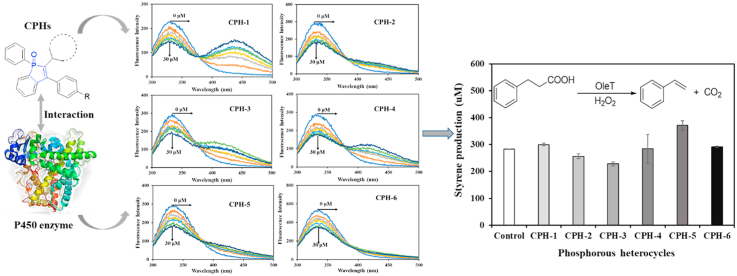
Abstract
P450 fatty acid decarboxylase OleT from Staphylococcus aureus (OleTSA) is a novel cytochrome P450 enzyme that catalyzes the oxidative decarboxylation of fatty acids to yield primarily terminal alkenes and CO2 or minor α- and β-hydroxylated fatty acids as side-products. In this work, the interactions between a series of cycloalkyl phosphorus heterocycles (CPHs) and OleTSA were investigated in detail by fluorescence titration experiment, ultraviolet–visible (UV–vis) and 31P NMR spectroscopies. Fluorescence titration experiment results clearly showed that a dynamic quenching occurred when CPH-6, a representative CPHs, interacted with OleTSA with a binding constant value of 15.2 × 104 M−1 at 293 K. The thermodynamic parameters (ΔH, ΔS and ΔG) showed that the hydrogen bond and van der Waals force played major roles in the interaction between OleTSA and CPHs. The UV–vis and 31P NMR studies indicated the penetration of CPH-6 into the interior environment of OleTSA, which greatly affects the enzymatic activity of OleTSA. Therefore, our study revealed an effective way to use phosphorus heterocyclic compounds to modulate the activity of cytochrome P450 enzymes.
Keywords: Phosphorus heterocycles, Cytochrome P450, OleT, Interaction, Spectroscopy
Graphical abstract
Highlights
-
•
The interaction between cycloalkyl phosphorus heterocycles (CPHs) and P450 OleTSA were studied by spectroscopic methods.
-
•
Dynamic mode of quenching mechanism was noticed in OleTSA - CPHs interaction.
-
•
Hydrogen bond and van der Waals force were determined to contribute the conformation of OleTSA- CPHs complex.
-
•
The penetration of CPH-6 into the interior environment of OleTSA was proposed.
-
•
The effect of CPHs on OleTSA enzyme activity was studied.
1. Introduction
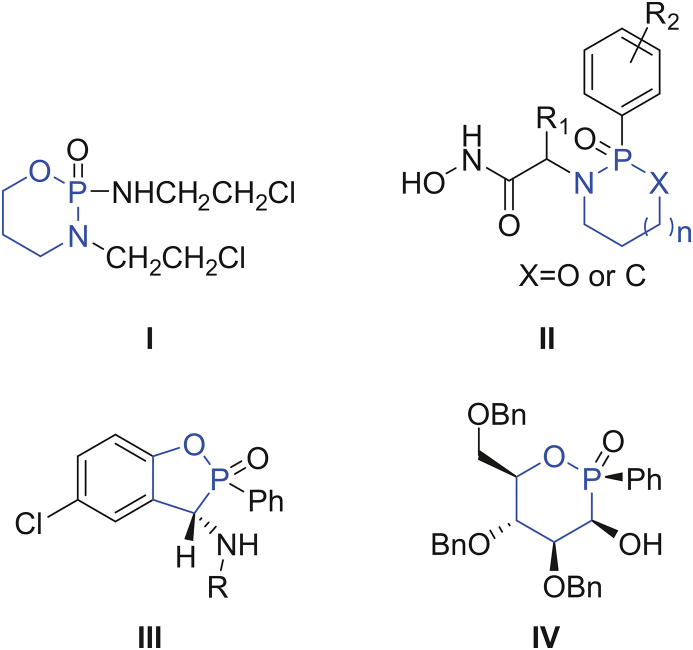
Organophosphorus compounds widely exist in biological systems and have drawn largely attentions in medicinal application [[1], [2], [3], [4], [5], [6], [7], [8], [9]]. They have been used for the treatment of human diseases including Alzheimer's, schistosomiasis and osteoporosis [10,11]. Phosphorus heterocycles are heterocyclic compounds containing hetero (O, N) and phosphorus atoms. They have obtained widespread interests because of their potential pharmaceutic applications [[12], [13], [14], [15], [16]]. For example, the oxazaphosphorine ifosfamide (I, Fig. 1) is commonly used as an alkylating chemotherapeutic agent in oncology such as lung, cervical, testicular cancers, and bone or soft tissue sarcomas [12]. A group of new nonpeptidic cyclic phosphon-based hydroxamic acids (II, Fig. 1) have been employed as inhibitors of metalloproteinases [14]. In another example, benzoxaphosphol-2-ones (III, Fig. 1) were designed and prepared, which show significant antibacterial activities [15]. [1,2]oxaphosphinane (IV, Fig. 1) was reported to have an antiproliferative activity against a large panel of NCI cancer cell lines [16]. In addition, interactions of phosphorus heterocyclic compounds with biomacromolecules have been reported, suggesting important biological and pharmaceutical values of these derivatives [17,18].
