Abstract
Background
Confocal laser microscopy (CLM) is one of the optical techniques that are promising methods of intraoperative in vivo real-time tissue examination based on tissue fluorescence. However, surgeons might struggle interpreting CLM images intraoperatively due to different tissue characteristics of different tissue pathologies in clinical reality. Deep learning techniques enable fast and consistent image analysis and might support intraoperative image interpretation. The objective of this study was to analyze the diagnostic accuracy of newly trained observers in the evaluation of normal colon and peritoneal tissue and colon cancer and metastasis, respectively, and to compare it with that of convolutional neural networks (CNNs).
Methods
Two hundred representative CLM images of the normal and malignant colon and peritoneal tissue were evaluated by newly trained observers (surgeons and pathologists) and CNNs (VGG-16 and Densenet121), respectively, based on tissue dignity. The primary endpoint was the correct detection of the normal and cancer/metastasis tissue measured by sensitivity and specificity of both groups. Additionally, positive predictive values (PPVs) and negative predictive values (NPVs) were calculated for the newly trained observer group. The interobserver variability of dignity evaluation was calculated using kappa statistic. The F1-score and area under the curve (AUC) were used to evaluate the performance of image recognition of the CNNs' training scenarios.
Results
Sensitivity and specificity ranged between 0.55 and 1.0 (pathologists: 0.66–0.97; surgeons: 0.55–1.0) and between 0.65 and 0.96 (pathologists: 0.68–0.93; surgeons: 0.65–0.96), respectively. PPVs were 0.75 and 0.90 in the pathologists' group and 0.73–0.96 in the surgeons' group, respectively. NPVs were 0.73 and 0.96 for pathologists' and between 0.66 and 1.00 for surgeons' tissue analysis. The overall interobserver variability was 0.54. Depending on the training scenario, cancer/metastasis tissue was classified with an AUC of 0.77–0.88 by VGG-16 and 0.85–0.89 by Densenet121. Transfer learning improved performance over training from scratch.
Conclusions
Newly trained investigators are able to learn CLM images features and interpretation rapidly, regardless of their clinical experience. Heterogeneity in tissue diagnosis and a moderate interobserver variability reflect the clinical reality more realistic. CNNs provide comparable diagnostic results as clinical observers and could improve surgeons' intraoperative tissue assessment.
Keywords: Machine learning strategies, Deep learning, Colon cancer, Confocal laser microscopy, Convolutional neural networks, Medical engineering, Minimal invasive surgery
Introduction
Cancer will be a leading cause of death in a few decades due to the aging world population and better treatment of cardiovascular disease [1]. In 2018, 18.1 million new cancer cases and 9.6 million cancer deaths occurred [2]. The economic impact of cancer is considerable with a total annual cost estimated at approximately USD 1.16 trillion in 2010 [3]. Since the introduction of minimally invasive surgical techniques in oncological surgery with reduced tactility and, in some cases, a limited intraoperative overview, surgeons request an intraoperative tissue navigation system enabling individual cancer resection while protecting healthy tissue [4].
Confocal laser microscopy (CLM) is one of the imaging techniques that are promising methods for intraoperative in vivo real-time tissue examination based on tissue fluorescence [5]. Especially during minimally invasive surgery, optical biopsy tissue navigation by CLM might support surgeons' resection strategies. Several studies are evaluating the feasibility of CLM during surgery [6, 7]. In recent studies, we evaluated a newly developed CLM device for minimally invasive surgery that does not need any fluorescent staining and showed that this CLM system enables to differentiate between benign and malignant colon and peritoneal tissue, respectively [8, 9].
Despite its promising diagnostic potential, newly trained users might struggle interpreting CLM images. In particular, large amounts of data may create an information overload for surgeons during surgery. Also, the diagnosis is examiner-dependent, leading to considerable interobserver variability. Furthermore, surgeons will need computer-aided tissue examination tools to overcome the limitations of tissue interpretation and classification.
Recently, using deep learning strategies such as convolutional neural networks (CNN), automatic tissue characterization has been successfully addressed for semantic segmentation and classification [10]. Several studies showed that deep learning strategies are powerful tools for automatic tissue characterization, for example, in dermatology [11]. However, the number of medical images is typically rather small. This can be problematic as insufficient data might lead to overfitting and limited generalization when training machine learning methods. This is particularly important for deep learning models, which are prone to overfitting due to the large number of trainable parameters. To overcome this issue, transfer learning methods have been proposed where a deep learning model is first pretrained on a different, large dataset, and afterward the information from the source domain can be transferred to the (medical) target domain [12]. In a recent study, we showed that CNNs and transfer learning strategies can be used to identify and classify the benign and malignant colon and peritoneal tissue, respectively [12]. However, there has not been any comparison of diagnostic accuracy between observers and CNN-based CLM image interpretation yet.
Therefore, a first purpose of this study was the evaluation of diagnostic accuracy and interobserver variability of newly trained observers interpreting CLM images of the benign and malignant colon and peritoneal tissue, respectively. Second, we compared the diagnostic accuracy of newly trained observers with that of deep learning image analysis strategies.
