (si)RNA for Sec10, Sec3, or Exo70 protein. PCNA, proliferating cell nuclear antigen. (A) Representative images of western blotting. (B) The summary data of western blotting compared with siCtrl. (C) The summary data of CCK-8 compared with those of siCtrl. CCK-8, cell counting kit-8. ∗p < 0.05, ∗∗∗p < 0.001 by t test. (D) Quantitative CCK-8 assay showing proliferation of HN4 cells after treatment with equal volumes of cell culture medium from HN4 cells transfected with siCtrl or with Exo70, Sec3, or Sec10 siRNA. ∗p < 0.05 versus siCtrl by t test. (E) Quantitative CCK-8 assay showing proliferation of HN4 cells after treatment with equal amounts of exosomes from HN4 cells transfected with siCtrl or with Exo70, Sec3, or Sec10 siRNA. (F) Quantitative CCK-8 assay showing proliferation of HN4 cells after treatment with an equal volume of exosome separated from HN4 cells transfected with siCtrl or with Exo70, Sec3, or Sec10 siRNA. OD, optical density.
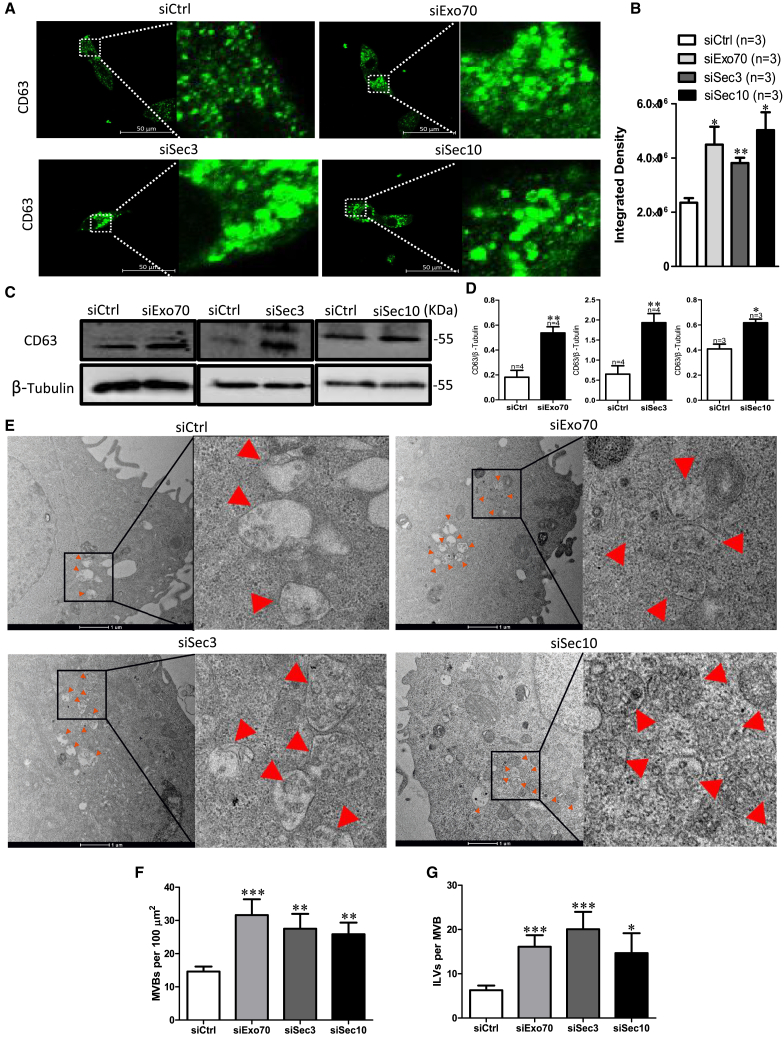
Exocyst depletion induces the accumulation of CD63-positive MVBs
To reveal the mechanism of regulation of exosome biogenesis by exocyst, we analyzed the subcellular expression of CD63, a marker of late endosomes and MVBs (LEs/MVBs) by immunofluorescence staining under exocyst depletion. As shown in Figures 3A and 3B, exocyst depletion resulted in the enlargement of CD63-positive MVBs and the formation of larger and stronger fluorescent spots. The results of western blotting confirmed that CD63 expression was significantly increased in HN4 cells after Sec10, Sec3, or Exo70 knockdown compared with that in cells transfected with control siRNA (Figures 3C and 3D). To gain further insight into the role of the exocyst in exosome biogenesis, we performed electron microscopy to investigate the number and morphology of ILVs and MVBs. In contrast to a significantly decreasing number of extracellular exosomes, the number of MVBs per 100 μm2 and ILVs per MVB markedly increased in HN4 cells with the knockdown of the expression of exocyst subunits compared with those in control cells (Figures 3E–3G). These results suggested that exocyst depletion abated exosome secretion because MVBs could not be trafficked to the plasma membrane for releasing and then accumulating in cells.
Figure 3.
