Abstract
The Samar Cobra, Naja samarensis, is endemic to the southern Philippines and is a WHO-listed Category 1 venomous snake species of medical importance. Envenomation caused by N. samarensis results in neurotoxicity, while there is no species-specific antivenom available for its treatment. The composition and neutralization of N. samarensis venom remain largely unknown to date. This study thus aimed to investigate the venom proteome of N. samarensis for a comprehensive profiling of the venom composition, and to examine the immunorecognition as well as neutralization of its toxins by a hetero-specific antivenom. Applying C18 reverse-phase high-performance liquid chromatography (RP-HPLC) and tandem mass spectrometry (LC-MS/MS), three-finger toxins (3FTx) were shown to dominate the venom proteome by 90.48% of total venom proteins. Other proteins in the venom comprised snake venom metalloproteinases, phospholipases A2, cysteine-rich secretory proteins, venom nerve growth factors, L-amino acid oxidases and vespryn, which were present at much lower abundances. Among all, short-chain alpha-neurotoxins (SαNTX) were the most highly expressed toxin within 3FTx family, constituting 65.87% of the total venom proteins. The SαNTX is the sole neurotoxic component of the venom and has an intravenous median lethal dose (LD50) of 0.18 μg/g in mice. The high abundance and low LD50 support the potent lethal activity of N. samarensis venom. The hetero-specific antivenom, Philippine Cobra Antivenom (PCAV, raised against Naja philippinensis) were immunoreactive toward the venom and its protein fractions, including the principal SαNTX. In efficacy study, PCAV was able to cross-neutralize the lethality of SαNTX albeit the effect was weak with a low potency of 0.20 mg/ml (defined as the amount of toxin completely neutralized per milliliter of the antivenom). With a volume of 5 ml, each vial of PCAV may cross-neutralize approximately 1 mg of the toxin in vivo. The findings support the potential para-specific use of PCAV in treating envenomation caused by N. samarensis while underscoring the need to improve the potency of its neutralization activity, especially against the highly lethal alpha-neurotoxins.
Keywords: Southern Philippine Cobra, spitting cobra, venomics, alpha-neurotoxin, immunoreactivity
Introduction
Each year, about 5.4 million snakebites occur worldwide, resulting in 1.8–2.7 million cases of envenomation. Consequently, 81,000 to 138,000 people die due to the toxic effects of snake venom, with three times as many continue to suffer long-term complications of various physical and mental sequelae (Kasturiratne et al., 2008; World Health Organization [WHO], 2019). South Asia, sub-Saharan Africa and Southeast Asia are the regional trio that features the highest prevalence and mortality of snakebite envenomation. This public health crisis heavily affects the impoverished populations in the rural areas, where health resources are scarce, venomous snakes are abundant and human-snake interaction is common due to extensive agricultural practice (World Health Organization [WHO], 2016; Gutiérrez et al., 2017; Ralph et al., 2019).
In Asia, cobras (Naja spp.) are the quintessential venomous snakes, typified by their lethal and incapacitating bite. Cobra bite envenomation causes neuromuscular paralysis that leads to respiratory failure, multiple organ failure and death, if proper treatment is unavailable (Tan and Tan, 2016; Thatoi et al., 2016; World Health Organization [WHO], 2016). Also, extensive tissue necrosis can take place at the bite site, resulting in amputation or severe physical deformity and compromising the victim’s quality of life (Mehta and Sashindran, 2002; Jayawardana et al., 2018). The clinical syndrome observed is largely attributed to the neurotoxins (causing paralysis) and cytotoxins (causing tissue necrosis) found abundantly in cobra venoms (Tan et al., 2017c; Wong et al., 2018). Nevertheless, cobra venoms can vary substantially between different species (inter-species venom variation) and even within the same species (intra-species venom variation) due to factors of geographical distribution, ontogenic development and sex of the snake (Mackessy, 2009; Tan et al., 2015d; Huang et al., 2015; Wong et al., 2016; Dutta et al., 2017). Variation in the composition and antigenicity of snake venom has important medical implications as it can result in variable venom toxicity and inconsistent effectiveness of antivenom therapy. Moreover, species-specific antivenoms are still lacking in many regions.
It is therefore crucial to characterize the venom properties of different snake species, addressing the unique protein composition and antigenicity therein. In the Philippines, there are two endemic cobra species in the country: the Philippine cobra (Naja philippinensis) which is distributed in the northern Philippines, and the Samar cobra (Naja samarensis) that can be found in the southern Philippines (Wüster and Thorpe, 1990; Sanguila et al., 2016; Sy and Mangkabong, 2018). Unlike the Philippine Cobra, the Samar Cobra (N. samarensis, also known as Visayan cobra or Peters’ cobra) is lesser known perhaps owing to insufficient epidemiological data and the lack of study on its venom properties. The Samar Cobra is distributed in the southern part of the Philippines, as in the islands of Visayas (Bohol, Leyte and Samar), Mindanao, Camiguin, Dinagat, Basilan and Siquijor (Sanguila et al., 2016) (Figure 1). N. samarensis can grow up to 1.4 m in length, and it displays striking body coloration of black and bright yellow, in contrast to N. philippinensis which is usually light to olive brown (Avilonzoo, 2015) (Figure 1). Envenomation caused by N. samarensis has been anecdotally reported to cause severe neurotoxicity somewhat similar to that caused by N. philippinensis, manifested by rapid neuromuscular paralysis that leads to respiratory failure and death (Watt et al., 1988; World Health Organization [WHO], 2018). As a spitting cobra, it is also capable of spitting (or spraying) venom into the eyes of aggressors, resulting in venom ophthalmia (Chu et al., 2010; Chang et al., 2020).
FIGURE 1.
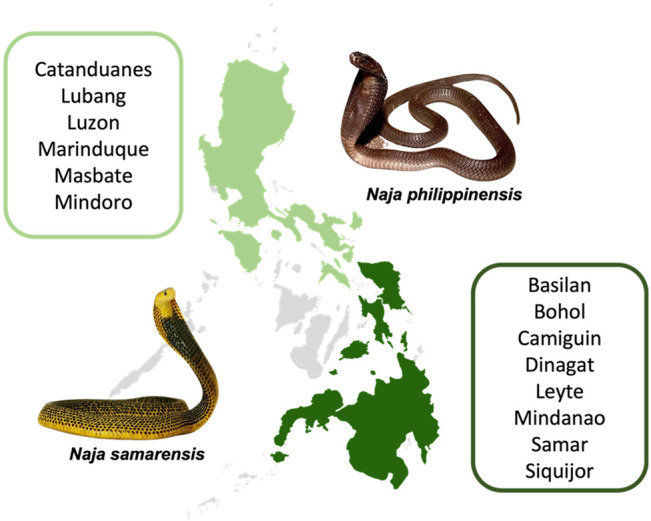
Geographical distribution of the Northern Philippine Cobra (Naja philippinensis) and Samar Cobra or Southern Philippine Cobra (Naja samarensis). Regions shaded in green are where the cobras are native to: N. philippinensis (light green); N. samarensis (dark green). Words in green boxes next to the shaded areas refer to islands in the Philippines where the respective cobra species can be found (Sy and Mangkabong, 2018; Sy and Bucol, 2020; Uetz et al., 2021a; Uetz et al., 2021b).
Although antivenom is the only definitive treatment for snakebite envenomation, there is however, no antivenom specific for N. samarensis available currently. The Research Institute for Tropical Medicine (RITM) in the Philippines does produce an antivenom raised against N. philippinensis, i.e., the Philippine Cobra Antivenom (PCAV) for domestic use, mainly in the northern region of the country (Dimaano, 2018). The supply of PCAV is, however, limited and its clinical use for N. samarensis envenomation has not been well established. In a preclinical study, Tan et al. (2021) recently showed that PCAV weakly cross-neutralized the lethal effect of N. samarensis venom in mice with a low potency. Presumably, the cross-neutralization efficacy of PCAV was limited by variation in the N. samarensis venom composition and toxin antigenicity. Hence, applying a protein decomplexation strategy, the present study investigated the venom proteome of N. samarensis to unravel the venom composition and toxins implicated in envenomation. The immunoreactivity and neutralization efficacy of PCAV against the toxins were also examined for insights into the optimization of antivenom production in the region.
Materials and Methods
Venom and Antivenoms
The venom sample of Naja samarensis was pooled from the milking of multiple adult specimens (n = 5) from the southern Philippines. The N. philippinensis and Calloselasma rhodostoma venoms were supplied by Latoxan Venom Supply (Valence, France) and Queen Saovabha Memorial Institute (QSMI, Thailand), respectively. All venom samples were kept in lyophilized form at −20°C until use. The antivenom used in this work was the Philippine Cobra Antivenom (PCAV; batch no.: 201804; expiry date: April 2021). PCAV is a monovalent antivenom raised against the venom of N. philippinensis, a product from Research Institute for Tropical Medicine (RITM), the Philippines. PCAV was used before its expiration date.
Determination of Protein Concentration of Venoms and Antivenom
Protein concentration of venoms and toxins was determined using Nanodrop™ 2000 spectrophotometer (Waltham, MA, United States). Protein concentration of PCAV was determined with Thermo Scientific™ Pierce™ Bicinchoninic Acid (BCA) Protein Assay kit (Rockford, IL, United States) with bovine serum albumin used as protein standard in calibration.
Chemicals and Materials
Ammonium bicarbonate, dithiothreitol (DTT) and iodoacetamide (IAA) were purchased from Sigma-Aldrich (St. Louis, MO, United States). MS grade trypsin protease and HPLC grade solvents were purchased from Thermo Scientific™ Pierce™ (Rockford, IL, United States). LiChrospher® WP300 RP-18 HPLC column (5 μm particle size) and Millipore ZipTip® C18 Pipette Tips were procured from Merck (Burlington, MA, United States). Other chemicals and solvents used were of analytical grade and purchased from Sigma-Aldrich (St. Louis, Missouri, United States).