Materials and Methods
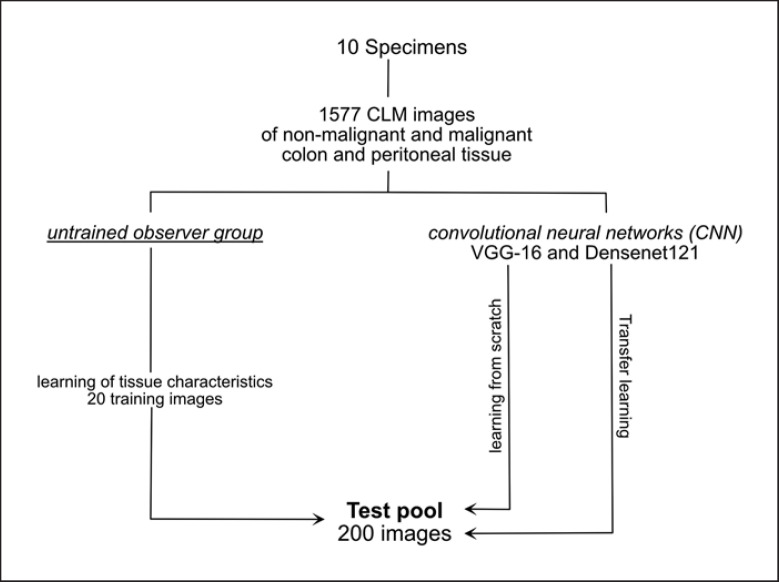
The study was designed to evaluate the diagnostic accuracy and interobserver variability of CLM image interpretation by newly trained observers compared to deep learning-based CNN image analysis. All procedures were approved by the Medical Ethics Committee (register number: 19-427A). We used CLM images of the normal colon and peritoneum and colon cancer, and its peritoneal metastasis, respectively, reported by our study group in a previous study [9]. Two hundred (50 each of normal colon, normal peritoneum, colon cancer, and peritoneal metastases) out of 1,577 CLM images were selected by the study investigators for diagnostic accuracy tests of newly trained participants and CNNs (VGG-16 and Densenet121), respectively. Afterward, we performed the training of newly trained participants and the training of the CNNs. The newly trained observer group consists of surgeons and pathologists because we assumed that pathologists would perform better in CLM interpretation due to their histopathological expertise. Presenting randomized CLM images online, the participants had to distinguish between the benign and malignant tissue and colon, peritoneum, and malignant tissue, respectively. The CNNs had to distinguish between the normal and malignant tissue during the test (Fig. 1).
Fig. 1.
Study design.
CC531 Adenocarcinoma Model
We used the CC531 colon adenocarcinoma model in rats developed by Marquet et al. [13]. Maintenance and care of all animals used in this study were carried out according to the direction of national animal protection law and according to the directories of European Community Council (2010/63/E4). The animals were housed in groups of 3 up to 4 animals and had free access to food and water. They got accustomed to their new surrounding and to the investigator for at least 10 days before the first surgical intervention.
CC531 moderately differentiated colon adenocarcinoma cells (Cell Lines Services GmbH, Eppelheim, Germany) were cultured in 20 mL complete RMPI 1640 medium (Cell Lines Services GmbH, Eppelheim, Germany) with 10% heat-inactivated fetal bovine serum (Cell Lines Services GmbH, Eppelheim, Germany) and 1% penicillin/streptomycin at 37°C and 5% CO2 in monolayer cultures. To prepare a tumor suspension for intraperitoneal application, the complete medium was removed; the cells were washed with 20 mL phosphate-buffered saline and were detached by 4 mL Accutase (Cell Lines Services GmbH, Eppelheim, Germany). After incubating for 10 min at 37°C, 6 mL complete medium was added. The cells were harvested, suspended in phosphate-buffered saline, and centrifuged at 300 g for 5 min. Vital counting was performed in a Burker hematocytometer.
All rats were anesthetized with 2% xylazinum (4–6 mg/kg body weight) and 10% S-ketamine (2–3 mg/kg body weight) by intraperitoneal injection. After opening the abdomen via a midline incision, 200 µL of the tumor suspension (density: 2.5 × 106 vital cell/200 µL) was implanted in the colon and the peritoneum of the right abdomen. The midline incision was closed by subcutaneous polyfilament 4–0 continuous suture (polyglectin 910, Vicryl® Ethicon, Germany) and cutaneous monofilament 3–0 interrupted suture (polyamide, Vicryl® Ethicon, Germany). After the surgical procedure, animals were allowed to recover from anesthesia before returning to the animal facility. They were weighted and examined for side effects (wound infection, loss of appetite, fatigue syndrome, and lethargy).
