Abstract
Introduction
Triple-negative breast cancer (TNBC) is considered the most aggressive type of breast cancer (BC) with limited options for therapy. TNBC is a heterogeneous disease and tumors have been classified into TNBC subtypes using gene expression profiling to distinguish basal-like 1, basal-like 2, immunomodulatory, mesenchymal, mesenchymal stem-like, luminal androgen receptor (LAR), and one nonclassifiable group (called unstable).
Objectives
The aim of this study was to verify the clinical relevance of molecular subtyping of TNBCs to improve the individual indication of systemic therapy.
Patients and Methods
Molecular subtyping was performed in 124 (82%) of 152 TNBC tumors that were obtained from a prospective, multicenter cohort including 1,270 histopathologically confirmed invasive, nonmetastatic BCs (NCT 01592825). Treatment was guideline-based. TNBC subtypes were correlated with recurrence-free interval (RFI) and overall survival (OS) after 5 years of observation.
Results
Using PAM50 analysis, 87% of the tumors were typed as basal with an inferior clinical outcome compared to patients with nonbasal tumors. Using the TNBCtype-6 classifier, we identified 23 (15%) of TNBCs as LAR subtype. After standard adjuvant or neoadjuvant chemotherapy, patients with LAR subtype showed the most events for 5-year RFI (66.7 vs. 80.6%) and the poorest probability of 5-year OS (60.0 vs. 84.4%) compared to patients with non-LAR disease (RFI: adjusted hazard ratio [aHR] = 1.87, 95% confidence interval [CI] 0.69–5.05, p = 0.211; OS: aHR = 2.74, 95% CI 1.06–7.10, p = 0.037).
Conclusion
Molecular analysis and subtyping of TNBC may be relevant to identify patients with LAR subtype. These cancers seem to be less sensitive to conventional chemotherapy, and new treatment options, including androgen receptor-blocking agents and immune checkpoint inhibitors, have to be explored.
Keywords: Early breast cancer, Triple-negative breast cancer, Molecular triple-negative breast cancer subtypes, Luminal androgen receptor, Prognosis
Introduction
Triple-negative breast cancer (TNBC) comprises 10–15% of newly diagnosed invasive breast cancers (BCs). TNBC is characterized by lack of expression of estrogen receptor (ER), lack of progesterone receptor (PR), and lack of expression or amplification of the human epidermal growth factor receptor 2 (HER2). The steroid hormone receptor status and HER2 status of tumors are routinely determined at the time of diagnosis by histopathology. Therapeutic decisions are based on status information according to current guidelines. Compared to patients with hormone receptor-positive cancers, TNBC patients are often younger, their tumors are larger in size, and triple-negative cancers are more likely of undifferentiated grading [1].
Both the biology of TNBC and the clinical course of the disease are characterized by a strong heterogeneity [2]. This heterogeneity is probably caused by a rather high rate of somatic mutations and genetic instabilities followed by differential gene expression [3]. Patients with no residual invasive disease in breast and lymph nodes after conventional neoadjuvant chemotherapy (NACT), defined as pathological complete responders, have a high likelihood of cure. In contrast, for those with a generally poor prognosis, TNBC patients without pathological complete response (pCR) can be offered additional postneoadjuvant therapies after surgery, such as capecitabine, but with limited effect [4]. Hence, there is an urgent medical need to identify those TNBC patients who are at very high risk of recurrence and who may benefit from additional therapy [5].
Roughly 75% of TNBCs are classified as basal tumors based on respective gene expression signatures [6, 7]. Most of the remaining TNBC tumors represent a nonbasal intrinsic expression pattern, mostly HER2-enriched, followed by Luminal B and Luminal A [8]. Focusing on TNBC tumors as a clinically heterogeneous group, Lehmann et al. [9] used gene expression profiles from 14 public data sets and identified 18% of cases with TNBCs. Furthermore, the authors defined six stable subtypes, all of which could be identified by specific biological pathways, and one unstable group (UNS), which included all samples that could not be allocated to one of the six stable subtypes [10]. The basal-like 1 (BL1) subtype is marked by genes involved in cell cycle and cell division pathways as well as in DNA damage response. Basal-like 2 (BL2) is associated with growth factor enrichment. These two basal-like subtypes show a high rate of mutations. The mesenchymal (M) subtype is characterized by genes involved in cell motility, extracellular receptor interaction, and cell differentiation. The luminal androgen receptor (LAR) subtype could be identified by genes involved in hormonally regulated pathways, including steroid synthesis as well as androgen and estrogen metabolism. The remaining two subtypes are the mesenchymal stem-like (MSL) subtype that expresses genes involved in cell motility and processes of epithelial to mesenchymal transition, and the immunomodulatory (IM) subtype characterized by immune cell signaling. Both subtypes were excluded from the subtyping TNBCtype-6 classifier, arguing that their signatures may originate from cancer-associated stromal and immune cells (e.g., tumor-infiltrating lymphocytes [TILs]) rather than from cancer cells directly. This led to the condensed TNBCtype-4 classifier with four stable subtypes (BL1, BL2, M, and LAR) [11]. The web-based TNBC subtyping tool (https://cbc.app.vumc.org/tnbc/) can be used for calculation of subtype-specific coefficients and p values which can be used for classification of TNBC samples according to the TNBCtype-6 and TNBCtype-4 classifier [12].
Applying these classifications, TNBC subtypes show different responses to NACT and are associated with different clinical outcomes [11, 13]. The BL1 subtype has the highest pCR rate, BL2 and LAR the lowest [14]. As there are several ways to split TNBC into subgroups, currently molecular approaches may help to overlook the diversity of this heterogeneous disease [15].
We designed a multicenter study (n = 1,270; NCT 01592825) to evaluate the clinical, histopathological, and biological markers of BC in routine clinical care [16, 17, 18]. Here, we focused on patients with TNBC (n = 152, 12% of the total BC cohort) to assess whether molecular subtyping would uncover the heterogeneity of TNBC in our patient cohort as a prerequisite to potentially integrate this information into clinical routine. To this end, we applied recurrence-free interval (RFI) and overall survival (OS) as endpoints and found that the classifier was indeed helpful in subtyping TNBC tumors and identifying patients with LAR tumors at particularly high risk. The ultimate goal of this approach is to hopefully improve the tailoring of treatment.
