Abstract
Background/purpose
Genetics plays a role in the susceptibility to periodontitis and tooth loss. Several studies examined the involvement of polymorphisms in candidate genes. We hypothesize that bone metabolism-related polymorphisms could be associated with the number of remaining teeth.
Materials and methods
Participants in the Pro.V.A. longitudinal Study: 3099 Italians (aged 65+ at baseline), 2196 at follow-up 1 (5yrs), 1641 at follow-up 2 (7yrs) underwent detailed interview and clinical-instrumental examination. Subjects, grouped by remaining teeth number (0, 1–7, 8–19, 20+), were genotyped for six different bone-related polymorphisms: collagen type Iα1 (COL1A1, Sp1, Ss alleles, n = 1068), vitamin D receptor (VDR, Fok I, Ff alleles, n = 300), calcitonin receptor (CALCR, Alu I, CT alleles, n = 1430), estrogen receptor alpha (ESR1, Pvu II and Xba I, Pp and Xx alleles, n = 1335 and n = 1324).
Results
COL1A1 associated with dental status: ss carriers had reduced incident tooth loss (p < 0.05). The low frequency of this genotype, 3.6% in the whole population, didn't grant sufficient statistical power to other findings, such as the lower prevalence of edentulism, consistent throughout the study. In men, CC genotype of CALCR was associated with higher tooth loss between follow ups (p < 0.05). Biochemical markers of inflammation didn't differ by genotype. Confounders such as diabetes, neoplasms, and smoking hampered the detrimental effect of S allele in the logistic regression analysis (OR = 0.67, 95% CI 0.4–1.0, p = 0.06).
Conclusion
The present study, demonstrating an association between tooth loss and COL1A1 and -in men- CALCR, contributes to the identification of genes involved in tooth loss and, possibly, susceptibility to periodontitis.
Keywords: Bone, Genetic polymorphisms, Oral health, Tooth loss, Type I collagen
Introduction
Edentulism is a multifactorial condition whose prevalence among the elderly of western countries ranges from less than 20 to 60%.1, 2, 3 Many studies have shown that the main cause of tooth loss in advanced age is chronic periodontitis, an inflammatory disease of periodontal tissues characterized by gingival swelling, loss of alveolar bone, and tooth movement.4, 5, 6, 7, 8 While gingivitis is generally reversible, periodontitis destroys the periodontium, thus determining loss of tooth attachment.9 The etiology of periodontitis is complex and related to multiple environmental and clinical risk factors as well as to individual differences in the structure of periodontal tissues. It has been advanced that genetics might play a prominent role in tooth loss and susceptibility to periodontal disease, but the disorder is likely polygenic and the identification of genetic risk factors is further complicated by its possible multiple --chronic and aggressive— presentation, while the difference in the number of remaining teeth among individuals may reflect genetic variation within the population.10, 11, 12 Several studies have examined the association between polymorphisms in candidate genes and periodontitis. Since Gram negative anaerobic bacteria can induce the loss of attachment observed in periodontitis, candidate genes have been initially selected among those codifying for inflammatory mediators extending thereafter to various others, involved in infection susceptibility and association with chronic diseases, with conflictual results.13, 14, 15, 16, 17, 18 Several aspects of the relationship between oral indicators and bone mineral density have also been investigated.19,20 As the rate of local bone loss and structural factors of periodontal tissues may influence also the rate of tooth loss, the hypothesis that polymorphisms in genes of the host bone metabolism could be associated with the number of remaining teeth and with incident tooth loss relies on a robust rationale. We went on investigating this topic and the findings are reported in the present study. We used the number of remaining teeth and the loss of teeth over two follow ups of 5 and 7 years as an index of periodontal disease susceptibility, in an elderly Italian population that was genotyped for a panel of polymorphisms described in bone metabolism-associated genes, namely collagen 1 type I (COL1A1), vitamin D (VDR), calcitonin receptor (CALCR), and estrogen receptor α (ESR1).
Materials and methods
Study population
The Pro.V.A. (Progetto Veneto Anziani) Study is a large observational community-based cohort survey on 3099 subjects aged 65 and older (1245 men and 1854 women), residing in the Veneto Region (Rovigo and Camposampiero areas, in Northeast Italy), designed to assess the functional status and the prevalence of disability in a representative sample of the population.21 The cross-sectional phase began in 1995 and ended in 1998, while the longitudinal one consisted of two follow ups at 5 (FU1) and 7 years (FU2), respectively. The Pro.V.A. Study design and protocol were approved by the two Institutional Review Boards of the Veneto Region Health Care Agencies. All the participants signed an informed written consent form, were interviewed at their homes, and examined by specially trained physicians and nurses through a protocol consisting of an extensive battery of clinical, instrumental, biochemical tests at the clinic facilities provided for the study's implementation.