In vivo CLM
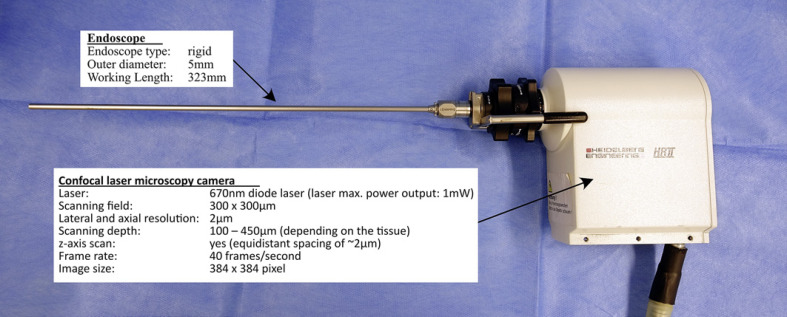
After 7–10 days of tumor growth, we performed a re-laparotomy using anesthesia, as described above, and CLM was performed in both the normal and malignant tissues in the colon and peritoneum. The CLM system consists of a modified Rostock Cornea module of the Heidelberg Engineering Retina Tomograph II HRT II (Heidelberg Engineering GmbH, Heidelberg, Germany) and a specially developed rigid endoscope (KARL STORZ SE & Co. KG, Tuttlingen, Germany) (Fig. 2). The light source is a 670-nm diode laser. A beam splitter deflects the collimated laser beam, the laser light passes the rigid endoscope and the backscattered light travels the same way back. A second beam splitter separates the reflected light from the illumination path, and the signal is detected by the photo diode with pinhole in the confocal plane. The circular scan field is 300 µm × 300 µm with a maximum lateral and axial calculated resolution of 1–2 µm. The penetration depth is between 100–450 µm depending on the tissue. The scan rate of 40 frames provides a real-time assessment.
Fig. 2.
Features of the eCLM device. eCLM, endoscopic confocal laser microscopy.
Newly Trained Observers and Online Questionnaire
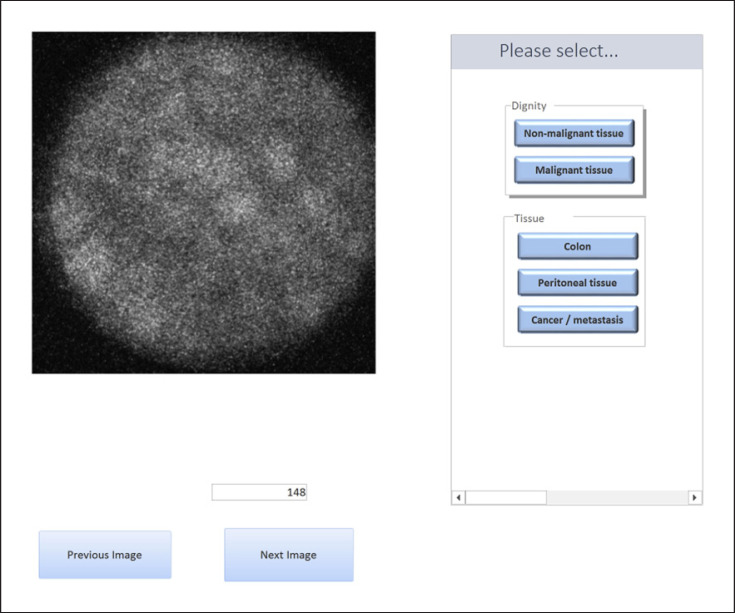
Six surgeons and 2 pathologists participated in this study. The surgeons involved in this study were experts in the field of visceral surgery but not in the field of CLM imaging. The pathologists were experts in the field of gastrointestinal pathology. However, they had no experience with CLM imaging. The participants were explained the features of each tissue type in a short presentation (Fig. 3 and Table 1). The CLM images used in the presentation were not part of the diagnostic accuracy test pool. No histological images were used for training or during the questionnaire. After 1 week, they were allowed to perform the test. An online questionnaire was developed with MicrosoftTM Access, with questions concerning tissue dignity (normal or malignant) and type of tissue (normal colon, normal peritoneum, and colon cancer/metastasis) (Fig. 4). The CLM images were presented randomly to the observers.
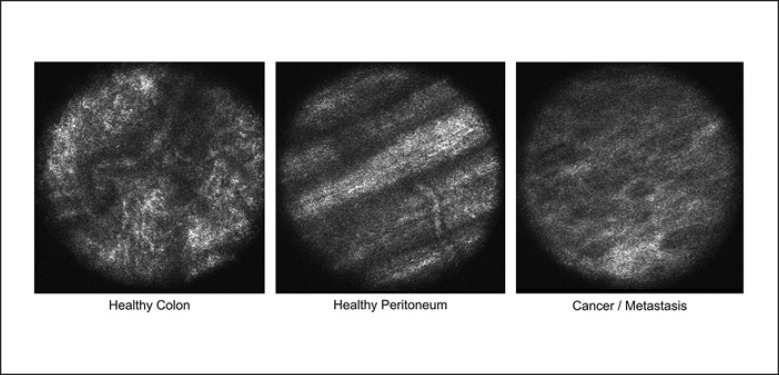
Fig. 3.
