Abstract
MicroRNAs (miRNAs) have been shown to affect expression of several genes contributing in important biological processes. miR-1290 a member of this family with crucial roles in the carcinogenesis. This miRNA is transcribed from MIR1290 gene on chromosome 1p36.13. This miRNA has interactions with a number of mRNA coding genes as well as non-coding RNAs SOCS4, GSK3, BCL2, CCNG2, KIF13B, INPP4B, hMSH2, KIF13B, NKD1, FOXA1, IGFBP3, CCAT1, FOXA1, NAT1, SMEK1, SCAI, ZNF667-AS1, ABLIM1, Circ_0000629 and CDC73. miR-1290 can also regulate activity of JAK/STAT3, PI3K/AKT, Wnt/β-catenin and NF-κB molecular pathways. Most evidence indicates the oncogenic roles of miR-1290, yet controversial evidence also exists. In the present review, we describe the results of in vitro, animal and human investigations about the impact of miR-1290 in the development of malignancies.
Keywords: miR-1290, cancer, biomarker, miRNA, expression
Introduction
MicroRNAs (miRNAs) are a group of small-sized transcripts with a wide range of regulatory roles. They are mostly produced through a multistep mechanism. These steps include transcription from DNA sequences into primary miRNAs and processing into precursor miRNAs and subsequently into mature miRNAs. The majority of bind with the 3′ untranslated region (3′ UTR) of target transcripts to either degrade mRNA or repress its translation. In some circumstances, miRNAs can induce translation or control transcription (O'Brien et al., 2018). Approximately 50% of all miRNAs are transcribed from intragenic regions. These miRNAs are mainly produced from introns and a number of exons of protein coding genes. Other miRNAs are intergenic and are produced in an independent manner from a host gene. Thus, these miRNAs have their own promoters (Kim and Kim, 2007; De Rie et al., 2017). miRNAs partake in the regulation of important biological functions, such as cell proliferation, differentiation and apoptosis, thus being involved in the pathoetiology of several disorders, particularly neoplastic disorders (Peng and Croce, 2016). These transcripts participate in the pathoetiology of diverse cancers (Abolghasemi et al., 2020).
miR-1290 is transcribed from MIR1290 gene on chromosome 1p36.13. The primary transcript (NR_031622.1) has 78 nucleotides (GAGCGUCACGUUGACACUCAAAAAGUUUCAGAUUUUGGAACAUUUCGGAUUUUGGAUUUUUGGAUCAGGGAUGCUCAA). The mature transcript of hsa-miR-1290 (MIMAT0005880) has 19 nucleotides (UGGAUUUUUGGAUCAGGGA). This miRNA has important functions in the carcinogenesis. Several in vitro studies have assessed function of miR-1290. Moreover, animal studies in lung, colon and liver cancer models have assessed functional consequences of up-regulation or silencing of this miRNA. However, some inconsistencies exist regarding the role of miR-1290. In the present manuscript, we describe the results of in vitro, animal and human assays about the influence of miR-1290 in the development of cancers.
In vitro Studies
Forced over-expression of miR-1290 in AsPC1 and Panc5.04 pancreatic cancer cell lines has led to enhancement of cell proliferation. Inhibition of miR-1290 in pancreatic cancer cells has the reverse effects. miR-1290 mimics have also enhanced invasive properties of these cells (Li et al., 2013).
Over-expression of miR-1290 has enhanced proliferation of proliferation of lung adenocarcinoma cells and induced cell cycle progression and invasiveness. Moreover, this miRNA has suppressed cell apoptosis in this cell line. miR-1290 has been found to downregulate expression of SOCS4 to activate JAK/STAT3 and PI3K/AKT pathways (Xiao et al., 2018).
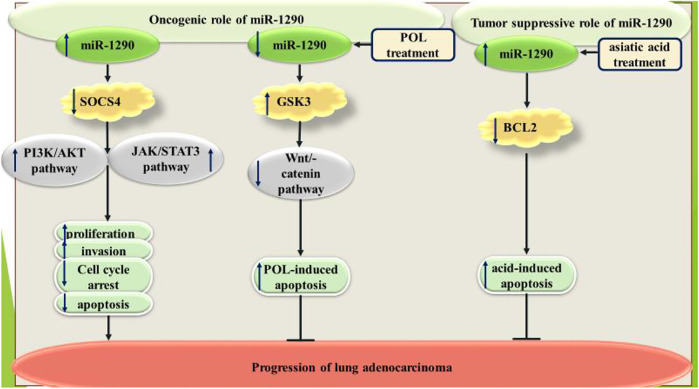
The anti-proliferative and apoptosis-inducing agent polygonatum odoratum lectin (POL) has been shown to decrease miR-1290 levels in A549 lung adenocarcinoma cells. Down-regulation of miR-1290 has been shown to increase POL-associated apoptosis in these cells. GSK3β has been found as the direct target of miR-1290 in A549 cells (Wu et al., 2016). Conversely, miR-1290 has been shown to sensitize A549 cells to the apoptosis-inducing agent asiatic acid through negatively regulating expression of BCL2. Expression of miR-1290 has been up-regulated by asiatic acid. Most notably, the apoptosis-inducing effect of asiatic acid relies on miR-1290 activity. Taken together, miR-1290 has been shown to suppress viability and cell cycle progression of A549 cells (Kim et al., 2014). Figure 1 summarizes the effects of miR-1290 in the pathoetiology of lung cancer.
FIGURE 1.
Dual roles of miR-1290 in the pathoetiology of lung cancer.