Reverse-Phase High-Performance Liquid Chromatography
The crude sample in the form of lyophilized venom (3 mg) was reconstituted in 200 μl ultrapure water and subjected to C18 reverse-phase high-performance liquid chromatography (RP-HPLC) at a flow rate of 1 ml/min. The LiChrospher® WP300 RP-18 HPLC column (250 × 4 mm, 5 μm particle size) was pre-equilibrated with Solvent A (0.1% TFA in water) and eluted with Solvent B (0.1% TFA in ACN) in linear gradient setting (5% B for 10 min, 5–15% B for 20 min, 15–45% B for 120 min and 45–70% for 20 min). The eluted peaks were detected at 215 nm and fractions were manually collected, lyophilized and stored at −20°C until use.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was conducted as per Laemmli (Laemmli, 1970). SmoBio PM2700 ExcelBand™ 3-color Broad Range Protein Marker (5–245 kDa) (Hsinchu, Taiwan) was utilized for molecular weight calibration. 25 μg of crude venoms (N. samarensis and N. philippinensis) and 10 μg of RP-HPLC fractions (eluted proteins) were redissolved in ultrapure water and separated electrophoretically via 15% SDS-PAGE under reducing conditions, at 90 V for 2.5 h. Proteins were visualized using Coomassie Brilliant Blue R250.
In-Solution Tryptic Digestion
The fractions collected from RP-HPLC were subjected to in-solution tryptic digestion as per the protocol previously outlined (Tan et al., 2017a). The venom samples were reduced with DTT, alkylated with IAA, and digested with MS grade trypsin protease. The tryptic peptides were concentrated and desalted with Millipore ZipTip® C18 Pipette Tips in accordance with the manufacturer’s recommendations to augment the performance of mass spectrometry.
Protein Identification by Tandem Mass Spectrometry (Nano-ESI-LCMS/MS)
The trypsin-digested peptides were redissolved in 7 μl of 0.1% formic acid in water and exposed to nano-electrospray ionization MS/MS by adopting Agilent 1200 HPLC-Chip/MS Interface in conjunction with Agilent 6550 Accurate-Mass Q-TOF LC/MS system. Samples were primed in a large capacity chip Zorbax 300 Å, C18, 160 nl enrichment column, 75 μm × 150 mm analytical column and 5 μm particles (Agilent part no. G4240–62010). Running parameters were configured to 1 μl injection volume per sample, 0.4 μl/min flow rate, and the peptides were separated by running Solution B (0.1% formic acid in acetonitrile) in configured linear gradient setting (5–50% B for 11 min, 50%–70% B for 4 min and 70% B for 3 min) using Agilent 1200 series nano-flow LC pump. Positive ionization mode was selected for ion polarity. Flow rate and temperature of drying gas were set to 11 l/min and 290°C, respectively. Fragmentor voltage was configured to 175 V and the capillary voltage was set to 1800 V. Spectra was obtained in a MS/MS mode with a MS scan range of 200–3,000 m/z and MS/MS scan range of 50–3,200 m/z. Precursor charge selection was set as doubly charge state and above with the exclusion of precursors 1221.9906 m/z (z = 1) and 299.2944 (z = 1) set as reference ions. Data were generated with MH (protonated peptide ion) mass span between 50 and 3,200 Da and sorted with Agilent Spectrum Mill MS Proteomics Workbench software packages. Carbamidomethylation of cysteine residues was set as a fixed modification and oxidation of methionine residues as variable modification. The data derived from mass spectrometry was searched against a non-redundant NCBI database of Serpentes (taxid: 8570) integrated with an in-house transcript database containing related elapids (cobras and king cobra) as previously described (Tan et al., 2015a; Tan et al., 2017a; Chong et al., 2019). Protein identification was verified with the following filters: protein score >15 and peptide score >5. Identified proteins were filtered at <1% false discovery rate (FDR).
Estimation of Relative Abundance of Proteins
The abundance of each individual protein in a fraction was estimated based on the mean spectral intensity (MSI) of its peptides and the peak area under curve (AUC) of its corresponding chromatographic fraction, as described in the formula below:
The MSI of protein A in HPLC-derived fraction B is the mean spectral intensity of the peptide ions assigned to protein A eluted from fraction B. The peak area under curve (AUC) was determined from the chromatogram using the Shimadzu LC Solution Software (Shimadzu, Kyoto, Japan).
ELISA Immunoprofiling of Venom Fractions
The immunological binding properties between antivenom and venom fractions were tested with an indirect enzyme-linked immunosorbent assay (ELISA) as described by Tan et al. (2015c). First, 96-well immunoplate were pre-coated overnight with 10 ng of whole venoms and RP-HPLC-derived fractions of N. samarensis at 4°C. N. philippinensis and C. rhodostoma venoms were served as positive and negative controls, respectively. Next day, the wells were flicked dried and repeatedly washed four times with phosphate-buffered saline containing 0.5% Tween®20 (PBST). Antivenom (PCAV) was prepared at a stock concentration of 10 mg/ml and further diluted as needed. 100 μl of the properly diluted antivenom (1:450) in PBST was added into venom-coated wells, followed by incubation for 1 h at room temperature. The wells were then added with horseradish peroxidase-conjugated anti-horse-IgG in PBST (1:10000) and incubated for another hour at room temperature. The wells were washed four times with PBST before adding 50 μl freshly prepared 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate. The plate was left for 25 min at room temperature in the dark for enzymatic reaction to take place. The reaction was then terminated by adding 50 μl of 2 M sulfuric acid per well, and the absorbance was read at 450 nm against a blank. Values presented were the means ± S.E.M. of triplicates.
Determination of Lethality of Venom Toxins and the Neutralization by Antivenom
The median lethal doses (LD50) of the principal toxins in N. samarensis venom (Fraction 1 from the RP-HPLC, which was found lethal following the lethality screening of various fractions) and in N. philippinensis venom (Fraction 1, as identified according to Tan et al. (2019)) were determined in ICR albino mice (n = 4 per dose, 20–25 g), supplied by the Animal Experimental Unit, University of Malaya. The experiment procedure was approved by the Institutional Animal Care and Use Committee of the university (reference: 2019-220108/PHAR/R/TCH).
In brief, various doses of the toxins were prepared in 100 µl and injected intravenously into the mice via caudal vein. The mice were closely monitored and allowed free access to food and water ad libitum. The survival ratio was recorded 24 h post-injection for determination of LD50, which is defined as the toxin dose (µg of venom/g of mice) at which 50% of the mice were dead. In antivenom neutralization study, different doses of PCAV were mixed with the respective toxins at a challenge dose of 5 LD50 each, dissolved in a total volume of 250 µl and incubated at 37°C for 30 min. The pre-incubated mixture was then injected intravenously into the mice (n = 4 per dose, 20–25 g). The neutralization was determined by median effective dose (ED50), defined as the volume dose of antivenom (µl) at which 50% of the mice survived. Both LD50, ED50 (Morais et al., 2010) and the 95% confidence intervals were calculated by the Probit analysis (Finney and Tattersfield, 1952), using BioStat 2009 analysis software (AnalystSoft Inc., Canada).
The neutralizing capacity of PCAV was further expressed as median effective ratio (ER50), defined as the ratio of venom or toxin (mg) to the volume of antivenom (ml) at which 50% of mice survived) and potency (P), defined as the amount of venom or toxin (mg) completely neutralized per unit volume (ml) of antivenom (thus 100% survival). The potency (P) was normalized by the PCAV concentration to obtain normalized potency (n-P), defined as the amount of venom or toxin (mg) completely neutralized per unit amount of antivenom protein (g) (Tan and Tan, 2019).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository (https://www.iprox.org) (Ma et al., 2019) with the dataset identifier PXD026270, subproject ID: IPX0003097001.
Results and Discussion
Chromatographic and Electrophoretic Profiles of Naja samarensis Venom
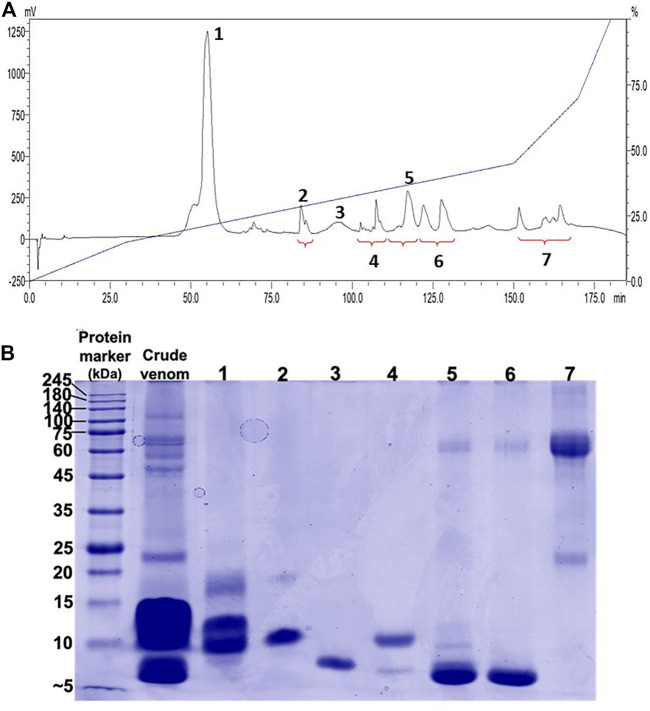
Reverse-phase HPLC (RP-HPLC) resolved N. samarensis venom into multiple peaks that were grouped into seven fractions (Figure 2). As depicted by SDS-PAGE in Figure 2, the majority of venom proteins eluted from fractions 1 to 6 amid chromatography (corresponding to the elution time between 50 and 130 min) were low molecular weight proteins (<20 kDa), consistent with low hydrophobicity of protein. The medium (>25–35 kDa) and high (>35 kDa) molecular weight proteins were present more predominantly in fraction seven which was eluted between 150 and 170 min. Low molecular weight proteins constituted more than 95% of the total venom proteins based on the calculation of chromatographic peak area.
FIGURE 2.
Chromatographic and electrophoretic profiles of Naja samarensis venom. (A) Upper panel: C18 reverse-phase high-performance liquid chromatography (RP-HPLC) of venom. (B) Lower panel: 15% polyacrylamide gel electrophoresis (SDS-PAGE) profile of RP-HPLC eluted fractions electrophoresed under reducing conditions.