Subjects and Methods
Study Design, Patients, and Tumors
We designed a prospective and unselected cohort of early BC patients in accordance with the REMARK recommendations for prognostic markers [19]. Between 2009 and 2011, 1,270 patients from five certified breast cancer centers in Germany were enrolled in the PiA study (Prognostic Assessment in Routine Application; NCT 01592825). Patients were consecutively included at the time of diagnosis and received either primary surgery (n = 1,070) or neoadjuvant treatment before surgery (n = 200). All procedures for diagnosis and treatment were applied according to the annually updated German AGO Guideline valid at the respective time (https://www.ago-online.de/leitlinien-empfehlungen/leitlinien-empfehlungen/kommission-mamma).
Patients were recruited if the following inclusion criteria were met: female, invasive, nonmetastasized BC; aged 18 years or older, any tumor size, lymph node involvement, grading or receptor status (ER, PR, HER2) available. Patients with nonbreast second cancers were excluded. Tumors were staged according to the Union for International Cancer Control TNM rules [20] and graded according to Elston and Ellis [21]. TNBC was diagnosed using formalin-fixed, paraffin-embedded (FFPE) tumor material from surgical excision on primary surgery or from core needle biopsies before neoadjuvant treatment. Local pathology results were used. According to respective guideline recommendations, tumors staining for ER and PR protein expression in <1% of tumor cells or having an immune reactivity score <2 [22, 23] were considered hormone receptor-negative. HER2 status was determined applying the DAKO score and, if necessary, by in situ hybridization (ISH) [24]. Applying the recommendations of the International TILs Working Group [25], the tumors were classified using H&E staining according to the percentage of tumor stroma covered by TILs, including the invasive margin (<10%, 10–60%, >60%). TILs outside the tumor border, necrosis, and tertiary lymphoid structures were not considered [25].
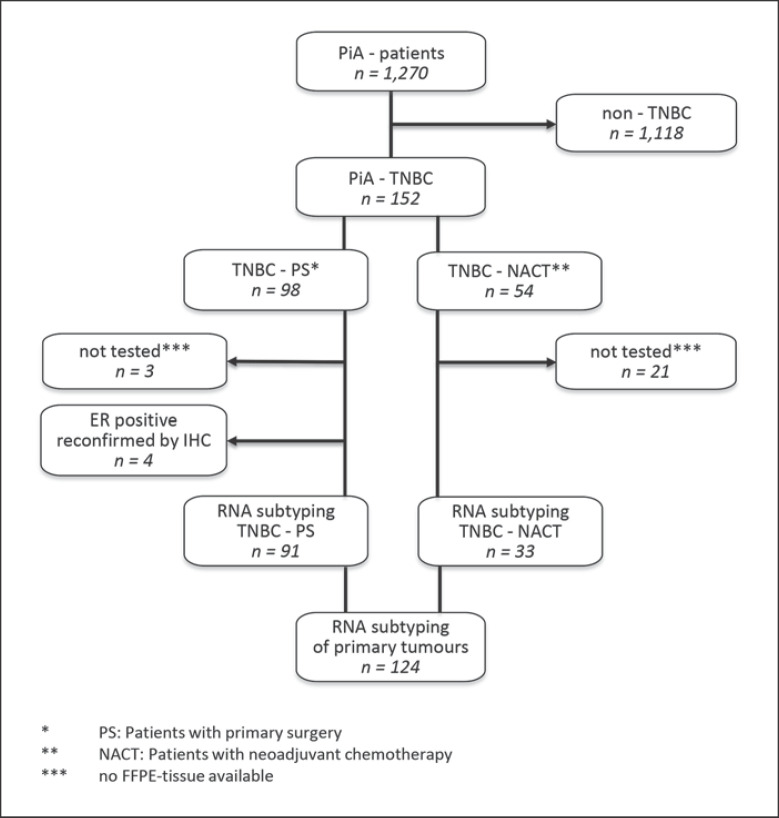
In the total cohort of 1,270 patients, TNBC was diagnosed in 152 patients, and molecular subtyping was performed with 124 tumors (Fig. 1, GEO accession number GSE167213). Due to old age or frailty, 10 TNBC patients (6.6%) did not receive any chemotherapy. These patients were included into the subtyping analysis, but excluded from all survival analyses.
Fig. 1.
Enrolment of TNBC patients (n = 152) of the PiA cohort (n = 1,270) and RNA samples for molecular subtyping (n = 124). TNBC, triple-negative breast cancer.
Following NACT, pCR was defined as the absence of invasive cancer (ypT0/Tis ypN0) [26], and “total pCR” was used for the stricter definition of no cancer cells at all (ypT0 ypN0, total pCR) in breast and axilla at the time of surgery.
Patient and Tumor Characteristics
The median age of patients in the TNBC cohort was 56 years; 37% were younger than 51 years at the time of diagnosis compared to 24% in the non-TNBC group. Seventy percent of the TNBC tumors were larger than 2 cm compared to 47% in non-TNBC cases. Axillary involvement was 49 and 37%, respectively, and tumors were undifferentiated (G3) in 60 and 20%, respectively.
The distribution of patient characteristics, as well as the histopathological parameters of those TNBC tumors that were studied for molecular subtypes, was not statistically different (χ2) from the entire TNBC cohort. With regard to TIL evaluation, the TNBC cohort contained more frequent high infiltration than the non-TNBC cohort [27], and over half of the TNBC tumors (53%) were assigned to the intermediate group with 10–60% TILs, 28% to the low group, and 19% to the high group.
While two-thirds of TNBC patients received primary surgery (n = 98), 54 patients had surgery after NACT, with significant differences for age, tumor size, and nodal status: 28 and 54% were younger than 51 years, respectively; 56 and 94% had tumors larger than 2 cm, respectively; and 41 and 65% were node-positive, respectively (important parameters are summarized in Table 1). Most of the TNBC patients treated with chemotherapy (n = 142) received taxane-anthracycline combination (67%, n = 96), 13% (n = 18) anthracycline-free taxane-based chemotherapy, and 20% (n = 28) anthracycline-based chemotherapy.
Table 1.