Oral examination
During the physical work up at the study center, trained clinicians performed an oral examination to assess each patient's general oral status and number of teeth. Examinations were performed in a well-calibrated manner, using a standard dental mirror under a sharp artificial lighting. All teeth whether sound, decayed, or treated with any kind of restoration, were considered when the total number of remaining teeth was being calculated. The presence of an upper and/or lower jaw prosthesis also was recorded. Data about oral conditions were obtained from 3083 subjects (1237 M, 1846 F) at baseline, from 2196 survivors (805 M, 1391 F) at the first FU examination, and from 1640 (576 M, 1064 F) at the second one.
Laboratory data
Serum inflammation indexes: white blood cells (WBC), platelet count, fibrinogen, erythrocyte sedimentation rate (ESR) were determined during routinary procedures at the study centers. Serum 25-OH calciferol (vit.D) levels were measured at the central University facilities by radioimmunoassay (DiaSorin, Saluggia, Italy). The intra-assay and inter-assay coefficients of variations (CV) for vit.D were 8.1% and 10.2%, respectively.
Genotype analysis
Genomic DNA was extracted from peripheral blood of 1443 participants (582 M; 861 F) using the salting out procedure described by Miller.22 DNA was amplified in a programmable thermal cycler (MJ Research Inc, Waltham, MA, USA) using specific primers and programs for each gene analyzed, as specified below. All of the amplifications were performed in a final volume of 50 μL, using 0.2–0.4 μg of genomic DNA. All programs included an initial denaturation at 94 °C for 3 min prior to the first cycle, and a final extension step of 7 min at 72 °C. For the analyses of RFLPs (restriction fragment length polymorphisms) PCR (polymerase chain reaction) products were digested with gene-specific restriction enzymes. All digestions were followed by electrophoresis in a 1.5–3% agarose gel containing ethidium bromide. The primers used for amplification were obtained from Primm (Primm Biotech, Napoli, Italy), Taq DNA polymerase, restriction enzymes and other molecular biology reagents were from Celbio (Celbio-Euroclone, Pero, Italy) while chemicals were from Sigma (Sigma–Aldrich, Milano, Italy), unless otherwise specified. Disposable materials were from Corning (Corning GmbH, Wiesbaden, Germany).
COL1A1
Type I collagen, the major protein of bone, consists of a triple chain made up of two alpha 1 chains and one alpha 2 chain encoded respectively by COL1A1 and COL1A2 genes. A single nucleotide polymorphism characterized by a G→T transition in the binding site for the transcription factor Sp1 in the COL1A1 (S/s allelic variants) has been described and demonstrated to be related with susceptibility to osteoporotic fractures. Allelic variants for COL1A1-Sp binding site were detected by mean of ARMS-PCR (amplification refractory mutation system–polymerase chain reaction), cleaving the product with Bal I/Mlu NI as described by Grant.23 COL1A1 genotyping was completed in 1068 subjects.
VDR
The vitamin D-derived steroid hormone 1,25 (OH)2vitD plays a major role in bone metabolism. Its receptor (VDR) has been described in a variety of cell types, thus broadening the sphere of influence of the hormone from mineral metabolism and calcium homeostasis to the immune system and host response. It has been also found, and shown to have regulatory activity, in the periodontal ligament.24 Several single nucleotide polymorphisms (SNPs), variously associated with bone loss/risk of fractures and periodontal disease, have been described within this gene.25,26 VDR analysis was performed as described by Gross, digesting the PCR product with Fok I (F/f allelic variants).27 VDR genotyping was completed in 300 subjects.
CALCR
Calcitonin is a polypeptide hormone produced by parafollicular C cells of the thyroid gland. It is known to inhibit osteoclastic bone resorption and to stimulate urinary calcium excretion. These actions are exerted through the binding to a specific G-protein-associated membrane receptor encoded by the CALCR gene, cloned in 1992.28 A base mutation T→C (allelic variants T/C) in the third intracellular C-terminal domain changing a proline (CCG) at position 447 to a leucine (CTG), might influence the intracellular signal pathway. CALCR analysis was performed as described by Nakamura, digesting the PCR product with the restriction enzyme Alu I. CALCR genotyping was completed in 1430 subjects.29
ESR1
Estrogens play a central role in bone metabolism and polymorphisms of estrogen receptor α (ESR1) gene have been reported to be associated with the occurrence of osteoporosis.30 Analyses of the Pvu II and Xba I restriction enzyme sites of the first intron (allelic variants P/p and X/x respectively) were performed as previously described.31 ESR1-Pvu genotyping was completed in 1335 subjects and ESR1-Xba in 1324.
