Abstract
Circular RNAs (circRNAs) are classified as noncoding RNAs because they are devoid of a 5’ end cap and a 3’ end poly (A) tail necessary for cap-dependent translation. However, increasing numbers of translated circRNAs identified through high-throughput RNA sequencing overlapping with polysome profiling indicate that this rule is being broken. CircRNAs can be translated in cap-independent mechanism, including IRES (internal ribosome entry site)-initiated pattern, MIRES (m6A internal ribosome entry site) -initiated patterns, and rolling translation mechanism (RCA). CircRNA-encoded proteins harbour diverse functions similar to or different from host proteins. In addition, they are linked to the modulation of human disease including carcinomas and noncarcinomas. CircRNA-related translatomics and proteomics have attracted increasing attention. This review discusses the progress and exclusive characteristics of circRNA translation and highlights the latest mechanisms and regulation of circRNA translatomics. Furthermore, we summarize the extensive functions and mechanisms of circRNA-derived proteins in human diseases, which contribute to a better understanding of intricate noncanonical circRNA translatomics and proteomics and their therapeutic potential in human diseases.
Keywords: Translation, Cap-independent, Circular RNA, Protein, Carcinomas
Introduction
Protein-coding genes annotated in the Human Genome Project account for only approximately 2.5% of the human genome [1]. The remaining transcripts in the genome are considered noncoding RNAs (ncRNAs) comprising circular RNAs (circRNAs), microRNAs, long noncoding RNAs, PIWI-interacting RNAs, and others [2]. CircRNAs are a unique subclass of ncRNAs characterized by covalently closed loops without 5’ and 3’ terminals [3, 4]. Generated from back splicing (head-to-tail splicing) events or noncolinear splicing reactions of precursor mRNAs (pre-mRNAs) in the nuclear and mitochondrial genomes of mammals [5–14], circRNAs have exclusive signs termed “back splicing junctions (BSJs)” to distinguish them from cognate linear RNAs.
Sanger HL and coauthors first discovered peculiar single-stranded covalently closed molecules in viroids by utilizing electron microscopy and created the term “circular RNA” [15]. Subsequently, these strange molecules were observed in pathogens and eukaryotic cells [16], but they have received little attention since for a long time they were considered to be functionless products of exon aberrant splicing. In 2013, a breakthrough in two articles published in NATURE revealed the sponge-like function of circRNAs in microRNAs [6, 17], transforming them from “waster to treasure (research hotspots)”. Recently, vast numbers of functional circRNAs were discovered in the eukaryotic tree of life from fungi to mammals, conserved across varied species [18–21]. These molecules are enriched in specific tissues/or cells or particularly developmental stages owing to their abundance or longer half-life [21–24]. The circular structure provides circRNAs with resistance against attacks by most ribonucleases except RNases encompassing RNase A, RNase T1, and RNase T2; as a result, circRNAs are more stable than their linear counterparts [21, 25, 26].
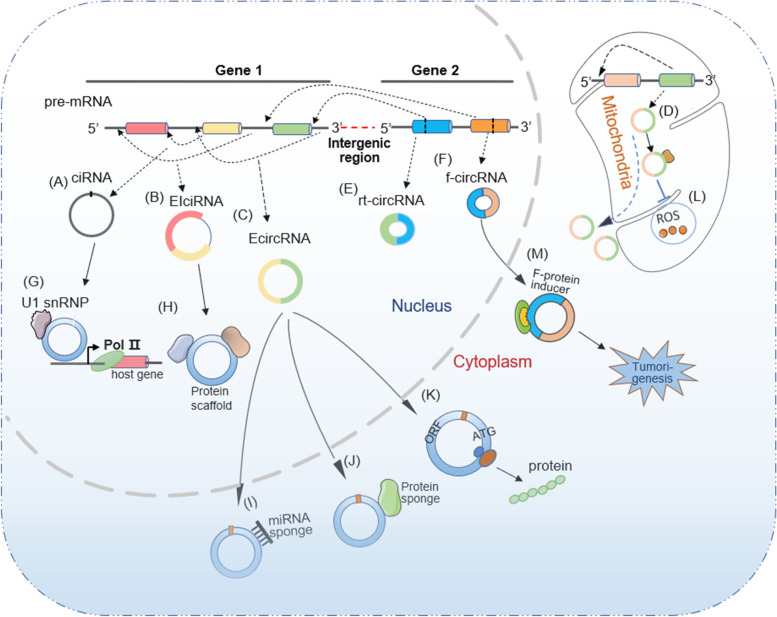
Circular RNA classification is based on various origination and circularization modes and consists of intronic circRNAs (ciRNAs) circularized from introns (Fig. 1A), exon-intron circRNAs (EIciRNAs) from exons covering intronic regions (Fig. 1B), exon circRNAs (EcircRNAs) formed from mono- or multi-exons in nuclear genomes and mitochondrial genomes (mecciRNAs), certain exonic mitochondrial circRNAs may contain intron retained sequences (Fig. 1C-D) [8, 11–14, 27–29], read-through circRNA (rt-circRNA) from exons between neighbouring genes on the same strand (intrachromosomal chimaeras) (Fig. 1E), and fused circRNAs (f-circRNAs) that are fused exons between two distant genes (interchromosomal chimaeras) in the translocation process (Fig. 1F) [30–32]. Intriguingly, EIciRNAs and ciRNAs are sequestered in the nucleus [10, 29], while most EcircRNAs derived from the nuclear genome are exported to the cytoplasm [27, 30], and f-circRNAs may be localized in the nucleus and cytoplasm. The resultant discrepancy in localization is closely linked to their diverse functions. Nuclear ciRNAs and EIciRNAs are capable of enhancing the Pol II transcription rate of its host gene via interaction with the U1 small nuclear ribonucleoprotein (snRNP), as exemplified by circEIF3J and circPAIP2 (Fig. 1G), or as scaffolds to recruit functional molecules (circ-Amotl1, circ-Foxo3) (Fig. 1H) [7, 33–35]. F-circRNA derived from cancer-associated chromosomal translocations can be an oncogenic molecule involved in tumorigenesis (Fig. 1M) [31]. MecciRNAs are linked to the regulation of ROS in mitochondria (Fig. 1L) [12, 30–32], and the read-through circRNA function remains unclear [31]. Mature exonic circRNAs are exported to the cytoplasm as sponges of microRNAs and proteins (Fig. 1I-J) or as templates to be translated into novel proteins/peptides (Fig. 1K) [17, 36–39].
Fig. 1.
Biogenesis and function of various circular RNAs. Circular RNAs are the product of back splicing of pre-messenger RNAs (pre-mRNAs), comprising intronic circRNA (ciRNA) from intron (A), Exonic and intronic circRNA (EIciRNA) from exon covering intronic regions (B), exon circRNA (EcircRNA) from exons in nuclear (C) and mitochondrial genomes (mecciRNAs) (D), read-through circRNA (rt-CircRNA) from exons between neighboring genes on the same strand (E), and fused circRNA (f-circRNA) fuse from exons between two distant genes (F). CiRNA interacts with small nuclear ribonucleoprotein (snRNP) to enhance transcription rate of its host gene (G). EIciRNA can be a scaffold to recruit functional molecules (H). EcircRNAs are exported to cytoplasm as the sponges of micro-RNA (I) and protein (J), or as the templates to be translated into novel protein (K). MecciRNA may be linked to an inhibition of Reactive oxygen species-ROS (L). F-circRNA accompanied with fused protein to promote tumorigenesis (M)
Previously, the translation of circRNAs was under speculation because canonical theory indicated that protein synthesis of eukaryotic mRNA in ribosome scanning mechanism requires the 5’ m7GpppN (m7G) cap and 3’ poly-A tail [40–43]. Due to the absence of the 5’ end and 3’ end for landing the m7G cap and poly-A tail, respectively, circRNAs were presumed to be untranslated. Moreover, traditional annotation of protein-coding genes only covers proteins more than 100 amino acids; thus, those shorter than 100 amino acids translated from circRNAs were ignored [44]. Recently, numerous translated circRNAs have been detected with the advent of advanced high-throughput technology, including RNA sequencing (RNA-seq) combined with polysome profiling and circRNA-specific bioinformatics algorithms [38, 39, 45, 46]. Natural circRNA translation is independent of the m7G cap but is dependent on internal ribosome entry (IRES) elements, including IRES or short IRES-like A/U enriched sequences [38, 45, 47–54], MIRES (m6A-IRES) [55, 56]. Moreover, circRNA can be translated in rolling circle amplification (RCA) mechanism (Fig. 2) [46, 57]. CircRNA-translated proteins (circ-proteins) modulate various physiopathologic processes ranging from carcinomas, including glioblastoma, colorectal and colon cancers, gastric cancer, hepatocellular carcinoma, and multiple myeloma [47, 56–62], and noncarcinomas, such as cardiac remodelling and Alzheimer’s disease (Fig. 3, Table 1) [66, 67].