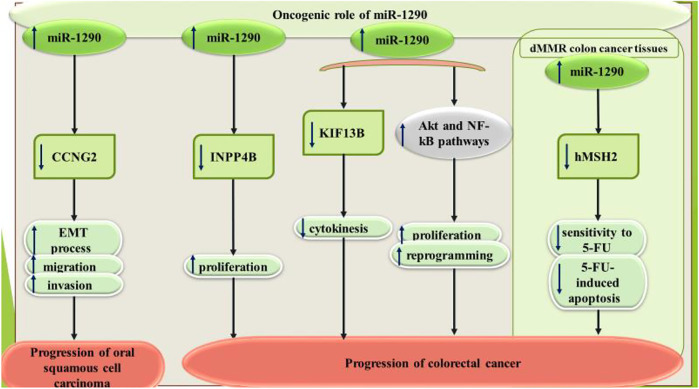
In oral squamous cell carcinoma, miR-1290 has been shown to be up-regulated parallel with downregulation of CCNG2. miR-1290 silencing has inhibited metastatic ability and epithelial-mesenchymal transition (EMT). CCNG2 has been identified as the direct target of miR-1290 (Qin et al., 2019). Functional studies in laryngeal squamous cell carcinoma has shown that miR-1290 targets two tumor suppressor genes, namely ITPR2 and MAF (Janiszewska et al., 2015).
miR-1290 has oncogenic roles in colorectal cancer. miR-1290 silencing has suppressed proliferation of colorectal cancer cells. miR-1290 up-regulation has decreased expression of p27 and enhanced transcript and protein amounts of cyclin D1. NPP4B has been recognized as the target of miR-1290 (Ma et al., 2018). Moreover, miR-1290 silencing has improved cytotoxic effects of 5-fluouracil in colorectal cancer cells through targeting hMSH2 (Ye et al., 2017). Figure 2 shows the oncogenic role of miR-1290 in squamous cell carcinoma and colorectal cancer.
FIGURE 2.
Oncogenic influence of miR-1290 in squamous cell carcinoma and colorectal cancer.
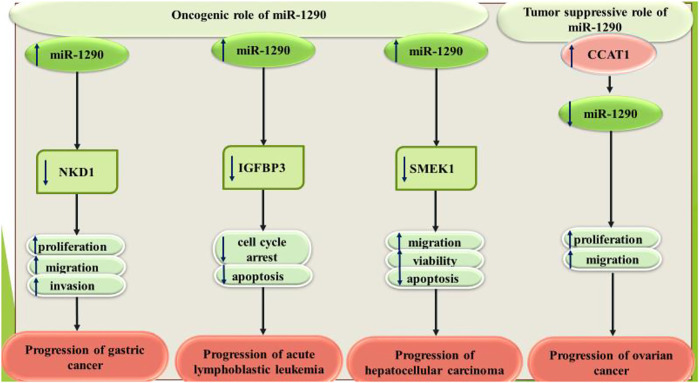
Exosomal miR-1290 has been found to be high in gastric cancer cell lines. miR-1290-containing exosomes could promote proliferation, migratory aptitude, and invasiveness of gastric cancer cells. NKD1 has been identified as the direct target of miR-1290 in these cells (Huang et al., 2019). Moreover, miR-1290 has been revealed to increase proliferation and migratory aptitude of gastric cancer cells through targeting FOXA1 (Lin et al., 2016).
miR-1290 has also been shown to be overexpressed in B-acute lymphoblastic leukemia (ALL) cell line SUP-B15. The anticancer agent resveratrol has been found to down-regulate expression of miR-1290 and enhance IGFBP3 levels in the ALL cells. miR-1290 can target 3′ UTR of IGFBP3 (Zhou et al., 2017). Besides, exosomal miR-1290 has been demonstrated to promote angiogenic processes in hepatocellular carcinoma through influencing expression of SMEK1 (Wang et al., 2021c).
On the other hand, miR-1290 has been shown to exert tumor suppressive role in ovarian cancer. In fact, the oncogenic long non-coding RNA (lncRNA) CCAT1 facilitates ovarian carcinogenesis through decreasing miR-1290 levels (Lai and Cheng, 2018).
Figure 3 shows the roles of miR-1290 in gastric cancer, ALL, hepatocellular carcinoma and ovarian cancer.
FIGURE 3.
miR-1290 has oncogenic roles in gastric cancer, acute lymphoblastic leukemia and hepatocellular carcinoma, while it has tumor suppressive roles in ovarian cancer.
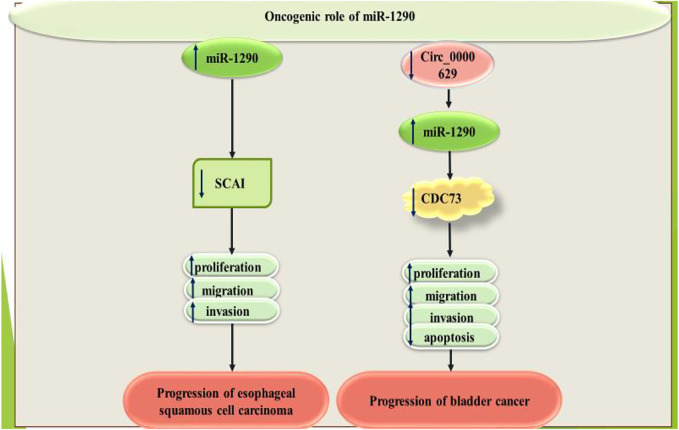
Over-expression of miR-1290 has enhanced esophageal squamous cell carcinoma growth, migration and invasiveness through decreasing SCAI levels (Li et al., 2015). In bladder cancer cells, tumor suppressor Circular RNA circ_0000629 has been shown to exert its effects through suppressing miR-1290 levels and up-regulating CDC73 expression (Wang et al., 2021b). Figure 4 shows the oncogenic role of miR-1290 in esophageal and bladder cancers.
FIGURE 4.
Oncogenic role of miR-1290 in esophageal and bladder cancers.
Summary of in vitro studies regarding the role of miR-1290 in the carcinogenesis is provided in Table 1.
TABLE 1.