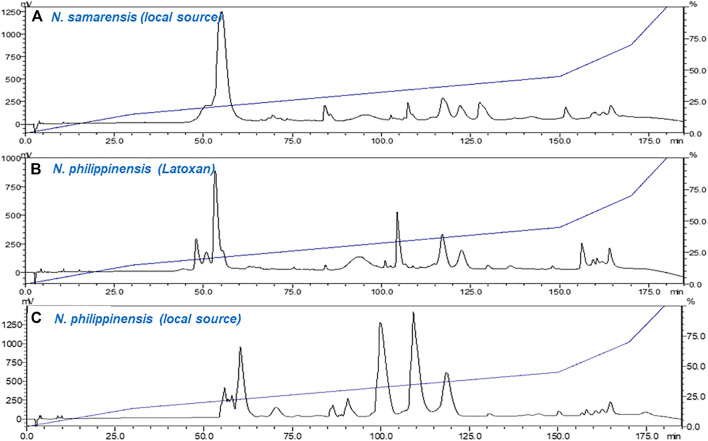
The venom chromatogram (C18 RP-HPLC) of N. samarensis in this study was further compared with that of N. philippinensis from a captive source (Latoxan) and a wild-caught specimen. Under the same experimental conditions, the venoms of N. samarensis and N. philippinensis showed similar patterns of protein separation (Figure 3). RP-HPLC profiling of the venoms showed the elution of predominantly low molecular weight neurotoxins between 45 and 80 min (Figure 3). Between 80 and 140 min, the elution profiles of N. samarensis venom and the commercially supplied N. philippinensis venom (Latoxan) were comparable, whereas variation was noticed in the wild-caught N. philippinensis venom in which proteins eluted had a higher abundance (based on area under curve). The intra-specific venom variability in N. philippinensis observed is probably related to habitat and availability of specific preys (dietary conditions), as previously it has been established that venom compositions can be greatly influenced by the diets of snake (Daltry et al., 1996; Pahari et al., 2007; Barlow et al., 2009; Harris et al., 2020). Although an earlier study suggested the expression of venom proteins is unlikely to be affected by their external housing enclosures (captive and wild-caught) (McCleary et al., 2016), minor differences in their expression abundances was notable. In N. samarensis venom, the dominance of low molecular weight proteins is similar to the venom of N. philippinensis (Tan et al., 2019b), and several other medically important Asiatic cobras reported including N. kaouthia (Tan et al., 2015d; Xu et al., 2017), Naja siamensis (Liu et al., 2017), Naja sputatrix (Tan et al., 2017c), Naja atra (Huang et al., 2015) and Naja naja (Sintiprungrat et al., 2016; Chanda et al., 2018; Wong et al., 2018). Consistently, high molecular weight proteins constituted minor components in these cobra venoms, with a relative protein abundance of less than 10%.
FIGURE 3.
Chromatograms of crude venom samples fractionated using C18 reverse-phase high-performance liquid chromatography (RP-HPLC). (A) Naja samarensis (local source); (B) Naja philippinensis (captive source from Latoxan); and (C) Naja philippinensis (local source). Red dotted lines highlight the elution between 80 and 140 min.
Venom Proteome of Naja samarensis
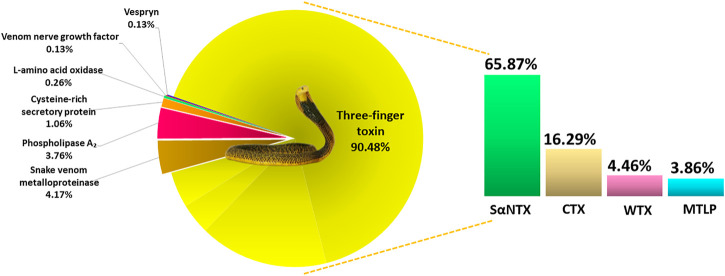
Using tandem mass spectrometry, proteins in the RP-HPLC fractions of N. samarensis venom were identified and shown as in Table 1. In total, 31 non-redundant proteins were identified from the seven RP-HPLC fractions. These proteins were further classified into seven protein families of typical snake venom toxins at varying relative protein abundances (Figure 4). Three-finger toxins (3FTx) were the most abundantly and diversely expressed proteins, comprising 16 distinct proteins that constitute 90.48% of total venom proteins, and followed by snake venom metalloproteinases (SVMP, 4.17%) and phospholipases A2 (PLA2, 3.76%). The remaining minor toxins collectively accounted for <2% of total venom proteins; these were toxin families of cysteine-rich secretory protein (CRiSP, 1.06%), L-amino acid oxidase (LAAO, 0.26%), venom nerve growth factor (vNGF, 0.13%) and vespryn (VES, 0.13%) (Figure 4). The proteins identified were consistent with the chromatographic and electrophoretic profiles of the venom, where low MW proteins including three-finger toxins (3FTx) and phospholipases A2 (PLA2) were eluted mainly from fraction 1 to fraction 6 of the RP-HPLC, followed by medium and high MW proteins from fraction 7 (Figure 2). The data of mass spectrometry analysis including the spectral mass/charge values, protein scores and peptide sequences were provided in Supplemental File.
TABLE 1.
Protein identification from Naja samarensis venom fractions isolated by reverse-phase high-performance liquid chromatography using nano-ESI-LCMS/MS.
| RP-HPLC derived fractions/Protein name | Database accession | Species annotated | Protein score | Relative abundance (%) |
| Fraction 1 | 65.87 | |||
| Short neurotoxin 1 | P60774 | N. samarensis | 96.24 | 27.93 |
| Short neurotoxin 1 | P01424 | N. melanoleuca | 42.31 | 34.87 |
| Cobrotoxin | P60770 | N. atra | 22.22 | 3.07 |
| Fraction 2 | 3.86 | |||
| Muscarinic toxin-like protein 2 | P82463 | N. kaouthia | 46.73 | 3.86 |
| Fraction 3 | 4.46 | |||
| Weak toxin CM-2 | P01415 | N. haje | 17.79 | 4.46 |
| Fraction 4 | 3.98 | |||
| Acidic phospholipase A2 natratoxin | A4FS04 | N. atra | 68.39 | 1.93 |
| Acidic phospholipase A2 D | Q9I900 | N. sputatrix | 57.64 | 1.83 |
| Cytotoxin 1 | P01467 | N. mossambica | 40.65 | 0.11 |
| Cytotoxin 3 | P01470 | N. mossambica | 37.27 | 0.11 |
| Fraction 5 | 6.93 | |||
| Cytotoxin 2 | P01441 | N. oxiana | 162.93 | 1.86 |
| Cytotoxin 10 | P86541 | N. naja | 78.78 | 0.65 |
| Cytotoxin 2 | P01445 | N. kaouthia | 74.29 | 0.37 |
| Cytotoxin SP15c | P60308 | N. atra | 73.38 | 3.26 |
| Cytotoxin homolog | P14541 | N. kaouthia | 59.57 | 0.15 |
| Cytotoxin 1 | P01467 | N. mossambica | 50.54 | 0.27 |
| Venom nerve growth factor 2 | Q5YF89 | N. sputatrix | 30.51 | 0.13 |
| Hemextin A | P0DQH3 | H. haemachatus | 18.59 | 0.23 |
| Cysteine-rich venom protein ophanin | Q7ZT98 | O. hannah | 16.61 | 0.01 |
| Fraction 6 | 9.36 | |||
| Cytotoxin 2 | P01441 | N. oxiana | 108.69 | 1.00 |
| Cytotoxin 2a | P86538 | N. naja | 62.12 | 1.74 |
| Cytotoxin 1 | P01447 | N. naja | 52.31 | 5.87 |
| Cytotoxin 1 | P01467 | N. mossambica | 30.39 | 0.70 |
| Zinc metalloproteinase-disintegrin-like atrase-B | D6PXE8 | N. atra | 24.83 | 0.07 |
| Fraction 7 | 5.54 | |||
| L-amino-acid oxidase | P81383 | O. hannah | 111.89 | 0.26 |
| Natrin | *CL2736.Contig1_NsM2 | N. atra | 100.47 | 0.42 |
| Cysteine-rich venom protein natrin-1 | Q7T1K6 | N. atra | 78.40 | 0.33 |
| Cysteine-rich venom protein ophanin | Q7ZT98 | O. hannah | 65.76 | 0.16 |
| Cysteine-rich venom protein ophanin-like | XP_026568636.1 | P. textilis | 48.83 | 0.14 |
| Zinc metalloproteinase-disintegrin-like atrase-A | D5LMJ3 | N. atra | 79.58 | 0.21 |
| Metalloproteinase atrase A | *CL115.Contig7_NkT | N. atra | 41.01 | 0.20 |
| Metalloproteinase | B5G6F7 | D. vestigiata | 49.02 | 0.15 |
| Zinc metalloproteinase-disintegrin-like MTP4 | F8RKW1 | D. coronoides | 19.50 | 1.22 |
| Zinc metalloproteinase-disintegrin-like atrase-B | D6PXE8 | N. atra | 22.21 | 2.22 |
| Hemorrhagic metalloproteinase-disintegrin-like kaouthiagin | P82942 | N. kaouthia | 22.92 | 0.10 |
| Ohanin | P83234 | O. hannah | 20.66 | 0.13 |
Relative abundance (%) refers to the abundance of individual proteins in a fraction, which was estimated based on the mean spectral intensity (MSI) of the peptide ions and the peak area under curve of its corresponding chromatographic fractions. Bold values are subtotals of protein abundances per fraction.
*Protein codes with suffix “_NsM2” and “_NkT” indicate venom proteins identified based on tryptic peptides matched to sequence from an in-house transcript database containing RNAseq, specific for the following: NsM: Naja sumatrana, Malaysia and NkT: Naja kaouthia, Thailand. Mass spectrometric data and peptide sequences are available in Supplemental file.
Abbreviations: D. coronoides, Drysdalia coronoides; D. vestigiata, Demansia vestigiata; H. haemachatus, Hemachatus haemachatus; N. atra, Naja atra; N. haje, Naja haje; N. kaouthia, Naja kaouthia; N. melanoleuca, Naja melanoleuca; N. mossambica, Naja mossambica; N. naja, Naja naja; N. oxiana, Naja oxiana; N. samarensis, Naja samarensis; N. sputatrix, Naja sputatrix; NCBI, National Centre for Biotechnology Information; O. hannah, Ophiophagus hannah; P. textilis, Pseudonaja textilis; RP-HPLC, reverse-phase high performance liquid chromatography.
FIGURE 4.
Venom proteome of Naja samarensis classified according to the toxin protein families, with relative abundances (%) based on HPLC profile and relative spectral intensity from mass spectrometry analysis (as described in the method section). Abbreviations: CTX, cytotoxin; MTLP, muscarinic toxin-like protein; SαNTX, short-chain alpha-neurotoxin; WTX, weak toxin.