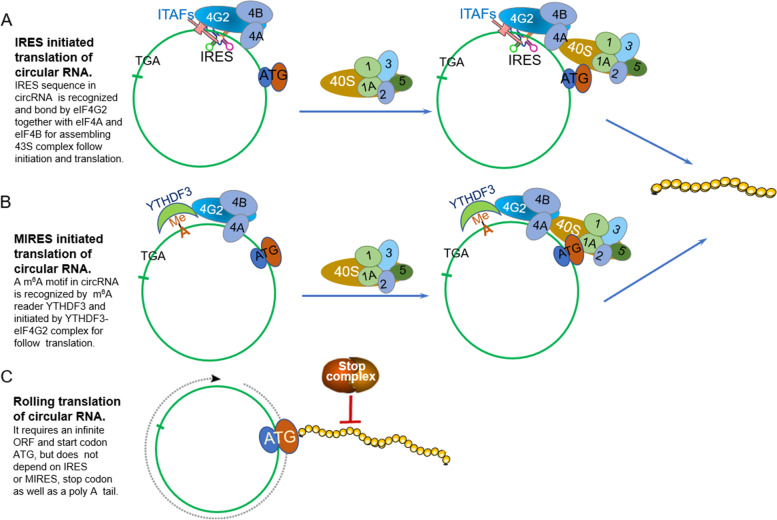
Fig. 2.
Translation mechanism of circular RNA. A IRES initiated translation of circular RNA. IRES is recognized and bond by eIF4G2 and acted as scaffold together with eIF4A and eIF4B for assembling 43S initiation complex including 40S ribosomal subunit and combination of eIF4G2 and eIFs complex to encounter start codon ATG for translation initiation and synthesis, which may be assisted with ITAFs (IRES trans-acting factors). B MIRES initiated translation of circular RNA. The m6A motif in circRNA is recognized by a m6A reader YTH domain family protein 3 (YTHDF3) to recruit eIF4G2 together with eIF4A and eIF4B for assembling 43S initiation complex including 40S ribosomal subunit and combination of eIF4G2 and eIFs complex to encounter ATG for translation initiation and synthesis. C circRNA harboring an infinite ORF and start codon ATG enables continuous translation termed rolling circle amplification without IRES element and stop codon, which can be terminated by a complex system named “programmed-1 ribosomal frameshifting”(-1PRF)-mediated out of-frame stop codon (OSC)
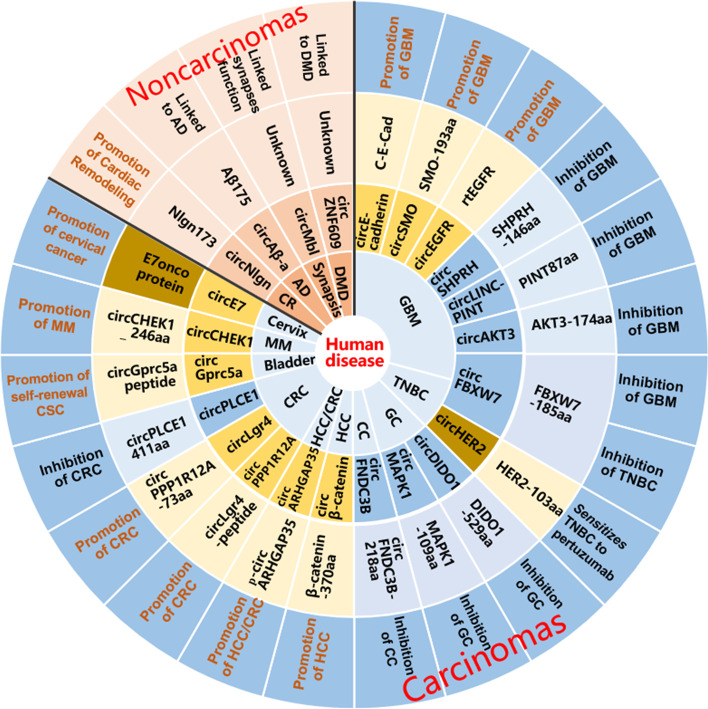
Fig. 3.
Translated circular RNAs and encoded proteins with various functions in carcinomas and noncarcinomas. Identified translated circular RNAs and encoded proteins with various functions in carcinomas containing glioblastoma (GBM), Triple-Negative Breast Cancer (TNBC), gastric cancer (GC), colon cancer (CC), colorectal cancer (CRC), hepatocellular carcinoma (HCC), multiple myeloma (MM), bladder cancer(Bladder), cervical cancer (Cervix), and in noncarcinomas diseases including Duchenne Muscular Dystrophy (DMD), synapsis function (Synapsis), Alzheimer’ s Disease (AD), cardiac remodeling (CR)
Table 1.
Overview of translated circRNAs and circ-proteins in carcinomas and noncarcinomas
| Diseases | Translated circular RNAs | Circ-proteins | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CircRNAs | Length (nt) | Ribosome binding | Translation mechanism | Circ-proteins | Length (aa) | Cellular location | Sequences covering domain of host protein | Expression (Up/Down) | Function and mechanism | ||
| Carcinomas | |||||||||||
| GBM | circE-cadherin (hsa_circ _0039992) | 733 |
reads across BSJ in ribosome-Seq |
IRES | C-E-Cad | 254 | Membrane, same to E-cadherin | Unique 14-aa tail at C-termini of C-E-Cad besides shared aa 282-522 at N-terminal of E-cadherin | Up | Unique 14-aa had new function that bond to the CR2 domain of EGFR, promoted STAT3 phosphorylation and nuclear translocation to interact with EGFRvIII, inducing GBM. | [46] |
| GBM | circSMO (hsa_circ _0001742) | 727 | In M and L polysomes fractions | IRES | SMO-193a.a | 193 | Cytoplasm and Membrane, same to SMO | Shared aa 230-421 at N-terminal of SMO covering most seven transmembrane domains besides one Glu at its C-termini | Up | Shared motif bond to N-terminal of SMO to translocate cholesterol, freed SMO from patched transmembrane receptors to maintain CSC self-renewal, inducing GBM. | [51] |
| GBM | circEGFR (hsa_circ_0080229) | 249 | In M and L polysomes fractions | Rolling translation | rtEGFR | 83 | Membrane, same to EGFR | shared aa 561-627 across extracellular domain IV of EGFR besides unique 19 amino acids, has similar function with EGFR | Up | Shared motif bond to extracellular Domain IV of EGFR to increase EGFR stability and membrane localization, attenuating its endocytosis and degradation, inducing GBM. | [57] |
| GBM | circSHPRH (hsa_circ_0001649) | 440 | undetected | IRES | SHPRH-146aa | 146 | unknown | shared aa1520–1651 covering the SNF2 domain of SHPRH besides 8-aa at its C-termini | Up | Shared motif as a decoy competitively bond to DTL to prevent its ubiquitination SHPRH which could ubiquitinate PCNA, inhibiting GBM. | [52] |
| GBM | circLINC-PINT (hsa_circ_0082389) | 1084 | two readers across BSJ by RNC-seq | IRES | PINT87aa | 87 | Nucleus, same to PINT | shared amino acids at N-terminal of PINT besides 10-aa at its C-termini | Down | Bond to the domain of PAF1 as an anchor keeping PAF1 complex on target genes’ promoter to repress transcriptional elongation and GBM. | [63] |
| GBM | circAKT3 (hsa_circ_0017250) | 524 | undetected | IRES | AKT3-174aa | 174 | Cytoplasm, similar to AKT3 | shared aa 62-232 covering PH-domain and thr-308 of AKT3 besides 3-aa at its C-termini | Down | Shared motif bond to PDK1 to activate it, blocking AKT phosphorylation, inhibiting GBM. | [54] |
| GBM | circFBXW7 (hsa_circ_022705) | 620 | undetected | IRES | FBXW7-185aa | 185 | unknown | shared 167-aa with FBXW7a besides 18-aa at its C-termini | Down | Shared motif as decoy competitively bond to USP28, releasing FBXW7a to degrade c-Myc, inhibiting GBM. | [53] |
| TNBC | 620 | undetected | IRES | FBXW7-185aa | 185 | unknown | shared 167-aa with FBXW7a besides 18-aa at its C-termini | Down | Increased the abundance of FBXW7 and inducing c-Myc degradation, inhibiting TNBC. | [53] | |
| TNBC | circHER2 (hsa_circ_0007766) | 676 | In M and L polysomes fractions | IRES | HER2-103 | 103 | Membrane, same to HER2 | shared 103 amino acids at N-terminal (aa 198-300) of HER2 covering most CR I domain of HER2 for EGFR/HER3 homo/heterodimer. | Up | Bond to CR I domain of HER2, stimulated EGFR/HER3 homo/heterodimer formation, phosphorylation and activation of AKT to sensitize Pertuzumab treatment of HER-103+ TNBC. | [64] |
| GC | circDIDO1 (hsa_circ_0061137) | 1787 | undetected | IRES | DIDO1-529aa | 529 | Nucleus, different from DIDO1-1a | shared 529 amino acids with NLS and PHD domain, lack of nuclear export sequence of DIDO1-1a | Down | Contrary to DIDO1-1a. bond to DNA binding domain and catalytic domain of PARP1 to block its activity, repressing GC, | [60] |
| GC | circMAPK1 (hsa_circ_0004872) | 490 | undetected | IRES | MAPK1-109aa | 109 | Cytoplasm, similar to MAPK1 | shared aa 98-203 covering phosphorylated sites of MAPK1 besides 3-aa at its C-termini | Down | Contrary to MAPK1, competitively bond to MEK1 to block extracellular signals to intracellular signals for MAPK phosphorylation, repressing GC. | [59] |
| CC | circFNDC3B (hsa_circ_0006156) | 526 | undetected | IRES | circFNDC3B-218aa | 218 | Cytoplasm, similar to FNDC3B | shared 509 amino acids with FNDC3B besides 17-aa at its C-termini | Down | Attenuated Snail expression, enhanced FBP1-induced OXPHOS to repress CC. | [58] |
| HCC | circβ-catenin (hsa_circ_0004194) | 1129 | undetected | IRES | β-catenin-370aa | 370 | Cytoplasm, similar to β-catenin | shared 361 amino acids at N-terminal of β-catenin besides 9-aa at its C-termini | Up | As a decoy binding to GSK3β to prevent it degrade β-catenin, freed β-catenin activated Wnt/β-catenin pathway, promoting HCC. | [48] |
| HCC and CRC | circARHGAP35 (hsa_circ_0109744) | 3867 | in M polysomes fractions | m6A modification | p-circARHGAP35 | 1289 | Nucleus, opposite to ARHGAP35 | shared most amino acids at N-terminal of ARHGAP35 containing four FF domains lack of Rho GAP domain | Up | Contrary to ARHGAP35, shared motif interacted with nuclear TFII-I protein, promoting progression of HCC and CRC. | [56] |
| CRC | circPPP1R12A (has_circ_0000423) | 1138 | undetected | undetected | circPPP1R12A-73aa | 73 | unknown | shared 55 amino acids besides 17-aa unique tail | Up | Activated Hippo-YAP pathway to promote CRC, which is different from PPP1R12A. | [49] |
| CRC | circLgr4 (hsa_circ_02276) | unknown | undetected | undetected | circLgr4-peptide | 19 | Cytoplasm and nucleus | has 19-aa | Up | Interacted with extracellular domain LGR4 to activate Wnt/β-catenin pathway, promoting self-renewal and metastasis of CSC, driving CRC. | [50] |