Expression pattern of miR-1290 in cancer cell lines (∆: knock-down or deletion, POL: Polygonatum odoratum lectin, 5-FU: 5-Fluorouracil).
| Tumor type | Targets/Regulators and signaling pathways | Cell line | Function | References |
|---|---|---|---|---|
| Pancreatic cancer | — | Panc5.04, Panc8.13, Panc10.05, Panc198, HPDE | ↑ miR-1290: ↑ proliferation, ↑ invasion | Li et al. (2013) |
| Lung cancer | SOCS4, JAK/STAT3 signaling pathway, PI3K/AKT signaling pathway | BEAS-2B, A549, SPC-A1 | ↑ miR-1290: ↑ proliferation, ↑ invasion, ↓ G1/G0 phase arrest, ↓ apoptosis | Xiao et al. (2018) |
| GSK3, Wnt/-catenin pathway | A549 | POL treatment: ↓ miR-1290 | Wu et al. (2016) | |
| ↑ miR-1290 + POL treatment: ↓ POL-induced apoptosis | ||||
| ∆ miR-1290 + POL treatment: ↑ POL-induced apoptosis | ||||
| ↑ miR-1290: did not affect proliferation, did not affect autophagy | ||||
| ∆ miR-1290: did not affect proliferation, did not affect autophagy | ||||
| BCL2 | A549 | asiatic acid treatment: ↑ miR-1290 | Kim et al. (2014) | |
| ↑ miR-1290: ↑ acid-induced apoptosis | ||||
| Oral squamous cell carcinoma | CCNG2 | NHOK, Cal-27, SCC-9, SCC-25, Tca-8113 c | ∆ miR-1290: ↓ migration, ↓ invasion | Qin et al. (2019) |
| ↑ miR-1290: ↑ EMT process | ||||
| Laryngeal squamous cell carcinoma | KIF13B | UT-SCC-34 | — | Janiszewska et al. (2015) |
| Colorectal cancer | INPP4B | FHC, and CRC cells SW480, HT-29, COLO205, SW403, KM202L, SW620 | ↑ miR-1290: ↑ proliferation | Ma et al. (2018) |
| ∆ miR-1290: ↓ proliferation | ||||
| — | Caco2, DLD1, HT29, LoVo, SW480 | ∆ miR-1290: ↓ proliferation, ↓ migration, ↓ invasion | Imaoka et al. (2016) | |
| hMSH2 | RKO, SW480, HCT116, and LoVo | ↑ miR-1290: ↑ viability, ↓ sensitivity to 5-FU | Ye et al. (2017) | |
| ∆ miR-1290: ↑ sensitivity to 5-FU, ↑ apoptosis | ||||
| KIF13B, Akt and NF-kB pathways | SW620, 293T, SGC7901 c | ↑ miR-1290: ↑ proliferation, ↑ reprogramming, ↓ cytokinesis | Wu et al. (2013) | |
| Gastric cancer | NKD1 | SGC7901, AGS, and BGC823, GES | ↑ miR-1290: ↑ proliferation, ↑ invasion, ↑ migration | Huang et al. (2019) |
| FOXA1 | GES-1, SGC-7901 | ∆ miR-1290: ↓ proliferation, ↓ migration, no significant difference in apoptosis | Lin et al. (2016) | |
| Acute lymphoblastic leukemia | IGFBP3 | PBMCs | ∆ miR-1290: ↑ cell cycle arrest, ↑ apoptosis | Zhou et al. (2017) |
| Ovarian cancer | CCAT1 | OVCAR-8, SKOV-3 w, IOSE386, OMC685 | ∆ lncRNA CCAT1 (which sponges miR-1290): ↓ proliferation, ↓ migration | Lai and Cheng, (2018) |
| Breast cancer | FOXA1, NAT1 | T47D, MCF-7 | ↑ miR-1290: ↓ expression levels of FOXA1 and NAT1 in ER-positive breast cancer cells | Endo et al. (2013) |
| Hepatocellular carcinoma | SMEK1 | HUVECs, Hep3 B, HepG2, SMMC-7721, PLC/PRF/5, L-02 | ↑ miR-1290: ↑ migration, ↑ viability, ↑ capacity of HUVECs to form tube-like structures | Wang et al. (2021c) |
| ∆ miR-1290: ↓ migration, ↓ viability, ↑ apoptosis | ||||
| Esophageal squamous cell carcinoma | SCAI | Eca109, TE13 | ↑ miR-1290: ↑ proliferation, ↑ invasion, ↑ migration | Li et al. (2015) |
| Chordoma | NONHSAT024778, Robo1 | U-CH1 | ↑ NONHSAT024778 (which sponges miR-1290): ↑ proliferation, ↑ invasion, ↑ migration | Wang et al. (2021a) |
| Nasopharyngeal carcinoma | ZNF667-AS1, ABLIM1 | NP69, c666-1, CNE-1, CNE-2, HNE1 | ↑ miR-1290: ↑ proliferation, ↑ invasion, ↑ migration, ↓ apoptosis | Chen et al. (2020) |
| Bladder cancer | Circ_0000629, CDC73 | T24, SW780 | ↑ miR-1290: ↑ growth, ↑ invasion, ↑ migration, ↓ apoptosis | Wang et al. (2021b) |
Studies in animal models.
miRNA-1290 has important roles in determination of response of cancer cells to 5-fluouracil. miR-1290 silencing has improved cytotoxic effects of 5-fluouracil in xenografts models of this cancer via targeting hMSH2 (Ye et al., 2017). Other studies have shown oncogenic roles of miR-1290 in animal models of lung cancer (Xiao et al., 2018), hepatocellular carcinoma (Wang et al., 2021c) and nasopharyngeal carcinoma (Chen et al., 2020) (Table 2). On the other hand, animal studies have shown that the oncogenic lncRNA NONHSAT024778 acts through sponging miR-1290, thus revealing a tumor suppressor role for miR-1290 (Wang et al., 2021a).
TABLE 2.