Exocyst depletion induces the accumulation of CD63-positive MVBs
(A–D) CD63 expression in HN4 cells after the cell were transfected with siCtrl or with Exo70, Sec3, or Sec10 siRNA. (A) CD63 immunofluorescence in HN4 cells accumulates after transfection with Exo70 small interfering (si)RNA, Sec3 siRNA, or Sec10 siRNA versus with scrambled siRNA. Scale bar, 50 μm. (B) Quantification analysis of fluorescence intensity of CD63. (C) Representative western blotting images of CD63 expression. (D) Quantification of CD63. β-Tubulin was used for normalization. ∗p < 0.05, ∗∗p < 0.01 versus siCtrl by t test. (E–G) The number of MVBs and ILVs after transfection with scrambled siRNA, Exo70 siRNA, Sec3 siRNA, or Sec10 siRNA. (E) Representative electron microscopy images of HN4 cells after transfection with scrambled siRNA, Exo70 siRNA, Sec3 siRNA, or Sec10 siRNA. Scale bar, 1 μm (F) The number of MVBs per 100 μm2. (G) The number of ILVs per MVB. ∗p < 0.05, ∗∗p < 0.01, ∗∗p < 0.01 versus siCtrl by t test.
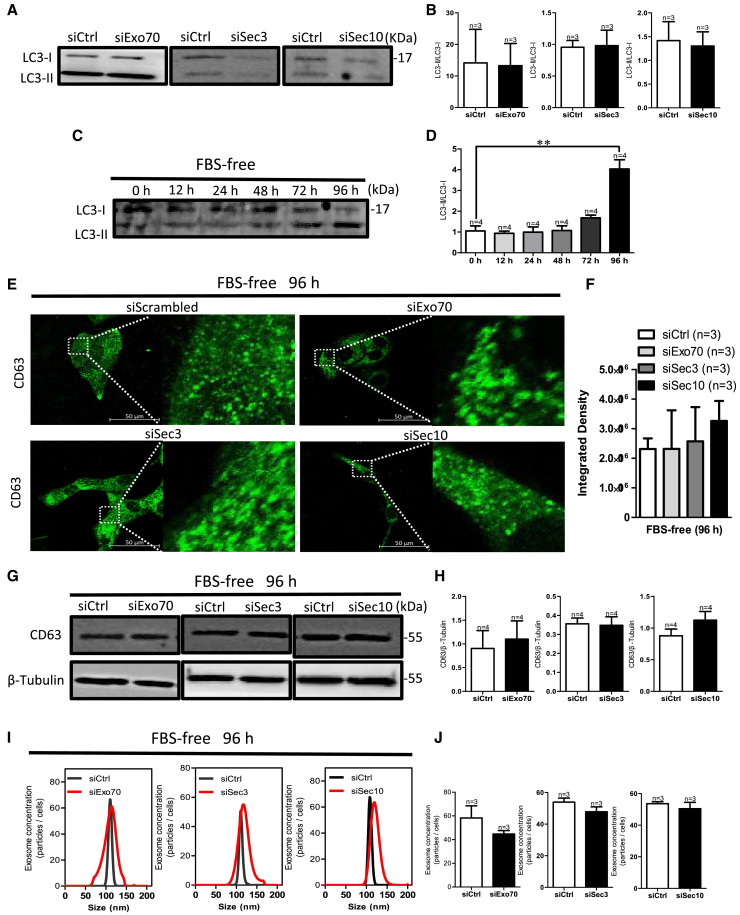
Autophagy activation consumes accumulated ILVs
Autophagy is a key mechanism for exosome degradation.17 To determine whether accumulated MVBs induced by knockdown of exocyst subunits was attributed to a reduction of autophagy, we assessed the expression level of the microtubule-associated protein 1A/1B-light chain 3 (LC3-II/LC3-I) ratio after Sec10, Sec3, or Exo70 knockdown. The results indicated that exocyst depletion did not affect the LC3-II/LC3-I ratio compared with control HN4 cells (Figures 4A and 4B), suggesting that the accumulated MVBs did not activate autophagy after knocking down the exocyst. ILVs may be degraded through fusion of MVBs with autophagosomes.30 Thus, we performed immunofluorescence assays to detect the co-localization of CD63 with the autophagy marker LC3 or Beclin1 or with the lysosomal marker LAMP1 in HN4 cells after transfection with scrambled siRNA or specific Sec10, Sec3, or Exo70 knockdown. The results suggested that CD63 was not co-localized with LC3, Beclin1, or LAMP1 in any of these four groups (Figures S4A–S4R), suggesting that accumulated MVBs cannot co-localize with autophagosomes and lysosomes after knocking down the exocyst. Next, we found that activated autophagy under serum starvation could rescue the enhancement of CD63 expression and accumulation of LE/MVBs induced by exocyst depletion (Figures 4C–4H). To assess whether the accumulation of LE/MVBs in cells was degraded rather than released after autophagy activation, we tested exosome secretion by HN4 cells transfected with scrambled, Sec10 siRNA, Sec3 siRNA, or Exo70 siRNA. As shown in Figures 4I and 4J, exosome concentration did not differ among HN4 cells with knockdown of the expression of any of the exocyst subunits and control cells when autophagy was activated with serum starvation. These results suggested that, although intracellular exosomes, namely ILVs, can be accumulated after knockdown of the exocyst and cannot activate autophagy, autophagy activation consumes accumulated ILVs.