| CRC | circPLCE1 (hsa_circ_0019223) | 1570 | undetected | IRES | circPLCE1 411 | 411 | Cytoplasm | shared aa 1-403 at N-terminal of PLCE1 protein with distinct function with PLCE1, besides own 8-aa tail | Down | Bond to ATP binding domain of HSP90α to accelerate RPS3 to dissociate from the HSP90α/RPS3 complex, leading to the HSP70-induced ubiquitin-dependent degradation of RPS3 and suppression of NF-κB pathway, blocking CRC. | [61] |
| Bladder cancer | circGprc5a (hsa_circ_02838) | unknown | undetected | undetected | circGprc5a-peptide | 11 | unknown | unknown | Up | bind to Gprc5a to activate the GPCR signalling pathway and promote self-renewal and metastasis of cancer stem cells | [47] |
| MM | circCHEK1 hsa_circ_0024792 | 738 | undetected | IRES | circCHEK1_246aa | 246 | unknown | shared N-terminus of CHEK1, has same function with CHEK1 | Up | induced Chromosomal Instability and bone lesion formation by interaction with and decrease mutant CEP170 | [62] |
| Cervical cancer | circE7 | 472 | in polysomes fractions | m6A modification | E7 oncoprotein | 98 | Cytoplasm | Shared most amino acids with E7 protein harbouring unique sequences | Up | E7 oncoprotein is independent for the transforming activity of circE7 promoting progress of cervical cancer | [65] |
| Noncarcinomas | |||||||||||
| CR | circNlgn (hsa_circ_0003046) | 813 | In heavy polysomes fractions | IRES | Nlgn173 | 173 | Nucleus, different from Nlgn | 9-aa tail at Nlgn173 C-termini for nuclear localization besides 164-aa at N- terminal of Nlgn | Up | Unique 9-aa motif interacts with the LaminB1 forcing nuclear localization of Nlgn173 to promote SGK3 and inhibit ING4, inducing Cardiac Remodelling, which is different Nlgn. | [66] |
| AD | circAβ-a (hsa_circ_0007556) | 524 | undetected | IRES-like A/U-rich sequences | Aβ175 | 175 | unknown | remaining 158-aa approaching to C-terminal of Amyloid β peptide | Up | Its expression raised in brain tissues of AD patients | [67] |
| Synaptic function | circMbl | unknown | Ribosome binding | IRES-like UTR | none | unknown | Cytoplasm | unknown | Up | maybe linked to regulation of synaptic function | [45] |
| DMD | circZNF609 | unknown | M and L polysomes | IRES-like UTR | none | 753 | unknown | unknown | Up | Linked to myoblast proliferation | [38] |
| Lifespan extending | circSfl | unknown | Ribosome binding | unknown | none | unknown | unknown | Sharing N-terminus with cytoplasmic and transmembrane domain | Up | extending lifespan of fruit flies | [68] |
ORF Open Reading Framework, BSJ back splice junction, IRES Internal Ribosome Entry Site, GBM glioblastoma, rtEGFR rolling translation EGFR, TNBC Triple-Negative Breast Cancer, GC gastric cancer, CC colon cancer, CRC colorectal carcinoma, HCC hepatocellular carcinoma, MM multiple myeloma, CR Cardiac Remodeling, AD Alzheimer’s Disease, DMD Duchenne Muscular Dystrophy, m6A, N6-methyladenosine, UTR Untranslated Region
The current work summarizes the progress and exclusive characteristics of circRNA translation, highlighting the latest regulation mechanisms of circRNA translation and extensive function of circRNA-encoded proteins in human disease, which contributes to a better understanding of noncanonical circRNA-oriented translatomics and related therapeutic potential in human disease.
Profiling of circRNA translation
History of circRNA translation
In 1986, Wang and coauthors observed that a hepatitis delta viral genome-derived single-stranded circular RNA harboured an open reading framework (ORF), start codon AUG and stop codon, enabling translation of a polypeptide with 215 amino acids, indicating the potential of translation [69]. Similar translated circular RNAs were subsequently identified in plant viroids, virusoids, and the Sry gene of adult mouse testes [70, 71]. Such pioneering work failed to be accepted at that time because canonical theory assumed that protein translation of eukaryotic mRNA requires the m7G cap and poly A tail, both of which are absent in circRNAs [40, 42]. In 1995, one decade later, Chen and Sarnow showed an exception to this rule. They successfully created a circRNA carrying an ORF and IRES of encephalomyocarditis virus. This molecule generated an approximately 23 kDa protein product, suggesting that circRNAs can be translated in an IRES-dependent pattern independent of the m7G cap [72]. Nevertheless, they failed to demonstrate that natural circRNAs can be translated.
This challenge was overcome by AbouHaidar et al. in 2014. These scholars revealed that the covalently closed RNA from a virusoid produced a 16 kDa protein in an uncapped IRES-dependent mechanism [73]. Intriguingly, this translation used overlapping initiation-termination codons (UGAUGA). They speculated that overlapping codons may be generated by overlapping reading frames registered in the genome where the UGAUGA sequence becomes two consecutive opal termination UGA codons, causing overlapping start and stop codons [73]. In 2015, Wang et al. utilized minigenes containing GFP fragments and IRES sequences and directly demonstrated that in human and Drosophila cells, natural exonic circRNAs with putative ORFs could be translated into functional proteins in uncapped IRES-dependent pathways [74].
Abe et al. discovered an unexpected protein synthesis mechanism termed “rolling circle amplification (RCA)”. They created an exonic circRNA bearing an infinite ORF (iORF) and a start codon AUG and introduced it into rabbit reticulocyte lysate or human living cells, producing the expected protein bands despite the lack of a given IRES element, 5’m7G cap structure and a 3’poly-A tail [75, 76]. These findings support that circRNA can be translated in a cap-independent pathway with or without IRES, even the 3’polyA tail. However, circRNA translation remains debatable due to a lack of evidence of polysome involvement and any evidence of the biological functions of circ-proteins in human disease.
Through 2017, several exciting studies confirmed natural circRNA translation ability with proof of polysome involvement [38, 45, 55, 77]. Using ribosome footprinting (RFP) datasets, Pamudurti et al. identified 192 polysome-bound circRNAs (ribo-circRNAs) in rat/mouse tissues and 151 ribo-circRNAs in Drosophila. In particular, circMbl3 derived from the muscleblind (mbl) gene yielded a 37.03 kDa protein band detected by Western blot and MS analysis [45]. The IRES activity was determined by conducting a double luciferase assay and it was catalysed by overexpression of 4E-BP, an inhibitor of cap-dependent translation [45, 78]. Similarly, using a polysome sucrose gradient fractionation assay, Legnini et al. revealed that circ-ZNF609 was combined with heavy polysomes in human and mouse tissues. This ribo-circRNA generated an approximately 30 kDa protein in an IRES-like sequence (UTR)-directed mechanism [38].
Yang et al. discovered MIRES-dependent circRNA translation [55, 77]. They observed that numerous circRNAs harboured a higher density of m6A sites by m6A-RIP assay and an RRACH consensus motif of m6A modification approaching the start codon (R = purine, H = pyrimidine or A). One or two m6A sites in the circRNA were adequate to induce its translation [55]. Zhou et al. employed a genome-wide map of m6A circRNAs in human embryonic stem cells and observed hundreds of m6A methylation-modified circRNAs [77], implying the existence of MIRES-dependent circRNA translation. These data strongly confirm the capability of natural circRNA translation.
In 2018, Zhang and coauthors revealed that natural circRNA-translated proteins (SHPRH-146aa, FBXW7-185aa) perform inhibitory functions in the tumorigenesis of GBM [52, 53], defining the roles of circRNA-derived proteins (circ-proteins) in human disease. To date, dozens of functional circ-proteins have been found to be involved in various cancers and other diseases, which unveils just the beginning of the hidden translation omics of circRNA [46–54, 56–62, 66, 67].