In vivo CLM of the normal tissue in the colon and peritoneum and cancer/metastasis regions. In normal colon regions, regular round or oval structures were seen. Additionally, small vessels can be detected. In colon cancer regions, regular patterns were deregulated and irregular cell arrangements and morphology were detected. Observing the normal peritoneum, abdominal wall muscles in areas with the healthy tissue can be identified due to scanning depth of the CLM camera system. Peritoneal metastasis regions showed the same pattern as primary colon cancer regions in the CLM scan. CLM, confocal laser microscopy.
Table 1.
CLM criteria for tissue interpretation
| CLM criteria | |
|---|---|
| Normal colon |
Regular round or oval crypts Linear crypts Vessels detecting possible |
| Normal peritoneum |
Abdominal wall muscles Striated pattern with white and black bands Indistinct longitudinal stripes |
| Colon cancer and metastasis | Irregular cell arrangements Large, elongated crypts Diffuse edge Necrotic debris No vessel detection |
CLM, confocal laser microscopy.
Fig. 4.
Online questionnaire presented to the study participants.
CNN Model Architecture and Training Strategies
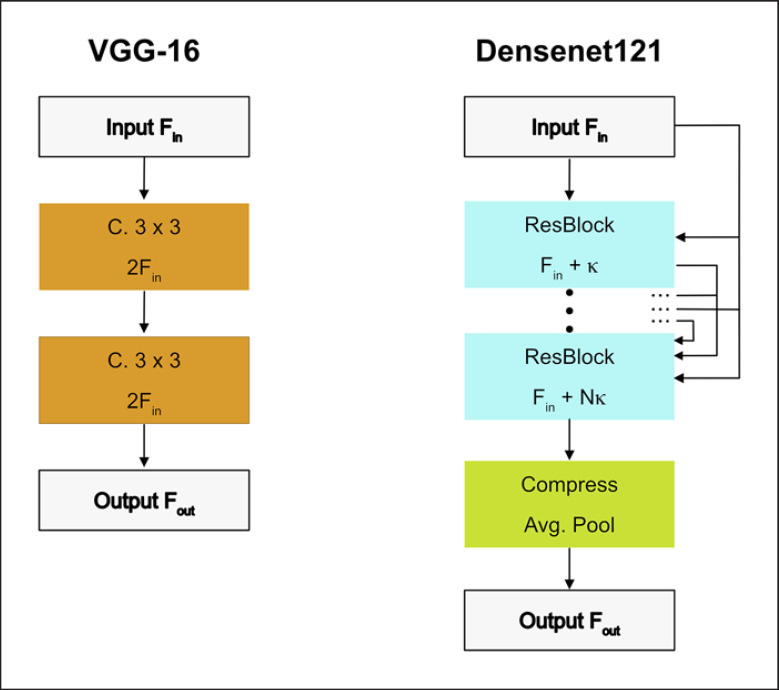
The VGG-16 model is simple as it consists of several stacked convolutional layers that gradually reduce spatial dimensions while increasing the feature map size. Convolutional layers use kernel sizes 3 × 3 and 1 × 1. Spatial reduction is performed with max pooling. After max pooling, the next layer doubles the number of feature maps. We added batch normalization for faster training of the model by reducing the internal covariate shift (Fig. 5a) [14].
Fig. 5.
Building blocks of CNN architectures. We employ VGG-16 (a) and Densenet121 (b). F denotes the number of feature maps in each block. The Conv blocks also contain ReLU activations and batch normalization for VGG-16. C is an abbreviation for convolutional layers. CNN, convolutional neural network.
The Densenet121 is a state-of-the-art architecture that is more efficient than VGG-16 as it reuses its features frequently [15]. The features computed in a Dense Block also use features that have been computed by previous layers. To keep feature map sizes moderate, compression blocks reduce the number of features maps between Dense Blocks (Fig. 5b).
As a baseline, we considered training from scratch, that is, all weights were randomly initialized. We also investigated a transfer learning strategy with partial freezing, where an initial part of the network remains frozen, and the part closer to the classifier is retrained. In this way, generalizable features that have been learned on a generic and large dataset (ImageNet) can be reused. Due to the small dataset size, we chose a cross-validation scheme where images from 1 subject were left for evaluation, and training was performed on the remaining ones. We considered the differentiation of normal and malignant tissue with the binary classification.
To further improve generalization, we employed online data augmentation with random image flipping and random changes in brightness and contrast. Furthermore, we used random cropping with crops of size 224 × 224) taken from the full images of size 384 × 384. We applied the Adam algorithm for optimization and adapted learning rates and the number of training epochs for the different transfer scenarios. We used a cross-entropy loss function with additional weighting to account for the slight class imbalance. In detail, we multiplied the loss of a training example by N/ni, where N is the total number of training examples in the current fold and ni is the number of examples belonging to class i in the current fold. In this way, underrepresented classes received a higher weighting in the loss function. During evaluation, we used multi-crop evaluation with Nc = 36 evenly spread crops over the images. This ensured that all image regions were covered with large overlaps between crops. The final predictions were averaged over the Nc crops. We implemented our models in PyTorch. Training was performed on an NVIDIA GTX 1080TI.