Impact of miR-1290 in carcinogenesis based on investigations in animal models (∆: knock-down or deletion).
| Tumor type | Animal models | Results | References |
|---|---|---|---|
| Lung cancer | BALB/c-nu/nu nude mice | ↑ miR-1290: ↑ tumor volume, ↑ tumor weight, ↑ invasion, ↑ metastasis | Xiao et al. (2018) |
| Colon cancer | male BALB/c nude mice | ∆ miR-1290: ↑ 5-FU-induced apoptosis | Ye et al. (2017) |
| Hepatocellular carcinoma | male BALB/c and NOD-SCID mice | ∆ miR-1290: ↓ tumor volumes, ↓ tumor weights, ↓ proliferation, ↑ apoptosis | Wang et al. (2021c) |
| Chordoma | male Balb/c NOD nude mice | ∆ NONHSAT024778 (which sponges miR-1290): ↓ tumor volumes, ↓ tumor weights, ↓ tumor growth | Wang et al. (2021a) |
| Nasopharyngeal carcinoma | BALB/c nude mice | ∆ miR-1290: ↓ tumor volumes, ↓ tumor weights | Chen et al. (2020) |
Studies in Clinical Samples
In lung adenocarcinoma tissues, expression of miR-1290 has been negatively correlated with SOCS4 levels. Expression of SOCS4 has been inversely correlated with higher clinical stages and lymph node metastases (Xiao et al., 2018). Moreover, miR-1290 levels have been associated with clinicopathological landscapes and poor prognosis of patients with oral squamous cell carcinoma (Qin et al., 2019). In laryngeal squamous cell carcinoma, a high throughput miRNA profiling experiment has shown up-regulation of 33 miRNAs, among them being miR-1290 (Janiszewska et al., 2015).
Comparison of miRNA profiles between deficient and proficient mismatch repair colon cancer tissues has shown up-regulation of miR-1290 in deficient mismatch repair colorectal cancer tissues. Expression of miR-1290 has been correlated with poor prognoses of colon cancer in stages II and III patients who took 5-fluouracil-based chemotherapeutics regimens (Ye et al., 2017).
miR-1290 has also been exhibited to be up-regulated in serum exosomes of gastric cancer patients compared with healthy people (Huang et al., 2019). Another study in gastric cancer patients has shown correlation between miR-1290 over-expression and clinical stage, deepness of invasion and lymph node positivity (Lin et al., 2016).
miR-1290 has also been shown to be upregulated in esophageal squamous cell carcinoma tissues compared with unaffected neighboring samples. Over-expression of miR-1290 has been associated with level of differentiation, N classification TNM stage in this type of esophageal cancer (Li et al., 2015).
On the other hand, in oral squamous cell carcinoma, levels of this miRNA has been reported to be decreased in blood samples of patients compared with control samples (Nakashima et al., 2019). Moreover, expression of miR-1290 has been reported to be decreased in chordoma samples (Wang et al., 2021a). Table 3 summarizes the results of studies that reported dysregulation of miR-1290 in clinical samples.
TABLE 3.
Dysregulation of miR-1290 in clinical specimens (DC: benign pancreatic disease controls, PFS: progression free survival, LUAD: Lung adenocarcinoma, ANCTs: adjacent non-cancerous tissues, OS: Overall survival, DFS: disease-free survival, TNM: tumor-node-metastasis, NSCLC: non-small-cell lung cancer, CRA: colorectal adenoma, HGSOC: high grade serous ovarian cancer, EOC: epithelial ovarian cancer, HGSOC: high grade serous ovarian carcinoma.).
| Tumor type | Samples | Expression (tumor vs. Normal) | Kaplan-Meier analysis (impact of miR-1290 up-regulation) | Univariate/Multivariate cox regression | Association of miR-1290 expression with clinicopathologic characteristics | References |
|---|---|---|---|---|---|---|
| Prostate cancer | 23 CRPC patients | up | Poor OS | — | — | Huang et al. (2015) |
| Pancreatic cancer (PC) | GEO datasets: (GSE113486 and GSE106817) | up | — | — | — | Wei et al. (2020) |
| 120 PC patients, 40 DC patients, and 40 healthy controls | up | — | miR-1290 expression was independent risk factors for PC. | gender (male), and stage III and IV | ||
| 167 PC patients and 267 healthy subjects | up | shorter OS and DFS | miR-1290 was not found to be an independent negative prognostic factor for OS and DFS in PC patients | PC aggressiveness | Tavano et al. (2018) | |
| 81 PDAC patients, 28 PNETs patients, 20 IPMN patients, 45 chronic pancreatitis patients, and 39 healthy controls | higher in patients with IPMNs than healthy controls, higher in patients with invasive pancreatic cancer than patients with IPMNs, higher in intermediate- and high-grade dysplasia than those with low-grade dysplasia | — | — | — | Li et al. (2013) | |
| Lung cancer | 70 LUAD patients and 40 healthy controls | up | shorter PFS | The level of miR-1290 was an independent prognostic factor in LUAD patients | gender (male), advanced TNM stage, tumor size, lymph node metastasis, distant metastasis, smoking, and drinking | Wu et al. (2020) |
| 32 pairs of LUAD tissues and ANCTs | up | — | — | — | Xiao et al. (2018) | |
| 33 pairs of NSCLC tissues and ANCTs | up | shorter OS | — | stage IIIa, lymph node metastasis | Mo et al. (2015) | |
| serum samples from 73 NSCLC patients, 19 patients with various benign lung disease, 34 healthy controls | up | shorter OS | TNM stage and lymph node metastasis status and serum miR-1290 expression were found to be the independent prognostic factors for OS. | TNM stage, lymph node metastasis | ||
| Oral squamous cell carcinoma (OSCC) | 47 pairs of OSCC tissues and ANCTs | up | shorter OS | — | TNM stage and the lymph node metastasis | Qin et al. (2019) |
| 10 OSCC patients and 10 healthy volunteers | down | — | — | — | Nakashima et al. (2019) | |