CircRNA translatomics characteristics
ORFs spanning back-splicing junctions
An open reading frame is a nucleic acid sequence starting with AUG (or CUG, GUG, ACG) and continuing in three-base sets for protein synthesis until it hits a stop codon [79]. The most significant difference in the ORF structure between mRNAs and circRNAs is that circRNA ORF requires a spanning back-splice junction with a minimum read-junction overlap of 9 nt on either side of the junction, and it recycles more than once [39]. The identified ORFs of circRNAs contain ORFs smaller than 100 aa, ORFs greater than 100 aa, and noncanonical infinite ORFs with start codons lacking stop codons (circE-cadherin, circEGFR) [46, 57, 75, 76], indicating the ORF complexity of circRNAs.
Start and stop codons in circRNA translation
Codons are nucleotide triplets within RNA encoding specific amino acids incorporated into a polypeptide chain, including the start codons and stop codons [80]. The start codon, consisting of a canonical AUG (ATG for RNA) or noncanonical start codons covering CUG, GUG and ACG, is the classic signal that tells ribosomes where to initiate eukaryotic mRNA translation [81, 82]. A stop codon refers to the signal to terminate polypeptide chain synthesis, involving one of three termination codons (UAG, UAA, or UGA) [80].
As the only start codon identified so far, ATG is indispensable for circRNA-related translation. However, stop codons are not necessary for rolling translation of circRNA except IRES/MIRES-dependent circRNA translation, which is distinct from mRNA-related translation [38, 39, 46, 57, 75, 76]. Intriguingly, overlapping initiation-termination codons (UGAUGA or “UAAUGA”) have been observed in certain translated circRNAs (circSHPRH, circPPP1R12A, and circ-AKT3) [49, 52, 54, 73], demonstrating a discrepancy between circRNA and mRNA translation.
Internal ribosome entry elements for circRNA translation
IRES is a crucial cis-acting RNA regulatory element that initiates factor complexes to recruit 40S ribosomal subunits for cap-independent protein translation [83]. Identified IRESs of circRNAs include typical IRES sequences or IRES-like sequences (A/U enriched sequences). Although circRNA IRESs harbour varied localizations between or behind the ORF or within the UTR of circRNA, these molecules exert the same activity to drive circRNA translation [38, 45, 67, 72, 84, 85]. MIRES, an m6A-modified structure (“RRACH”), is another internal element for initiating factor complexes to bind 40S ribosomal subunits to drive cap-independent protein translation [55]. MIRES in the 5’ UTR approaching to the start codon ATG of circRNA can be recognized by YTHDF3-eIF4G2 complex, which induces circRNA translation (Fig. 2) [86–89].
Ribosome-associated circRNA translation
Ribosomes are necessary for protein synthesis. They provide the synthesis environment, serve as a molecular scaffold to promote the interactions of codons contained in RNA and the anticodons in tRNA, and present peptidyl transferase activity, permitting the formation of peptide bonds between adjacent amino acids. Mono-/multiribosome-bound RNA is a signal for protein translation, which can be captured by advanced ribosome sequencing techniques [39, 90–92].
Ribosome-associated circRNAs can be predicted by ribosome profiling sequencing (Ribo-seq), ribosome footprint (RFP) dataset profiling, ribosome nascent-chain complex profiling sequencing (RNC-seq) and the Ribosome Atlas, or be detected by ribosome enrichment assays [38, 39, 46, 91]. To screen ribosome-bound circRNAs and diminish errors, filtering criteria in circRNA Ribo-seq require that more than 3 unique ribosome-binding reads and at least 5 total junction-spanning back-splice junction reads in circRNAs overlap to ensure the potential of ribo-circRNAs. Indeed, 40 ribosome-associated circRNAs in human heart tissues have been identified, pointing to the potential of circRNA translation [39]. The ribosome footprint refers to the 3-nt codon movement to manifest ribosome translation activity [90]. To select ribosome-combined circRNAs, the RFP assay requires that at least one RFPread covers the back-splice junctions of the circRNAs. Combined with the translating ribosome affinity purification (TRAP) assay, 37 ribo-circRNAs, including circ-ZNF-609, were found to be associated with light polysomes [45]. Independent of high concentrations of sucrose sediment and unconsolidated binding with ribosomes, RNC-seq is able to enrich more ribo-circRNAs; sometimes, it may result in biased analyses and higher false-positive rates. Sucrose gradient isolation of ribosome assays can be conducted to determine single ribosome-bound circRNA (ribo-circRNA) and their translation efficiency. Puromycin or EDTA treatment enables the evaluation of the activity of ribosomes during translation. The identified ribosome-associated circRNAs are illustrated in Table 1 [66].
CircRNA translation mechanism and regulation
Protein synthesis in eukaryotes includes four phases: initiation, elongation, termination, and ribosome recycling [93, 94]. Translation initiation is the rate-limiting stage of translation. Canonical translation of eukaryotic mRNA depends on the m7G cap for recognition of the cap-binding protein initiation factor eIF4E complex, including eIF4E and eIF4G (a scaffold protein) and eIF4A (a helicase protein), and assembly of the 43S initiation complex to direct protein synthesis [42, 43]. In contrast, uncapped circRNA translation requires IRES or MIRES to combine with the initiation factor eIF4G2 or eIF3 complex containing eIF4G2, eIF4A and eIF4B, anchoring the 43S complex for protein translation (Fig. 2A-B) [72, 78], or it requires an infinite ORF and start codon to initiate translation in a rolling translation pathway (Fig. 2C) [46, 57, 75, 76], suggesting a discrepancy in the translation initiation pattern between cap-dependent and cap-independent translation.
IRES-dependent circRNA translation
During IRES-dependent circRNA translation, IRES acts as the RNA scaffold to interact with alternative initiation factors eIF4G (eIF4G2 or DAP-5) or the eIF3 complex including eIF4A and eIF3, instead of the 5’cap-eIF4E complex to recruit 40S ribosomal subunits, followed by assembly of the 43S initiation complex to initiate translation (Fig. 2A) [72, 82, 95, 96]. Unexpectedly, certain lines of evidence revealed that the IRES activity of the virus could be regulated by IRES trans-acting factors (ITAFs), such as heterogeneous nuclear ribonucleoproteins (hnRNPs) [84, 97, 98]. hnRNPI can bind to two IRES activity sites within the EMCV IRES upstream of the AUG codon, stabilizing the IRES conformation for ribosome recruitment [99, 100]. hnRNPQ accelerates secondary structure unwinding of the IRES to enhance its activity and translation efficiency. PABPC1 and hnRNP U enable the recognition of noncanonical IRES-like elements (A/U-rich sequences) to promote cap-independent translation of circRNA [85]. QKI indicates two-faced effects on IRES activity as an inhibitor or promoter [36, 84]. HNRNPL elevates the back-splicing of circARHGAP35 by reorganizing CA-rich elements in flanking of its gene locus, resulting in an increase in oncogenic circ-protein (p-ARHGAP35) [56]. These data demonstrate the ability of ITAFs to control IRES activity and circRNAs biogenesis. The mechanism of these ITAFs in regulating IRES activity in endogenous circRNA translation remains unknown.
MIRES-dependent circRNA translation
N 6-methyladenosine (m6A) is formed by methylated adenosine residues within RNA. m6A modification-triggered cap-independent translation mainly occurs in the 5’-UTR of certain circRNAs [55, 89]. CircRNAs bearing putative ORF spanning junctions and the m6A motif “RRACH” (R = G or A; H = A, C or U) in the 5’-UTR can be reorganized by YTH domain family protein 3 (YTHDF3, m6A reader), subsequently binding to the initiation factor eIF4G2 for anchoring and attachment of the 40S ribosomal subunit complex and the 43S complex to induce translation (Fig. 2B) [55, 87]. Interestingly, the eIF3-YTHDF1 or eIF3-METTL3 complex in the 3’UTR m6A sites and the single eIF3 in the 5’end of m6A sites have been found to be able to recruit the 40S ribosomal subunit for the translation of linear mRNA in response to stress [86–89]. The activity of these initiated complexes driving circRNA translation has not been determined.
Rolling translation of circRNA
Given the unique rolling translation of circRNA, an infinite ORF and start codon (ATG) are sufficient for continuous translation, independent of IRES or MIRES elements [57, 75, 76, 78]. The RCA pattern is similar to an isothermal and enzymatic process induced by a particular group of DNA polymerases for the ultrasensitive detection of DNA. During this reaction, longer nucleic acids can be generated using an infinitely repeating circular template in a given period [101]. In certain translatable circRNAs, because of the looped structure, their nucleotides are not matched with integral multiples of three, and stop codons fail to be engaged in all reading frames or out of in-frame, causing the formulation of an infinite ORF [73], which offers the possibility of circRNA translation using the RCA mechanism. Furthermore, rolling circRNA translation is a considerably efficient and simpler process compared to canonical protein synthesis. The latter follows the ribosome scanning mechanism and requires complex recycling and reinitiation processes, attenuating the synthesis efficiency and protein outputs [42, 72].
It appears that artificial circRNA rolling translation is ceaseless due to an infinite ORF [75, 76]; in contrast, rolling translation in certain natural circRNAs can be terminated. Liu and coauthors discovered that with an infinite ORF and the start codon ATG, cirEGFR encoded a polymetric protein complex termed rtEGFR (rolling translation EGFR) in a rolling translation mechanism, while this process was terminated by a complex system named “programmed-1 ribosomal frameshifting” (-1PRF)- mediated out-of-frame stop codon (OSC) to break the endless movement of the codons (Fig. 2C) [57, 102, 103].
IRES- and m6A-mediated translation initiation and rolling translation are crucial mechanisms for circRNA translation (Fig. 2). Its regulatory mechanism in eukaryotic cells is still unclear.