Based on the relative abundances of the proteins, 3FTxs in N. samarensis venom are composed of diverse isoforms of short-chain alpha-neurotoxin (SαNTX), cytotoxin (CTX), muscarinic toxin-like protein (MTLP) and weak toxin (WTX), accounting for 90.48% of the total venom proteins. Among these, SαNTX is the sole and most abundantly expressed subclass of alpha-neurotoxin (αNTX) identified in the venom, accounting for 65.87% of the total venom proteins. In total, three SαNTX forms were present (Table 2). Cobra venoms, in general, exhibit a common phenotype characterized by dominant expression of either short-chain or long-chain alpha-neurotoxins (SαNTX or LαNTX). Interestingly, the N. samarensis venom proteome has only SαNTX as its alpha-neurotoxins whereas LαNTX was absent—a feature similar to that observed in the venom proteome of N. philippinensis (Tan et al., 2019b), an allopatric cobra species distributed in the northern islands. The lack of LαNTX has also been noted in the venom proteomes of the monocled cobra (N. kaouthia) from Vietnam (Tan et al., 2015d) and the Chinese cobra (Naja atra) from mainland China (Shan et al., 2016). These cobras, representing far eastern dispersal of Naja species, probably evolved a specialized toxin arsenal that fully deploys SαNTX in predation. Nevertheless, the expression levels of SαNTX vary between these species, with N. samarensis recording the highest SαNTX abundance (65.87% of total venom proteins, current work), followed by N. philippinensis (44.55%) (Tan et al., 2019b), N. atra (southern China Mainland) (11.2%) (Shan et al., 2016) and N. kaouthia (Vietnam) (9.2%) (Tan et al., 2015d). The apparent variance in SαNTX content is reflected in the differential lethal potency of the venoms, as discussed below.
TABLE 2.
Relative abundance of different toxin families identified from Naja samarensis venom.
| Protein family/Name | Fraction(s) | Database accession | Species | Relative abundance (%) |
| Three-finger toxin (3FTx) | 90.48 | |||
| Short-chain alpha-neurotoxin (SαNTX) | 65.87 | |||
| Short neurotoxin 1 | 1 | P01424 | N. melanoleuca | 34.87 |
| Cobrotoxin | 1 | P60770 | N. atra | 3.07 |
| Short neurotoxin 1 | 1 | P60774 | N. samarensis | 27.93 |
| Cytotoxin (CTX) | 16.29 | |||
| Cytotoxin 1 | 6 | P01467 | N. mossambica | 0.70 |
| Cytotoxin 2 | 5,6 | P01441 | N. oxiana | 2.84 |
| Cytotoxin 2 | 5 | P01445 | N. kaouthia | 0.37 |
| Cytotoxin 1 | 6 | P01447 | N. naja | 5.87 |
| Cytotoxin 1 | 4,5 | P01467 | N. mossambica | 0.38 |
| Cytotoxin 3 | 4 | P01470 | N. mossambica | 0.11 |
| Hemextin A | 5 | P0DQH3 | H. haemachatus | 0.23 |
| Cytotoxin homolog | 5 | P14541 | N. kaouthia | 0.15 |
| Cytotoxin SP15c | 5 | P60308 | N. atra | 3.26 |
| Cytotoxin 2a | 6 | P86538 | N. naja | 1.74 |
| Cytotoxin 10 | 5 | P86541 | N. naja | 0.65 |
| Muscarinic toxin-like protein (MTLP) | 3.86 | |||
| Muscarinic toxin-like protein 2 | 2 | P82463 | N. kaouthia | 3.86 |
| Weak toxin (WTX) | 4.46 | |||
| Weak toxin CM-2 | 3 | P01415 | N. haje | 4.46 |
| Snake venom metalloproteinase (SVMP) | 4.17 | |||
| Metalloproteinase | 7 | B5G6F7 | D. vestigiata | 0.15 |
| Zinc metalloproteinase-disintegrin-like atrase-A | 7 | D5LMJ3 | N. atra | 0.21 |
| Zinc metalloproteinase-disintegrin-like atrase-B | 6,7 | D6PXE8 | N. atra | 2.29 |
| Zinc metalloproteinase-disintegrin-like MTP4 | 7 | F8RKW1 | D. coronoides | 1.22 |
| Hemorrhagic metalloproteinase-disintegrin-like kaouthiagin | 7 | P82942 | N. kaouthia | 0.10 |
| Metalloproteinase atrase A | 7 | *CL115.Contig7_NkT | N. atra | 0.20 |
| Phospholipase A2 (PLA2) | 3.76 | |||
| Acidic phospholipase A2 natratoxin | 4 | A4FS04 | N. atra | 1.93 |
| Acidic phospholipase A2 D | 4 | Q9I900 | N. sputatrix | 1.83 |
| Cysteine-rich secretory protein (CRiSP) | 1.06 | |||
| Cysteine-rich venom protein natrin-1 | 7 | Q7T1K6 | N. atra | 0.33 |
| Cysteine-rich venom protein ophanin | 5,7 | Q7ZT98 | O. hannah | 0.17 |
| Cysteine-rich venom protein ophanin-like | 7 | XP_026568636.1 | P. textilis | 0.14 |
| Natrin | 7 | *CL2736.Contig1_NsM2 | N. atra | 0.42 |
| L-amino-acid oxidase (LAAO) | 0.26 | |||
| L-amino-acid oxidase | 7 | P81383 | O. hannah | 0.26 |
| Venom nerve growth factor (vNGF) | 0.13 | |||
| Venom nerve growth factor 2 | 5 | Q5YF89 | N. sputatrix | 0.13 |
| Vespryn (VES) | 0.13 | |||
| Ohanin | 7 | P83234 | O. hannah | 0.13 |
The percentages (%) refers to the relative abundances of proteins identified from the HPLC-derived venom fractions. Bold values are subtotals of protein abundances per toxin family or subfamily.
*Protein accession numbers with suffixes “_NsM2” and “_NkT” indicate proteins matched to sequences from an in-house transcript database containing RNAseq, of the following cobras: NsM: Naja sumatrana, Malaysia and NkT: Naja kaouthia, Thailand. Mass spectrometric data and peptide sequences are available in Supplemental File.
Abbreviations: D. vestigiata, Demansia vestigiata; D. coronoides, Drysdalia coronoides; H. haemachatus, Hemachatus haemachatus; N. atra, Naja atra; N. haje, Naja haje; N. kaouthia, Naja kaouthia; N. melanoleuca, Naja melanoleuca; N. mossambica, Naja mossambica; N. naja, Naja naja; N. oxiana, Naja oxiana; N. samarensis, Naja samarensis; N. sputatrix, Naja sputatrix; O. hannah, Ophiophagus hannah; P. textilis, Pseudonaja textilis.
In cobra bite envenomation, the alpha-neurotoxins block post-synaptic nicotinic acetylcholine receptors (nAChR) at the neuromuscular junctions, thereby inhibiting neurotransmission and causing paralysis (Barber et al., 2013). These toxins are typically the principal lethal components in the venoms of most cobra and sea snake species (Tan et al., 2016b; Wong et al., 2016; Tan et al., 2017c; Silva et al., 2018; Tan et al., 2019a; Harris et al., 2020). Previous studies indicated that short neurotoxins (SαNTX), in contrast to LαNTX, bind less irreversibly to nAChR and are therefore, theoretically, “less lethal” with low medical importance (Chicheportiche et al., 1975; Utkin, 2013). In lethality test, however, both SαNTX and LαNTX induced comparable neurotoxicity in vivo, and are highly lethal to mice with median lethal doses (LD50) ranging between 0.05 and 0.25 μg/g (Tan et al., 2015b; Tan et al., 2016b; Wong et al., 2016; Tan et al., 2017a; Tan et al., 2019a). The lethal potency of cobra venom has been shown to correlate with the abundance of alpha-neurotoxins (both SαNTX and LαNTX) in the venom (Tan et al., 2019b). Comparing the aforementioned eastern cobras whose venom neurotoxicity is exclusively SαNTX-driven, we observed a trend that supports the correlation between the differential SαNTX expression, and the lethality induced. The highly lethal N. samarensis and N. philippinensis venoms (with intravenous LD50 between 0.1 μg/g and 0.2 μg/g) were dominated by SαNTX up to 50–70% (Tan et al., 2019b; Tan et al., 2021); while in the Chinese N. atra and Vietnamese N. kaouthia venoms (intravenous LD50 is ∼0.9 μg/g), the lethality is approximately 5-fold weaker, consistent with their much lower expression (by ∼6-fold) of SαNTX (∼9%) in the respective venoms (Tan et al., 2015d; Ratanabanangkoon et al., 2016). The finding in the present work indicates that the neurotoxicity of N. samarensis venom toxicity is principally driven by the abundant SαNTX and underscores the need for antivenom treatment that can neutralize the toxin effectively.
Cytotoxins (CTX, including cytotoxin-like homologs) made up 16.29% of the total venom proteins in N. samarensis (Figure 4). CTX is the second most abundantly expressed 3FTx, with 11 distinct subtypes identified in the proteome (Table 2). Contrary to alpha-neurotoxins, CTX is generally less lethal with intravenous LD50 values higher than 1.0 μg/g in mice (Leong et al., 2015; Tan et al., 2016b; Wong et al., 2016; Tan et al., 2017c). CTX with membrane-damaging disposition is implicated in the local envenoming effect of cobras for tissue necrosis and venom ophthalmia (Chu et al., 2010; Tan and Tan, 2016; Chang et al., 2020). CTX usually constitutes the major subtype of 3FTx in most cobra venoms, accounting for at least 20–72% of total venom proteins (Huang et al., 2015; Laustsen et al., 2015; Tan et al., 2015d; Shan et al., 2016; Sintiprungrat et al., 2016; Wong et al., 2016; Tan et al., 2017c). The lower abundance of CTX in N. samarensis venom (current work) is exceptional, as with the proteomic abundance of CTX in N. philippinensis venom (q.v. Table 3 for comparison). Together, the observation suggests that the two endemic cobra species in the Philippines exhibit a unique venom phenotype characterized by reduced expression of CTX and high abundance of SαNTX. The finding is also consistent with clinical envenomation caused by the Philippine cobras, in which local tissue necrotic effect is rare while neurotoxicity is prominent (Watt et al., 1988). On the other hand, other eastern Asiatic cobra species, exemplified by N. atra (China and Taiwan) and N. kaouthia (Vietnam), have venoms that contain high abundances of CTX but fewer alpha-neurotoxins (Tan et al., 2015d; Huang et al., 2015; Shan et al., 2016). Clinically, they produce envenomation effect marked by severe tissue necrosis but little or negligible neurotoxicity—a clinical phenotype opposing to that in envenomation caused by the Philippine cobras. The mechanisms that underlie the evolutionary divergence in their 3FTx compositions and the resultant dichotomous toxicity of the venoms remain unknown, but presumably are related to the paleo-geographical separation of the Philippine islands from Taiwan in the north (by Luzon Strait) and the Sunda Shelf (rest of Southeast Asia) in the west (by South China Sea). Hence, the overall management for envenomation caused by cobras in the region involving antivenom use and/or supportive treatment, cannot be generalized and should be strategized according to the venom’s principal toxicity of different species.