Figure 4.
Autophagy activation consumes accumulated ILVs
(A and B) Representative western blotting images (A) and summary data (B) of LC3-I and LC3-II expression after specific low expression of Exo70, Sec3, or Sec10. (C and D) Representative western blotting images (C) and summary data (D) of LC3-II and LC3-I in HN4 cells when cultured in fetal bovine serum (FBS)-free medium for 0, 12, 24, 48, 72, and 96 h. ∗∗p < 0.01 versus siCtrl by t test. (E–H) CD63 expression in HN4 cells after culture in FBS-free medium for 96 h and transfection with siCtrl or with Exo70 siRNA, Sec3 siRNA, or Sec10 siRNA. (E) CD63 immunofluorescence in HN4 cells. Scale bar, 50 μm. (F) Quantification analysis of fluorescence intensity of CD63. (G) Representative western blotting images of CD63 expression. (H) Quantification of CD63. β-Tubulin was used for normalization. (I and J) Exosome concentrations after HN4 cells were cultured in an FBS-free medium for 96 h and then transfected with scrambled, Exo70 siRNA, Sec3 siRNA, or Sec10 siRNA. (I) Representative nanoparticle tracking analysis traces. (J) The summary data of exosome concentration. Values are shown as the mean ± SD. Statistical analysis was performed using the t test.
Exocyst trafficks MVBs by interacting with Rab11a
To further determine the mechanism of exocyst-mediated exosome biogenesis, we examined known and predicted protein-protein interactions by analyzing data in the Search Tool for the Retrieval of Interacting Genes (STRING) database. The results showed that exocyst complex components interacted with Rab11a (Figure 5A). The results of a co-immunoprecipitation (co-IP) assay provided supporting evidence that the exocyst complex subunits Sec10, Sec3, and Exo70 interact with Rab11a (Figures 5B–5E). In addition, we found that Rab11a was highly expressed in LC tissue and that extracellular exosome secretion was significantly decreased in HN4 cells with knockdown of Rab11a expression (Figures 5F–5I, S2F, and S2H), suggesting that Rab11a can also mediate exosome secretion in HNC cells.
Figure 5.
Exocyst trafficks MVBs by interacting with Rab11a
(A) Exocyst interacts with Rab11a, as assessed using the STRING database. (B–E) Co-immunoprecipitation (co-IP) assay results showed that Sec10, Sec3, Exo70, and Rab11a interacted in HN4 and CNE2 cells. (F and G) Representative images of immunohistochemical staining (brown) (F) and summary data showing Rab11a expression in LC tissue versus adjacent healthy tissue (Ctrl) (G). ∗p < 0.05 versus siCtrl by t test. Scale bar, 50 μm (H and I) Exosome concentrations after the cells were transfected with scrambled or Rab11a siRNA in HN4 cells. (H) Representative nanoparticle tracking analysis traces. (I) The quantification of exosome concentrations in HN4 cells. ∗∗p < 0.01 versus siCtrl by t test. (J) Representative western blotting images show that CD9, CD63, and CD81 each interact with Rab11a, Exo70, Sec3, and Sec10 in HN4 and CNE2 cells. Lys, whole-cell lysates; Pre-immu, preimmune serum. (K–Z) The interactions between CD63 and Exo70, Sec3, Sec10, or Rab11a after siRNA-mediated of Exo70, Sec3, Sec10, or Rab11a knockdown. CD63 was used for normalization. ∗p < 0.05, ∗∗p < 0.01 versus siCtrl by t test.
To explore whether the exocyst mediates the docking of MVBs to the plasma membrane via Rab11a, we used the MVBs markers CD9, CD63, and CD81 to pull down each of the Rab11a and exocyst subunits in co-IP assays. The results demonstrated a physical interaction among MVBs, Rab11a, and exocysts (Figure 5J). With the knockdown of Rab11a expression, the interactions between the MVBs marker CD63 and Sec10, Sec3, or Exo70 were significantly weakened (Figures 5K–5N). After blocking the assembling of the exocyst via knockdown of Sec10 expression, the interactions between CD63 and membrane part units of Sec3 or Exo70 were significantly decreased; however, there was no effect on the interaction between CD63 and Rab11a (Figures 5O–5R). When we separately knocked down the expression of the membrane-targeting exocyst subunits, namely, Sec3 and Exo70, the interactions among CD63, Rab11a, and Sec 10 were unaffected (Figures 5S–5Z). These results suggested that Rab11a and the exocyst bind with MVBs for delivery to the cell membrane and assemble a complete exocyst with Exo70 and Sec3, located in the plasma membrane, which are the necessary steps before fusion of MVBs with the cell membrane and exosome secretion.