Data Analysis
The primary endpoint of the newly trained observer test was the correct diagnosis of normal and cancer/metastasis tissue. Sensitivity, specificity, positive predictive values (PPVs), and negative predictive values (NPVs) with corresponding 95% Wilson confidence intervals (CIs) were calculated. Furthermore, the interobserver variability of dignity evaluation was calculated using Cohen's kappa and multi-rater Fleiss's kappa statistics, respectively, with the following classification: poor <0.2, fair 0.21–0.4, moderate 0.41–0.6, substantial 0.61–0.8, and excellent 0.81–1. Due to the expected heterogeneity between observers, we calculated the observer-specific percentage of correct tissue ratings for each observer. All analyses were performed using the statistical software environment R [16].
For the diagnostic accuracy of the CNNs, we calculated the sensitivity and specificity. Additionally, we calculated the accuracy score, the recall ratio (F1-score), and the area under the curve for the performance interpretation of the CNN scenarios.
Results
Newly Trained Observers
In total, 6 surgeons and 2 pathologists performed the complete questionnaire. Pathologists' clinical experience was 20 and 10 years, respectively. In the surgeons group, the experience ranged between 3 and 21 years. Untrained observers' results of diagnostic accuracy are displayed in Table 2.
Table 2.
Newly trained observers' diagnostic accuracy
| Tester | Clinical experience, years | Sensitivity [95% CI] | Specificity [95% CI] | PPV [95% CI] | NPV [95% CI] |
|---|---|---|---|---|---|
| 1 | 20 | 0.66 [0.56; 0.75] | 0.93 [0.86; 0.97] | 0.90 [0.82; 0.95] | 0.73 [0.65; 0.80] |
| 2 | 10 | 0.97 [0.92; 0.99] | 0.68 [0.58; 0.76] | 0.75 [0.67; 0.82] | 0.96 [0.88; 0.99] |
| 3 | 20 | 0.55 [0.45; 0.64] | 0.89 [0.81; 0.94] | 0.83 [0.73; 0.90] | 0.66 [0.58; 0.73] |
| 4 | 21 | 0.93 [0.86; 0.97] | 0.65 [0.55; 0.74] | 0.73 [0.64; 0.80] | 0.90 [0.81; 0.95] |
| 5 | 9 | 0.93 [0.86; 0.97] | 0.89 [0.81; 0.94] | 0.89 [0.82; 0.94] | 0.93 [0.86; 0.96] |
| 6 | 5 | 0.90 [0.83; 0.94] | 0.94 [0.88; 0.97] | 0.93 [0.87; 0.97] | 0.90 [0.83; 0.95] |
| 7 | 3 | 0.96 [0.90; 0.98] | 0.94 [0.88; 0.97] | 0.94 [0.88; 0.97] | 0.96 [0.90; 0.98] |
| 8 | 4 | 1.00 [0.96; 1.00] | 0.96 [0.90; 0.98] | 0.96 [0.91; 0.98] | 1.00 [0.96; 1.00] |
CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
The results of sensitivity and specificity were heterogeneous. In detail, the pathologists' sensitivity and specificity were 0.66 (95% CI: 0.56; 0.75) and 0.97 (95% CI: 0.92; 0.99) and 0.93 (95% CI: 0.86; 0.97) and 0.68 (95% CI: 0.58; 0.76), respectively. The sensitivity ranged between 0.55 (95% CI: 0.45; 0.64) and 1.0 (95% CI: 0.81; 0.94) in the surgeon observer group. Also, the specificity showed a range of 0.65 (95% CI: 0.55; 0.74) to 0.96 (95% CI: 0.90; 0.98). PPVs and NPVs varied between 0.75 (95% CI: 0.67; 0.82) and 0.90 (95% CI: 0.82; 0.95) and 0.73 (95% CI: 0.63; 0.80) and 0.96 (95% CI: 0.88; 0.99), respectively, in the pathologists' group. The PPVs and NPVs range was between 0.73 (95% CI: 0.64; 0.80) and 0.96 (95% CI: 0.91; 0.98) and 0.66 (95% CI: 0.58; 0.73) and 1.00 (95% CI: 0.96; 1.00), respectively, in the surgeons' group. The surgeons obtained a substantial kappa value of 0.65, followed by a low value of 0.37 for the pathologists. The overall interobserver variability was moderate (0.54).
The correct tissue diagnosis ranged between 49% and 97% in both groups (pathologists: 76% and 82%; surgeons: 49–97%). Corresponding kappa values were moderate, with 0.47 for pathologists and surgeons, respectively, and 0.42 in total. The required time to complete the online questionnaire ranged between 10 and 30 min. This corresponds to an image interpretation time of 3–9 s per image.