Patient characteristics and histopathological parameters of the tumors used for subtyping
| Parameters | TNBC all (n = 152) | TNBC and subtyping (n = 124) | TNBC chemotherapy and subtyping (n = 115) | TNBC primary surgery (n = 98) | TNBC primary surgery and subtyping (n = 91) | TNBC NACT (n = 54) | TNBC NACT and subtyping (n = 33) |
|---|---|---|---|---|---|---|---|
| Age at diagnosis | |||||||
| <35 years | 11 (7%) | 8 (6%) | 8 (7%) | 5 (5%) | 5 (5%) | 6 (11%) | 3 (10%) |
| 35–50 years | 45 (30%) | 35 (28%) | 35 (30%) | 22 (23%) | 20 (22%) | 23 (43%) | 15 (45%) |
| 51–75 years | 74 (49%) | 60 (49%) | 58 (51%) | 50 (51%) | 45 (50%) | 24 (44%) | 15 (45%) |
| >75 years | 22 (14%) | 21 (17%) | 14 (12%) | 21 (21%) | 21 (23%) | 1 (2%) | 0 (0%) |
| p value, χ2 | 0.280a | 0.535a | 0.433b | 0.559b | |||
|
| |||||||
| Tumor histology | |||||||
| Ductal (NST) | 131 (86%) | 106 (86%) | 98 (85%) | 82 (84%) | 75 (83%) | 49 (91%) | 31 (94%) |
| Lobular | 7 (5%) | 4 (3%) | 4 (4%) | 2 (2%) | 2 (2%) | 5 (9%) | 2 (6%) |
| Others | 14 (9%) | 14 (11%) | 13 (11%) | 14 (14%) | 14 (15%) | 0 (0%) | 0 (0%) |
| p value, χ2 | 0.050a | 0.165a | 0.479b | 0.309b | |||
|
| |||||||
| Tumor size at time of diagnosis | |||||||
| <2 cm | 46 (30%) | 41 (33%) | 39 (34%) | 43 (44%) | 38 (42%) | 3 (6%) | 3 (9%) |
| 2–5 cm | 84 (55%) | 68 (55%) | 61 (53%) | 50 (51%) | 48 (53%) | 34 (63%) | 20 (61%) |
| >5 cm | 22 (15%) | 15 (12%) | 15 (13%) | 5 (5%) | 5 (5%) | 17 (31%) | 10 (30%) |
| p value, χ2 | 0.111a | 0.205a | 0.296b | 0.364b | |||
|
| |||||||
| Nodal status at time of diagnosis | |||||||
| Negative | 77 (51%) | 64 (52%) | 58 (51%) | 58 (59%) | 45 (59%) | 19 (35%) | 10 (30%) |
| Positive | 75 (49%) | 60 (48%) | 57 (49%) | 40 (41%) | 37 (41%) | 35 (65%) | 23 (70%) |
| p value, χ2 | 0.620a | 0.923a | 0.909b | 0.346b | |||
|
| |||||||
| Tumor differentiation | |||||||
| G1 | 1 (1%) | 1 (1%) | 1 (1%) | 1 (1%) | 1 (1%) | 0 (0%) | 0 (0%) |
| G2 | 60 (39%) | 50 (40%) | 46 (40%) | 36 (37%) | 32 (35%) | 24 (44%) | 18 (55%) |
| G3 | 91 (60%) | 73 (59%) | 68 (59%) | 61 (62%) | 58 (64%) | 30 (56%) | 15 (45%) |
| p value, χ2 | 0.794a | 0.820a | 0.500b | 0.061b | |||
| Parameters | TNBC all (n = 133) | TNBC and subtyping (n = 124) | TNBC chemotherapy and subtyping (n = 115) | TNBC primary surgery (n = 95) | TNBC primary surgery and subtyping (n = 91) | TNBC NACT (n = 38) | TNBC NACT and subtyping (n = 33) |
|---|---|---|---|---|---|---|---|
| TILs | |||||||
| <10% | 37 (28%) | 34 (28%) | 31 (27%) | 23 (24%) | 21 (23%) | 14 (37%) | 13 (39%) |
| 10–60% | 71 (53%) | 66 (53%) | 60 (52%) | 51 (54%) | 49 (54%) | 20 (53%) | 17 (52%) |
| >60% | 25 (19%) | 24 (19%) | 24 (21%) | 21 (22%) | 21 (23%) | 4 (10%) | 3 (9%) |
| p value, χ2 | 0.812a | 0.312a | 0.353b | 0.608b |
Values are n (%) unless indicated otherwise. NACT, neoadjuvant chemotherapy; TNBC, triple-negative breast cancer.
The p value describes the difference of parameters to all TNBCs (n = 152).
The p value describes the difference of parameters within the groups (n = 91 vs. n = 98 and n = 33 vs. n = 51).
Endpoints and Outcome Assessment
We chose RFI as the first endpoint applying the Standardized Definitions of Efficacy Endpoints criteria [28] which consider local invasive recurrence, distant recurrence, and death from BC as events. The second endpoint was OS (death from BC, non-BC cause, and unknown causes). The median observation time was 63 months (9–88 months). For all survival analyses, only patients treated with chemotherapy were considered (n = 115, Table 1).
Statistical Analysis
Statistical analyses were carried out by using SPSS 25 (IBM, Armonk, NY, USA) including the subtype-specific probability of RFI and OS of the patients. Survival curves were generated as Kaplan-Meier estimates, differences described by log-rank test and Cox regression model were applied to calculate hazard ratios for selected parameters with 95% confidence intervals (CIs), and p values <0.05 were defined as significant. Only cases with information for all covariates (including chemotherapy) were taken into account for all survival analyses.
Microdissection and RNA Extraction
After identification of the tumor areas on H&E-stained slides by the pathologist, 3–5 adjacent unstained tumor slices (10 μm) were processed for RNA isolation using the miRNeasy FFPE kit (Qiagen, Hilden, Germany). The DNA microarray analysis was started with a standard amount of 25 ng RNA.
DNA Microarray Analysis and Gene Expression Profiling
The GeneChip 3′IVT Pico Reagent Kit was used for sample preparation followed by hybridization on HG U133 Plus 2.0 microarrays (both Affymetrix, Thermo Fisher Scientific, Applied Biosystems, formerly known as Affymetrix, Santa Clara, CA, USA) applying standard protocols. Raw data were imported using the simpleaffy R package [29] using the “robust multi-array average” normalization [30]. From this point onwards, all further analyses were conducted with log2 expression values. For probe set annotation the bioconductor package hgu133plus2.db was used [31]. Gene expression was estimated by averaging the expression of all probe sets targeting the same gene. Please note that taking the arithmetic mean of the logarithms is equivalent to taking the geometric mean of the original expression values.