CircRNA-encoded proteins in carcinomas and noncarcinomas
CircRNA-encoded proteins in carcinomas
14-aa tail of C-E-Cad in response to GBM
A novel protein termed the E-cadherin protein variant (C-E-Cad) is translated from circE-cadherin (hsa_circ_0039992), and it is 254 amino acids in length. C-E-Cad harbours a unique 14 aa tail at the C-terminus due to a natural frameshift in the second-round translation of circE-cad, possessing a multiple-round ORF (Table 1) [46].
EGFR signal phosphorylation and activation are pivotal for the tumorigenicity of GBM [104]. C-E-Cad colocalized in the cell membrane with full-length EGFR and EGFRvIII, an active EGFR mutant frequently amplified and coexpressed with EGFR in GBM. The C-E-Cad 14-aa tail enables direct binding to the CR2 domain of full-length EGFR through salt bridge and hydrogen bond interactions, accompanied by EGFRvIII to promote STAT3 phosphorylation and nuclear translocation and AKT and ERK1/2 phosphorylation, resulting in the tumorigenicity of GBM [46, 105]. Consequently, C-E-Cad is an individual target for combined antibody treatment of glioblastoma because it facilitates malignant phenotypes, including proliferation, invasion, antiapoptosis, senescence resistance and cell stemness properties and sphere-forming frequency (Table 1).
SMO-193a.a, a scaffold for SMO cholesterol modification inducing GBM
A nascent protein with 193 amino acids termed SMO-193a.a. is generated from circSMO (hsa_circ_0001742). SMO-193a.a. shares the same 192 amino acids with the full-length SMO protein, which covers seven transmembrane domains responsible for cytoplasmic and membrane localization [51].
Hedgehog (HH) signalling is involved in the tumorigenesis of many cancers, including GBM, which is activated through the binding of HH to PTCH and derepressing SMO, releasing the Gli1 transcription factor for nuclear translocation and gene expression regulation [106, 107]. Cholesterol modification is indispensable for full-length SMO activation, which requires seven transmembrane domains for binding to cholesterol and PTCH1-blocked SMO cholesterol modification [107, 108]. SMO-193a.a. promotes cholesterol modification of full-length SMO by directly binding to N-terminus of SMO as a scaffold to translocate cholesterol to full-length SMO, functionally maintaining the cell stem cell (CSC) self-renewal ability and tumorigenicity of GBM [51].
rtEGFR, a platform enhancing EGFR stability to drive GBM
Rolling-translated EGFR (rtEGFR) is the polymetric protein complex translated from circEGFR (hsa_circ_0080229) in a rolling translation pattern due to an infinite ORF. Its core 83 amino acid sequence spans extracellular domain IV of the host protein EGFR, which comprises four crucial sites necessary for EGFR/rtEGFR interaction and EGFR activation. When colocalized in the cell membrane with full-length EGFR, rtEGFR can interact with extracellular domain IV of EGFR, maintaining EGFR stability and membrane localization, promoting the tumorigenicity of GBM [57]. Consequently, rtEGFR becomes an optimal target for monoclonal antibody or nimotuzumab-combined treatment of GBM to overcome nimotuzumab resistance [57, 109].
SHPRH-146aa, a decoy releasing SHPRH, blocking GBM
A newly discovered protein with 146 amino acids named SHPRH-146 aa is the product of circSHPRH (hsa_circ_0001649). It shares the same amino acids 1520-1651 at the C-terminus of full-length SHPRH containing two ubiquitination sites at K1562 and K1572 [52]. SHPRH is a well-characterized E3 ligase that targets proliferating cell nuclear antigen (PCNA) for degradation [110, 111]. Degradation induced by the E3 ligase DTL requires preferential interaction with the C-terminal sequence of SHPRH. Both SHPRH and SHPRH-146aa can be ubiquitin targets of DTL, while SHPRH-146aa possesses a stronger affinity. As a result, SHPRH-146aa acts as a decoy that competitively binds to DTL to release the host SHPRH from ubiquitination degradation. “Freed SHPRH” causes PCNA ubiquitination degradation to repress cell proliferation, reducing the tumorigenesis of GBM [52].
PINT87aa, a novel anchor for PAF1 to block GBM
A novel peptide named PINT87aa is translated from circLINC-PINT (hsa_circ_0082389), the unique circular form of a long intergenic noncoding RNA p53-induced transcript (LINC-PINT), mainly concentrated in the nucleus [63].
Although covering the same 77 amino acids as the N-terminus of the full-length PINT protein, PINT87aa exerts a unique tumour suppressive function that differs from that of the host PINT protein. It has been revealed that the PAF1 complex is involved in RNA II polymerase (Pol II) recruitment and regulation of the transcriptional elongation of downstream genes [112, 113]. PINT87 directly binds to the middle region (150-300 aa) of PAF1 and serves as an anchor, holding the PAF1 complex on the target gene promoter, hampering Pol II-induced mRNA elongation of multiple oncogenes (CPEB1, SOX-2) and abolishing cell cycle progression [63, 112–114].
AKT3-174aa, a blocker of PDK1-mediated AKT phosphorylation and GBM
A nascent protein with 174 amino acids termed AKT3-174 aa is generated from circAKT3 (hsa_circ_0017250). It covers the same amino acids 62-232 in the middle region of full-length AKT3 (PKBγ, protein kinase Bγ) containing a PH domain with the Thr308 site necessary for phosphorylation [54].
Notably, AKT3-174aa exerts an inhibitory function in GBM contrary to that of the host AKT protein [115]. PDK1-mediated AKT phosphorylation at Thr308 and activation are the initial key steps in activating the RTK/PI3K/AKT signalling pathway for GBM progression [116, 117]. Due to its higher binding affinity to phosphorylated PDK1 (p-PDK1), AKT3-174aa prefers to interact with activated PDK1, blocking Akt phosphorylation at Thr308 and negatively modulating PI3K/AKT signal intensity to reduce cell proliferation, resulting in the reduction of tumorigenicity and the radiation resistance of GBM [54].
FBXW7-185aa, a novel inhibiter of GBM and TNBC
FBXW7-185aa, a novel FBXW7a variant with 185 amino acids, is the product of circFBXW7 (hsa_circ_022705). It shares the same 165 amino acids at the N-terminus of the full-length FBXW7a protein, a crucial isoform of the E3 ligase FBXW7 [53]. The deubiquitinating enzyme USP28 reportedly binds to the N-terminus of FBXW7a for deubiquitination degradation and then induces c-Myc to promote GBM [118–120]. Since it possesses the same motif as FBXW7a but with a stronger affinity to USP28, FBXW7-185aa acts as a decoy and preferentially binds to USP28 to release FBXW7a. Freed FBXW7a induces c-Myc ubiquitin degradation, diminishing the cell proliferation and tumorigenesis of GBM [53].
Similarly, FBXW7-185aa demonstrates an inhibitory capability in triple-negative breast cancer owing to an enhancement of FBXW7 expression to induce c-Myc degradation (Table 1) [121].
HER2-103 sensitizes TNBC to pertuzumab
HER2-103, a new HER2 variant with 103 amino acids, is translated from circHER2 (hsa_circ_0007766). The sequence of these 103 amino acids is same as the CR I domain of full-length HER2, which is indispensable for EGFR/HER2-103 or HER3/HER2-103 dimerization and downstream signalling cascade activation [64, 122, 123] and it is localized close to the cell membrane, similar to the full-length HER2 protein. Accordingly, HER2-103 is able to promote TNBC cell proliferation and invasion by interacting with the CR I domain of HER2, stimulating EGFR/HER3 homo/heterodimer formation and phosphorylation, and sequential AKT activation, displaying obvious tumorigenicity [64]. Pertuzumab, an anti-HER2 antibody, is ineffective in TNBC patients lacking HER2 expression [124]. Notably, HER2-103-expressing TNBC cells and mouse xenografts are sensitive to pertuzumab treatment against certain TNBCs by acting on its shared CR I domain of HER2, which enables it to be an optimal target for anti-HER2 mono-antibodies such as pertuzumab or combined with trastuzumab [64, 124].
DIDO1-529aa, a new GC inhibitor
DIDO1-529aa translated from circDIDO1 (hsa_circ_0061137) is a new isoform of DIDO1-1a (death-inducer obliterator isoform 1-1a). It shares all 529 amino acids, similar to the DIDO1-1a protein, which contains NLS and PHD domains, while lacking the nuclear export sequence (NES) of the C-terminus, causing nuclear localization [60]. Interestingly, due to the absence of an NES, DIDO1-529aa demonstrates a new function independent of full-length DIDO1-1a; it inhibits GC through direct interaction of the DNA binding domain (DBD) and the catalytic domain (CAT) to block the activity of poly ADP-ribose polymerase 1 (PARP1). Moreover, it acts as a partner by binding to peroxiredoxin 2 (PRDX2) to induce an E3 ligase of the SCF ubiquitination complex for RBX1-mediated ubiquitination and degradation of PRDX2 and inactivation of its downstream signalling pathways, hampering the proliferation and invasion of GC [60, 125].
MAPK1-109aa, a novel blocker of MAPK1 signalling
A novel MAPK1 variant with 109 amino acids termed MAPK1-109 aa is encoded by circMAPK1 (hsa_circ_0004872). This variant shares the same sequences at amino acids 98-203 with the full-length MAPK1 protein, which includes indispensable MAPK1 phosphorylation sites [59]. MEK1 is a pivotal component of the Ras-Raf-MEK-MAPK cascade for the transmission of extracellular signals to intracellular signals and the phosphorylation of its downstream substrates, which is positively linked to cell growth and proliferation in numerous cancers [126, 127].