| plasma samples from 55 OSCC patients | down | higher OS and DFS | Expression OF miR-1290 was found to be a significant prognostic factor for OSCC patients | tumor differentiation and response to CRT | ||
| Laryngeal squamous cell carcinoma (LSCC) | 50 LSCC patients and 5 epithelial no tumor controls | up | — | — | — | Janiszewska et al. (2015) |
| 5 pairs of LSCC tissues and ANCTs | up | — | — | — | Sun et al. (2013) | |
| 48 LSCC patients | up | — | — | — | ||
| Colorectal cancer (CRC) | GEO datasets: (GSE108153, GSE81581, GSE55139 and GSE41655) | up | — | — | — | Liu et al. (2019) |
| 15 CRC patients, 15 adenoma cases and 15 healthy controls | up | — | — | — | ||
| 80 CRC patients, 50 adenoma cases, and 30 healthy controls | up | — | — | larger tumor size, advanced TNM stage, lymph node metastasis, and distant metastasis | ||
| 8 pairs of CRC tissues and ANCTs | up | — | — | — | Ma et al. (2018) | |
| 20 normal colon samples and 50 CRC samples | up | — | — | — | ||
| 12 pairs of CRC tissues and ANCTs, and 12 colorectal adenomas tissues | up | poorer OS | High miR-1290 expression, large tumor size, lymphatic invasion, venous invasion, high T stage, lymph node metastasis, distant metastasis, and high carcinoembryonic antigen levels were associated with poor OS. | — | Imaoka et al. (2016) | |
| serum samples from 12 CRC patients,12 adenoma patients, and 12 healthy persons | up | worse OS | Increased serum miR-1290 level, poor differentiation, lymphatic invasion, venous invasion, high T stage, lymph node metastasis, distant metastasis, and high CEA levels were associated with poor OS. | — | ||
| serum samples from 211 CRC patients, 56 colorectal adenoma patients, and 57 healthy controls | up | — | — | stage IV, tumor size, serosal invasion, lymphatic and venous invasion, and metastasis | ||
| GEO database: GSE39833 (88 CRC patients and 11 healthy controls) | up | — | — | — | Li et al. (2016) | |
| Colorectal cancer (CRC) | 54 CRA patients | up | — | — | adenoma size | Handa et al. (2021) |
| Colon cancer | 291 colon cancer tumor tissues | up | Lower OS and DFS | miR-1290 expression, N stage, AJCC stage, tumor differentiation, vascular invasion, miR-and MMR status were associated with decreased OS and DFS. | dMMR Status, tumor location, N stage, and tumor differentiation | Ye et al. (2017) |
| 25 pairs of colon cancer tissues and ANCTs | up | — | — | — | Wu et al. (2013) | |
| Gastric cancer (GC) | serum samples from 20 GC patients and 10 healthy controls | up | — | — | — | Huang et al. (2019) |
| 20 pairs of GC tissues and ANCTs | up | — | — | advanced clinical staging and depth of tumor invasion | Lin et al. (2016) | |
| Acute lymphoblastic leukemia (ALL) | 15 ALL patients and 15 healthy controls | IGFBP3 (a target of miR-1290) expression is decreased | — | — | — | Zhou et al. (2017) |
| Ovarian cancer (OC) | sera samples from 70 EOC patients and 13 healthy controls | no significant difference | — | — | — | Kobayashi et al. (2018) |
| 30 HGSOC patients and 13 healthy controls | up | — | — | tumor burden | ||
| 40 pairs of OC tissues and ANCTs | upregulation of lncRNA CCAT1 (which sponges miR-1290) | higher CCAT1 = shorter OS | — | tumor size and lymph node metastasis | Lai and Cheng, (2018) | |
| Breast cancer | blood samples from 60 breast cancer patients and 20 healthy controls | up | — | — | lymph node metastasis and Stage II/III | Li et al. (2021) |
| 4 ER-high Ki67-low tumor tissues and 4 ER-low Ki67-high tumor tissues | down in ER-high Ki67-low tumors | — | — | tumor grade | Endo et al. (2013) | |
| Hepatocellular carcinoma (HCC) | 49 pairs of HCC tissues and ANCTs | Up | — | — | — | Wang et al. (2021c) |
| serum samples of 49 HCC patients and serum samples of 28 healthy controls | Up | — | — | — | ||
| Esophageal squamous cell carcinoma (ESCC) | 24 pairs of ESCC tumor tissues and ANCTs | up | — | — | differentiation, N classification and tumor-node-metastasis stage | Li et al. (2015) |
| Chordoma | 20 chordoma tissues and 10 FNP tissues | down | — | — | — | Wang et al. (2021a) |
| Nasopharyngeal carcinoma (NPC) | GEO database: (GSE70970) | up | — | — | — | Chen et al. (2020) |
| Cutaneous squamous cell carcinoma (cSCC) | 8 cSCC patients and 8 controls | up | — | — | — | Geusau et al. (2020) |
| Cervical cancer | sera from 6 cervical cancer patients and 6 healthy persons | up | — | — | — | Nagamitsu et al. (2016) |
| Sera of 20 cervical cancer patients 10 healthy persons | up | — | — | — | ||
| serum samples from 100 cervical cancer patients and 31 healthy controls | up | — | — | — | ||
| microarray analysis | up in cells with HPV infection upon 5-AZA treatment | — | — | — | Yao et al. (2013) |
Serum levels of miR-1290 have been shown to be higher in patients with intraductal papillary mucinous pancreatic cancer compared with healthy subjects. The ability of serum levels of miR-1290 in separation of patients with low-stage pancreatic cancer from controls has been higher than CA19-9. Notably, higher levels of miR-1290 has been predictive of poor outcome following pancreaticoduodenectomy (Li et al., 2013). In this type of cancer, miR-1290 has been shown to appropriately distinguish neoplastic condition from both healthy condition and chronic pancreatitis (Wei et al., 2020). In colorectal cancer, levels of this miRNA could distinguish cancer status from healthy condition with up to ideal diagnostic power. Moreover, it can separate colorectal adenoma from healthy status with lower values (Imaoka et al., 2016). Table 4 shows the diagnostic value of miR-1290 in cancers.
TABLE 4.