Statistical analysis
Quantitative variables were summarized as mean ± standard deviation (median, minimum–maximum for non normal) and qualitative ones as frequency distributions. Analysis of variance or t test was used to compare mean values among groups for normally distributed variables, and non parametric Mann–Whitney test for non normal variables. Chi square or Fisher's exact tests were applied to compare categorical distributions. Logistic regression analysis was performed to estimate the independent contribution of each variable considered to tooth loss. For the study of the association of natural dentition with different genotypes, the number of remaining teeth was applied as dichotomous (none vs at least 1) or categorical (0; 1–7; 8–19; ≥20). For the analysis of longitudinal data, the difference between the number of teeth at baseline and that at follow ups (excluding from the analysis subjects who were edentulous at baseline), categorizing the loss at the 75th percentile, was also considered. Confounding factors were entered either as categorical - age (65–74, 75–84, ≥85 years), smoking habits (current, former or never) - or dichotomous - diagnosis of diabetes and neoplasm (yes/no).
A p level lower than 0.05 was considered significant. All statistical analyses were performed using SAS Statistical Software Package version 9.1 (SAS Institute, Cary, NC, USA).32
Results
Genotype distribution was not affected by sex and age for all the genes considered and all were in Hardy Weinberg equilibrium in the Pro.V.A. population (data not shown).
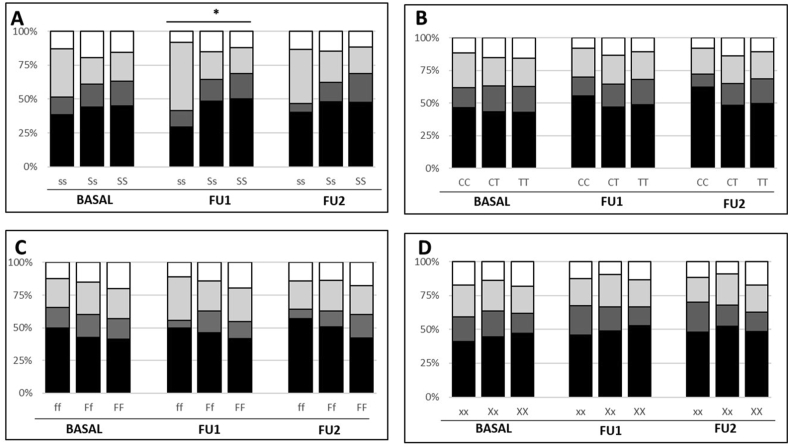
Tooth class frequencies by genotype at baseline and follow ups are detailed in Fig. 1. The prevalence of edentulism was high in the studied population (43.8%, 48.4%, and 48.2% at baseline, FU1 and FU2, respectively). The ss genotype of COL1A1 presented with the lowest values at all timepoints reaching 29% at FU1 (p < 0.02). As the prevalence of subjects retaining 20+ teeth was not high in the general population, further analyses were performed grouping the subjects into two tooth classes, with a cut off at 8. Again, the ss genotype resulted strongly associated with a better dentition at both follow ups (p < 0.02 at FU1 and p = 0.05 at FU2). All other genes did not show significant association with the number of remaining teeth.
Figure 1.
Prevalence of subjects by tooth class and genetic polymorphisms at baseline and follow ups. Panel A: COL1A1 (collagen type Iα1); Panel B: CALCR (calcitonin receptor); Panel C: VDR (vitamin D receptor); Panel D: ESR1-Xba (estrogen receptor alpha). ∗p < 0.02
As potential intermediates in the relationship, baseline levels of biochemical markers of inflammation and vit.D levels were evaluated according to genotype. As reported in Table 1, inflammation markers did not differ among genotypes, except for lower platelets in ESR1-Pvu genotype PP. As expected, vit.D levels were lower in ff genotype of VDR gene. In women, differences were found in ESR1-Pvu PP and ESR1-Xba XX, both presenting with higher erythrocyte sedimentation rate and platelets, and in VDR ff, with slightly higher WBC and confirmative lower vit.D. In men, reduced platelets and WBC were associated with genotypes ESR1-Pvu PP and ESR1-Xba XX, while the lower levels of vit.D, found in VDR ff, did not reach statistical significance.
Table 1.