TABLE 3.
Comparison of venom toxin families of Naja samarensis and Naja philippinensis.
| Toxin families | Naja samarensis | Naja philippinensis a |
| Localities | Southern Philippines | Northern Philippines |
| Venom source | Pooled sample (5 adults) b | Pooled sample (6 adults) c |
| Fractionation method | Reverse-phase HPLC | Reverse-phase HPLC |
| Toxin identification method | Nano-ESI-LCMS/MS | Nano-ESI-LCMS/MS |
| Toxin family abundance (%) | ||
| 3FTx | 90.48 | 66.64 |
| SαNTX | 65.87 | 44.55 |
| LαNTX | ND | ND |
| CTX | 16.29 | 21.31 |
| MTLP | 3.86 | 0.77 |
| WTX | 4.46 | 0.01 |
| PLA2 | 3.76 | 22.88 |
| SVMP | 4.17 | 3.93 |
| CRiSP | 1.06 | 1.49 |
| LAAO | 0.26 | 0.42 |
| NGF | 0.13 | 0.06 |
| VES | 0.13 | 0.05 |
| CVF | ND | 2.38 |
| 5′NUC | ND | 0.55 |
| PDE | ND | 0.45 |
| SVSP | ND | 0.35 |
Referenced from Tan et al. (2019b).
Wild caught.
Supplied by Latoxan Venom Supply, Valence, France.
Abbreviations: 3FTx, Three-finger toxin; 5′NUC, 5′ nucleotidase; CRiSP, cysteine-rich secretory protein; CTX, cytotoxin; CVF, cobra venom factor; HPLC, high-performance liquid chromatography; LAAO, L-amino acid oxidase; LαNTX, long-chain alpha-neurotoxin; MTLP, muscarinic toxin-like protein; ND, not detected; NGF, nerve growth factor; PDE, phosphodiesterase; PLA2, phospholipase A2; SαNTX, short-chain alpha-neurotoxin; SVMP, snake venom metalloproteinases; SVSP, snake venom serine protease; VES, vespryn; WTX, weak toxin.
Cumulatively, other toxin families made up less than 10% of the total proteins in N. samarensis venom (Figure 3). Of these, snake venom metalloproteinases (SVMP) constituted 4.17% of the total venom proteins, comprising six distinct protein forms (Table 2). The SVMP proteins identified belong to the P-III subclass with peptides containing the disintegrin and cysteine-rich domains (Fox and Serrano, 2008). In agreement with existing venom proteomic data of snakes (where details on protein subtypes and abundances were reported), the SVMP in cobra (Naja spp.) venoms are typically P-III subclass, and present in trace amount (Tan et al., 2015d; Sintiprungrat et al., 2016; Tan et al., 2017c; Dutta et al., 2017; Chanda et al., 2018; Wong et al., 2018; Tan et al., 2019b). Similarly, SVMP abundance in venom proteome is typically low in Asian elapids including the kraits (Bungarus spp.) (Oh et al., 2017; Oh et al., 2019; Hia et al., 2020) and sea snakes (Tan et al., 2015b; Tan et al., 2019a), with the exceptions of king cobra (Tan et al., 2015a; Liu et al., 2017) and Asiatic coral snakes (Tan et al., 2016a; Tan et al., 2019d). The pathophysiological role of SVMP in Naja cobra venoms may be related to inflammation and cytotoxicity, while the exact function remains to be elucidated.
The PLA2 identified in N. samarensis venom proteome (3.76% of total venom proteins) are homologous to the acidic PLA2 of N. kaouthia and N. naja. The acidic PLA2 annotated are non-lethal but may contribute to local envenomation effect (Tan et al., 2016b; Wong et al., 2016). In cobra venoms, we previously demonstrated that PLA2 follows a unique trend of distribution across the different cobra subgenera, in which the species of Uraeus subgenus (African non-spitting cobras) have none to negligible amount of snake venom PLA2, while the PLA2 abundance is variably higher in the other subgenera (Naja, Afronaja and Boulengerina), especially the spitting cobras presumably associated with algesic (pain-inducing) properties for defense (Tan et al., 2019c). A recent study showed that cobras of spitting lineages have upregulated PLA2 to potentiate CTX in activating mammalian sensory nerves, inferring that PLA2 expression is indicative of convergent evolution of pain-inducing defensive venoms in the spitting cobras (Kazandjian et al., 2021). The current and previous studies, as shown in Table 3, which analyzed the venom proteomes of the two Asiatic spitting cobras (N. philippinensis and N. samarensis), reveal an otherwise indication: the two Philippine spitting cobras greatly downregulated the production of PLA2 and, to a lesser extent the CTX, while conserving the venom-spitting or venom-spraying trait. The cobra species of far-eastern dispersal in the Philippines thus probably deploy various other toxin components, e.g., SVMP, as above, and L-amino acid oxidases (LAAO, 0.26%), for cytotoxic and pain-inducing properties in venom ophthalmia.
Other minor proteins in N. samarensis venom are cysteine-rich secretory protein (CRiSP) (1.06% of total venom protein), venom nerve growth factor (vNGF, 0.13%) and vespryn (0.13%). CRiSP can inhibit the growth of new blood vessels (angiogenesis), increase vascular permeability and thus promote inflammatory responses (leukocyte and neutrophil infiltration) (Yamazaki and Morita, 2004; Mackessy and Heyborne, 2009). Vespryn has been shown to facilitate immobilization of prey by inducing hypolocomotion (Pung et al., 2005; Pung et al., 2006), and vNGF can potentiate venom spread through the release of histamine in the vasculature around the bite site (Kostiza et al., 1995). These are, in general, proteins with higher molecular weights (contrary to the major toxin group, 3FTx) but trace amounts in the venom, implying that they serve ancillary functions without direct lethal implication.
Immunological Cross-Reactivity of Antivenom and Cross-Neutralization of Venom Toxicity
The cross-reactivity of PCAV toward N. samarensis venom and its protein fractions separated by RP-HPLC was examined on ELISA (Figure 5). PCAV exhibited comparable immunoreactivity toward the N. samarensis venom and the homologous N. philippinensis venom, implying the sharing of common toxin epitopes between the two phylogenetically related cobra species. The immunological binding activities of PCAV toward the different protein fractions of N. samarensis venom (tested at the same protein amount) were, in general, moderate to strong. High immunoreactivity was observed in fractions 1, 3, 5 and 6 (absorbance values: 2.0–3.0), while the immunoreactivity was moderate in fractions 2, 4 and 7 (absorbance values: 1.0–2.0). Essentially, fraction 1, which formed the major bulk of the venom proteins, showed the highest immunoreactivity with PCAV. Fraction 1 contained the principal toxins, i.e., SαNTX, which are highly neurotoxic and lethal with a low LD50 of 0.18 μg/g in mice (Table 4).
FIGURE 5.
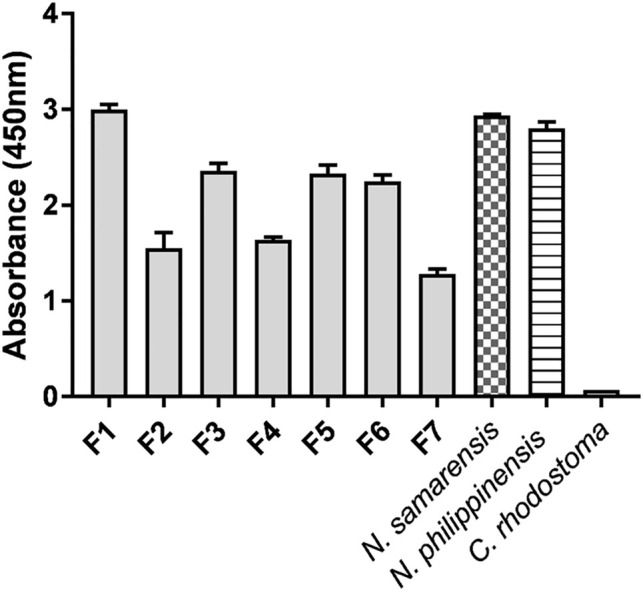
Immunoreactivity of Philippine Cobra Antivenom (PCAV) toward the reverse-phase high-performance liquid chromatography (RP-HPLC) protein fractions (F1-F7) of Naja samarensis venom. Values were means ± S.E.M. of triplicates. Naja philippinensis and Calloselasma rhodostoma venoms were used as positive and negative controls, respectively.
TABLE 4.
Neutralization of the alpha-neurotoxins and venoms of Philippine cobras, Naja samarensis and Naja philippinensis by antivenom.
| LD50 (μg/g) | Challenge dose | PCAV (16.2 ± 1.1 mg/ml) | |||||
| ED50 (μl) | ER50 (mg/ml) | P (mg/ml) | Normalized P (mg/g) | ||||
| Alpha-neurotoxins | NS-SαNTX | 0.18 (0.12–0.29) | 5 LD50 | 91.24 (58.29–142.80) | 0.25 (0.16–0.40) | 0.20 | 12.18 |
| NP-SαNTX | 0.10 (0.06–0.15) | 5 LD50 | 44.94 (29.60–68.23) | 0.28 (0.17–0.42) | 0.22 | 13.74 | |
| Venom | Naja samarensis* | 0.20 (0.16–0.25) | 5 LD50 | 120.86 (104.79–139.40) | 0.21 (0.17–0.26) | 0.17 | 10.21 |
| Naja philippinensis # | 0.18 (0.12–0.27) | 5 LD50 | 44.94 (29.60–68.23) | 0.50 (0.33–0.75) | 0.40 | 24.72 | |
LD50 (Lethal dose): Venom dose (µg/g) at which 50% of the mice were dead.