Fig. 1.
Structure of representative phosphorus heterocycles as novel pharmaceuticals.
Cytochromes P450 (P450s) are widely found in a variety of species throughout the biosphere [19]. As a large family of heme-containing redox enzymes, they play an important role in the synthesis of many molecules such as steroid hormones, cholesterol and other fats and acids. Additional P450s serve to metabolize and detoxify exogenous compounds presented to the organism [[20], [21], [22], [23], [24]]. Studying the interaction between drugs and P450s is significant not only for drugs’ pharmacological characteristic analysis but also for the enzyme inhibitor/activator discovery [25]. Identification of effective binders to P450s is crucial in distinguishing the risk of drug-drug interactions, where a compound potentially influences the metabolism of other compounds [26]. Moreover, investigations on the interactions of small molecules and P450s will provide insights into the mechanisms of drug action as well as the design of novel medicines.
Fluorescence spectroscopy is a powerful tool to explore the interactions between small molecules and P450s, which can reveal valuable information of the binding features of small molecules to proteins. Several studies have been investigated, indicating that the fluorescence quenching of P450 enzymes could be triggered by a variety of molecules [[27], [28], [29], [30]]. Mitra and co-workers [27] employed steady-state and time-resolved fluorescence spectroscopy to study the binding of camphor in the active side of cytochrome P450cam. Shumyantseva et al. [28] utilized similar fluorescence assays to detect the existence of riboflavin binding site on cytochrome P450 2B4. In addition, cytochrome P450 CYP1A2 and CYP2D6 were selected to study their interaction with puerarin by Shao et al. by using different spectroscopic studies [29]. Guengerich and co-workers [30] focused on the binding relationship of 7,8-benzoflavone to human cytochrome P450 3A4, revealing the complex fluorescence quenching and binding mechanism.
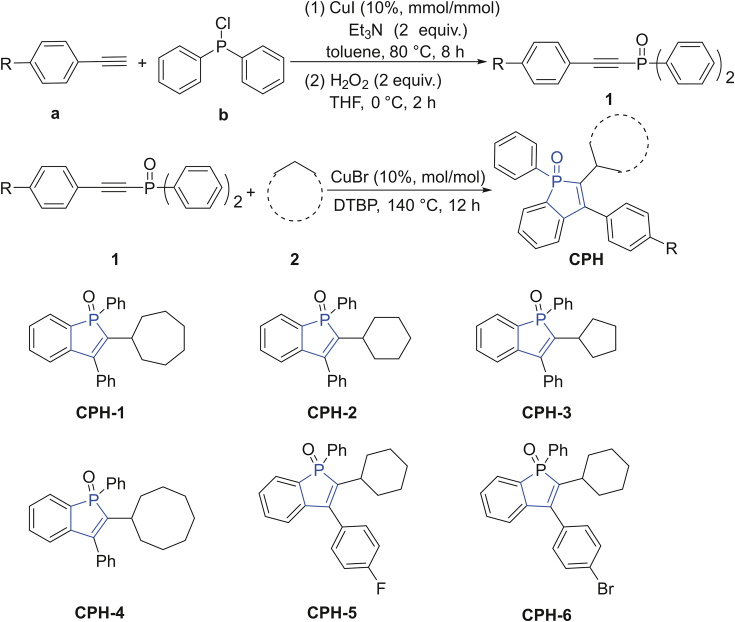
Recently, we reported a novel method to synthesize a series of cycloalkyl phosphorus heterocycles (CPHs) [31]. Their biological applications are yet to be discovered. Interestingly, these molecules display specific fluorescence and environment responsive properties (e.g. polarity, as shown in Fig. S1), making them excellent probes for small molecule/P450s binding study. OleT is a new P450 enzyme first reported by Rude et al. [32] in 2011. Collective effort has been made to improve the enzyme's catalytic efficiency including the use of small non-polar molecules to stimulate its activity [[33], [34], [35]]. Detailed binding mechanisms are needed to further understand the stimulation effect. Herein, we studied the interactions between CPHs and P450 fatty acid decarboxylase OleT from Staphylococcus aureus (OleTSA) by using fluorescence spectrum, UV–vis spectrum and 31P nuclear magnetic resonance (NMR) spectroscopies. The effect of CPHs on OleTSA enzymatic activity was also tested. To the best of our knowledge, it is the first work to describe the interaction of phosphorous heterocycles with P450s, which will provide useful information for phosphorus heterocycles' potential biological application. The binding mechanisms illustrated in this work also guide us to design novel stimulator to leverage OleT.