Main inflammation markers and vitamin D levels according to genotype in the total population.
| Gene | Genotype (n) | Age (yrs) | Fibrinogen (g/dL) | ESR (mm/h) | WBC (n103/mm3) | Platelets (n103/mm3) | 25-OH vit.D (nmol/L) |
|---|---|---|---|---|---|---|---|
| COL1A1 | |||||||
| ss (39) | 76.1 ± 7.0 | 373.5 ± 95.8 | 21.2 ± 20.2 | 6.1 ± 1.1 | 213.3 ± 57.5 | 78.9 ± 69.8 | |
| Ss (345) | 76.1 ± 7.5 | 360.5 ± 102.5 | 20.2 ± 19.7 | 5.8 ± 1.7 | 218.2 ± 62.8 | 81.5 ± 54.4 | |
| SS (688) | 76.5 ± 7.8 | 358.5 ± 87.7 | 21.7 ± 20.1 | 6.1 ± 2.7 | 219.4 ± 63.1 | 80.3 ± 52.6 | |
| VDR | |||||||
| ff (32) | 76.6 ± 8.1 | 386.3 ± 126.4 | 20.7 ± 23.9 | 6.4 ± 1.9 | 233.7 ± 59.7 | 62.2 ± 38.4∗ | |
| Ff (133) | 75.8 ± 7.2 | 378.9 ± 91.7 | 22.3 ± 21.9 | 5.6 ± 1.6 | 214.8 ± 60.2 | 73.5 ± 51.6 | |
| FF (136) | 75.9 ± 8.0 | 391.1 ± 89.8 | 21.2 ± 19.2 | 5.8 ± 1.7 | 225.7 ± 58.0 | 86.1 ± 54.7 | |
| CALCR | |||||||
| CC (86) | 75.7 ± 7.4 | 355.3 ± 82.3 | 22.9 ± 19.9 | 5.9 ± 1.7 | 218.4 ± 62.7 | 78.0 ± 44.6 | |
| CT (539) | 76.3 ± 7.7 | 346.7 ± 85.2 | 23.0 ± 23.6 | 6.2 ± 1.8 | 216.7 ± 61.6 | 87.2 ± 63.6 | |
| TT (813) | 76.0 ± 7.2 | 349.3 ± 153.5 | 21.3 ± 18.8 | 6.2 ± 2.3 | 218.2 ± 61.1 | 85.3 ± 59.6 | |
| ESR1-Pvu | |||||||
| pp (386) | 75.1 ± 7.1 | 357.5 ± 175.6 | 20.0 ± 16.4 | 6.2 ± 2.8 | 215.6 ± 58.1 | 82.1 ± 59.0 | |
| Pp (705) | 76.0 ± 7.6 | 357.0 ± 114.6 | 21.9 ± 21.1 | 6.2 ± 2.5 | 218.4 ± 58.8 | 84.5 ± 58.2 | |
| PP (249) | 76.3 ± 7.3 | 352.6 ± 104.3 | 22.4 ± 20.9 | 5.9 ± 1.5 | 211.0 ± 60.7 | 94.9 ± 63.1∗ | |
| ESR1-Xba | |||||||
| xx (476) | 75.4 ± 7.2 | 357.7 ± 161.7 | 20.7 ± 21.2 | 6.1 ± 2.6 | 213.2 ± 57.7 | 87.3 ± 61.3 | |
| Xx (684) | 76.1 ± 7.7 | 357.9 ± 117.5 | 22.0 ± 18.9 | 6.2 ± 2.5 | 220.0 ± 60.1 | 81.5 ± 56.3 | |
| XX (179) | 75.7 ± 6.8 | 346.7 ± 103.5 | 21.1 ± 19.8 | 6.0 ± 1.5 | 209.0 ± 56.5∗ | 98.7 ± 64.8∗ | |
Genes: CALCR = calcitonin receptor; COL1A = collagen type Iα1; ESR1 = estrogen receptor alpha; VDR = vitamin D receptor inflammation markers: ESR = erythrocyte sedimentation rate; WBC = white blood cells.
∗p < 0.05.
Remaining teeth number at baseline and follow ups by genotype are reported in Table 2. The prevalence of individuals retaining more teeth was always higher in those carrying the ss genotype, without reaching statistical significance possibly as a consequence of the low number of subjects presenting with a good dentition that further weakened the already low frequency of the ss genotype (3.6% in the general population). Analysis of the number of lost teeth at follow ups was performed excluding subjects who were edentulous at baseline. Difference in tooth number at the different timepoints confirmed the positive role played by ss genotype, associated with a lower number of lost teeth between the two follow ups (p = 0.017), the proportion of subjects loosing less than 2 teeth being consistently lower in ss compared to the other genotypes (23.5 vs 53.7% in SS basal-FU1, p = 0.043; 36.4% vs 67.3% basal-FU2, p = 0.048; 18.2 vs 24.4% FU1-FU2, p = 0.021). CALCR genotype CC resulted associated with more lost teeth between F1 and F2 only in men (p = 0.023). No association was found with the other genotypes. Further analyses performed to better explore the role of possible confounders in determining the number of teeth, showed no relationship between any genotype and diabetes, neoplasms, or smoking habits. In the logistic regression analysis, adjustment for confounders hampered the detrimental effect of S allele in tooth retention between FU1 and FU2 (OR = 0.67, 95% CI: 0.4–1.0, p = 0.064).
Table 2.