Interestingly, MAPK1-109aa exhibits the opposite function to full-length MAPK1. Due to sharing the same sequences, MAPK1-109a competitively binds to MEK1 to block the transmission of extracellular signals to intracellular signals to phosphorylate MAPK and downstream substrates encompassing p-c-Fos, p-c-JUN, and p-RSK, repressing the proliferation and invasion of GC cells [59].
CircFNDC3B-218aa, a novel CC inhibitor
CircFNDC3B-218aa is a novel protein with 218 amino acids translated from circFNDC3B (hsa_circ_0006156). It shares 201 amino acids with the N-terminal sequence of full length FNDC3B [58]. Tumorigenesis depends on an enhanced glycolytic phenotype switched from OXPHOS, termed the Warburg effect [128]. This metabolic shift can promote EMT progression, inducing tumour malignancy. The gluconeogenic enzyme fructose-1,6-bisphosphatase 1 (FBP1), one of the related-limiting enzymes in gluconeogenesis, exerts crucial function switching from glycolysis to oxidative phosphorylation (OXPHOS), which is necessary for tumour malignancy, blocking cancer progression via the Snail-FBP1 axis [129, 130]. CircFNDC3B-218aa presents an inhibitory ability to restrict tumorigenesis by attenuating Snail expression, enhancing FBP1-induced OXPHOS, as shown by the reduction of glucose uptake, pyruvate production and lactate production, and the promotion of metabolic reprogramming from glycolysis to oxidative phosphorylation [58].
β-catenin-370aa, a decoy of GSK3β releasing β-catenin to promote HCC
β-catenin-370aa, a nascent β-catenin isoform with 370 amino acids, is encoded by circβ-catenin (hsa_circ_0004194). It shares 361 amino acids at the N-terminus of the full-length β-catenin protein in addition to a 9-aa tail at the C-terminus. This novel molecule is found in the cytoplasm, unlike β-catenin, which is found in the nucleus [48]. As a vital component of the Wnt pathway, active β-catenin accumulates in the nucleus and participates in a variety of pathological events after being freed from glycogen synthase kinase 3β (GSK3β)-induced phosphorylation and proteasome-mediated ubiquitination degradation [131, 132]. Instead, β-catenin-370aa is localized in the cytoplasm and can be a decoy that preferentially binds to cytoplasmic GSK3β, protecting full-length β-catenin from GSK3β-directed degradation. Released β-catenin stimulates the Wnt/β-catenin pathway, promoting liver cancer progression [48].
p-circARHGAP35, an oncoprotein in HCC and CRC
P-circARHGAP35 is a novel oncoprotein with 1289 amino acids translated from circARHGAP35 (hsa_circ_0109744). This oncoprotein harbours the same amino acids with sequences at the N-terminus of the full-length ARHGAP35 protein, which encompasses four FF domains without the Rho GAP domain [56]. P-circARHGAP35 accumulates in the nucleus, unlike ARHGAP35 in the cytoplasm [133]. Accordingly, nuclear p-circARHGAP35 functions as an oncoprotein to facilitate tumour migration, invasion, and metastasis of cancer cells, including both hepatocellular carcinoma and colorectal cancer, owing to interaction with the transcriptional regulator TFII-I. This function is opposite to host ARHGAP35, a tumour inhibitor of RhoA activation in the cytoplasm [56, 133], demonstrating the complexity of the cancer transcriptome.
CircPPP1R12A-73aa and CircLgr4-peptide, novel CRC inducers
A novel protein with 73 amino acids named circPPP1R12A-73 aa is translated from circPPP1R12A (hsa_circ_0000423). It shares 56 amino acids with sequences at the N-terminus of full-length PPP1R12A. However, circPPP1R12A-73aa confers distinct functions to facilitate the proliferation and metastasis of CRC owing to its activation of the Hippo-YAP signalling pathway; nevertheless, full-length PPP1R12A does not show increased expression in CRC tissues [49].
The circLgr4 peptide is a small polypeptide with 19 amino acids encoded by circLgr4 (hsa_circ_02276) [50]. Lgr4, Lgr5 and Lgr6 are vital members of the Rspo/Lgr4/5 signalling pathway, which belongs to the bypass pathway of the Wnt/β-catenin signalling pathway [134]. CircLgr4 peptide function is closely dependent on its host protein Lgr4, one of the activated receptors in the noncanonical Wnt/β-catenin signalling pathway [135]. It interacts with LGR4 to efficiently activate Wnt/β-catenin signalling and promotes the self-renewal and metastasis of cancer stem cells [50].
CircPLCE1-411, a novel CRC inhibitor
CircPLCE1-411, a novel protein with 411 amino acids, is derived from circPLCE1 (hsa_circ_0019223). It shares the same amino acids 1-403 at the N-terminus of the full-length PLCE1 protein [61].
Ribosomal protein S3 (RPS3), a component of the 40S ribosomal subunit, harbours the ability to combine with the p65 subunit of NF-κB to drive nuclear translocation of the NF-κB complex [136]. HSP90α, a partner of RPS3, maintains RPS3 stability resistance to ubiquitin-dependent degradation [137, 138], while blocking the activation of the ATP-binding domain of HSP90 enables the release of RPS3 from the HSP90α/RPS3 complex [139, 140]. Freed RPS3 can preferentially bind and be degraded by the chaperone E3 ligase complex HSP70-CHIP [141]. Despite sharing the same sequences with the host PLCE1, circPLCE1-411 demonstrates a distinct capability to suppress colorectal cancer cell proliferation and migration via its interaction with the ATP-binding domain of HSP90α to accelerate the dissociation of RPS3 from the HSP90α/RPS3 complex, leading to HSP70-induced ubiquitin-dependent degradation of RPS3 and the suppression of NF-κB nuclear translocation and activation [61].
CircGprc5a-peptide, a bladder cancer promoter
CircGprc5a-peptide with 11 amino acids is the product of circGprc5a (hsa_circ_002838). G-protein-coupled receptor C family 5A (Gprc5a), a key membrane protein in the GPRC signalling pathway, is involved in the maintenance of self-renewal and metastasis of tumour stem cells [47, 142]. The function of circGprc5a-peptide is Gprc5a-dependent. It can drive bladder oncogenesis and metastasis accompanied by Gprc5a to activate the GPCR signalling pathway and promote the self-renewal and metastasis of cancer stem cells [47].
CircCHEK1_246aa induces MM
A nascent CHEK1 variant termed circCHEK1_246aa is translated from circCHEK1 (hsa_circ_0024792). Multiple myeloma (MM) cells expressed CircCHEK1_246aa is mainly in the CHEK1 kinase catalytic centre, and mature circCHEK1_246aa can be secreted into the bone marrow microenvironment. In this region it interacts with native centrosomal protein 170 (CEP170) to attenuate mutant CEP170 expression in MM cells, promoting multiple myeloma (MM) by inducing chromosomal instability and bone lesion formation, exerting a similar function to full-length CHEK1 [62].
E7 oncoprotein transforms the activity of HPVs
The E7 oncoprotein, with 98 amino acids, is the product of circE7 originating from human papillomavirus 16. Its translation can be facilitated by QKI and heat shock stress. The ability of the E7 oncoprotein to suppress cervical cancer cell growth indicates that virus-derived circRNA translation may be responsible for the transforming properties of some HPVs [65].
CircRNA-encoded proteins in noncarcinomas
Unique 9-aa of Nlgn173 confers cardiac remodelling
A nascent protein with 173 amino acids termed Nlgn173 translated from circNlgn (hsa_circ_0003046) harbours an exclusive 9 amino acid motif (GYRPAANWI) at the C-terminus responsible for nuclear localization, and the remaining 164 amino acids are the same as the full-length Nlgn protein [66].
Nlgn173 is highly expressed in hypertrophic and fibrotic hearts tissues, and its particular 9-amino-acid domain interacts with the structural protein LaminB1 to force Nlgn173 nuclear localization. Nuclear Nlgn173 combines with the helix-turn-helix (HTH) DNA binding motif at the promoters of glucocorticoid-inducible kinase-3 (SGK3) to induce cardiac fibroblast proliferation and inhibits growth protein 4 (ING4) to attenuate cardiomyocyte survival [66, 143, 144], finally leading to cardiac remodelling.
Aβ175 expression linked to AD
The novel polypeptide with 175 amino acids named Aβ175 is derived from circAβ-a (hsa_circ_0007556). This molecule shares the same 158 amino acids approaching the C-terminus of the full-length amyloid β peptide. Aβ175 expression is enhanced in brain tissues of Alzheimer’s disease patients, hinting at its potential to induce AD [66]. The detailed mechanism has not been investigated.
Other functional peptides of circRNAs
Other peptides from translated circRNAs have been verified to be biologically functional. Pamudurti et al. certified that a nascent protein encoded from circMbl3 tends to be linked to the regulation of synaptic function [45]. CircZNF609-translated peptide is prone to regulate myoblast proliferation since circZNF609 is strongly expressed in human myoblasts with Duchenne muscular dystrophy (DMD) [38]. Another circ-protein from circSfl bears the same N-terminal sequence as full-length Sfl, including its cytoplasmic and transmembrane domains. Although the enzyme active domain is absent, this nascent protein is capable of extending the lifespan of fruit flies, playing an important role in regulating ageing in vivo [68].
Conclusions and future perspectives
An accumulating number of circRNAs have been confirmed to be translated through uncapped translation mechanisms, including IRES- or MIRES-dependent pathways, o RCA mechanism (Fig. 2). The resultant proteins play various biological roles (Table 1). This noncanonical translatomics has attracted increasing attention, but several issues need to be addressed in the future.