Convolutional Neural Networks
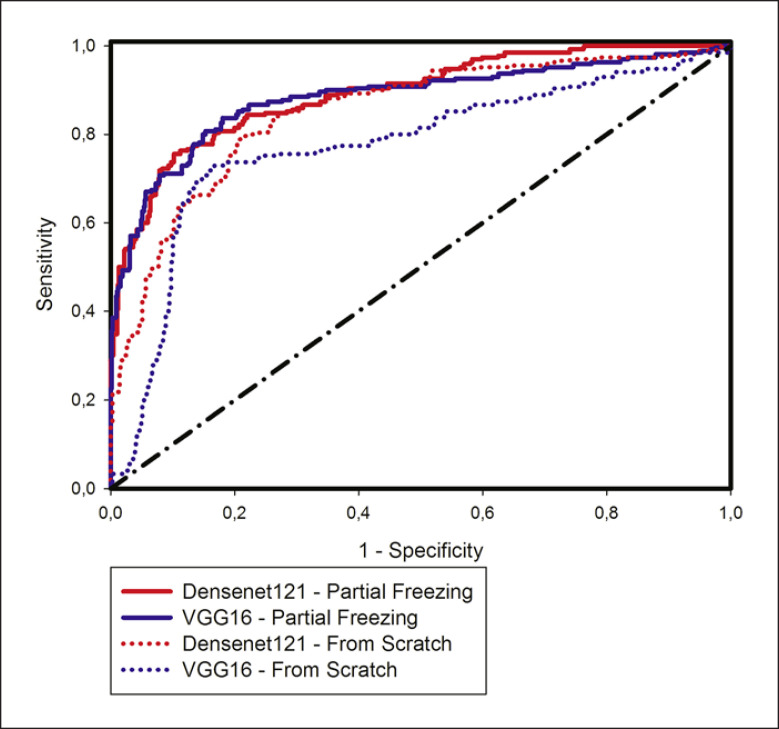
Both learning scenarios showed high values of differentiation of normal and cancer/metastasis tissue. Figure 6 shows the ROC curves for all models with transfer learning methods.
Fig. 6.
ROC curve for the different CNN architectures and the different training types. CNN, convolutional neural network.
For both VGG-16 and Densenet121, partial freezing performed better than learning from scratch. Comparing individual results for each architecture, Densenet121 performed slightly better than VGG-16 (Table 3). In contrast to the VGG-16 scenario, transfer learning with partial freezing improved the precision and recall values (F1-score) in the Densenet121 scenario.
Table 3.
CNNs' diagnostic accuracy
| Type | Accuracy | Sensitivity | Specificity | F1-score | AUC | |
|---|---|---|---|---|---|---|
| VGG-16 | From scratch | 0.72 | 0.67 | 0.87 | 0.81 | 0.77 |
| Partial freezing | 0.79 | 0.85 | 0.97 | 0.82 | 0.88 | |
| Densenet121 | From scratch | 0.72 | 0.88 | 0.64 | 0.72 | 0.85 |
| Partial freezing | 0.77 | 0.84 | 0.78 | 0.80 | 0.89 |
AUC, area under the curve; CNN, convolutional neural network.
The partial freezing transfer learning strategy affected sensitivity and specificity in different ways in the CNN scenarios. Due to partial freezing, sensitivity increased and specificity decreased in VGG-16. In contrast, partial freezing decreased the true positive rate and increased the true negative rate in the Densenet121 scenario. In both cases, the transfer learning strategy led to a better balance between sensitivity and specificity.
Training VGG-16 and Densenet121 from scratch for 90 epochs took 13 and 8 min, respectively. Partial freezing, where only a part of the CNN was retrained, took 8 and 4 min, respectively, for VGG-16 and Densenet121.
Discussion
In this study, we report the diagnostic accuracy of newly trained observers and compare the results to deep learning strategies. First, all newly trained observers were able to learn the CLM features of the normal colon and peritoneal tissue as well as colon cancer. However, the diagnostic accuracy data obtained in this study indicate that there is great heterogeneity and interobserver variability between the newly trained observer. We believe that these results reflect the clinical reality more realistically, especially at the beginning of CLM usage by newly trained investigators. In the field of CLM image usage during endoscopic procedures, some studies indicated that it is possible to reach a reliable diagnostic accuracy and interobserver variability [17, 18].
Although pathologists were experts in the field of gastrointestinal histology, we found comparable diagnostic accuracy in the surgeons' group. The results indicate that investigation of CLM features of the normal colon and peritoneal tissue as well as colon cancer/metastasis can be trained. Some endoscopic studies conducted state that a correct interpretation can be learned rapidly [16, 17, 18]. In this regard, diagnostic accuracy does not correlate with the clinical experience. The investigator with the shortest period of clinical experience performed best in the questionnaire. Thus, we believe that surgeons at the beginning of their clinical practice and senior surgeons are able to learn, interpreting CLM features for interoperative tissue assessment. Hoffmann et al. [16] mentioned that training success depends on the investigated tissue. They showed that the correct diagnosis of the benign and malignant tissue is more difficult in esophagus pathologies than in the gastric or colon tissue. In this case, pathology will be the backbone of tissue diagnosis and learning curve. Surgeons and pathologists will perform the tissue analysis intraoperatively together [7].