ED50 (Median effective dose): Antivenom dose (μl) at which 50% of tested mice survived.
ER50 (Median effective ratio): Antivenom dose (mg/ml) corresponding to ED50.
P: Potency of antivenom (mg venom neutralized per ml antivenom).
Normalized P: Potency of antivenom (mg venom neutralized per g antivenom).
Abbreviations: NP-SαNTX, Naja philippinensis short-chain alpha-neurotoxin; NS-SαNTX, Naja samarensis short-chain alpha-neurotoxin; PCAV, Philippine Cobra Antivenom.
The high immunoreactivity of PCAV toward the principal toxin fraction supports the previous observation of PCAV cross-neutralizing the whole venom of N. samarensis, though did not explain its differential neutralization efficacy against N. philippinensis and N. samarensis venoms, in which the neutralization of the former (homologous venom) was more efficacious (Tan et al., 2021). To elucidate the observation, we isolated the SαNTX from the venoms of N. samarensis and N. philippinensis based on the RP-HPLC profiles and tested the in vivo neutralization activity of PCAV against the toxins from the respective species. The neutralization effects of PCAV against the toxins of N. samarensis and N. philippinensis were compared in terms of potency, defined as the amount of the toxin completely neutralized per volume dose of the antivenom (mg/ml) (Table 4). It was found that PCAV neutralized the SαNTX of N. samarensis and N. philippinensis with equipotency (p = 0.20 and 0.22 mg/ml, respectively), implying that their SαNTX indeed shared conserved antigenicity that allows immunorecognition and neutralization by the same antivenom. The neutralization potency against the whole venom, however, was lower in N. samarensis than N. philippinensis by approximately 2-fold. Based on the venom proteomic findings, this could be explained by the higher abundance of SαNTx in the N. samarensis venom (65.87%) than in the N. philippinensis venom (44.55%) (Table 3), thus requiring a higher dose of antivenom for neutralizing the whole venom toxicity. On the whole, the neutralization potency of PCAV against the SαNTX is low; this is the limiting factor of PCAV efficacy in neutralizing the whole venom toxicity, as shown by the low potency of 0.20–0.40 mg/ml (Table 4). Considering that an adult cobra can readily deliver 50–100 mg dry weight of venom in a bite (author’s venom-milking experience), in the case of complete absorption of 50 mg venom into the systemic circulation, a theoretical dose of 125–250 ml of PCAV which is equivalent to tens of vials of antivenom would be required for complete neutralization in clinical treatment. High doses of antivenom increase the risk of hypersensitivity reactions (de Silva et al., 2016; Williams et al., 2018), incur exorbitant treatment cost and exhaust antivenom stock which is inadequately supplied to begin with (Williams et al., 2011). The limited neutralization capacity of PCAV, nonetheless, is similar to that reported in a number of cobra venom neutralization studies, in which the in vivo neutralization effects of antivenom were consistently low (potency <1 mg/ml) (Tan et al., 2016b; Wong et al., 2016; Tan et al., 2017c; Tan et al., 2020; Wong et al., 2021). The key toxins (alpha-neurotoxins) in cobra venoms are small, less immunogenic proteins, and this presumably compromises the production of effective antibody and the neutralization activity of antivenom (Clementi et al., 1991). The development of more efficacious antivenom products for cobra bite neurotoxic envenomation is therefore warranted. This is achievable through improving the immunogen formulation with strategies such as protein-adjuvant conjugation and enrichment with toxins from multiple species known as “diverse toxin repertoire” (Bermúdez-Méndez et al., 2018; Ratanabanangkoon et al., 2020; Ratanabanangkoon, 2021).
Conclusion
The present proteomic study unveiled a unique snake venom phenotype of N. samarensis, an Asiatic spitting cobra that represents the easternmost dispersal of Naja cobra species. Three-finger toxins (3FTx) constituted close to 90% of the total venom proteins, recording by far the highest 3FTx abundance in snake venom. The major 3FTx proteins in the venom were short-chain alpha-neurotoxins (SαNTX) which made up two-thirds of the total venom composition, while the long-chain alpha-neurotoxin (LαNTX) was absent. The venom proteomes of N. samarensis (current work) and N. philippinensis (Tan et al., 2019b) demonstrated that SαNTX are exclusively the neurotoxic components accountable for the venom lethality of these cobra species of high medical importance in Asia. Also, the abundance of cytotoxins (or cardiotoxins, and homologs) was considerably low (∼16%) in N. samarensis venom proteome compared to most other cobra species, and this is consistent with the lack of local tissue effect in envenomation caused by the Philippine cobras. Besides, the PLA2 content in the venom was exceptionally low (∼4%) for a spitting cobra, showing that the Philippine spitting cobras do not conform to the upregulating trend of PLA2 and CTX despite evolving the venom-spitting (spraying) trait. The hetero-specific antivenom PCAV was immunoreactive toward the N. samarensis venom and its toxin fractions including SαNTX, and the antivenom weakly cross-neutralized the toxin lethality albeit with a low potency. Together, the present study provides a deep insight into the venom composition and the pathophysiology of envenomation of this lesser-known spitting cobra species in Southeast Asia. The study also underscores the need for an antivenom product with improved efficacy in the region, which may be achieved by immunogen re-formulation to increase the neutralizing strength of the antibody against the principal neurotoxins as identified through venom proteomics and toxin-specific neutralization test.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, accession ID: PXD026270.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee, Faculty of Medicine, University of Malaya.
Author Contributions
Conceptualization: CHT and KYT; Methodology: CHT, KYT, and PP; Investigation: PP, YWC, and KYT; Validation: PP, KYT, and CHT; Data curation: CHT and KYT; Writing—original draft preparation: PP; Writing—review and editing: CHT, KYT, and PP; All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the special research funding from the University of Malaya (BKS003-2020) and the Fundamental Research Grant Scheme from the Ministry of Higher Education Malaysia (FRGS/1/2019/SKK08/UM/02/2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.727756/full#supplementary-material
References
- Avilonzoo. (2015). Samar Cobra. Available at: https://www.avilonzoo.ph/avilon-zoo/meet-our-animals/item/65-samar-cobra . [Accessed May 21, 2021].
- Barber C. M., Isbister G. K., Hodgson W. C. (2013). Alpha Neurotoxins. Toxicon 66, 47–58. 10.1016/j.toxicon.2013.01.019 [DOI] [PubMed] [Google Scholar]
- Barlow A., Pook C. E., Harrison R. A., Wüster W. (2009). Coevolution of Diet and Prey-specific Venom Activity Supports the Role of Selection in Snake Venom Evolution. Proc. Biol. Sci. 276 (1666), 2443–2449. 10.1098/rspb.2009.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-Méndez E., Fuglsang-Madsen A., Føns S., Lomonte B., Gutiérrez J., Laustsen A. (2018). Innovative Immunization Strategies for Antivenom Development. Toxins 10 (11), 452. 10.3390/toxins10110452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda A., Patra A., Kalita B., Mukherjee A. K. (2018). Proteomics Analysis to Compare the Venom Composition between Naja naja and Naja kaouthia from the Same Geographical Location of Eastern India: Correlation with Pathophysiology of Envenomation and Immunological Cross-Reactivity towards Commercial Polyantivenom. Expert Rev. Proteomics 15 (11), 949–961. 10.1080/14789450.2018.1538799 [DOI] [PubMed] [Google Scholar]
- Chang K. C., Huang Y. K., Chen Y. W., Chen M. H., Tu A. T., Chen Y. C. (2020). Venom Ophthalmia and Ocular Complications Caused by Snake Venom. Toxins (Basel) 12 (9). 10.3390/toxins12090576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicheportiche R., Vincent J. P., Kopeyan C., Schweitz H., Lazdunski M. (1975). Structure-function Relationship in the Binding of Snake Neurotoxins to the Torpedo Membrane Receptor. Biochemistry 14 (10), 2081–2091. 10.1021/bi00681a007 [DOI] [PubMed] [Google Scholar]
- Chong H. P., Tan K. Y., Tan N. H., Tan C. H. (2019). Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics. Toxins (Basel) 11 (2). 10.3390/toxins11020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E. R., Weinstein S. A., White J., Warrell D. A. (2010). Venom Ophthalmia Caused by Venoms of Spitting Elapid and Other Snakes: Report of Ten Cases with Review of Epidemiology, Clinical Features, Pathophysiology and Management. Toxicon 56 (3), 259–272. 10.1016/j.toxicon.2010.02.023 [DOI] [PubMed] [Google Scholar]
- Clementi M. E., Marini S., Condò S. G., Giardina B. (1991). Antibodies against Small Molecules. Ann. Ist Super Sanita 27 (1), 139–143. [PubMed] [Google Scholar]
- Daltry J. C., Wüster W., Thorpe R. S. (1996). Diet and Snake Venom Evolution. Nature 379 (6565), 537–540. 10.1038/379537a0 [DOI] [PubMed] [Google Scholar]
- de Silva H. A., Ryan N. M., de Silva H. J. (2016). Adverse Reactions to Snake Antivenom, and Their Prevention and Treatment. Br. J. Clin. Pharmacol. 81 (3), 446–452. 10.1111/bcp.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano A. A. (2018). PCAV: Saving Lives One Ampoule at a Time . Available at: https://ritm.gov.ph/pcav-saving-lives-one-ampoule-at-a-time/ . [Accessed May 6, 2021].