First, the precise regulatory mechanism of natural circRNA translation remains unclear. ITAFs such as hnRNPI, hnRNPQ reportedly dominated the activity of certain viral IRESs [99, 100]. Additionally, Chen certified that a stem-loop structured RNA element in IRES termed SuRE was located 40-60 nt from the first nucleotide of the IRES of circular RNAs, particularly promoting translation of circRNAs instead of linear RNAs [145]. The detailed mechanism of ITAFs and SuRE involved in regulating IRES activity in natural circRNA translation in cells requires elucidation.
Second, engineering exosome-delivered translated circRNAs and their functions in human disease has yet to be defined. Rolling translation is a novel mechanism driving circRNA translation to efficiently generate a large number of protein products beyond that of cognate mRNAs [146], offering the possibility to utilize circRNA-derived proteins for future gene therapy [146, 147]. Scholars have revealed the potential of circRNA translation engineering by designing a novel expression system (rAAV) to induce the initiation of artificial circRNA translation [148] or natural circRNA translation in eukaryotic cells in order to increase the output and expression duration of circRNAs [146], whereas suitable vehicles delivering translated circRNAs into target organs are unknown. Exosomal circRNAs are stable regulators or biomarkers involved in human diseases [149, 150]. Exosome-transferred lncRNA RN7SL1 is able to stimulate CAR-T cells to accelerate autonomous and endogenous immune function [151]. Engineering of exosome-delivered translated circRNAs aimed at individual diagnoses and treatments appears to be a promising field.
Third, the development of more sensitive approaches is required to discover the low-abundance peptides from translated circular RNAs.
Finally, mature translated circRNAs are exported to the cytoplasm as templates for protein synthesis, while the resultant proteins exhibit various localizations and functions similar to or different from host proteins. How are translated circRNAs trafficked to the cytoplasm from the nucleus? Why do these proteins have different localizations and functions compared to their host proteins? These issues remain under-addressed thus far. Future work will be aimed at the elucidation of the regulatory mechanism underlying translation and function of circRNAs and the resultant proteins in pathophysiological processes, as well as the exploration of the therapeutic potential of translated circRNAs and exosome-coated translated circRNAs to benefit individual diagnoses and human disease treatments.
Acknowledgements
Not applicable.
Abbreviations
- circRNA
Circular RNA
- IRES
Internal ribosomal entry sequence
- m6A
N6-methyladenosine
- RCA
Rolling translation mechanism
- ncRNAs
Non-coding RNAs
- pre-mRNA
Precursor mRNA
- BSJ
Back splicing junction
- ciRNA
Intronic circRNA
- EIciRNA
Exonic and intronic circRNA
- EcircRNA
Exon circRNA
- mecciRNA
Mitochondrial circRNA genomes
- rt-circRNAs
Read-through circRNA
- f-circRNA
Fused circRNA
- snRNP
Small nuclear ribonucleoprotein
- ROS
Reactive oxygen species
- m7G
5’ m7GpppN
- RNA-seq
RNA-sequencing
- ORF
Open reading frame
- iORF
Infinite ORF
- RFP
Ribosome footprinting
- ribo-circRNA
Ribosome-bond circRNA
- m6A-RIP
Methylated RNA immunoprecipitation sequencing
- GBM
Glioblastoma
- circ-protein
circRNA-derived protein
- sORF
Smaller ORF
- uORF
Upstream ORF
- UTR
Untranslated Region
- Ribo-seq
Ribosome profiling sequencing
- RNC-seq
The ribosome nascent-chain complex profiling sequencing
- eIF3
The eukaryotic translation initiation factor 3
- eIF4G2
The eukaryotic translation initiation factor4G2
- eIF4A
The eukaryotic translation initiation factor 4A
- eIF4E
The eukaryotic translation initiation factor4E
- eIF4B
The eukaryotic translation initiation factor4B
- ITAFs
Trans-acting factors
- hnRNPs
Heterogeneous nuclear ribonucleoproteins
- hnRNP I
Heterogeneous nuclear ribonucleoprotein I
- hnRNP Q
Heterogeneous nuclear ribonucleoprotein Q
- QKI
Quaking
- YTHDF3
YTH domain family protein 3
- METTL3
Methyltransferase-like 3
- rtEGFR
Rolling translation EGFR
- OSC
Out of-frame stop codon
- C-E-Cad
E-cadherin protein variant
- SMO
The G protein-coupled-like receptor smoothened
- PDK1
The 3-phosphoinositide-dependent protein kinase 1
- TNBC
Triple-Negative Breast Cancer
- GC
Gastric cancer
- MAPK
Mitogen-activated protein kinases
- OXPHOS
Oxidative phosphorylation
- HCC
Hepatocellular carcinoma
- GSK3β
Glycogen synthase kinase 3β
- CC
Colon cancer
- CRC
Colorectal cancer
- MM
Multiple myeloma
- CR
Cardiac remodeling
- AD
Alzheimer’ s Disease
- DMD
Duchenne Muscular Dystrophy
- CAR-T
Chimeric antigen receptor T-cell
- rAAV
Recombinant adeno-associated virus
Authors’ contributions
JB, JNZ, YW conceived manuscript. YW, CJW and YD collected relevant references, drafted manuscript and finished the figures. ZWL, MLL, PFH, ZGS and SFC offered crucial content revision and language polishing. JB and YW completed the final manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82072649, 81872304), the Outstanding Youth Foundation of Jiangsu Province (BK20200046), the Education Department of Jiangsu Province (No. 19KJA130001), the Natural Science Foundation of Jiangsu Province (BK20180989), the Natural Science Foundation of Xuzhou (KC21259), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX21_2663).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yan Wang, Chunjie Wu and Yu Du contributed equally to this work.
Contributor Information
Junnian Zheng, Email: jnzheng@xzhmu.edu.cn.
Jin Bai, Email: bj@xzhmu.edu.cn.
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141. doi: 10.1177/1535370217708978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adelman K, Egan E. Non-coding RNA: More uses for genomic junk. Nature. 2017;543(7644):183–185. doi: 10.1038/543183a. [DOI] [PubMed] [Google Scholar]
- 4.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA splicing. Adv Exp Med Biol. 2018;1087:41–52. doi: 10.1007/978-981-13-1426-1_4. [DOI] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Hu Z, Zhou J, Tian C, Tian G, He M, et al. Interior circular RNA. RNA Biol. 2020;17(1):87–97. doi: 10.1080/15476286.2019.1669391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mance LG, Mawla I, Shell SM, Cahoon AB. Mitochondrial mRNA fragments are circularized in a human HEK cell line. Mitochondrion. 2020;51:1–6. doi: 10.1016/j.mito.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Q, Liu J, Deng H, Ma R, Liao JY, Liang H, et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell. 2020;183(1):76–93.e22. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, Sun H, Wang C, Liu W, Liu M, Zhu Y, et al. Mitochondrial Genome-Derived circRNA mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol Ther Nucleic Acids. 2020;20:801–811. doi: 10.1016/j.omtn.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Wang X, Li J, Hu S, Deng Y, Yin H, et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63(10):1429–1449. doi: 10.1007/s11427-020-1631-9. [DOI] [PubMed] [Google Scholar]
- 15.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 17.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 18.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med. 2017;95(11):1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18(6):984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281(40):29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 27.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Vidal AF. Read-through circular RNAs reveal the plasticity of RNA processing mechanisms in human cells. RNA Biol. 2020;17(12):1823–1826. doi: 10.1080/15476286.2020.1805233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A Circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7(16):3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu D, et al. The Circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25(9):2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular rna that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, et al. The translational landscape of the human heart. Cell. 2019;178(1):242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 41.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 43.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonkin J, Rosenthal N. One small step for muscle: a new micropeptide regulates performance. Cell Metab. 2015;21(4):515–516. doi: 10.1016/j.cmet.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23(3):278–291. doi: 10.1038/s41556-021-00639-4. [DOI] [PubMed] [Google Scholar]
- 47.Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S, et al. circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol Ther Nucleic Acids. 2018;13:633–641. doi: 10.1016/j.omtn.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhi X, Zhang J, Cheng Z, Bian L, Qin J. circLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int J Cancer. 2019. [DOI] [PubMed]
- 51.Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li B, et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021;22(1):33. doi: 10.1186/s13059-020-02250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 Circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18(1):131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T, et al. HNRNPL Circularizes ARHGAP35 to Produce an Oncogenic Protein. Adv Sci (Weinh) 2021;8(13):2001701. doi: 10.1002/advs.202001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Li Z, Zhang M, Zhou H, Wu X, Zhong J, et al. Rolling-translated EGFR variants sustain EGFR signaling and promote glioblastoma tumorigenicity. Neuro-Oncology. 2021;23(5):743–756. doi: 10.1093/neuonc/noaa279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19(1):71. doi: 10.1186/s12943-020-01179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20(1):66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z, et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol Cancer. 2021;20(1):101. doi: 10.1186/s12943-021-01390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang ZX, Liu HS, Xiong L, Yang X, Wang FW, Zeng ZW, et al. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol Cancer. 2021;20(1):103. doi: 10.1186/s12943-021-01404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu C, Wang W, Tang X, Xu T, Zhang Y, Guo M, et al. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021;20(1):84. doi: 10.1186/s12943-021-01380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J, Ma M, Yang X, Zhang M, Luo J, Zhou H, et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to pertuzumab. Mol Cancer. 2020;19(1):142. doi: 10.1186/s12943-020-01259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10(1):2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du WW, Xu J, Yang W, Wu N, Li F, Zhou L, et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ Res. 2021;129(5):568–582. doi: 10.1161/CIRCRESAHA.120.318364. [DOI] [PubMed] [Google Scholar]
- 67.Mo D, Li X, Raabe CA, Rozhdestvensky TS, Skryabin BV, Brosius J. Circular RNA encoded amyloid beta peptides-A novel putative player in Alzheimer’s disease. Cells. 2020;9(10):2196. doi: 10.3390/cells9102196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weigelt CM, Sehgal R, Tain LS, Cheng J, Eßer J, Pahl A, et al. An insulin-sensitive circular RNA that regulates lifespan in Drosophila. Mol Cell. 2020;79(2):268–279.e5. doi: 10.1016/j.molcel.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, et al. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986;323(6088):508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 70.Makino S, Chang MF, Shieh CK, Kamahora T, Vannier DM, Govindarajan S, et al. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature. 1987;329(6137):343–346. doi: 10.1038/329343a0. [DOI] [PubMed] [Google Scholar]