At this point, we note that tissue interpretation might be more difficult in rats than in the human colon and peritoneal tissue, and the investigators might perform better questionnaires in human tissue interpretation. This assumption is based on the fact that we were able to demonstrate a significantly more detailed image resolution with the same eCLM device in a previous study in the human colon tissue [8]. Second, autofluorescence, due to aging products like lipofuscin, cannot be excluded. This might improve image quality in the human tissue as well.
As mentioned earlier, the laparoscopic CLM device does not need any fluorescent staining for comparable CLM image quality. The intraoperative usage of fluorescent dye will have advantages and disadvantages. On the one hand, fluorescent tissue staining might enable the improvement of correct image interpretation and diagnosis by highlighting features of the expected pathology. On the other hand, surgical procedures will be stalled awaiting the best image contrast, and repetitive tissue assessment might be unfeasible.
Due to its resolution, CLM tissue investigation is slower than OCT or macroscopic tissue investigation tools. Deep learning strategies will improve the assessment speed due to automatic tissue interpretation. In particular, deep learning methods provide consistent assessments. Our results showed that the newly trained observer needed 10–30 min for tissue interpretation of 200 CLM images. In contrast, longest duration of CNNs took 13 min for learning and test scenario together. Considering the interobserver variability, we found for CLM image interpretation, deep learning methods could reduce variability across clinics and departments. Still, we have to keep in mind that the results of tissue diagnosis are likelihoods, which the surgeon has to confirm.
The diagnostic accuracy of CNNs allows for 2 conclusions. First, CNNs enable comparable diagnostic accuracy, although we had to use significantly fewer CLM images for training the CNNs than other CNN training scenarios dealing with CLM image interpretation, for example, in neurosurgery [19]. In order to compensate for this disadvantage, we used a cross-validation scheme where 1 patient was held out for testing in each validation step. The CNN training scenario differed from the test pool for the newly trained observers as it contained images from several patients. A patient-stratified training pool would be too small for meaningful training, and mixing patients in the training and test pool would cause data leakage [20]. Nevertheless, our CNN-based performance metrics provided indications for the feasibility of automated tissue analysis. Moreover, we used transfer learning strategies to improve the CNNs' diagnostic accuracy. The partial freezing, which performed best in the precedent study [12], enhanced the diagnostic accuracy in both CNN scenarios. The idea of transfer learning strategies is to reuse a model that has already been trained on a large dataset. This model should have learned generic features that can be reused for CLM image classification. With partial freezing, we reused a part of these features while also learning new, CLM-specific features in the higher level CNN layers. This strategy has been successful in different medical image classification tasks [21]. However, until now, it is unclear which CNN performs best in different pathologies, and further studies are necessary to evaluate CNN and transfer learning strategies assessing different pathologies intraoperatively.
This study has several limitations. First, we consider only 2 CNN models and 2 transfer learning strategies. We chose Densenet121 with partial freezing and compared it to a standard CNN (VGG-16) because it appeared more consistent with interpreting colon and peritoneal CLM images [12]. Further studies are necessary to confirm Densenet121 and partial freezing for the best automatic tissue diagnosis of the colon and peritoneal tissue. Moreover, due to the small dataset size, the training set for CNN training was small. A larger training set will likely improve diagnostic accuracy. However, we used a cross-validation scheme to minimize the bias of diagnostic accuracy. Also, the number of newly trained observers is small. Yet, even the small observer number shows a heterogeneity, which reflects the real clinical situation.
In conclusion, newly trained investigators are able to learn CLM image features and interpretation rapidly, regardless of their clinical experience. However, there is heterogeneity in tissue diagnosis and a moderate interobserver variability, which reflects the clinical reality more realistic. Deep learning strategies provide comparable diagnostic results like the observers and could improve surgeons' intraoperative diagnostic tissue assessment as they can provide consistent estimates. However, further studies are necessary to evaluate the best deep learning strategies for automatic tissue diagnosis.
Statement of Ethics
Ethical approval for this study was provided by the University of Luebeck Ethics Committee (Register number: 19-427A). The images used in this study were obtained from study “Confocal laser microscopy as novel approach for real-time and in-vivo tissue examination during minimal-invasive surgery in colon cancer,” which was approved by the National Ethical Committee for Animal Studies.
Conflict of Interest Statement
The authors have no conflicts of interest or financial ties to disclose.
Funding Sources
Dr. David B. Ellebrecht got a junior research grant of the University of Lübeck for the study “Confocal laser microscopy as novel approach for real-time and in-vivo tissue examination during minimal-invasive surgery in colon cancer” (J02-2015).
Author Contributions
Study concept and design: David B. Ellebrecht, and N. Gessert. Acquisition of data: David B. Ellebrecht. Analysis and interpretation: David B. Ellebrecht, Nicole Heßler, A. Schlaefer, and N. Gessert. Study supervision: David B. Ellebrecht and N. Gessert. All authors have provided final approval of the version submitted.