TNBCtype and PAM50 Subtyping
Gene expression values were entered into the TNBCtype online analysis tool [12], and correlation results were used to compute TNBCtype-6 classifications [9]. Considering stromal tissue including TILs [32], we used TNBCtype-6 for the main analyses [9]. Additional analyses were made for TNBCtype-4. To do so, we determined the highest correlation of the subtypes and requested according to the TNBCtype classification (1) a minimum correlation coefficient of 0.1, (2) a p value <0.05, and (3) a difference of at least 0.05 from the best correlation coefficient to the second-best coefficient if this value also conformed to (2). PAM50 intrinsic subtypes [33] were calculated in R. Bias correction for our pure TNBC study cohort was performed according to Zhao et al. [34].
Results
Molecular Subtyping and Clinical and Histopathological Differences in the TNBC Subtypes and PAM50 Intrinsic Types
Microarray analysis was feasible in 84% of the TNBC tumors (128 of 152) (Fig. 1). Molecular subtyping was performed for 97% of the FFPE RNAs from surgical and for 61% from core needle biopsy material. Most failures were caused by missing FFPE blocks or no remaining tumor tissue in the FFPE blocks (mostly affecting the core needle biopsies). Four tumors were reclassified as ER-positive based on the gene expression results and were excluded from further analyses after confirmation by immunohistochemistry (IHC). Finally, 124 tumors (82%) were successfully classified by TNBCtype-6 into BL1 (17%, n = 21), BL2 (13%, n = 16), M (14%, n = 17), MSL (4%, n = 5), IM (22%, n = 27), LAR (15%, n = 18), and UNS (16%, n = 20) (online suppl. Table 2; see www.karger.com/doi/10.1159/000519255 for all online suppl. material). The same tumors were classified according to TNBCtype-4 into BL1 (29%, n = 36), BL2 (20%, n = 25), M (16%, n = 20), LAR (20%, n = 24), and UNS (15%, n = 19) (online suppl. Table 2).
BL1 tumors were significantly less differentiated, and in this group were the youngest patients. Most of the LAR tumors (80%) were significantly better differentiated compared to non-LAR tumors (10–60%) and the intrinsic nonbasal group better than the basal group (73 vs. 36%). Although not significant, there was an enrichment of lymph node metastases in LAR and BL2 patients. As for the evaluation of TILs, the IM group was the subtype showing significantly the highest TIL proportion with >60% TILs. Conversely, LAR was the subtype with the significantly fewest TILs. We further classified the TNBC tumors into the PAM50 intrinsic types, classifying the majority of TNBCs as basal (n = 109 of 124, 88%). This is in accordance with previous reports [11] also reported from our group [35]. The distribution of the parameters in the distinct molecular subtypes and in the intrinsic types is summarized in Table 2, considering patients who underwent chemotherapy.
Table 2.
Patient characteristics and histopathological parameters of the tumors distributed into subtypes by TNBCtype-6 and PAM50 classification (n = 115)
| Parameters | TNBC (IHC) |
TNBC molecular subtypes |
PAM50 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TNBC | BL1 | BL2 | IM | M | MSL | LAR | UNS | basal | nonbasal | |
| chemotherapy & subtyping (n = 115) (100%) | (n = 20) (17%) | (n = 14) (12%) | (n = 27) (24%) | (n = 15) (13%) | (n = 5) (4%) | (n = 15) (13%) | (n = 19) (17%) | (n = 100) (87%) | (n = 15) (13%) | |
| Age at diagnosis | ||||||||||
| ≤50 years | 41 (100%) | 12 (30%)a | 5 (12%) | 9 (22%) | 3 (7%) | 0 (0%) | 5 (12%) | 7 (17%) | 37 (90%) | 4 (10%) |
| >50 years | 74 (100%) | 8 (11%) | 9 (12%) | 18 (24%) | 12 (16%) | 5 (7%) | 10 (14%) | 12 (16%) | 63 (85%) | 11 (15%) |
|
| ||||||||||
| Tumor size at time of diagnosis | ||||||||||
| <2 cm | 39 (100%) | 6 (15%) | 3 (8%) | 10 (26%) | 5 (13%) | 4 (10%) | 6 (15%) | 5 (13%) | 32 (82%) | 7 (18%) |
| >2 cm | 76 (100%) | 14 (19%) | 11 (14%) | 17 (22%) | 10 (13%) | 1 (1%) | 9 (12%) | 14 (19%) | 68 (89%) | 8 (11%) |
|
| ||||||||||
| Nodal status at time of diagnosis | ||||||||||
| Negative | 58 (100%) | 12 (21%) | 6 (10%) | 16 (27%) | 9 (16%) | 5 (9%) | 6 (10%) | 4 (7%) | 50 (86%) | 8 (14%) |
| Positive | 57 (100%) | 8 (14%) | 8 (14%) | 11 (19%) | 6 (11%) | 0 (0%) | 9 (16%) | 15 (26%) | 50 (88%) | 7 (12%) |
|
| ||||||||||
| Tumor differentiation | ||||||||||
| G1, G2 | 47 (100%) | 2 (4%)b | 6 (13%) | 10 (21%) | 7 (15%) | 3 (6%) | 12 (26%)b | 7 (15%) | 36 (77%)b | 11 (23%)b |
| G3 | 68 (100%) | 18 (26%) | 8 (12%) | 17 (25%) | 8 (12%) | 2 (3%) | 3 (4%) | 12 (18%) | 64 (94%) | 4 (6%) |
|
| ||||||||||
| TILs | ||||||||||
| <10% | 31 (100%) | 5 (16%) | 2 (67%) | 1 (3%)c | 9 (29%) | 0 (0%) | 9 (29%)c | 5 (16%) | 23 (74%)c | 8 (26%)c |
| 10–60% | 60 (100%) | 11 (18%) | 12 (20%) | 10 (17%) | 6 (10%) | 5 (8%) | 5 (8%) | 11 (18%) | 54 (90%) | 6 (10%) |
| >60% | 24 (100%) | 4 (17%) | 0 (0%) | 16 (66%) | 0 (0%) | 0 (0%) | 1 (4%) | 3 (13%) | 23 (96%) | 1 (4%) |
Values are n (%). Bold indicates statistical significance. BL1, basal-like 1; BL2, basal-like 2; IHC, immunohistochemistry; IM, immunomodulatory; LAR, luminal androgen receptor; M, mesenchymal; MSL, mesenchymal stem-like; TILs, tumor-infiltrating lymphocytes; TNBC, triple-negative breast cancer; UNS, unstable.