Remaining teeth at baseline and follow ups, differences at follow ups according to genotypes.
| Gene | Genotype | B |
FU1 |
FU2 |
Delta n teeth median (min–max) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Median min–max |
IQR | n | Median min–max |
IQR | n | Median min–max |
IQR | B-FU1 | B-FU2 | FU1-FU2 | ||
| COL1A1 | |||||||||||||
| ss | 39 | 7 0–31 |
0–14 | 24 | 8.5 0–25 |
0–13.25 | 15 | 9 0–25 |
0–15.5 | 0# 0–6 |
0 0–9 |
0∗ 0–6 |
|
| Ss | 345 | 3 0–30 |
0–14.25 | 234 | 1.5 0–30 |
0–15 | 161 | 1 0–22 |
0–14 | 2 0–22 |
3 0–22 |
1 0–16 |
|
| SS | 688 | 2 0–31 |
0–14 | 468 | 0 0–30 |
0–11 | 312 | 1 0–24 |
0–10 | 2 0–27 |
2.5 0–24 |
0 0–24 |
|
| VDR | |||||||||||||
| ff | 32 | 0.5 0–32 |
0–9.75 | 18 | 1.5 0–32 |
0–12 | 14 | 0 0–30 |
0–11.5 | 1 0–5 |
2 0–14 |
0 0–10 |
|
| Ff | 133 | 3 0–30 |
0–14 | 84 | 2 0–30 |
0–15 | 59 | 0 0–28 |
0–14 | 1 0–15 |
3 0–16 |
0 0–16 |
|
| FF | 135 | 5 0–31 |
0–18 | 86 | 4.5 0–30 |
0–16 | 50 | 4 0–29 |
0–12.75 | 2 0–27 |
2 0–20 |
0 0–11 |
|
| CALCR | |||||||||||||
| CC | 86 | 2 0–28 |
0–13 | 63 | 0 0–28 |
0–8 | 50 | 0 0–25 |
0–8 | 1 0–10 |
2 0–21 |
0 0–21 |
|
| CT | 536 | 3 0–31 |
0–14 | 380 | 1 0–30 |
0–12 | 284 | 1 0–31 |
0–13 | 1 0–27 |
2 0–18 |
0 0–15 |
|
| TT | 808 | 3 0–30 |
0–14 | 613 | 1 0–30 |
0–11 | 508 | 1 0–30 |
0–10 | 2 0–21 |
3 0–21 |
0 0–15 |
|
| ESR1-Pvu | |||||||||||||
| pp | 382 | 5 0–29 |
0–14 | 281 | 2 0–29 |
0–10 | 212 | 2 0–29 |
0–10 | 2 0–22 |
3 0–22 |
0 0–14 |
|
| Pp | 705 | 2 0–31 |
0–13 | 509 | 1 0–30 |
0–11 | 367 | 0 0–29 |
0–10 | 1 0–22 |
3 0–21 |
0 0–21 |
|
| PP | 248 | 2 0–31 |
0–14 | 182 | 0 0–28 |
0–11 | 130 | 2 0–28 |
0–13 | 1 0–20 |
2 0–20 |
0 0–10 |
|
| ESR1-Xba | |||||||||||||
| xx | 472 | 4 0–31 |
0–14.25 | 351 | 2 0–30 |
0–10 | 252 | 1 0–29 |
0–10 | 2 0–22 |
3 0–22 |
0 0–14 |
|
| Xx | 683 | 2 0–31 |
0–13 | 485 | 1 0–30 |
0–11 | 359 | 0 0–29 |
0–11 | 1 0–22 |
3 0–21 |
0 0–21 |
|
| XX | 179 | 2 0–28 |
0–15.5 | 135 | 0 0–28 |
0–10.5 | 97 | 1 0–28 |
0–14 | 2 0–20 |
2 0–20 |
0 0–10 |
|
B = baseline; FU = follow up; IQR = interquartile range; genes: CALCR = calcitonin receptor; COL1A = collagen type Iα1; ESR1 = estrogen receptor alpha; VDR = vitamin D receptor.
∗p = 0.017; #p = 0.057.
Discussion
The purpose of the present study was to examine whether polymorphisms of genes involved in processes other than inflammation (i.e. structural factors of periodontal tissue) are associated with tooth loss. All the genes analyzed are known to play a role in bone metabolism and may therefore be concerned in the impaired attachment that characterizes the process leading to tooth loss. While a common genetic factor has not been identified for all forms of periodontal disease, at an individual level it comes as the interplay between innate genetic susceptibility and various environmental factors. Gene polymorphisms represent a way by which individuals may exhibit variations within the range of what is considered biologically normal.12 As most of them are located on introns and do not cause amino-acid substitution, the mechanism by which these polymorphisms affect gene function cannot be inferred in a straightforward manner and large studies are needed to verify the contribution of candidate genes to periodontitis and subsequent tooth loss.