References
- 1.Dagenais GR, Leong DP, Rangarajan S, Lanas F, Lopez-Jaramillo P, Gupta R, et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. Lancet. 2019 Sep 3;395:785–94. doi: 10.1016/S0140-6736(19)32007-0. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68((6)):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.WHO . World Health Organizsation (WHO); 2019. Cancer. [Google Scholar]
- 4.Crawshaw B, Delaney CP. Gastrointestinal surgery: real-time tissue identification during surgery. Nat Rev Gastroenterol Hepatol. 2013 Nov;10((11)):624–5. doi: 10.1038/nrgastro.2013.176. [DOI] [PubMed] [Google Scholar]
- 5.Ellebrecht DB, Latus S, Schlaefer A, Keck T, Gessert N. Towards an optical biopsy during visceral surgical interventions. Visc Med. 2020;36:70–9. doi: 10.1159/000505938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goetz M, Kiesslich R, Dienes HP, Drebber U, Murr E, Hoffman A, et al. In vivo confocal laser endomicroscopy of the human liver: a novel method for assessing liver microarchitecture in real time. Endoscopy. 2008 Jul;40((7)):554–62. doi: 10.1055/s-2008-1077296. [DOI] [PubMed] [Google Scholar]
- 7.Fuks D, Pierangelo A, Validire P, Lefevre M, Benali A, Trebuchet G, et al. Intraoperative confocal laser endomicroscopy for real-time in vivo tissue characterization during surgical procedures. Surg Endosc. 2019 May;33((5)):1544–52. doi: 10.1007/s00464-018-6442-3. [DOI] [PubMed] [Google Scholar]
- 8.Ellebrecht DB, Gebhard MP, Horn M, Keck T, Kleemann M. Laparoscopic confocal laser microscopy without fluorescent injection: a pilot ex vivo study in colon cancer. Surg innovation. 2016 Aug;23((4)):341–6. doi: 10.1177/1553350616637690. [DOI] [PubMed] [Google Scholar]
- 9.Ellebrecht DB, Kuempers C, Horn M, Keck T, Kleemann M. Confocal laser microscopy as novel approach for real-time and in-vivo tissue examination during minimal-invasive surgery in colon cancer. Surg Endosc. 2019 Jun;33((6)):1811–7. doi: 10.1007/s00464-018-6457-9. [DOI] [PubMed] [Google Scholar]
- 10.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017 Feb 2;542((7639)):115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gessert N, Bengs M, Wittig L, Drömann D, Keck T, Schlaefer A, et al. Deep transfer learning methods for colon cancer classification in confocal laser microscopy images. Int J Comput Assist Radiol Surg. 2019 May 25;14((11)):1837–45. doi: 10.1007/s11548-019-02004-1. [DOI] [PubMed] [Google Scholar]
- 13.Marquet RL, Westbroek DL, Jeekel J. Interferon treatment of a transplantable rat colon adenocarcinoma: importance of tumor site. Int J Cancer. 1984 May 15;33((5)):689–92. doi: 10.1002/ijc.2910330521. [DOI] [PubMed] [Google Scholar]
- 14.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016 June 26 - 2016 July 1; Las Vegas: Conference on Computer Vision and Pattern Recognition (CVPR) 2016:2818–26. [Google Scholar]
- 15.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017 July 22 - July 25; Honolulu: Conference on Computer Vision and Pattern Recognition (CVPR) 2017:4700–08. [Google Scholar]
- 16.Hoffman A, Rey JW, Mueller L, Hansen T, Goetz M, Tresch A, et al. Analysis of interobserver variability for endomicroscopy of the gastrointestinal tract. Dig Liver Dis. 2014 Feb;46((2)):140–5. doi: 10.1016/j.dld.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Lim LG, Yeoh KG, Salto-Tellez M, Khor CJ, Teh M, Chan YH, et al. Experienced versus inexperienced confocal endoscopists in the diagnosis of gastric adenocarcinoma and intestinal metaplasia on confocal images. Gastrointest Endosc. 2011 Jun;73((6)):1141–7. doi: 10.1016/j.gie.2011.01.068. [DOI] [PubMed] [Google Scholar]
- 18.Kuiper T, Kiesslich R, Ponsioen C, Fockens P, Dekker E. The learning curve, accuracy, and interobserver agreement of endoscope-based confocal laser endomicroscopy for the differentiation of colorectal lesions. Gastrointest Endosc. 2012 Jun;75((6)):1211–7. doi: 10.1016/j.gie.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 19.Izadyyazdanabadi M, Belykh E, Martirosyan N, Eschbacher J, Nakaji P, Yang Y, et al. Improving utility of brain tumor confocal laser endomicroscopy: objective value assessment and diagnostic frame detection with convolutional neural networks. SPIE. 2017;10134 [Google Scholar]
- 20.Goodfellow I, Bengio Y, Courville A. Deep learning. MIT press; 2016. [Google Scholar]
- 21.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35((5)):1285–98. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]