Diagnostic value of miR-1290 in cancers (PC: pancreatic cancer; DC: benign pancreatic disease control; LUAD: Lung adenocarcinoma, EOC: epithelial ovarian cancer, HGSOC: high grade serous ovarian carcinoma).
| Tumor type | Samples | Distinguish between | Area under curve | Sensitivity (%) | Specificity (%) | References |
|---|---|---|---|---|---|---|
| Pancreatic cancer (PC) | 120 PC patients and 40 healthy controls | PC patients vs. healthy controls | 0.93 | 75.0 | 97.5 | Wei et al. (2020) |
| 120 PC patients and 40 DC | PC patients vs. DC | 0.89 | 88.3 | 72.5 | ||
| 120 PC patients and controls | PC patients vs. all controls | 0.91 | 74.2 | 91.2 | ||
| 81 PDAC patients and 39 healthy controls | PDAC patients vs. healthy controls | 0.96 | — | — | Li et al. (2013) | |
| 81 PDAC patients and 45 chronic pancreatitis samples | PDAC patients vs. chronic pancreatitis samples | 0.81 | — | — | ||
| 81 PDAC patients and 28 PNETs patients | PDAC patients vs. PNET samples | 0.80 | — | — | ||
| 81 PDAC patients and all controlls | PDAC patients vs. all controls | 0.85 | — | — | ||
| Lung cancer | 70 LUAD patients and 40 healthy controls | LUAD patients vs. controls | 0.937 | 80.0 | 96.7 | Wu et al. (2020) |
| Colorectal cancer (CRC) | 15 CRC patients, 15 colorectal adenoma patients and 15 healthy controls | CRC patients vs. healthy controls | 0.96 | 78.79 | 93.33 | Liu et al. (2019) |
| colorectal adenoma patients vs. healthy controls | 0.92 | 79.66 | 86.67 | |||
| 12 CRC patients,12 colorectal adenoma patients, and 12 healthy controls | CRC patients vs. healthy controls | 1.000 | 100 | 100 | Imaoka et al. (2016) | |
| colorectal adenoma patients vs healthy controls | 0.722 | 50 | 100 | |||
| 211 CRC patients, 56 colorectal adenoma patients, and 57 healthy controls | CRC patients vs. healthy controls | 0.830 | 70.1% | 91.2 | ||
| colorectal adenoma patients vs. healthy controls | 0.718 | 46.4 | 91.2 | |||
| Ovarian cancer (OC) | sera samples from 70 EOC patients and 13 healthy controls | EOC patients vs. healthy controls | 0.48 | 0.51 | 0.57 | Kobayashi et al. (2018) |
| 30 HGSOC patients and 13 healthy controls | HGSOC patients vs. healthy controls | 0.71 | 0.63 | 0.85 |
Discussion
Several miRNAs have been found to influence the carcinogenesis. miR-1290 is an example of oncomiRs based on the bulk of relevant evidence. This miRNA has interactions with several cancer-related mRNAs such as SOCS4, GSK3, BCL2, CCNG2, KIF13B, INPP4B, hMSH2, KIF13B, NKD1, FOXA1, IGFBP3, FOXA1, NAT1, SMEK1, SCAI, ZNF667-AS1, ABLIM1, and CDC73.
Moreover, miR-1290 has interactions with a number of non-coding RNAs such as Circ_0000629, CCTA1 and NONHSAT024778. The interaction between lncRNAs/circRNAs and miRNAs has important implications in pathoetiology of cancers, thus future studies are needed to identify other non-coding RNAs that interact with miR-1290 in the context of neoplastic conditions. In fact, these lncRNAs and circRNAs can act as sponge for miRNAs to decrease its bioavalability, thus enhancing expression of targets of miR-1290. Therefore, they construct a competing endogenous RNA (ceRNA) network.
In addition to its role in the regulation of gene expression, miR-1290 can regulate activity of JAK/STAT3, PI3K/AKT, Wnt/β-catenin and NF-κB signaling pathways, thus influencing several cancer-related routes.
Most evidence indicates the oncogenic roles of miR-1290, yet controversial evidence also exists. Particularly, in the lung cancer, both oncogenic and tumor suppressor roles have been reported for miR-1290.
A number of anticancer agents such as POL, asiatic acid and resveratrol has been shown to affect expression of miR-1290. Moreover, this miRNA can influence response of neoplastic cells to the chemotherapeutic agent 5-fluouracil. Thus, one can deduce that miR-1290-targeting strategies can modulate response of cancer cells to a wide variety of antineoplastic modalities.
In addition to its therapeutic implications, the existence of miR-1290 in cancer-derived exosomes not only indicates its application in diagnostic approaches, but also shows the effect of these vehicles in conferring neoplastic features inside the tumor bulk.
The ceRNA networks constructed by circRNAs, miR-1290 and target mRNAs can be used as prognostic biomarkers and therapeutic targets in different cancers. These ceRNA networks are superior to single transcripts since they reflect a more comprehensive overview of dysregulated pathways. Theoretically, the ceRNA regulatory networks including lncRNAs or circRNAs-miR-1290-mRNAs can be applied as prognostic biomarkers and therapeutic targets in different cancers. High throughput sequencing methods have facilitated applicability of these networks in diagnostic, prognostic and therapeutic fields. Moreover, these techniques have facilitated design of personalized therapeutic options based on the identified dysregulated networks in samples obtained from each patient. Application of this data can enhance survival of patients.
Cumulatively, miR-1290 is a cancer-related miRNA with possible application as diagnostic and prognostic marker in diverse types of cancers. Therapeutic applications of anti-miR-1290 modalities should be assessed in future. Moreover, future studies should address the possibility of targeting the miR1290-containg ceRNA networks.