The most relevant finding of our work is that the S/s polymorphism in the transcription factor Sp1 in the COL1A1 gene is related with dental status. Specifically, the ss genotype resulted associated with reduced tooth loss and with a lower prevalence of edentulism at the first follow up, particularly in men. Only limited literature reports address the relationship between COL1A1 Sp1 polymorphism and oral status, with non-univocal results. Suzuki et al. in a study conducted in Japanese subjects, in which were investigated 310 SNPs in 125 genes encoding both inflammatory and structural factors of periodontal disease, reported a positive correlation between aggressive periodontitis and SNPs in the COL1A1, COL4A1, and IL-6 signal transducer (IL6ST) genes.33 On the other hand, Sakellari did not find any correlation investigating a Greek population, but the sample size was small.34
The VDR gene has been investigated by many Authors as a possible candidate for the susceptibility to periodontitis but only a few studies focused on the Fok I polymorphism.24, 25, 26 We found a higher prevalence of edentulous subjects carrying the ff genotype. This finding, though not supported by statistical significance, is in agreement with a study conducted in a Japanese population, in which middle-aged men F-carriers were less likely to develop severe chronic periodontitis.27 The rationale may rely in the lower levels of vitamin D in the ff genotype, confirmed in our population. Unfortunately, we characterized for VDR only a relatively small number of subjects, with a negative impact on the statistical power of our analyses. In the literature, the most frequent association was found analyzing the Taq I polymorphism T/t, but, again, studies were conducted in diverse populations and it must be considered that different ethnicities as well as periodontitis type may account for discrepancies in the results.35, 36
The polymorphism identified by Alu I (C/T) in CALCR gene generates an amino acid substitution that changes the net charge of the receptor and may influence the affinity for its ligand calcitonin. A role in the determinism of bone mass has been demonstrated in postmenopausal women.37 We found a significantly higher tooth loss from baseline to FU1 in men. Suzuki and co-workers, investigating in a small sample the correlation between severe periodontitis and a SNP positioned in an intron sequence that does not change the translated protein, also found a significant association. Their findings, though not easy to compare with ours, confirm a possible role of CALCR at dental level.38
Our research, conducted in an elderly population, suggests that the relationship between tooth loss and some bone metabolism-related genetic polymorphisms may be either direct as it appears in men or mediated by bone loss/osteoporosis as it is more likely in women. This observation has a rationale in that osteoporosis and tooth loss are multifactorial and polygenic conditions that are both established in women over 65 yrs of age while in men the onset is delayed of 10–15 yrs and therefore the genetic contribution is more evident in the same age group.
Tooth loss/chronic periodontitis and osteoporosis/osteopenia have some etiological factors in common. Although the elucidation of a mechanistic association awaits future study, these evidences support the involvement of shared genetic factors in tooth loss/chronic periodontitis and osteoporosis.39,40
Dental status in the elderly is a multifactorial condition in which genetics plays a modest role. The present study demonstrating an association between tooth loss and COL1A1 and CALCR (only in men), contributes to the identification of genes involved in tooth loss and possibly in susceptibility to periodontitis. Indeed, the research concerning the genetics of this field should not aim to single out one gene but rather to identify a combination of genes that predisposes to the disease.
Conflicts of interest
The authors have no conflicts of interest relevant to this article.
Acknowledgements
The only funding source of the Pro.V.A. Study was Fondazione Cassa di Risparmio di Padova e Rovigo. The Institutions: University of Padova, ULSS n. 15 and ULSS n. 18 were active partners in the Study and provided some facilities, services, and personnel.