Discussion
Tumor cells actively secrete large amounts of exosomes that form important components of the tumor microenvironment and are considered to be the main contributors to tumor progression and metastasis.31, 32, 33, 34, 35 Surgical treatments induce a dramatic reduction in the plasma levels of exosomes expressing CD63 in the early period, which shows that the tumor mass is responsible for the high levels of circulating exosomes detected in patients with cancer.36 In this study, we found that the expression level of exosome-specific markers (CD9, CD63, and CD81) was higher in HNC cells than in adjacent healthy tissue. Therefore, investigating the molecular mechanisms of tumor exosome biogenesis may be of great significance for improving the outcomes of HNC treatment. The exocyst, a conserved octameric complex, localizes to dynamic areas of the plasma membrane where it mediates the delivery of Golgi-derived secretory vesicles through its role in vesicular trafficking, tethering, and fusion, which are similar to the docking and fusion process of MVBs in exosome biogenesis. Studies have also reported on the involvement of exocyst components in exosome biogenesis. Chacon-Heszele et al. reported that the exocyst is involved in urinary exosomes.37 However, how the exocyst modulates exosome biogenesis in cancers, especially in HNCs, is not well understood. Focusing on the exocyst as a participator of exosome biogenesis, we aimed to investigate the mechanism underlying exosome biogenesis in HNC.
Exosomes contain many molecular proteins, including, mRNA, miRNA, lncRNA, and fatty acids.8 Various in vitro assays have demonstrated that cancer-derived exosomes, which act as cancer’s little army, promote tumor malignant biological behavior.38 The stomach adenocarcinoma cell SGC-7901 promoted tumor cell proliferation by exosome secretion in a dose-dependent manner.39 Exosomes secreted by human liver cancer cell HCC-LM3 significantly promoted the proliferation of liver cancer cells.40 Our results suggested that exocyst depletion significantly decreased the proliferation of HN4 cells but had no effect on apoptosis compared with control cells, as measured by assessing proliferating cell nuclear antigen protein expression using western blotting. This inhibition in cell proliferation might have been the result of reducing the amounts of exosomes and changing exosome contents.
A link between autophagy and exosomes has been proposed.41 Autophagy eliminates damaged organelles, aggregated proteins, and invading pathogens, and supplies nutrients during starvation.41 MVBs can fuse with autophagosomes or directly with lysosomes after autophagy activation.42 Fader et al. proposed that autophagy increases the fusion of MVBs with autophagosomes, which directly increases the degradation of MVBs and eventually reduces exosome release.43 Recent studies have suggested that cellular homeostasis decides the degradation or secretion of exosomes.44,45 In our study, we blocked the transport of MVBs and then induced the accumulation of MVBs in the cells without activating autophagy. We then activated autophagy through cell starvation using fetal bovine serum (FBS)-free medium and found that autophagy depleted the accumulated exosomes in HN4 cells. We hypothesized that the cells consumed the accumulated exosomes through activated autophagy. Although knockdown of exocyst expression to cause exosome accumulation was initially insufficient to activate autophagy, Sec10, Sec3, or Exo70 knockdown and their culturing in FBS for a prolonged time, activated autophagy to consume the accumulated exosomes.
The exocyst is regulated by many different small GTPases and has important roles in exocytosis, cytokinesis, cell growth, primary ciliogenesis, cell migration, and tumorigenesis.46 In exocytosis, the exocyst functions to mediate the transport of secretory vesicles to the plasma membrane before SNARE-mediated fusion.46 However, its role in the delivery of MVBs to the plasma membrane has not yet been reported. In this study, we found that the exocyst also participates in exosome biogenesis by mediating the docking of MVBs in the plasma membrane in HN4 cells. To gain insights into how the exocyst transports MVBs, we further investigated previous results indicating that the Rab family promotes vesicle transport and docking as well as the fusion of transport vesicles with membrane.19 Consistent with those results, we found that Rab11a was highly expressed in LC tissue and affected exosome release in HN4 cells. In addition, we found that the exocyst interacted with Rab11a in HN4 cells. There are at least two possible scenarios for this interaction: (1) the exocyst delivers Rab11a to the plasma membrane, or (2) the exocyst delivers MVBs to Rab11a, eventually resulting in the fusion of MVBs with the plasma membrane. To explore these possibilities, we knocked down Rab11a expression and assessed the binding of MVBs to Rab11a or exocyst subunits. The results showed that low Rab11a expression inhibited the interaction of MVBs with the exocyst, whereas decreasing the expression of the exocyst had no effect on the interaction of MVBs and Rab11a. Thus, we concluded that the exocyst was a connector that mediates the role of Rab11a in the transport of MVBs to the membrane, where MVBs eventually fused with the plasma membrane. Moreover, reduced expression of the exocyst subunit Sec10 affected the interaction of MVBs with the exocyst subunits Sec3 and Exo70, which are located in the plasma membrane. Conversely, reducing the expression of the exocyst subunits Sec3 or Exo70 did not affect the interaction of exosomes with exocyst subunit Sec10. These results provided additional evidence supporting the formation of the exocyst in cells, with Sec3 and Exo70 located at the plasma membrane to promote the attachment of the exocyst to the membrane and with Sec10 acting as part of the central hub of the exocyst.41,42 Sec10 knockdown inhibited the delivery of vesicles to the membrane, causing Sec3 and Exo70 on the membrane to have fewer vesicles to bind to. In contrast, Sec3 or Exo70 knockdown did not affect the formation of the exocyst, except with vesicles tethered to the plasma membrane. These findings are consistent with those of earlier reports describing the formation of exocyst.47
Globally, researchers are examining methods to use or control exosome secretion to limit the development of disease. Our results showed that exocysts play an important role in HNC tissues before the occurrence of Rab11a-mediated MVB fusion with the plasma membrane. Thus, inhibiting the expression of exocyst proteins can induce the accumulation of MVBs and limit exosome secretion, which may inhibit the proliferation of cancer cells in patients with HNC. Thus, restricting the biogenesis of tumor cell exosomes that play a pivotal role in tumor angiogenesis and promote growth and metastasis by knockdown of exocyst expression may not only inhibit tumor cell proliferation but also prevent other malignant biological behaviors.