Pearson's χ2 test with Yates continuity correction.
Fisher's exact test (cell frequency <5).
Pearson's χ2 test.
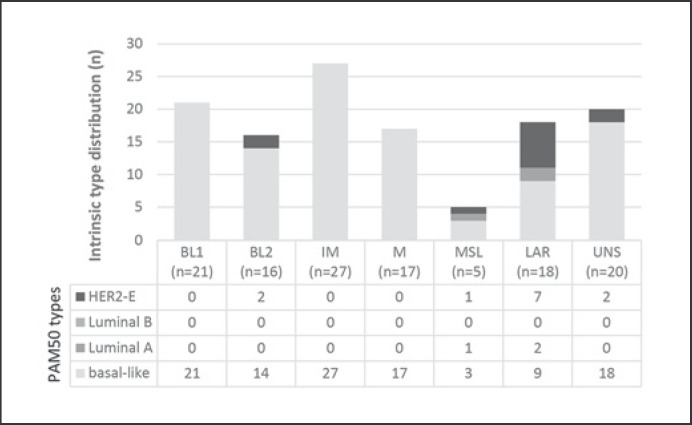
Details of the distribution of the nonbasal subtypes are shown in Figure 2. All BL1, IM, and M subtypes were PAM50 basal; amongst the BL2, MSL, and LAR types a variable proportion of nonbasal tumors was detected. Importantly, most of these nonbasal tumors were significantly well-differentiated, had fewer TILs, and expressed the LAR profile (9 of 15). Two of nonbasal LAR tumors (n = 9) were Luminal A and 7 were HER2-enriched. For 6 of the 7 HER2-enriched tumors, by IHC low HER2 expression (DAKO 1 or 2, chromogenic ISH negative) was reported. In comparison, in non-LAR tumors we found weak HER2 protein expression less frequently (71% in LAR vs. 20% in non-LAR tumors). Neither tumor size nor nodal status were associated with the PAM50 intrinsic types.
Fig. 2.
TNBC subtypes correlated with PAM50 intrinsic molecular types (n = 124). TNBC, triple-negative breast cancer.
Clinical Outcome and LAR Subtype
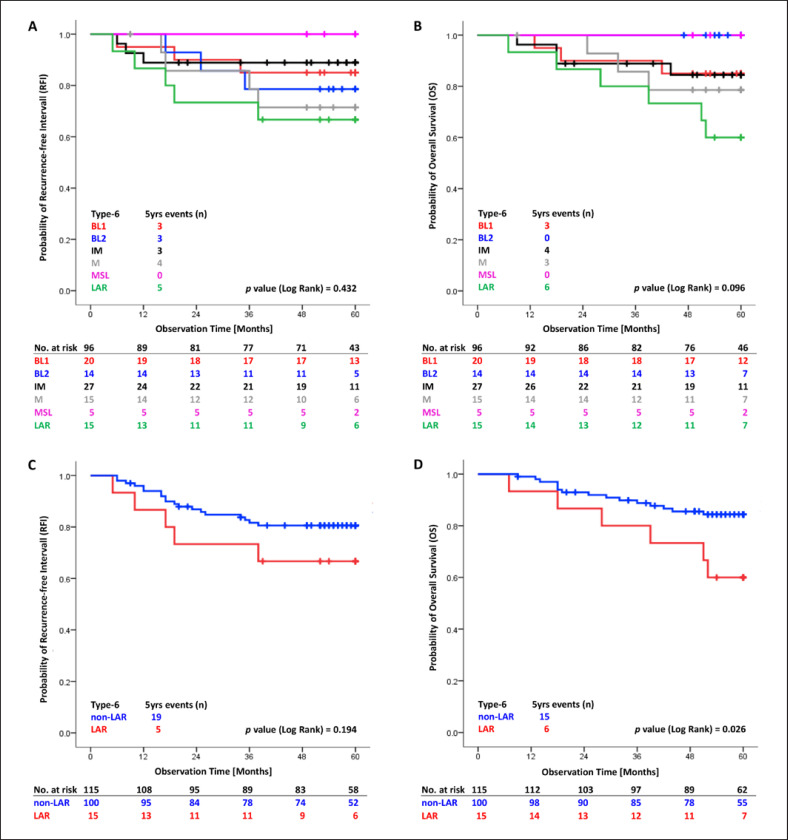
With a median observation time of 61 months, 107 patients of all TNBCs with chemotherapy were recurrence-free (RFI 74.9%, 95% CI 67.6–82.2) and 77.1% were still alive (95% CI 70.0–84.2).
Patients with LAR subtype, although treated with chemotherapy (n = 15), experienced the worst clinical outcome, visualized by Kaplan-Meier plots in Figure 3A and B. Free of an RFI event were 66.7% patients with LAR subtype (95% CI 42.8–90.6) compared to 80.6% (95% CI 72.8–88.4) in the non-LAR TNBC group (n = 100). These results were not significant for RFI but significant for OS (Fig. 3C, D). We showed a nearly doubled risk for RFI of the LAR versus the non-LAR group even if adjusted for tumor size and nodal status (adjusted hazard ratio [aHR] 1.87, 95% CI 0.60–5.05, p = 0.211). The LAR subtype was a significant, independent predictor for OS (aHR 2.74, 95% CI 1.06–7.10, p = 0.037) (Table 3). These subtype results demonstrate a distinct clinical course of patients within the TNBC group. Patients with a LAR nonbasal tumor showed a better outcome than patients with a LAR basal tumor, significant for OS (online suppl. Fig. 1).
Fig. 3.
TNBCtype-6 Kaplan-Meier plots. The tables present the effective sample size for each interval (number at risk). A, B All subdivided stable subtypes RFI (A) and OS (B). C, D Summarized groups LAR versus non-LAR subtypes RFI (C) and OS (D). LAR, luminal androgen receptor; OS, overall survival; RFI, recurrence-free interval.
Table 3.