References
- 1.WHO . Vol. 47. 1994. Oral Health. World Health Stat Q; pp. 42–94.http://www.who.int/mediacentre/factsheets/fs318/en/ WHO 2012. [Google Scholar]
- 2.Total tooth loss among persons aged > or =65 years--selected states, 1995-1997. MMWR Morb Mortal Wkly Rep. 1999;48:206–210. [PubMed] [Google Scholar]
- 3.Musacchio E., Perissinotto E., Binotto P., et al. Tooth loss in the elderly and its association with nutritional status, socio-economical and lifestyle factors. Acta Odontol Scand. 2007;65:78–86. doi: 10.1080/00016350601058069. [DOI] [PubMed] [Google Scholar]
- 4.Reich E., Hiller K.A. Reasons for tooth extraction in the western states of Germany. Community Dent Oral Epidemiol. 1993;21:379–383. doi: 10.1111/j.1600-0528.1993.tb01103.x. [DOI] [PubMed] [Google Scholar]
- 5.Cahen P.M., Frank R.M., Turlot J.C. A survey of the reasons for dental extractions in France. J Dent Res. 1985;64:1087–1093. doi: 10.1177/00220345850640081401. [DOI] [PubMed] [Google Scholar]
- 6.Morita M., Kimura T., Kanegae M., Ishikawa A., Watanabe T. Reasons for extraction of permanent teeth in Japan. Community Dent Oral Epidemiol. 1994;22:303–306. doi: 10.1111/j.1600-0528.1994.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 7.Hull P.S., Worthington H.V., Clerehugh V., Tsirba R., Davies R.M., Clarkson J.E. The reasons for tooth extractions in adults and their validation. J Dent. 1997;25:233–237. doi: 10.1016/s0300-5712(96)00029-2. [DOI] [PubMed] [Google Scholar]
- 8.McCaul L.K., Jenkins W.M., Kay E.J. The reasons for extraction of permanent teeth in Scotland: a 15-year follow-up study. Br Dent J. 2001;190:658–662. doi: 10.1038/sj.bdj.4801068. [DOI] [PubMed] [Google Scholar]
- 9.Armitage G.C. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Kinane D.F., Shiba H., Hart T.C. The genetic basis of periodontitis. Periodontol. 2000;39(2005):91–117. doi: 10.1111/j.1600-0757.2005.00118.x. [DOI] [PubMed] [Google Scholar]
- 11.Brett P.M., Zygogianni P., Griffiths G.S., et al. Functional gene polymorphisms in aggressive and chronic periodontitis. J Dent Res. 2005;84:1149–1153. doi: 10.1177/154405910508401211. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki A., Ji G., Numabe Y., et al. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004;317:887–892. doi: 10.1016/j.bbrc.2004.03.126. [DOI] [PubMed] [Google Scholar]
- 13.Jakovljevic A., Nikolic N., Jacimovic J., et al. Tumor necrosis factor alpha -308 G/A singlenucleotide polymorphism and apical periodontitis: an updated systematic review and metaanalysis. J Endod. 2021;47:1061–1069. doi: 10.1016/j.joen.2021.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Chatzopoulos G., Doufexi A.E., Wolff L., Kouvatsi A. Interleukin-6 and interleukin-10 gene polymorphisms and the risk of further periodontal disease progression. Braz Oral Res. 2018;32:e11. doi: 10.1590/1807-3107bor-2018.vol32.0011. [DOI] [PubMed] [Google Scholar]
- 15.Mashhadiabbas F., Dastgheib S.A., Hashemzehi A., et al. Association of IL-10 -1082A>G, -819C>T, and -592C>A polymorphisms with susceptibility to chronic and aggressive periodontitis: a systematic review and meta-analysis. Inflamm Res. 2021;70:509–524. doi: 10.1007/s00011-021-01448-z. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W., Xu P., Chen Z., Cheng Y., Li X., Mao Q. IL-13 -1112 polymorphism and periodontitis susceptibility: a meta-analysis. BMC Oral Health. 2018;18:21. doi: 10.1186/s12903-018-0481-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia X.W., Yuan Y.D., Yao Z.X., et al. Association between IL-4 and IL-4R polymorphisms and periodontitis: a meta-analysis. Dis Markers. 2017;2017:8021279. doi: 10.1155/2017/8021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Silva M.K., de Carvalho A.C.G., Alves E.H.P., da Silva F.R.P., Pessoa L.D.S., Vasconcelos D.F.P. Genetic factors and the risk of eriodontitis development: findings from a systematic review composed of 13 studies of meta-analysis with 71,531 participants. Int J Dent. 2017;2017:1914073. doi: 10.1155/2017/1914073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki K., Kurosu Y., Kamiya T., et al. Low metacarpal bone density, tooth loss, and periodontal disease in Japanese women. J Dent Res. 2001;80:1818–1822. doi: 10.1177/00220345010800090901. [DOI] [PubMed] [Google Scholar]
- 20.Hanai Y., Sugita N., Wang Y., et al. Relationships between IL-6 gene polymorphism, low BMD and periodontitis in postmenopausal women. Arch Oral Biol. 2015;60:533–539. doi: 10.1016/j.archoralbio.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Corti M.C., Guralnik J.M., Sartori L., et al. The effect of cardiovascular and osteoarticular diseases on disability in older Italian men and women: rationale, design, and sample characteristics of the Progetto Veneto Anziani (PRO.V.A.) study. J Am Geriatr Soc. 2002;50:1535–1540. doi: 10.1046/j.1532-5415.2002.50409.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant S.F.A., Reid D.M., Blake G., Herd R., Fogelman I., Ralston S.H. Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I a 1 gene. Nat Genet. 1996;14:203–205. doi: 10.1038/ng1096-203. [DOI] [PubMed] [Google Scholar]