Materials and methods
Cell culture and cell transfection
HN4, an HNC cell line, was derived from patients with HNSCC.48 HN4 cells were cultured in Dulbecco’s modified Eagle’s medium (4.5 g/L glucose) (BI, Israel) supplemented with 10% FBS (VivaCell, Shanghai, China) and antibiotics (100 kU/L penicillin and 100 mg/L streptomycin) (BI, Israel) in an incubator at 37°C with 5% CO2. The human nasopharyngeal carcinoma cell line, CNE2, and the normal NP69 cell lines were obtained from Fenghui Biological (ChangSha, Hunan).49 RPMI 1640 cell culture medium was used, which also contained FBS (VivaCell, Shanghai, China) (10%) and appropriate antibiotics (streptomycin 100 μg/mL and penicillin G 100 U/mL). The culture was maintained in an atmosphere containing 5% CO2 at 37°C. Exosome-free medium (SBI, USA) was prepared for isolation of exosomes from the cell culture supernatant.
HN4 cells were transiently transfected with specific siRNA against human Exo70 (the oligoribonucleotide sequence was: 5′-GGUUAAAGGUGACUGAUUA-3′; for Sec3, 5′-AGAUGAAUACCAAGAGUUA-dTdT-3′; for Sec10, 5′-GCAACAAUGUCAGAAAGAA-dTdT-3′; for Rab11a, 5′-AAGAGUAAUCUCCUGUCUCGA-3′), which were designed and obtained from Biomics using Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific, USA) following the manufacturer’s instructions, and then cultured for another 48 h before the following experiment.
PPI network analysis
The PPI network was predicted using the STRING (http://string-db.org) online database. In this study, the PPI network of Rab11a, EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7, and EXOC8 was constructed using the STRING database, and interaction with a combined score >0.4 was considered statistically significant.
Correlation analysis
The correlation between exocyst complex (EXOC1-8) and exosome-specific markers, CD9, CD63, and CD81, was analyzed using the Head and Neck Cancer Database (HNCDB: http://hncdb.cancerbio.info). The “ANALYSIS” component allows users to conduct correlation analysis using 2,403 samples from 78 HNC gene expression datasets.
Western blotting
For western blotting, cells or exosomes were lysed in RIPA buffer (150 mM NaCl; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% SDS; 50 mM Tris [pH 8.0]; 1× protease inhibitor cocktail [Biosharp]). Protein concentration was measured using a bicinchoninic acid (BCA) kit. Loads normalized to 20 μg protein were resolved to 10% or 15% homemade polyacrylamide gels, transferred onto PVDF membrane, and processed as described elsewhere.50 The following primary antibodies were used: anti-CD9 (1:250, Santa Cruz, USA), anti-CD63 (1:250, Santa Cruz), anti-CD81 (1:250, Santa Cruz), anti-Exo70 (1:200, Affinity Biosciences, USA), anti-Sec3 (1:200, Affinity Biosciences), anti-Sec10 (1:200, Affinity Biosciences), anti-Rab11a (1:200, Affinity Biosciences), and anti-vimentin (1:250, ZENBIO, China). Immunoreactive bands were detected using ECL (Tanon, Shanghai, China) and images were acquired using a chemiluminescence imaging system (P&Q Science & Technology, Shanghai, China).
Co-IP
Co-IP assays were performed using procedures described previously.51 In brief, HN4 cells were lysed with protein lysis buffer (1% Nonidet P-40, 150 mM NaCl, 20 mM Tris-HCl [pH 8.0], with the addition of a protease inhibitor cocktail), sonicated, and centrifuged at 10,000 × g for 20 min at 4°C. CD9, CD63, CD81, Exo70, Sec3, Sec10, or Rab11a proteins were immunoprecipitated by incubating 800 μg extracted proteins with 5 μg anti-CD6, anti-CD63, anti-CD81, anti-Exo70, anti-Sec3, anti-Sec10, or anti-Rab11a antibody, or preimmune IgG, on a rocking platform overnight at 4°C. Protein A agarose (Millipore, Germany) was then added and incubated for an additional 3 h at 4°C. The immunoprecipitates were washed three times with phosphate-buffered saline (PBS). Finally, the samples were tested using by western blotting.