- Dutta S., Chanda A., Kalita B., Islam T., Patra A., Mukherjee A. K. (2017). Proteomic Analysis to Unravel the Complex Venom Proteome of Eastern India Naja naja: Correlation of Venom Composition with its Biochemical and Pharmacological Properties. J. Proteomics 156, 29–39. 10.1016/j.jprot.2016.12.018 [DOI] [PubMed] [Google Scholar]
- Finney D. J., Tattersfield F. (1952). Probit Analysis. By D. J. Finney, M.A., Sc.D., [2nd Ed. Pp. Xiv + 318. Cambridge University Press, 1952. 35s.]. J. Inst. Actuar. 78 (3), 388–390. 10.1017/S0020268100052938 [DOI] [Google Scholar]
- Fox J. W., Serrano S. M. (2008). Insights into and Speculations about Snake Venom Metalloproteinase (SVMP) Synthesis, Folding and Disulfide Bond Formation and Their Contribution to Venom Complexity. Febs J 275 (12), 3016–3030. 10.1111/j.1742-4658.2008.06466.x [DOI] [PubMed] [Google Scholar]
- Gutiérrez J. M., Calvete J. J., Habib A. G., Harrison R. A., Williams D. J., Warrell D. A. (2017). Snakebite Envenoming. Nat. Rev. Dis. Primers 3 (1), 17063. 10.1038/nrdp.2017.63 [DOI] [PubMed] [Google Scholar]
- Harris R. J., Zdenek C. N., Harrich D., Frank N., Fry B. G. (2020). An Appetite for Destruction: Detecting Prey-Selective Binding of α-Neurotoxins in the Venom of Afro-Asian Elapids. Toxins (Basel) 12 (3). 10.3390/toxins12030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hia Y. L., Tan K. Y., Tan C. H. (2020). Comparative Venom Proteomics of Banded Krait (Bungarus fasciatus) from Five Geographical Locales: Correlation of Venom Lethality, Immunoreactivity and Antivenom Neutralization. Acta Trop. 207, 105460. 10.1016/j.actatropica.2020.105460 [DOI] [PubMed] [Google Scholar]
- Huang H. W., Liu B. S., Chien K. Y., Chiang L. C., Huang S. Y., Sung W. C., et al. (2015). Cobra Venom Proteome and Glycome Determined from Individual Snakes of Naja atra Reveal Medically Important Dynamic Range and Systematic Geographic Variation. J. Proteomics 128, 92–104. 10.1016/j.jprot.2015.07.015 [DOI] [PubMed] [Google Scholar]
- Jayawardana S., Arambepola C., Chang T., Gnanathasan A. (2018). Long-term Health Complications Following Snake Envenoming. J. Multidiscip. Healthc. 11, 279–285. 10.2147/JMDH.S126648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturiratne A., Wickremasinghe A. R., de Silva N., Gunawardena N. K., Pathmeswaran A., Premaratna R., et al. (2008). The Global burden of Snakebite: A Literature Analysis and Modelling Based on Regional Estimates of Envenoming and Deaths. Plos Med. 5 (11), e218. 10.1371/journal.pmed.0050218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazandjian T. D., Petras D., Robinson S. D., van Thiel J., Greene H. W., Arbuckle K., et al. (2021). Convergent Evolution of Pain-Inducing Defensive Venom Components in Spitting Cobras. Science 371 (6527), 386–390. 10.1126/science.abb9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostiza T., Dahinden C. A., Rihs S., Otten U., Meier J. (1995). Nerve Growth Factor from the Venom of the Chinese Cobra Naja naja atra: Purification and Description of Non-neuronal Activities. Toxicon 33 (10), 1249–1261. 10.1016/0041-0101(95)00086-2 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 227 (5259), 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Laustsen A. H., Gutiérrez J. M., Lohse B., Rasmussen A. R., Fernández J., Milbo C., et al. (2015). Snake Venomics of Monocled Cobra (Naja kaouthia) and Investigation of Human IgG Response against Venom Toxins. Toxicon 99, 23–35. 10.1016/j.toxicon.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Leong P. K., Fung S. Y., Tan C. H., Sim S. M., Tan N. H. (2015). Immunological Cross-Reactivity and Neutralization of the Principal Toxins of Naja sumatrana and Related Cobra Venoms by a Thai Polyvalent Antivenom (Neuro Polyvalent Snake Antivenom). Acta Trop. 149, 86–93. 10.1016/j.actatropica.2015.05.020 [DOI] [PubMed] [Google Scholar]
- Liu C. C., You C. H., Wang P. J., Yu J. S., Huang G. J., Liu C. H., et al. (2017). Analysis of the Efficacy of Taiwanese Freeze-Dried Neurotoxic Antivenom against Naja kaouthia, Naja siamensis and Ophiophagus hannah through Proteomics and Animal Model Approaches. PLOS Negl. Trop. Dis. 11 (12), e0006138. 10.1371/journal.pntd.0006138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., et al. (2019). iProX: an Integrated Proteome Resource. Nucleic Acids Res. 47 (D1), D1211–D1217. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy S., Heyborne W. (2009). “Cysteine-Rich Secretory Proteins in Reptile Venoms,” in Handbook of Venoms and Toxins of Reptiles (Boca Raton, FL: CRC Press; ), 325–336. 10.1201/9781420008661.ch16 [DOI] [Google Scholar]
- Mackessy S. P. (2009). Handbook of Venoms and Toxins of Reptiles. Boca Raton, FL: CRC Press. [Google Scholar]
- McCleary R. J., Sridharan S., Dunstan N. L., Mirtschin P. J., Kini R. M. (2016). Proteomic Comparisons of Venoms of Long-Term Captive and Recently Wild-Caught Eastern Brown Snakes (Pseudonaja textilis) Indicate Venom Does Not Change Due to Captivity. J. Proteomics 144, 51–62. 10.1016/j.jprot.2016.05.027 [DOI] [PubMed] [Google Scholar]
- Mehta S. R., Sashindran V. K. (2002). Clinical Features and Management of Snake Bite. Med. J. Armed Forces India 58 (3), 247–249. 10.1016/S0377-1237(02)80140-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais V., Ifran S., Berasain P., Massaldi H. (2010). Antivenoms: Potency or Median Effective Dose, Which to Use. J. Venom. Anim. Toxins Incl. Trop. Dis. 16, 191–193. 10.1590/S1678-91992010000200002 [DOI] [Google Scholar]
- Oh A. M. F., Tan C. H., Ariaranee G. C., Quraishi N., Tan N. H. (2017). Venomics of Bungarus caeruleus (Indian Krait): Comparable Venom Profiles, Variable Immunoreactivities Among Specimens from Sri Lanka, India and Pakistan. J. Proteomics 164, 1–18. 10.1016/j.jprot.2017.04.018 [DOI] [PubMed] [Google Scholar]
- Oh A. M. F., Tan C. H., Tan K. Y., Quraishi N. H., Tan N. H. (2019). Venom Proteome of Bungarus sindanus (Sind Krait) from Pakistan and In Vivo Cross-Neutralization of Toxicity Using an Indian Polyvalent Antivenom. J. Proteomics 193, 243–254. 10.1016/j.jprot.2018.10.016 [DOI] [PubMed] [Google Scholar]
- Pahari S., Bickford D., Fry B. G., Kini R. M. (2007). Expression Pattern of Three-finger Toxin and Phospholipase A2 Genes in the Venom Glands of Two Sea Snakes, Lapemis curtus and Acalyptophis peronii: Comparison of Evolution of These Toxins in Land Snakes, Sea Kraits and Sea Snakes. BMC Evol. Biol. 7, 175–179. 10.1186/1471-2148-7-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pung Y. F., Kumar S. V., Rajagopalan N., Fry B. G., Kumar P. P., Kini R. M. (2006). Ohanin, a Novel Protein from King Cobra Venom: Its cDNA and Genomic Organization. Gene 371 (2), 246–256. 10.1016/j.gene.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Pung Y. F., Wong P. T., Kumar P. P., Hodgson W. C., Kini R. M. (2005). Ohanin, a Novel Protein from King Cobra Venom, Induces Hypolocomotion and Hyperalgesia in Mice. J. Biol. Chem. 280 (13), 13137–13147. 10.1074/jbc.M414137200 [DOI] [PubMed] [Google Scholar]
- Ralph R., Sharma S. K., Faiz M. A., Ribeiro I., Rijal S., Chappuis F., et al. (2019). The Timing Is Right to End Snakebite Deaths in South Asia. BMJ 364, k5317. 10.1136/bmj.k5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanabanangkoon K. (2021). A Quest for a Universal Plasma-Derived Antivenom against All Elapid Neurotoxic Snake Venoms. Front. Immunol. 12 (1363), 668328. 10.3389/fimmu.2021.668328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanabanangkoon K., Tan K. Y., Eursakun S., Tan C. H., Simsiriwong P., Pamornsakda T., et al. (2016). A Simple and Novel Strategy for the Production of a Pan-specific Antiserum against Elapid Snakes of Asia. Plos Negl. Trop. Dis. 10 (4), e0004565. 10.1371/journal.pntd.0004565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanabanangkoon K., Tan K. Y., Pruksaphon K., Klinpayom C., Gutiérrez J. M., Quraishi N. H., et al. (2020). A Pan-specific Antiserum Produced by a Novel Immunization Strategy Shows a High Spectrum of Neutralization against Neurotoxic Snake Venoms. Sci. Rep. 10 (1), 11261. 10.1038/s41598-020-66657-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguila M. B., Cobb K. A., Siler C. D., Diesmos A. C., Alcala A. C., Brown R. M. (2016). The Amphibians and Reptiles of Mindanao Island, Southern Philippines, II: the Herpetofauna of Northeast Mindanao and Adjacent Islands. ZooKeys 624, 1–132. 10.3897/zookeys.624.9814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L. L., Gao J. F., Zhang Y. X., Shen S. S., He Y., Wang J., et al. (2016). Proteomic Characterization and Comparison of Venoms from Two Elapid Snakes (Bungarus multicinctus and Naja atra) from China. J. Proteomics 138, 83–94. 10.1016/j.jprot.2016.02.028 [DOI] [PubMed] [Google Scholar]
- Silva A., Cristofori-Armstrong B., Rash L. D., Hodgson W. C., Isbister G. K. (2018). Defining the Role of post-synaptic α-neurotoxins in Paralysis Due to Snake Envenoming in Humans. Cell Mol. Life Sci. 75 (23), 4465–4478. 10.1007/s00018-018-2893-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sintiprungrat K., Watcharatanyatip K., Senevirathne W. D., Chaisuriya P., Chokchaichamnankit D., Srisomsap C., et al. (2016). A Comparative Study of Venomics of Naja naja from India and Sri Lanka, Clinical Manifestations and Antivenomics of an Indian Polyspecific Antivenom. J. Proteomics 132, 131–143. 10.1016/j.jprot.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Sy E. Y., Bucol A. A. (2020). First Record of the Southern Philippine Cobra Naja samarensis on Siquijor Island, Philippines. Southeast Asia Vertebrate Records. ISSN: 2424‐8525. [Google Scholar]