- 24.Tang X., Meng H. Osteogenic induction and 1,25-dihydroxyvitamin D3 oppositely regulate the proliferation and expression of RANKL and the vitamin D receptor of human periodontal ligament cells. Arch Oral Biol. 2009;54:625–633. doi: 10.1016/j.archoralbio.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Tachi Y., Shimpuku H., Nosaka Y., et al. Vitamin D receptor gene polymorphism is associated with chronic periodontitis. Life Sci. 2003;73:3313–3321. doi: 10.1016/j.lfs.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Sun J.L., Meng H.X., Cao C.F., et al. Relationship between vitamin D receptor gene polymorphism and periodontitis. J Periodontal Res. 2002;37:263–267. doi: 10.1034/j.1600-0765.2002.01605.x. [DOI] [PubMed] [Google Scholar]
- 27.Gross C., Eccleshall T., Malloy P., Villa M., Marcus R., Feldman D. The presence of a polymorphism at the translation initiation site of the vitamin D receptor is associated with low bone mineral density in postmenopausal Mexican-American women. J Bone Miner Res. 1996;11:1850–1855. doi: 10.1002/jbmr.5650111204. [DOI] [PubMed] [Google Scholar]
- 28.Gorn A.H., Lin H.Y., Yamin M., et al. Cloning, characterization, and expression of a human calcitonin receptor from an ovarian carcinoma cell line. J Clin Invest. 1992;90:1726–1735. doi: 10.1172/JCI116046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura M., Zhang Z.Q., Shan L., et al. Allelic variants of human calcitonin receptor in the Japanese population. Hum Genet. 1997;99:38–41. doi: 10.1007/s004390050307. [DOI] [PubMed] [Google Scholar]
- 30.Langdahl B.L., Løkke E., Carstens M., Stenkjaer L.L., Eriksen E.F. A TA repeat polymorphism in the estrogen receptor gene is associated with osteoporotic fractures but polymorphisms in the first exon and intron are not. J Bone Miner Res. 2000;15:2222–2230. doi: 10.1359/jbmr.2000.15.11.2222. [DOI] [PubMed] [Google Scholar]
- 31.Yaich L., Dupont W.D., Cavener D.R., Parl F.F. Analysis of the PvuII restriction fragment-length polymorphism and exon structure of the estrogen receptor gene in breast cancer and peripheral blood. Cancer Res. 1992;52:77–83. [PubMed] [Google Scholar]
- 32.R Core Team . 2018. R: a language and environment for statistical computing.https://www.r-project.org Vienna, Austria URL: [Google Scholar]
- 33.Suzuki A., Ji G., Numabe Y., Ishii K., Muramatsu M., Kamoi K. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004;92:43–47. doi: 10.1007/s10266-004-0035-4. [DOI] [PubMed] [Google Scholar]
- 34.Sakellari D., Katsares V., Georgiadou M., Kouvatsi A., Arsenakis M., Konstantinidis A. No correlation of five gene polymorphisms with periodontal conditions in a Greek population. J Clin Periodontol. 2006;33:765–770. doi: 10.1111/j.1600-051X.2006.00983.x. [DOI] [PubMed] [Google Scholar]
- 35.Naito M., Miyaki K., Naito T., et al. Association between vitamin D receptor gene haplotypes and chronic periodontitis among Japanese men. Int J Med Sci. 2007;4:216–222. doi: 10.7150/ijms.4.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennig B.J., Parkhill J.M., Chapple I.L., Heasman P.A., Taylor J.J. Association of a vitamin D receptor gene polymorphism with localized early-onset periodontal diseases. J Periodontol. 1999;70:1032–1038. doi: 10.1902/jop.1999.70.9.1032. [DOI] [PubMed] [Google Scholar]
- 37.Masi L., Becherini L., Colli E., et al. Polymorphisms of the calcitonin receptor gene are associated with bone mineral density in postmenopausal Italian women. Biochem Biophys Res Commun. 1998;248:190–195. doi: 10.1006/bbrc.1998.8880. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki A., Ji G., Numabe Y., et al. Single nucleotide polymorphisms associated with aggressive periodontitis and severe chronic periodontitis in Japanese. Biochem Biophys Res Commun. 2004;317:887–892. doi: 10.1016/j.bbrc.2004.03.126. [DOI] [PubMed] [Google Scholar]
- 39.von Wowern N., Klausen B., Kollerup G. Osteoporosis: a risk factor in periodontal disease. J Periodontol. 1994;65:1134–1138. doi: 10.1902/jop.1994.65.12.1134. [DOI] [PubMed] [Google Scholar]
- 40.Hong S.J., Yang B.E., Yoo D.M., Kim S.J., Choi H.G., Byun S.H. Analysis of the relationship between periodontitis and osteoporosis/fractures: a cross-sectional study. BMC Oral Health. 2021;21:125. doi: 10.1186/s12903-021-01496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]