Immunofluorescence
HN4 cells treated with siRNAs were cultured on a coverslip (CITOTEST Scientific, Jiangsu, China) for 24 h and then fixed with 4% paraformaldehyde for 20 min at 25°C. HN4 cells were incubated with 5% BSA (Beyotime Biotechnology, Shanghai, China) and 0.2% Triton X-100 (Biosharp, Hefei, China) in PBS for 30 min at 25°C for blocking unspecific bindings. Then, rabbit anti-Beclin 1, anti-LC3, and anti-LAMP1 and mouse anti-CD63 were incubated in cells overnight at 4°C in a humidified chamber. The cells were incubated with donkey anti-rabbit IgG Alexa Fluor 594 (1:500, Thermo Fisher Scientific) and donkey anti-mice IgG Alexa Fluor 488 (1:500, Thermo Fisher Scientific) for 2 h at room temperature, respectively, after rinsing four times with PBS. Fluorescence images were obtained under a confocal laser-scanning microscope (ZEISS, Germany).
Immunohistochemistry
This study was approved by the Clinical Research Ethics Committee at Anhui Medical University. Each specimen was obtained from a patient who provided written informed consent. The tissues were fixed in 4% paraformaldehyde for 2–3 days, dehydrated, embedded in paraffin, and sliced into 5-μm-thick sections. The sections were then deparaffinized and rehydrated using routine methods. Antigen retrieval was accomplished by heating the sections in citrate buffer solution (0.01 M, pH 6.0) in a microwave oven. Then, an immunohistochemistry kit (ZsBio, Beijing, China) was used to continue the subsequent experiments. The primary antibodies (1:100–1:500) were incubated overnight at 4°C. Brown particles stained in cells were regarded as positive for the presence of antibodies. We used Image-Pro-plus software to analyze the image density.
TUNEL assay
Cell apoptosis was detected using the TUNEL assay. Experiments were performed according to the manufacturer’s protocol. In brief, HN4 cells on the coverslip were fixed with 4% paraformaldehyde solution and then incubated with Triton X-100 (0.1% in PBS) for 5 min at room temperature. Next, the cells were treated with an equilibration buffer for 30 min at room temperature. After washing with PBS three times, the cells were incubated in TdT buffer for another hour at 37°C. Following washing with PBS, HN4 cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology) for 10 min. Finally, the cells were dried and analyzed under a fluorescence microscope. Each cell was visualized by blue fluorescence (DAPI) at a wavelength of 460 nm, and the apoptotic cells were identified by green fluorescence at a wavelength of 520 nm. The percentage of apoptosis was calculated as apoptotic cell number/total cell number × 100%.
Wound healing assay
Cells were seeded at 2.5 × 104 cells per well with transfection-related siRNA. The cells were scratched using a pipette tip and cultured in an FBS-free medium. Images were taken again after 24 h.
CCK-8 assay
The proliferation of HN4 cells was determined using the CCK-8 assay. HN4 cells were trypsinized and seeded in 96-well plates at an equal density of 6 × 103 cells/well after knocking down the expression of Sec10, Sec3, or Exo70. On the day after the treatment, based on the experimental design, 10 μL of CCK-8 was added to each well and incubated for 4 h in an incubator at 37°C with 5% CO2. The absorbance was recorded at a wavelength of 450 nm. Cell viability is proportional to the value of the absorbance value.
Isolation of exosome and cell count
Exosomes were collected from exosome-free medium supplements over a period of 48–72 h. The culture supernatant was centrifuged at 2,000 × g for 30 min at 4°C to remove dead cells and cell debris. Then, we used the Total Exosome Isolation Reagent (Thermo Fisher Scientific) to isolate the exosome for western blotting and used a 0.22-μm PES membrane filter for diameter detection. HN4 cells were digested by 0.25% pancreatin and resuspended in PBS, then tested using an automated cell counter (Invitrogen, USA). The number of exosomes was measured using a BCA protein assay kit (Beyotime Biotechnology) and exosomes (20 μg/mL) were added to the cell culture medium for 48 h.52
Transmission electron microscopy of exosomes
To identify the presence of exosomes, the samples were analyzed using transmission electron microscopy (TEM). In brief, the extracted exosomes samples were dropped onto the plasmonic hydrophilic Formvar/carbon-coated grids for 90 s. Unabsorbed samples were absorbed on filter paper and stained twice with uranyl acetate (UA) solution on the surface of the EM grid. After rinsing, the samples were dyed on mesh copper with 5 μL UA for 90 s. The excess UA solution on the grid was removed. After drying, cells were observed under the FEICM 200 TEM (FEI, USA) at 120 kV, and the images were recorded using an Ouemesa CCD digital camera (Olympus Soft Imaging Solutions).