- 71.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 72.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 73.AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A. 2014;111(40):14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Eng. 2013;52(27):7004–7008. doi: 10.1002/anie.201302044. [DOI] [PubMed] [Google Scholar]
- 76.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prats AC, David F, Diallo LH, Roussel E, Tatin F, Garmy-Susini B, et al. Circular RNA, the key for translation. Int J Mol Sci. 2020;21(22):8591. doi: 10.3390/ijms21228591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20(23):8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong HR. Translation. Crit Care Med. 2005;33(12 Suppl):S404–S406. doi: 10.1097/01.ccm.0000186075.26921.d2. [DOI] [PubMed] [Google Scholar]
- 81.Mackiewicz P. AUG as the translation start codon in circular RNA molecules: a connection between protein-coding genes and transfer RNAs? Bioessays. 2020;42(6):e2000061. doi: 10.1002/bies.202000061. [DOI] [PubMed] [Google Scholar]
- 82.Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. 2020;22(12):2162–2169. doi: 10.1007/s12094-020-02371-1. [DOI] [PubMed] [Google Scholar]
- 83.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 84.Godet AC, David F, Hantelys F, Tatin F, Lacazette E, Garmy-Susini B, et al. IRES trans-acting factors, key actors of the stress response. Int J Mol Sci. 2019;20(4):924. doi: 10.3390/ijms20040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fan XJ, Yang Y, Chen CY, Wang ZF. Pervasive translation of circular RNAs driven by short IRES-like elements. BioRxiv. 2020;473207. 10.1101/473207. [DOI] [PMC free article] [PubMed]
- 86.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyer KD. m(6)A-mediated translation regulation. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):301–309. doi: 10.1016/j.bbagrm.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Calviello L, Ohler U. Beyond read-counts: Ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet. 2017;33(10):728–744. doi: 10.1016/j.tig.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 91.He L, Man C, Xiang S, Yao L, Wang X, Fan Y. Circular RNAs’ cap-independent translation protein and its roles in carcinomas. Mol Cancer. 2021;20(1):119. doi: 10.1186/s12943-021-01417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao J, Qin B, Nikolay R, Spahn CMT, Zhang G. Translatomics: the global view of translation. Int J Mol Sci. 2019;20(1):12. doi: 10.3390/ijms20010212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crick FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 94.Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14(8):1007–1017. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4(12):1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roberts L, Wieden HJ. Viruses, IRESs, and a universal translation initiation mechanism. Biotechnol Genet Eng Rev. 2018;34(1):60–75. doi: 10.1080/02648725.2018.1471567. [DOI] [PubMed] [Google Scholar]
- 97.Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, et al. Combinatorial control of drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29(20):2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rossbach O, Hung LH, Khrameeva E, Schreiner S, König J, Curk T, et al. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol. 2014;11(2):146–155. doi: 10.4161/rna.27991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Romanelli MG, Diani E, Lievens PM. New insights into functional roles of the polypyrimidine tract-binding protein. Int J Mol Sci. 2013;14(11):22906–22932. doi: 10.3390/ijms141122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kafasla P, Morgner N, Pöyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell. 2009;34(5):556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 101.Zhao W, Ali MM, Brook MA, Li Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew Chem Int Ed Eng. 2008;47(34):6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 102.Tse H, Cai JJ, Tsoi HW, Lam EP, Yuen KY. Natural selection retains overrepresented out-of-frame stop codons against frameshift peptides in prokaryotes. BMC Genomics. 2010;11:491. doi: 10.1186/1471-2164-11-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang X, Xuan Y, Han Y, Ding X, Ye K, Yang F, et al. Regulation of HIV-1 gag-pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting. Cell. 2019;176(3):625–635.e14. doi: 10.1016/j.cell.2018.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15(5):302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4(3):226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, et al. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature. 2019;571(7764):284–288. doi: 10.1038/s41586-019-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, et al. Cellular cholesterol directly activates smoothened in hedgehog signaling. Cell. 2016;166(5):1176–1187.e14. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Solomón MT, Selva JC, Figueredo J, Vaquer J, Toledo C, Quintanal N, et al. Radiotherapy plus nimotuzumab or placebo in the treatment of high grade glioma patients: results from a randomized, double blind trial. BMC Cancer. 2013;13:299. doi: 10.1186/1471-2407-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175(5):703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Unk I, Hajdú I, Fátyol K, Szakál B, Blastyák A, Bermudez V, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103(48):18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, et al. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell. 2015;162(5):1003–1015. doi: 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Strikoudis A, Lazaris C, Trimarchi T, Galvao Neto AL, Yang Y, Ntziachristos P, et al. Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat Cell Biol. 2016;18(11):1127–1138. doi: 10.1038/ncb3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karmakar S, Dey P, Vaz AP, Bhaumik SR, Ponnusamy MP, Batra SK. PD2/PAF1 at the crossroads of the cancer network. Cancer Res. 2018;78(2):313–319. doi: 10.1158/0008-5472.CAN-17-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, et al. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17(18):5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gagliardi PA, Puliafito A, Primo L. PDK1: at the crossroad of cancer signaling pathways. Semin Cancer Biol. 2018;48:27–35. doi: 10.1016/j.semcancer.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 117.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 118.Popov N, Herold S, Llamazares M, Schülein C, Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6(19):2327–2331. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- 119.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294(5540):173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 121.Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, et al. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, et al. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J Cell Sci. 2007;120(Pt 16):2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 123.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hurvitz SA, Martin M, Symmans WF, Jung KH, Huang CS, Thompson AM, et al. Neoadjuvant trastuzumab, pertuzumab, and chemotherapy versus trastuzumab emtansine plus pertuzumab in patients with HER2-positive breast cancer (KRISTINE): a randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2018;19(1):115–126. doi: 10.1016/S1470-2045(17)30716-7. [DOI] [PubMed] [Google Scholar]
- 125.Feng AL, Han X, Meng X, Chen Z, Li Q, Shu W, et al. PRDX2 plays an oncogenic role in esophageal squamous cell carcinoma via Wnt/β-catenin and AKT pathways. Clin Transl Oncol. 2020;22(10):1838–1848. doi: 10.1007/s12094-020-02323-9. [DOI] [PubMed] [Google Scholar]
- 126.Yang SH, Sharrocks AD, Whitmarsh AJ. MAP kinase signalling cascades and transcriptional regulation. Gene. 2013;513(1):1–13. doi: 10.1016/j.gene.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 127.Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23(3):316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lien WH, Fuchs E. Wnt some lose some: transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 2014;28(14):1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nejak-Bowen KN, Monga SP. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21(1):44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40(6):1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 134.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108(28):11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 136.Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131(5):927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 137.Hodgson A, Wier EM, Fu K, Sun X, Yu H, Zheng W, et al. Metalloprotease NleC suppresses host NF-κB/inflammatory responses by cleaving p65 and interfering with the p65/RPS3 interaction. PLoS Pathog. 2015;11(3):e1004705. doi: 10.1371/journal.ppat.1004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim TS, Jang CY, Kim HD, Lee JY, Ahn BY, Kim J. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol Biol Cell. 2006;17(2):824–833. doi: 10.1091/mbc.E05-08-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90(1):65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 140.Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17(16):4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hwang I, Cho SW, Ahn JY. Chaperone-E3 ligase complex HSP70-CHIP mediates ubiquitination of ribosomal protein S3. Int J Mol Sci. 2018;19(9):2723. doi: 10.3390/ijms19092723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 143.Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323(6088):560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 144.Narasimhan G, Bu C, Gao Y, Wang X, Xu N, Mathee K. Mining protein sequences for motifs. J Comput Biol. 2002;9(5):707–720. doi: 10.1089/106652702761034145. [DOI] [PubMed] [Google Scholar]
- 145.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 146.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meganck RM, Liu J, Hale AE, Simon KE, Fanous MM, Vincent HA, et al. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol Ther Nucleic Acids. 2021;23:821–834. doi: 10.1016/j.omtn.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meganck RM, Borchardt EK, Castellanos Rivera RM, Scalabrino ML, Wilusz JE, Marzluff WF, et al. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol Ther Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shang BQ, Li ML, Quan HY, Hou PF, Li ZW, Chu SF, et al. Functional roles of circular RNAs during epithelial-to-mesenchymal transition. Mol Cancer. 2019;18(1):138. doi: 10.1186/s12943-019-1071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johnson LR, Lee DY, Eacret JS, Ye D, June CH, Minn AJ. The immunostimulatory RNA RN7SL1 enables CAR-T cells to enhance autonomous and endogenous immune function. Cell. 2021;184(19):4981–4995.e14. doi: 10.1016/j.cell.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.