Author Contributions
SG-F wrote the draft and revised it. MT designed and supervised the study. TK and MS collected the data and designed the figures and tables. All the authors read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abolghasemi M., Tehrani S. S., Yousefi T., Karimian A., Mahmoodpoor A., Ghamari A., et al. (2020). MicroRNAs in Breast Cancer: Roles, Functions, and Mechanism of Actions. J. Cell Physiol 235, 5008–5029. 10.1002/jcp.29396 [DOI] [PubMed] [Google Scholar]
- Chen X., Huang Y., Shi D., Nie C., Luo Y., Guo L., et al. (2020). LncRNA ZNF667-AS1 Promotes ABLIM1 Expression by Adsorbing microRNA-1290 to Suppress Nasopharyngeal Carcinoma Cell Progression. Ott 13, 4397–4409. 10.2147/ott.s245554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rie D., Abugessaisa I., Abugessaisa I., Alam T., Arner E., Arner P., et al. (2017). An Integrated Expression Atlas of miRNAs and Their Promoters in Human and Mouse. Nat. Biotechnol. 35, 872–878. 10.1038/nbt.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Toyama T., Takahashi S., Yoshimoto N., Iwasa M., Asano T., et al. (2013). miR-1290 and its Potential Targets Are Associated with Characteristics of Estrogen Receptor α-positive Breast Cancer. Endocrine-related cancer 20, 91–102. 10.1530/erc-12-0207 [DOI] [PubMed] [Google Scholar]
- Geusau A., Borik-Heil L., Skalicky S., Mildner M., Grillari J., Hackl M., et al. (2020). Dysregulation of Tissue and Serum microRNAs in Organ Transplant Recipients with Cutaneous Squamous Cell Carcinomas. Health Sci. Rep. 3, e205. 10.1002/hsr2.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa T., Kuroha M., Nagai H., Shimoyama Y., Naito T., Moroi R., et al. (2021). Liquid Biopsy for Colorectal Adenoma: Is the Exosomal miRNA Derived from Organoid a Potential Diagnostic Biomarker? Clin. Transl Gastroenterol. 12, e00356. 10.14309/ctg.0000000000000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Shen M., Yan M., Cui Y., Gao Z., Meng X. (2019). Exosome-mediated Transfer of miR-1290 Promotes Cell Proliferation and Invasion in Gastric Cancer via NKD1. Acta Biochim. Biophys. Sinica 51, 900–907. 10.1093/abbs/gmz077 [DOI] [PubMed] [Google Scholar]
- Huang X., Yuan T., Liang M., Du M., Xia S., Dittmar R., et al. (2015). Exosomal miR-1290 and miR-375 as Prognostic Markers in Castration-Resistant Prostate Cancer. Eur. Urol. 67, 33–41. 10.1016/j.eururo.2014.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaoka H., Toiyama Y., Fujikawa H., Hiro J., Saigusa S., Tanaka K., et al. (2016). Circulating microRNA-1290 as a Novel Diagnostic and Prognostic Biomarker in Human Colorectal Cancer. Ann. Oncol. 27, 1879–1886. 10.1093/annonc/mdw279 [DOI] [PubMed] [Google Scholar]
- Janiszewska J., Szaumkessel M., Kostrzewska-Poczekaj M., Bednarek K., Paczkowska J., Jackowska J., et al. (2015). Global miRNA Expression Profiling Identifies miR-1290 as Novel Potential oncomiR in Laryngeal Carcinoma. PLoS One 10, e0144924. 10.1371/journal.pone.0144924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. B., Kim K., Bae S., Choi Y., Cha H. J., Kim S. Y., et al. (2014). MicroRNA-1290 Promotes Asiatic Acid-Induced Apoptosis by Decreasing BCL2 Protein Level in A549 Non-small Cell Lung Carcinoma Cells. Oncol. Rep. 32, 1029–1036. 10.3892/or.2014.3319 [DOI] [PubMed] [Google Scholar]
- Kim Y.-K., Kim V. N. (2007). Processing of Intronic microRNAs. Embo J. 26, 775–783. 10.1038/sj.emboj.7601512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Sawada K., Nakamura K., Yoshimura A., Miyamoto M., Shimizu A., et al. (2018). Exosomal miR-1290 Is a Potential Biomarker of High-Grade Serous Ovarian Carcinoma and Can Discriminate Patients from Those with Malignancies of Other Histological Types. J. Ovarian Res. 11, 81–10. 10.1186/s13048-018-0458-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X. J., Cheng H. F. (2018). LncRNA colon Cancer-Associated Transcript 1 (CCAT1) Promotes Proliferation and Metastasis of Ovarian Cancer via miR-1290. Eur. Rev. Med. Pharmacol. Sci. 22, 322–328. 10.26355/eurrev_201801_14175 [DOI] [PubMed] [Google Scholar]
- Li A., Yu J., Kim H., Wolfgang C. L., Canto M. I., Hruban R. H., et al. (2013). MicroRNA Array Analysis Finds Elevated Serum miR-1290 Accurately Distinguishes Patients with Low-Stage Pancreatic Cancer from Healthy and Disease Controls. Clin. Cancer Res. 19, 3600–3610. 10.1158/1078-0432.ccr-12-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhang H., Lu G., Li Q., Gu J., Song Y., et al. (2016). Mechanism Analysis of Colorectal Cancer According to the microRNA Expression Profile. Oncol. Lett. 12, 2329–2336. 10.3892/ol.2016.5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., He X.-Y., Zhang Z.-M., Li S., Ren L.-H., Cao R.-S., et al. (2015). MicroRNA-1290 Promotes Esophageal Squamous Cell Carcinoma Cell Proliferation and Metastasis. Wjg 21, 3245–3255. 10.3748/wjg.v21.i11.3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang M., Xu F., Wang Y., Leng D. (2021). Detection Significance of miR-3662, miR-146a, and miR-1290 in Serum Exosomes of Breast Cancer Patients. J. Can. Res. Ther. 17, 749. 10.4103/jcrt.jcrt_280_21 [DOI] [PubMed] [Google Scholar]