NanoSight measurements
Analyses were performed by the same operator using NanoSight NS300 (Malvern, UK) obtained from different laboratories. Cell supernatant was collected in 1.5-mL centrifuge tubes and diluted in particle-free PBS to obtain a concentration within the recommended measurement range (1–10 × 108 particles/mL); subsequently, the above diluent was filtered using a 0.22-μm filter (Millipore, Germany). Experimental videos were analyzed using nanoparticle tracking analysis (NTA) 2.3 build 17 software (Malvern) after capturing them in the script control mode (three videos of 30 s per measurement) using a 1-mL injection syringe (Becton Dickinson, USA). A total of 900 frames were examined per sample. The samples were captured and analyzed by applying either an identical or instrument-optimized setting.49
Statistical analysis
Statistical analyses were performed using Prism software (GraphPad) with a t test. Data are presented as means ± SEM.
Acknowledgments
The study was supported by grants from the National Natural Science Foundation of China (grant nos. 81972539, U1732157, 31701162, and 82003048), the Anhui Provincial Natural Science Foundation (1908085QC131), the Doctoral research fund of the first affiliated hospital of Anhui Medical University (no. 1307) and grants for Scientific Research of BSKY (XJ201726) from Anhui Medical University.
Author contributions
J.D. conceived the project and designed and supervised the research. S.B. designed, performed, and analyzed most of the experiments. W.Hou performed and analyzed the NTA data and exosome isolation. Y.Y. performed the TEM experiments. J.M. helped analyze online data and guide the design of some experiments. R.X. and Y.W. gave insightful suggestions on the manuscript. F.H. and X.H. performed some of the IHC staining. N.Z. helped with cell cultures. J.W. and Y.L. provided HNC patient tissues. F.T. and B.W. revised the manuscript. B.S. and H.L. provided suggestions for revised manuscript. Y.D., H.F., and W.H. provided technical guidance during the experiment.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.023.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Koh J., Walsh P., D’Costa I., Bhatti O. Head and neck squamous cell carcinoma survivorship care. Aust. J. Gen. Pract. 2019;48:846–848. doi: 10.31128/AJGP-08-19-5032. [DOI] [PubMed] [Google Scholar]
- 3.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Ang K.K., Harris J., Wheeler R., Weber R., Rosenthal D.I., Nguyen-Tan P.F., Westra W.H., Chung C.H., Jordan R.C., Lu C., et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Gu Y., Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4:e1027472. doi: 10.1080/2162402X.2015.1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J., Wasa M., Chan K.S.L., Shao Q., Yu J.Q. Palladium-catalyzed transformations of alkyl C–H bonds. Chem. Rev. 2017;117:8754–8786. doi: 10.1021/acs.chemrev.6b00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Niel G., D’Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 8.Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 9.Guo W., Gao Y., Li N., Shao F., Wang C., Wang P., Yang Z., Li R., He J. Exosomes: new players in cancer (review) Oncol. Rep. 2017;38:665–675. doi: 10.3892/or.2017.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien K., Rani S., Corcoran C., Wallace R., Hughes L., Friel A.M., McDonnell S., Crown J., Radomski M.W., O'Driscoll L. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur. J. Cancer. 2013;49:1845–1859. doi: 10.1016/j.ejca.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Sharma A., Khatun Z., Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine. 2016;11:421–437. doi: 10.2217/nnm.15.210. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Chen J.Q., Liu J.L., Tian L. Exosomes in tumor microenvironment: novel transporters and biomarkers. J. Transl. Med. 2016;14:297. doi: 10.1186/s12967-016-1056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., Li C., Wang S., Wang Z., Jiang J., Wang W., Li X., Chen J., Liu K., Li C., et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- 14.Xiao M., Zhang J., Chen W., Chen W. M1-like tumor-associated macrophages activated by exosome-transferred THBS1 promote malignant migration in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2018;37:143. doi: 10.1186/s13046-018-0815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui X., Xiao D., Cui Y., Wang X. Exosomes-derived long non-coding RNA HOTAIR reduces laryngeal cancer radiosensitivity by regulating microRNA-454-3p/E2F2 Axis. Onco Targets Ther. 2019;12:10827–10839. doi: 10.2147/OTT.S224881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messenger S.W., Woo S.S., Sun Z., Martin T.F.J. A Ca(2+)-stimulated exosome release pathway in cancer cells is regulated by Munc13-4. J. Cell Biol. 2018;217:2877–2890. doi: 10.1083/jcb.201710132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. Sup pp 11-13. [DOI] [PubMed] [Google Scholar]
- 20.Song L., Tang S., Han X., Jiang Z., Dong L., Liu C., Liang X., Dong J., Qiu C., Wang Y., et al. KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nat. Commun. 2019;10:1639. doi: 10.1038/s41467-019-09720-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.TerBush D.R., Maurice T., Roth D., Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 22.Heider M.R., Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue M., Chang L., Hwang J., Chiang S.H., Saltiel A.R. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Guo W. The exocyst complex in exocytosis and cell migration. Protoplasma. 2012;249:587–597. doi: 10.1007/s00709-011-0330-1. [DOI] [PubMed] [Google Scholar]