Multivariate analysis of RFI and OS for TNBC patients with chemotherapy and available TNBCtype-6 subtyping (n = 115)
| Parameters | RFI (24 events) |
OS (21 events) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| univariate analysis |
multivariate analysis |
univa |
riate analysis |
multivariate analysis |
|||||||||||
| HR | 95% CI | p value | HR | 95% CI | p value | HR | 95% CI | p value | HR | 95% CI | p value | ||||
| Age at diagnosis ≤50 years 51–75 years >75 years | 1.24 ref. 1.16 |
0.53–2.93 0.32–4.15 | 0.618 0.082 | 1.22 3.48 | 0.44–3.36 1.20–10.04 | 0.705 0.021 | |||||||||
|
| |||||||||||||||
| Tumor size at time of diagnosis <2 cm ≥2 cm | ref. 4.03 | 1.20–13.50 | 0.024 | 3.30 | 0.95–11.41 | 0.059 | 5.46 | 1.27–23.44 | 0.022 | 3.95 | 0.89–17.54 | 0.071 | |||
|
| |||||||||||||||
|
Nodal status at time of diagnosis Negative Positive |
ref. 2.74 | 1.13–6.60 | 0.025 | 2.05 | 0.83–5.08 | 0.119 | 4.73 | 1.59–14.07 | 0.005 | 3.32 | 1.08–10.14 | 0.035 | |||
|
| |||||||||||||||
|
Tumor differentiation G1, G2 G3 |
ref 0.49 | 0.22–1.09 | 0.082 | 0.63 | 0.27–1.49 | 0.293 | |||||||||
|
| |||||||||||||||
|
TNBCtype-6 Non-LAR LAR |
ref. 1.89 | 0.70–5.07 | 0.204 | 1.87 | 0.69–5.05 | 0.211 | 2.79 | 1.08–7.20 | 0.033 | 2.74 | 1.06–7.10 | 0.037 | |||
CI, confidence interval; HR, hazard ratio; LAR, luminal androgen receptor; OS, overall survival; ref., reference group; RFI, recurrence-free interval; TNBC, triple-negative breast cancer.
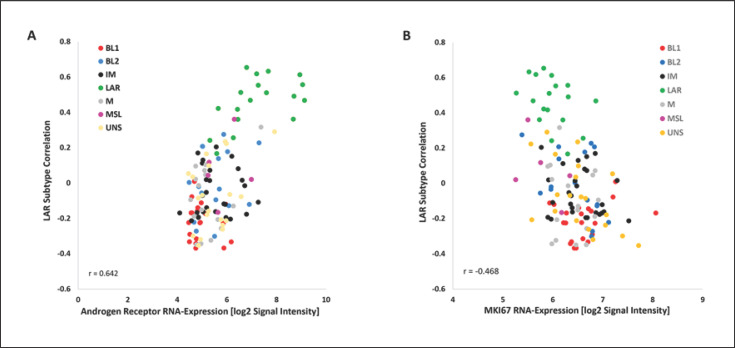
Androgen receptor mRNA levels were correlated to subtypes with the highest expression in LAR subtype [36] and associated with low MKI67 transcript levels (Fig. 4A, B).
Fig. 4.
Correlation of androgen receptor (A) and MKI67 (Ki-67) (B) RNA expression to LAR subtype (n = 124). LAR, luminal androgen receptor.
pCR and Subtypes
The pCR rate (ypT0/Tis ypN0) was 48% (26 of 54 patients) (online suppl. Table 1); 80.0% (95% CI 64.3–95.7) of them had no disease-specific event. Due to the small number of core needle biopsies with subtyping results before NACT (n = 33), no significant association could be found between TNBC subtypes and pCR. We emphasize that the best chemosensitivity as demonstrated by no residual invasive tumor tissue in the breast and axilla was observed in the IM subtype (4 of 6). No residual disease after NACT was diagnosed in 45% (13 of 29) of the patients with non-LAR subtype and in only 25% (1 of 4) of the patients with LAR subtype.
Discussion
In this study, we evaluated the distribution of TNBCtype-6 subtypes according to Lehmann et al. [9], also displaying the heterogeneity of the outcome (RFI, OS) of the TNBC patients.
Firstly, we successfully classified 84% of triple-negative tumors of an unselected clinical cohort (n = 124 of 152 TNBCs, total BC cohort n = 1,270). The array experiments and the subsequent subtyping was performed with each prepared RNA. None of the immunohistochemically detected TNBC tumors were classified as mRNA-ER-positive, as reported elsewhere [37], indicating high quality of the IHC/chromogenic ISH tests performed in the respective peripheral pathology centers. The reliable group segregation might be a consequence of the application of a lower cutoff for hormone receptor negativity (<1% stained cancer cells, immune reactivity score <2). The four reconfirmed ER-positive samples (Fig. 1) were excluded from all subtyping and survival analyses.
Secondly, using both TNBC classifiers (TNBCtype-6, TNBCtype-4), the subtype distribution was very similar to reports by other investigators [9, 38]. The IM subtype was most frequent (26% by excluding the UNS subtype; online suppl. Table 2). These tumors are enriched in genes linked to immune processes, also seen in our cohort with 57% with a high TIL score (16 of 28 of the IM samples; online suppl. Fig. 2A). Applying the condensed TNBCtype-4 classifier (online suppl. Fig. 2B), 66% of the IM samples (by TNBCtype-6) switched to the BL1 group (online suppl. Fig. 2C), suggesting a more favorable prognosis. Excluding the unclassifiable tumors by definition (see Subjects and Methods), we found that by TNBCtype-6 analysis more than one-third were BL1 and BL2 (combined 35%), 16% M, 17% LAR, and MSL was underrepresented (5%) (online suppl. Table 2), which is in line with other studies [38, 39].
Thirdly, each TNBC subtype showed a distinct clinical outcome for RFI and OS. As mentioned before, 15% of the TNBC samples of our study expressed the LAR profile by the TNBCtype-6 and 20% by the TNBCtype-4 classifier, representing 2% of all BCs in our cohort, as also shown by other studies [38, 39, 40]. Compared to the non-LAR group, patients with LAR subtype suffered the highest number of events with an adjusted 1.87-fold trend (aHR) for local recurrence, distant metastasis, and death caused by BC. We further emphasize a significant aHR of 2.74 of death for the LAR group compared to the non-LAR group classified by TNBCtype-6. Applying the TNBCtype-4 classifier we determined an aHR of 1.74 for RFI and a significant aHR of 2.62 for OS for the LAR subtype (online suppl. Table 3 and Fig. 3).