- Lin M., Shi C., Lin X., Pan J., Shen S., Xu Z., et al. (2016). sMicroRNA-1290 Inhibits Cells Proliferation and Migration by Targeting FOXA1 in Gastric Cancer Cells. Gene 582, 137–142. 10.1016/j.gene.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Liu X., Xu X., Pan B., He B., Chen X., Zeng K., et al. (2019). Circulating miR-1290 and miR-320d as Novel Diagnostic Biomarkers of Human Colorectal Cancer. J. Cancer 10, 43–50. 10.7150/jca.26723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Wang H., Sun J., Liu H., Zheng C., Zhou X., et al. (2018). LINC00152 Promotes Cell Cycle Progression in Hepatocellular Carcinoma via miR-193a/b-3p/CCND1 axis. Cell cycle 17, 974–984. 10.1080/15384101.2018.1464834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo D., Gu B., Gong X., Wu L., Wang H., Jiang Y., et al. (2015). miR-1290 Is a Potential Prognostic Biomarker in Non-small Cell Lung Cancer. J. Thorac. Dis. 7, 1570–1579. 10.3978/j.issn.2072-1439.2015.09.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamitsu Y., Nishi H., Sasaki T., Takaesu Y., Terauchi F., Isaka K. (2016). Profiling Analysis of Circulating microRNA Expression in Cervical Cancer. Mol. Clin. Oncol. 5, 189–194. 10.3892/mco.2016.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima H., Yoshida R., Hirosue A., Kawahara K., Sakata J., Arita H., et al. (2019). Circulating miRNA-1290 as a Potential Biomarker for Response to Chemoradiotherapy and Prognosis of Patients with Advanced Oral Squamous Cell Carcinoma: A Single-center Retrospective Study. Tumour Biol. 41, 1010428319826853. 10.1177/1010428319826853 [DOI] [PubMed] [Google Scholar]
- O'Brien J., Hayder H., Zayed Y., Peng C. (2018). Overview of microRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 9, 402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Croce C. M. (2016). The Role of MicroRNAs in Human Cancer. Signal. Transduct Target. Ther. 1, 15004–15009. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W. J., Wang W. P., Wang X. B., Zhang X. T., Du J. D. (2019). MiR-1290 Targets CCNG2 to Promote the Metastasis of Oral Squamous Cell Carcinoma. Eur. Rev. Med. Pharmacol. Sci. 23, 10332–10342. 10.26355/eurrev_201912_19671 [DOI] [PubMed] [Google Scholar]
- Sun X., Song Y., Tai X., Liu B., Ji W. (2013). MicroRNA Expression and its Detection in Human Supraglottic Laryngeal Squamous Cell Carcinoma. Biomed. Rep. 1, 743–746. 10.3892/br.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavano F., Gioffreda D., Valvano M. R., Palmieri O., Tardio M., Latiano T. P., et al. (2018). Droplet Digital PCR Quantification of miR-1290 as a Circulating Biomarker for Pancreatic Cancer. Sci. Rep. 8, 16389. 10.1038/s41598-018-34597-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Zhang K., Meng S., Shao X., Zhou Z., Mao H., et al. (2021a). LncRNA-NONHSAT024778 Promote the Proliferation and Invasion of Chordoma Cell by Regulating miR-1290/Robo1 axis. Int. J. Biol. Sci. 17, 796–806. 10.7150/ijbs.54091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Luo J., Wu X., Gao Z. (2021b). Circular RNA_0000629 Suppresses Bladder Cancer Progression Mediating MicroRNA-1290/CDC73. Cmar 13, 2701–2715. 10.2147/cmar.s292863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang G., Niu L., Zhao S., Li J., Zhang Z., et al. (2021). Exosomal MiR-1290 Promotes Angiogenesis of Hepatocellular Carcinoma via Targeting SMEK1. J. Oncol., 2021, 1–13. 10.1155/2021/6617700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Yang L., Wu Y.-N., Xu J. (2020). Serum miR-1290 and miR-1246 as Potential Diagnostic Biomarkers of Human Pancreatic Cancer. J. Cancer 11, 1325–1333. 10.7150/jca.38048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Ji X., Zhu L., Jiang Q., Wen Z., Xu S., et al. (2013). Up-regulation of microRNA-1290 Impairs Cytokinesis and Affects the Reprogramming of colon Cancer Cells. Cancer Lett. 329, 155–163. 10.1016/j.canlet.2012.10.038 [DOI] [PubMed] [Google Scholar]
- Wu L., Liu T., Xiao Y., Li X., Zhu Y., Zhao Y., et al. (2016). Polygonatum Odoratum Lectin Induces Apoptosis and Autophagy by Regulation of microRNA-1290 and microRNA-15a-3p in Human Lung Adenocarcinoma A549 Cells. Int. J. Biol. macromolecules 85, 217–226. 10.1016/j.ijbiomac.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Wu Y., Wei J., Zhang W., Xie M., Wang X., Xu J. (2020). Serum Exosomal miR-1290 Is a Potential Biomarker for Lung Adenocarcinoma. Ott 13, 7809–7818. 10.2147/ott.s263934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Yang D., Gong X., Mo D., Pan S., Xu J. (2018). miR-1290 Promotes Lung Adenocarcinoma Cell Proliferation and Invasion by Targeting SOCS4. Oncotarget 9, 11977–11988. 10.18632/oncotarget.24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Rao Q., Liu L., Zheng C., Xie Q., Liang J., et al. (2013). Exploration of Tumor-Suppressive microRNAs Silenced by DNA Hypermethylation in Cervical Cancer. Virol. J. 10, 175–177. 10.1186/1743-422X-10-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Jiang T., Shao H., Zhong L., Wang Z., Liu Y., et al. (2017). miR-1290 Is a Biomarker in DNA-Mismatch-Repair-Deficient colon Cancer and Promotes Resistance to 5-fluorouracil by Directly Targeting hMSH2. Mol. Ther. - Nucleic Acids 7, 453–464. 10.1016/j.omtn.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Wang S., Ying Y., Zhou R., Mao P. (2017). miR-196b/miR-1290 Participate in the Antitumor Effect of Resveratrol via Regulation of IGFBP3 Expression in Acute Lymphoblastic Leukemia. Oncol. Rep. 37, 1075–1083. 10.3892/or.2016.5321 [DOI] [PubMed] [Google Scholar]