- 25.Polgar N., Lee A.J., Lui V.H., Napoli J.A., Fogelgren B. The exocyst gene Sec10 regulates renal epithelial monolayer homeostasis and apoptotic sensitivity. Am. J. Physiol. Cell Physiol. 2015;309:C190–C201. doi: 10.1152/ajpcell.00011.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeffer S.R. Transport vesicle docking: SNAREs and associates. Annu. Rev. Cell Dev. Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- 27.Whyte J.R., Munro S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 28.Wu H., Turner C., Gardner J., Temple B., Brennwald P. The Exo70 subunit of the exocyst is an effector for both Cdc42 and Rho3 function in polarized exocytosis. Mol. Biol. Cell. 2010;21:430–442. doi: 10.1091/mbc.E09-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteside T.L. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Goot F.G., Gruenberg J. Intra-endosomal membrane traffic. Trends Cell Biol. 2006;16:514–521. doi: 10.1016/j.tcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Hood J.L., San R.S., Wickline S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 32.King H.W., Michael M.Z., Gleadle J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C., Yu S., Zinn K., Wang J., Zhang L., Jia Y., Kappes J.C., Barnes S., Kimberly R.P., Grizzle W.E., et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J. Immunol. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 34.Peinado H., Lavotshkin S., Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin. Cancer Biol. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Webber J., Steadman R., Mason M.D., Tabi Z., Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez Zorrilla S., Perez-Sayans M., Fais S., Logozzi M., Gallas Torreira M., Garcia Garcia A. A pilot clinical study on the prognostic relevance of plasmatic exosomes levels in oral squamous cell carcinoma patients. Cancers. 2019;11:429. doi: 10.3390/cancers11030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chacon-Heszele M.F., Choi S.Y., Zuo X., Baek J.I., Ward C., Lipschutz J.H. The exocyst and regulatory GTPases in urinary exosomes. Physiol. Rep. 2014;2:e12116. doi: 10.14814/phy2.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Sowayan B.S., Al-Shareeda A.T., Al-Hujaily E.M. Exosomes, cancer's little army. Stem Cell Investig. 2019;6:9. doi: 10.21037/sci.2019.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C., Liu D.R., Li G.G., Wang H.H., Li X.W., Zhang W., Wu Y.L., Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J. Gastroenterol. 2015;21:6215–6228. doi: 10.3748/wjg.v21.i20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju T., Wang S., Wang J., Yang F., Song Z., Xu H., Chen Y., Zhang J., Wang Z. A study on the effects of tumor-derived exosomes on hepatoma cells and hepatocytes by atomic force microscopy. Anal. Methods. 2020;12:5458–5467. doi: 10.1039/d0ay01730b. [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Yamagata A., Kubota K., Sato Y., Goto-Ito S., Fukai S. Crystal structure of Sec10, a subunit of the exocyst complex. Sci. Rep. 2017;7:40909. doi: 10.1038/srep40909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung K.P., Lau W.C.Y. Isolation of the plant exocyst complex. Methods Mol. Biol. 2017;1662:243–255. doi: 10.1007/978-1-4939-7262-3_22. [DOI] [PubMed] [Google Scholar]
- 43.Fader C.M., Sanchez D., Furlan M., Colombo M.I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 44.Baixauli F., Lopez-Otin C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desdin-Mico G., Mittelbrunn M. Role of exosomes in the protection of cellular homeostasis. Cell Adh. Migr. 2017;11:127–134. doi: 10.1080/19336918.2016.1251000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei K., Guo W. The exocyst complex. Curr. Biol. 2018;28:R922–R925. doi: 10.1016/j.cub.2018.06.042. [DOI] [PubMed] [Google Scholar]
- 47.Boyd C., Hughes T., Pypaert M., Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S.Y., Chu K.C., Lee H.R., Lee K.S., Carey T.E. Establishment and characterization of nine new head and neck cancer cell lines. Acta Otolaryngol. 1997;117:775–784. doi: 10.3109/00016489709113477. [DOI] [PubMed] [Google Scholar]
- 49.Guo Y., Hao Y., Guan G., Ma S., Zhu Z., Guo F., Bai J. Mukonal inhibits cell proliferation, alters mitochondrial membrane potential and induces apoptosis and autophagy in human CNE1 nasopharyngeal carcinoma cells. Med. Sci. Monit. 2019;25:1976–1983. doi: 10.12659/MSM.913915. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Bachurski D., Schuldner M., Nguyen P.H., Malz A., Reiners K.S., Grenzi P.C., Babatz F., Schauss A.C., Hansen H.P., Hallek M. Extracellular vesicle measurements with nanoparticle tracking analysis - an accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles. 2019;8:1596016. doi: 10.1080/20013078.2019.1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du J., Wong W.Y., Sun L., Huang Y., Yao X. Protein kinase G inhibits flow-induced Ca2+ entry into collecting duct cells. J. Am. Soc. Nephrol. 2012;23:1172–1180. doi: 10.1681/ASN.2011100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He L., Zhu W., Chen Q., Yuan Y., Wang Y., Wang J., Wu X. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9:8206–8220. doi: 10.7150/thno.37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.