Fourthly, as expected, the PAM50 classification predominantly clustered intrinsic basal type (88%), followed by HER2-enriched (10%) and Luminal A (2%) (for a review, see Lehmann and Pietenpol [10]). Focusing on nonbasal tumors (n = 15), 9 of them belonged to the LAR subtype, which is in line with published data, e.g., Masuda et al. [13]. Most of them (78%, 7 of 9) showed HER2-enriched profiling (Fig. 2) as also reported by Prat et al. [37] (20 of 28, 71%). For clinical practice, it could be of relevance that immunohistochemically clinical HER2-negative (DAKO 1 or 2, chromogenic ISH negative) tumors might still be sensitive to anti-HER2 therapy. These special TNBC groups can be identified by TNBC subtyping and additional PAM50 intrinsic classification using the same RNA expression data of the tumor.
Combining molecular TNBC subtyping and PAM50 classification, we identified six of 15 LAR tumors as LAR-basal and the patients as the group with most events for RFI or OS (50%, 95% CI 10–90, 3 events and 33.3%, 95% CI <1–70.9, 4 events, respectively). These patients have a three times (3.28) higher risk of inferior RFI (p = 0.172) and a 5.39 times higher risk of inferior OS (p = 0.034).
As previously reported by other authors, LAR tumors are characterized by low MKI67 mRNA expression (Fig. 4B) and intermediate grading as an indicator for low proliferation and pronounced chemoresistance [41]. Also, Witzel et al. [42] figured out that low chemotherapy response can be predicted by high androgen receptor mRNA expression that is overexpressed in the LAR subtype (Fig. 4A). In a phase I/II study, metastasized patients with a positive androgen receptor status at mRNA level benefited from anti-androgen receptor-hormonal therapy, so that may be an option to bypass chemoresistance and to de-escalate chemotherapy [36]. Considering the unfavorable prognosis of patients with LAR subtype, a combination of immune checkpoint inhibitors and androgen receptor-inhibiting treatment should also be studied [43].
Last, residual disease after NACT predicts inferior outcome [44]. Also, patients in our study without pCR suffered from worse RFI or OS compared to patients with pCR.
Even taking into account the limitations of low case numbers with subtyping data, a better pCR rate can be predicted for the non-LAR group than for the LAR group. Hence, patients with LAR subtype and residual disease showed the worst outcome for RFI and OS.
Some notable strengths of our study should be mentioned. In comparison to other studies, we analyzed a clinical cohort of patients rather than data from a web-based data repository. TNBC patients were consecutively and routinely enrolled in our prospective, unselected multicenter cohort evaluating prognostic factors safeguarding a well-documented follow-up. Our TNBC group showed characteristics representative of a typical TNBC population with respect to age, tumor size, nodal status, and grading [45]. For all survival analyses, we exclusively included patients treated with chemotherapy.
There are some limitations. Patients were treated at the discretion of the local tumor boards of the participating certified breast centers, but decisions were based on current national guideline recommendations. There is only a small sample size of patients treated with NACT, so no meaningful response and survival analyses for this subgroup could be performed. Due to the retrospective analysis of TNBC subtyping, only 82% of the FFPE blocks for RNA preparation were still available for this retrospective analysis.
Conclusion
TNBC subtyping using RNA profiling data that were generated in a clinical routine setting is feasible and differentiates TNBC patients into groups with distinct clinical outcomes. We found a clear trend for the LAR subtype as a predictor for worse RFI and OS. Since these cancers appear to be less sensitive to standard adjuvant chemotherapy new treatment options, androgen receptor-blocking agents and immune checkpoint inhibitors have to be explored.
Outlook
TNBC remains the challenge of BC treatment. There is further merit in prospective validation of pCR rates amongst TNBC subtypes and correlation with clinical outcomes of these neoadjuvantly treated patients. It appears clinically relevant to identify patients with the LAR subtype, since these cancers are less sensitive to chemotherapy and require other options.
Statement of Ethics
The study complied with the guidelines for human studies and was conducted in accordance with the World Medical Association Declaration of Helsinki. The study protocol was approved by the Ethical Board of the Medical Faculty of Martin Luther University Halle-Wittenberg (#214/16.12.09/11). All patients provided written informed consent.
Conflict of Interest Statement
C. Hartung, M. Porsch, K. Stückrath, S. Kaufhold, M.S. Staege, V. Hanf, S. Peschel, J. John, M. Pöhler, E. Weigert, J. Buchmann, K.-F. Bürrig, K. Schüler, D. Bethmann, I. Große, and E.J. Kantelhardt have no conflicts of interest to declare. T. Lantzsch reports personal honoraria from Pfizer, Astra Zeneca, Lilly, Roche, and Novartis, all outside the submitted work. C. Uleer reports personal honoraria from Astra Zeneca, Novartis, Onkowis, Pierre Fabre, Roche, and Seagen, all outside the submitted work. C. Thomssen reports personal honoraria from Amgen, Astra Zeneca, Celgene, Daiichi-Sankyo, Eisai, Lilly, MEDA-Pharma, MSD, Novartis, Pfizer, Pierre-Fabre, Roche, and Vifor, all outside the submitted work. M. Vetter got travel support from NanoString.
Funding Sources
This work was supported by the Wilhelm Roux Program of the Medical Faculty, Martin Luther University Halle-Wittenberg (FKZ 25/36), and the German Federal Ministry of Education and Research (e:Med FKZ: 031A429).
Author Contributions
E.J. Kantelhardt and M. Vetter designed the PiA study. C. Hartung, C. Thomssen, and M. Vetter conceived the TNBC analysis. C. Hartung and M. Vetter wrote the manuscript. V. Hanf, T. Lantzsch, C. Uleer, S. Peschel, J. John, M. Pöhler, E. Weigert, J. Buchmann, and K.-F. Bürrig contributed substantial input to the conception and acquisition of the work. K. Stückrath and S. Kaufhold carried out the DNA microarray experiments. M. Porsch, M.S. Staege, and I. Große performed the bioinformatics analyses of the DNA microarray data. D. Bethmann and K. Schüler defined and analyzed the TIL proportion. E.J. Kantelhardt was responsible for statistical evaluation. M. Vetter, C. Hartung, and C. Thomssen performed data interpretation. All authors revised the manuscript critically.