- Sy E. Y., Mangkabong S. G. (2018). First Record of Southern Philippine Cobra Naja samarensis on Basilan Island, Philippines. Southeast Asia Vertebrate Records. ISSN: 2424‐8525. [Google Scholar]
- Tan C. H., Fung S. Y., Yap M. K., Leong P. K., Liew J. L., Tan N. H. (2016a). Unveiling the Elusive and Exotic: Venomics of the Malayan Blue Coral Snake (Calliophis bivirgata flaviceps). J. Proteomics 132, 1–12. 10.1016/j.jprot.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Tan C. H., Palasuberniam P., Blanco F. B., Tan K. Y. (2021). Immunoreactivity and Neutralization Capacity of Philippine Cobra Antivenom against Naja philippinensis and Naja samarensis Venoms. Trans. R. Soc. Trop. Med. Hyg. 115 (1), 78–84. 10.1093/trstmh/traa087 [DOI] [PubMed] [Google Scholar]
- Tan C. H., Tan K. Y. (2019). Functional Application of Snake Venom Proteomics in In Vivo Antivenom Assessment. Methods Mol. Biol. 1871, 153–158. 10.1007/978-1-4939-8814-3_11 [DOI] [PubMed] [Google Scholar]
- Tan C. H., Tan K. Y., Fung S. Y., Tan N. H. (2015a). Venom-gland Transcriptome and Venom Proteome of the Malaysian King Cobra (Ophiophagus hannah). BMC Genomics 16, 687. 10.1186/s12864-015-1828-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. H., Tan K. Y., Lim S. E., Tan N. H. (2015b). Venomics of the Beaked Sea Snake, Hydrophis schistosus: A Minimalist Toxin Arsenal and its Cross-Neutralization by Heterologous Antivenoms. J. Proteomics 126, 121–130. 10.1016/j.jprot.2015.05.035 [DOI] [PubMed] [Google Scholar]
- Tan C. H., Tan K. Y., Ng T. S., Sim S. M., Tan N. H. (2019a). Venom Proteome of Spine-Bellied Sea Snake (Hydrophis curtus) from Penang, Malaysia: Toxicity Correlation, Immunoprofiling and Cross-Neutralization by Sea Snake Antivenom. Toxins (Basel) 11 (1). 10.3390/toxins11010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. H., Tan N. H., Tan K. Y., Kwong K. O. (2015c). Antivenom Cross-Neutralization of the Venoms of Hydrophis schistosus and Hydrophis curtus, Two Common Sea Snakes in Malaysian Waters. Toxins (Basel) 7 (2), 572–581. 10.3390/toxins7020572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. H., Wong K. Y., Tan K. Y., Tan N. H. (2017a). Venom Proteome of the Yellow-Lipped Sea Krait, Laticauda colubrina from Bali: Insights into Subvenomic Diversity, Venom Antigenicity and Cross-Neutralization by Antivenom. J. Proteomics 166, 48–58. 10.1016/j.jprot.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Tan C. H., Wong K. Y., Tan N. H., Ng T. S., Tan K. Y. (2019c). Distinctive Distribution of Secretory Phospholipases A₂ in the Venoms of Afro-Asian Cobras (Subgenus: Naja, Afronaja, Boulengerina and Uraeus). Toxins (Basel) 11 (2), 116. 10.3390/toxins11020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. H., Wong K. Y., Chong H. P., Tan N. H., Tan K. Y. (2019b). Proteomic Insights into Short Neurotoxin-Driven, Highly Neurotoxic Venom of Philippine Cobra (Naja philippinensis) and Toxicity Correlation of Cobra Envenomation in Asia. J. Proteomics 206, 103418. 10.1016/j.jprot.2019.103418 [DOI] [PubMed] [Google Scholar]
- Tan K. Y., Liew J. L., Tan N. H., Quah E. S. H., Ismail A. K., Tan C. H. (2019d). Unlocking the Secrets of Banded Coral Snake (Calliophis intestinalis, Malaysia): A Venom with Proteome Novelty, Low Toxicity and Distinct Antigenicity. J. Proteomics 192, 246–257. 10.1016/j.jprot.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Tan K. Y., Tan C. H., Chanhome L., Tan N. H. (2017b). Comparative Venom Gland Transcriptomics of Naja kaouthia (Monocled Cobra) from Malaysia and Thailand: Elucidating Geographical Venom Variation and Insights into Sequence Novelty. PeerJ 5 (4), e3142. 10.7717/peerj.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K. Y., Tan C. H., Fung S. Y., Tan N. H. (2016b). Neutralization of the Principal Toxins from the Venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into Toxin-specific Neutralization by Two Different Antivenoms. Toxins (Basel) 8 (4), 86. 10.3390/toxins8040086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K. Y., Tan C. H., Fung S. Y., Tan N. H. (2015d). Venomics, Lethality and Neutralization of Naja kaouthia (Monocled Cobra) Venoms from Three Different Geographical Regions of Southeast Asia. J. Proteomics 120, 105–125. 10.1016/j.jprot.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Tan K. Y., Wong K. Y., Tan N. H., Tan C. H. (2020). Quantitative Proteomics of Naja annulifera (Sub-saharan Snouted Cobra) Venom and Neutralization Activities of Two Antivenoms in Africa. Int. J. Biol. Macromolecules 158, 605–616. 10.1016/j.ijbiomac.2020.04.173 [DOI] [PubMed] [Google Scholar]
- Tan N. H., Wong K. Y., Tan C. H. (2017c). Venomics of Naja sputatrix, the Javan Spitting Cobra: A Short Neurotoxin-Driven Venom Needing Improved Antivenom Neutralization. J. Proteomics 157, 18–32. 10.1016/j.jprot.2017.01.018 [DOI] [PubMed] [Google Scholar]
- Tan N. H., Tan C. H. (2016). “Cytotoxicity of Snake Venoms and Toxins: Mechanisms and Applications,” in Snake Venoms and Envenomation. Editor Utkin Y.N. (New York: Nova Science Publishers; ). [Google Scholar]
- Thatoi P., Acharya R., Malla A. (2016). Acute Respiratory Failure Following Neurotoxic Snake Bite - A Study of 101 Cases of Neurotoxic Snake Bite from Eastern India. Eur. Respir. J. 48, 2140. 10.1183/13993003.congress-2016.PA2140 [DOI] [Google Scholar]
- Uetz P., Freed P., Aguilar R., Hošek J. (2021a). Naja philippinensis: The Reptile Database Available at: http://www.reptile-database.org [Accessed May 12, 2021].
- Uetz P., Freed P., Aguilar R., Hošek J. (2021b). Naja samarensis: The Reptile Database Available at: http://www.reptile-database.org [Accessed May 12, 2021].
- Utkin Y. N. (2013). Three-finger Toxins, a Deadly Weapon of Elapid Venom-Milestones of Discovery. Toxicon 62, 50–55. 10.1016/j.toxicon.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Watt G., Padre L., Tuazon L., Theakston R. D., Laughlin L. (1988). Bites by the Philippine Cobra (Naja naja philippinensis): Prominent Neurotoxicity with Minimal Local Signs. Am. J. Trop. Med. Hyg. 39 (3), 306–311. 10.4269/ajtmh.1988.39.306 [DOI] [PubMed] [Google Scholar]
- Williams D. J., Gutiérrez J. M., Calvete J. J., Wüster W., Ratanabanangkoon K., Paiva O., et al. (2011). Ending the Drought: New Strategies for Improving the Flow of Affordable, Effective Antivenoms in Asia and Africa. J. Proteomics 74 (9), 1735–1767. 10.1016/j.jprot.2011.05.027 [DOI] [PubMed] [Google Scholar]
- Williams D. J., Habib A. G., Warrell D. A. (2018). Clinical Studies of the Effectiveness and Safety of Antivenoms. Toxicon 150, 1–10. 10.1016/j.toxicon.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Wong K. Y., Tan C. H., Tan K. Y., Quraishi N. H., Tan N. H. (2018). Elucidating the Biogeographical Variation of the Venom of Naja naja (Spectacled Cobra) from Pakistan through a Venom-Decomplexing Proteomic Study. J. Proteomics 175, 156–173. 10.1016/j.jprot.2017.12.012 [DOI] [PubMed] [Google Scholar]
- Wong K. Y., Tan C. H., Tan N. H. (2016). Venom and Purified Toxins of the Spectacled Cobra (Naja naja) from Pakistan: Insights into Toxicity and Antivenom Neutralization. Am. J. Trop. Med. Hyg. 94 (6), 1392–1399. 10.4269/ajtmh.15-0871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. Y., Tan K. Y., Tan N. H., Tan C. H. (2021). A Neurotoxic Snake Venom without Phospholipase A2: Proteomics and Cross-Neutralization of the Venom from Senegalese Cobra, Naja senegalensis (Subgenus: Uraeus). Toxins 13 (1), 60. 10.3390/toxins13010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2016). Guidelines for the Management of Snakebites. Geneva: WHO. [Google Scholar]
- World Health Organization [WHO] (2018). Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins. Geneva: WHO. [Google Scholar]
- World Health Organization [WHO] (2019). Snakebite Envenoming: A Strategy for Prevention and Control: Executive Summary. Geneva: WHO. [Google Scholar]
- Wüster W., Thorpe R. S. (1990). Systematics and Biogeography of the Asiatic Cobra (Naja naja) Species Complex in the Philippine Islands. Vertebrates in the Tropics, 333–344. ISNI. 0000 0001 3573 482X. [Google Scholar]
- Xu N., Zhao H. Y., Yin Y., Shen S. S., Shan L. L., Chen C. X., et al. (2017). Combined Venomics, Antivenomics and Venom Gland Transcriptome Analysis of the Monocoled Cobra (Naja kaouthia) from China. J. Proteomics 159, 19–31. 10.1016/j.jprot.2017.02.018 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Morita T. (2004). Structure and Function of Snake Venom Cysteine-Rich Secretory Proteins. Toxicon 44 (3), 227–231. 10.1016/j.toxicon.2004.05.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository (https://www.iprox.org) (Ma et al., 2019) with the dataset identifier PXD026270, subproject ID: IPX0003097001.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, accession ID: PXD026270.