Supplementary Material
Supplementary data
Acknowledgements
We thank all patients participating the PiA study as well as the study nurses Katrin Losse (St. Elisabeth and St. Barbara Halle), Birgit Mauder (Fürth), and Andrea Meyer-Kühn (Hildesheim) for excellent documentation. We thank the core facility Cellular Analysis of our medical faculty, specifically Vesselin Christov providing the Affymetrix chip platform.
References
- 1.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–87. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–72. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 5.Rida P, Ogden A, Ellis IO, Varga Z, Wolff AC, Traina TA, et al. First international TNBC conference meeting report. Breast Cancer Res Treat. 2018;169:407–12. doi: 10.1007/s10549-018-4692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–50. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: a subtyping tool for triple-negative breast cancer. Cancer Inform. 2012;11:147–56. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19:5533–40. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann BD, Pietenpol JA. Clinical implications of molecular heterogeneity in triple negative breast cancer. Breast. 2015;24((Suppl 2)):S36–40. doi: 10.1016/j.breast.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrera Juarez M, Tolosa Ortega P, Sanchez de Torre A, Ciruelos Gil E. Biology of the triple-negative breast cancer: immunohistochemical, RNA, and DNA features. Breast Care (Basel) 2020;15:208–16. doi: 10.1159/000508758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernhardt S, Bayerlová M, Vetter M, Wachter A, Mitra D, Hanf V, et al. Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors. Breast Cancer Res. 2017;19:112. doi: 10.1186/s13058-017-0905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breunig C, Erdem N, Bott A, Greiwe JF, Reinz E, Bernhardt S, et al. TGFβ1 regulates HGF-induced cell migration and hepatocyte growth factor receptor MET expression via C-ets-1 and miR-128-3p in basal-like breast cancer. Mol Oncol. 2018;12:1447–63. doi: 10.1002/1878-0261.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer M, Kantelhardt EJ, Stiewe T, Nist A, Mernberger M, Politt K, et al. Specific allelic variants of SNPs in the MDM2 and MDMX genes are associated with earlier tumor onset and progression in Caucasian breast cancer patients. Oncotarget. 2019;10:19751992. doi: 10.18632/oncotarget.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Eur J Cancer. 2005;41:1690–6. doi: 10.1016/j.ejca.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 20.Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours. 7th ed. Somerset: Wiley; 2011. [Google Scholar]
- 21.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19((5)):403–10. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 22.Remmele W, Stegner HE. Vorschlag zur einheitlichen Definition eines immunreaktiven Score (IRS) für den immunhistochemischen Östrogenrezeptor-Nachweis (ER-ICA) im Mammakarzinomgewebe. Pathologe. 1987;8:138–40. [PubMed] [Google Scholar]
- 23.Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thürlimann B, Senn HJ. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20:1319–29. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 25.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. International TILs Working Group 2014. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–71. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 27.Kuhlmann JD, Bachmann HS, Link T, Wimberger P, Kröber E, Thomssen C, et al. Association of caspase 8 polymorphisms −652 6N InsDel and Asp302His with progression-free survival and tumor infiltrating lymphocytes in early breast cancer. Sci Rep. 2019;9:12594. doi: 10.1038/s41598-019-47601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25:2127–32. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 29.Miller CJ. simpleaffy: very simple high level analysis of Affymetrix data. 2007. [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Carlson M. hgu133plus2.db: Affymetrix Human Genome U133 Plus 2.0 Array annotation data (chip hgu133plus2) 2016 [Google Scholar]
- 32.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–98. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X, Rødland EA, Tibshirani R, Plevritis S. Molecular subtyping for clinically defined breast cancer subgroups. Breast Cancer Res. 2015;17:29. doi: 10.1186/s13058-015-0520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berchtold E, Vetter M, Gündert M, Csaba G, Fathke C, Ulbrich SE, et al. Comparison of six breast cancer classifiers using qPCR. Bioinformatics. 2019;35:3412–20. doi: 10.1093/bioinformatics/btz103. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann BD, Abramson VG, Sanders ME, Mayer EL, Haddad TC, Nanda R, et al. TBCRC 032 IB/II multicenter study: molecular insights to AR antagonist and PI3K inhibitor efficacy in patients with AR+ metastatic triple-negative breast cancer. Clin Cancer Res. 2020;26:2111–23. doi: 10.1158/1078-0432.CCR-19-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prat A, Fan C, Fernández A, Hoadley KA, Martinello R, Vidal M, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015;13:303. doi: 10.1186/s12916-015-0540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santonja A, Sánchez-Muñoz A, Lluch A, Chica-Parrado MR, Albanell J, Chacón JI, et al. Triple negative breast cancer subtypes and pathologic complete response rate to neoadjuvant chemotherapy. Oncotarget. 2018;9:26406–16. doi: 10.18632/oncotarget.25413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Echavarria I, López-Tarruella S, Picornell A, García-Saenz JÁ, Jerez Y, Hoadley K, et al. Pathological response in a triple-negative breast cancer cohort treated with neoadjuvant carboplatin and docetaxel according to Lehmann's refined classification. Clin Cancer Res. 2018;24:1845–52. doi: 10.1158/1078-0432.CCR-17-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer − deciphering the heterogeneity. Clin Cancer Res. 2014;20:782–90. doi: 10.1158/1078-0432.CCR-13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bareche Y, Venet D, Ignatiadis M, Aftimos P, Piccart M, Rothe F, et al. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol. 2018;29:895–902. doi: 10.1093/annonc/mdy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witzel I, Loibl S, Wirtz R, Fasching PA, Denkert C, Weber K, et al. Androgen receptor expression and response to chemotherapy in breast cancer patients treated in the neoadjuvant TECHNO and PREPARE trial. Br J Cancer. 2019;121:1009–15. doi: 10.1038/s41416-019-0630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Y, Lee JS, Yost SE, Frankel PH, Ruel C, Egelston CA, et al. A phase II clinical trial of pembrolizumab and enobosarm in patients with androgen receptor-positive metastatic triple-negative breast cancer. Oncologist. 2021;26((2)):99–e217. doi: 10.1002/onco.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC, et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161:279–87. doi: 10.1007/s10549-016-4059-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data