2. Materials and methods
2.1. Materials
The CPHs used here were prepared according to the synthetic method reported previously by our group [31]. The substrate diaryl (arylethynyl)phosphine oxide 1 was reacted with commercially available cycloalkane 2 in the presence of di-tert-butyl peroxide (DTBP) (Fig. 2), delivering excellent yield of CPHs. The crude product was purified by chromatographic techniques. The stock solutions of synthesized molecules were prepared by dissolving them in dimethyl sulfoxide (DMSO) to make the concentration as 30 mM. A phosphate buffer (200 mM K2HPO4, 100 mM NaCl, pH 7.4) was used unless otherwise noted. Other chemicals used were of analytical reagent grade and double-distilled water was used throughout.
Fig. 2.
Synthesis of cycloalkyl phosphorus heterocycles (CPHs).
OleTSA expression and purification were performed by following the published paper [36]. Briefly the kanamycin-resistant plasmid T5 containing the OleTSA-His6 gene was transformed to BL21 (DE3) containing the chloramphenicol-resistant pTF2 plasmid which has GroEL/GroES/Tig chaperones encoded. A modified terrific broth (TB) (24 g/L yeast extract, 12 g/L tryptone, and 4 g/L peptone) supplemented with 50 μg/mL kanamycin and 20 μg/mL chloramphenicol was used for culturing. 125 mg/L thiamine, and trace metals, 100 μM isopropyl β-d-1-thiogalactopyranoside (IPTG), 10 ng/mL tetracycline and 10 mg/L 5-aminolevulinic acid were added for protein expression. The expression was made at 18 °C for 16–18 h. Nickel-nitriloacetic acid (Ni-NTA) affinity chromatography was used for the purification of the protein, followed by Butyl-S-Sepharose column [37]. Fractions with an Rz value (Abs418 nm/Abs280 nm) above 1.2 were pooled and dialyzed against 200 mM KPi buffer (pH 7.4). Proteins were stored at −80 °C until further use.
2.2. Apparatus
Fluorescence data were collected using Cary-Eclipse Fluorescence Spectrophotometer (Palo Alto, CA, USA) equipped with an attached Cary Temperature Controller. UV–vis spectra were carried on HP 8453 spectrophotometer (Palo Alto, CA, USA). The 31P NMR spectra were obtained with a Bruker Avance 400 spectrometer at 298 K (Bruker, Faellanden, Switzerland).
2.3. Methods
2.3.1. Fluorescence spectroscopy
Fluorescence spectra of OleTSA (∼10 μM) were detected with varying the CPHs concentration (0, 5, 10, 15, 20, 25, 30 μM). With 280 nm excitation, the emission spectra were collected in the wavelength range from 300 to 500 nm at 293 K, 298 K, and 303 K, respectively. The different initial fluorescence intensity of the enzyme in all kinds of CPHs system were caused by the different concentration of OleTSA. In addition, the fluorescence spectra of CPH-6 were carried out by keeping the concentration of CPH-6 at 30 μM, meanwhile varying the OleTSA concentration (0, 0.2, 0.6, 2, 4, 6, 8 and 10 μM). The spectra were measured in the range of 270–500 nm at the excitation wavelength (λem) of 250 nm at 298 K. All the experiments were carried out in a conventional quartz cell using phosphate buffered saline (PBS).
2.3.2. UV–vis spectroscopy
Optical spectra were performed using 10 μM OleTSA and the sequential addition of CPH-6 from the 30 mM stock solution. The absorption spectra were collected from 300 nm to 700 nm at 303 K.
2.3.3. 31P NMR study
CPH-6 (50 or 100 μM) in 600 μL D2O was placed in a 10 mm NMR tube, followed by adding OleTSA (50 μM) and the 31P NMR spectrum was recorded. Bovine serum albumin (BSA) was chosen as a protein control.
2.3.4. OleTSA assay
The reaction mixtures (500 μL) containing 5 μM OleTSA, 2 mM hydrocinnamic acid, 2 mM H2O2 in the presence and absence of 200 μM phosphorus heterocyclic compounds were incubated at room temperature for 30 min. The reactions were quenched by addition of 1% acetic acid followed by 1,2,3,4-tetramethylbenzene as the internal standard. After centrifuging at 12,000 r/min for 20 min, the supernatant was extracted and tested on HPLC.
3. Results and discussion
3.1. Fluorescence quenching analysis
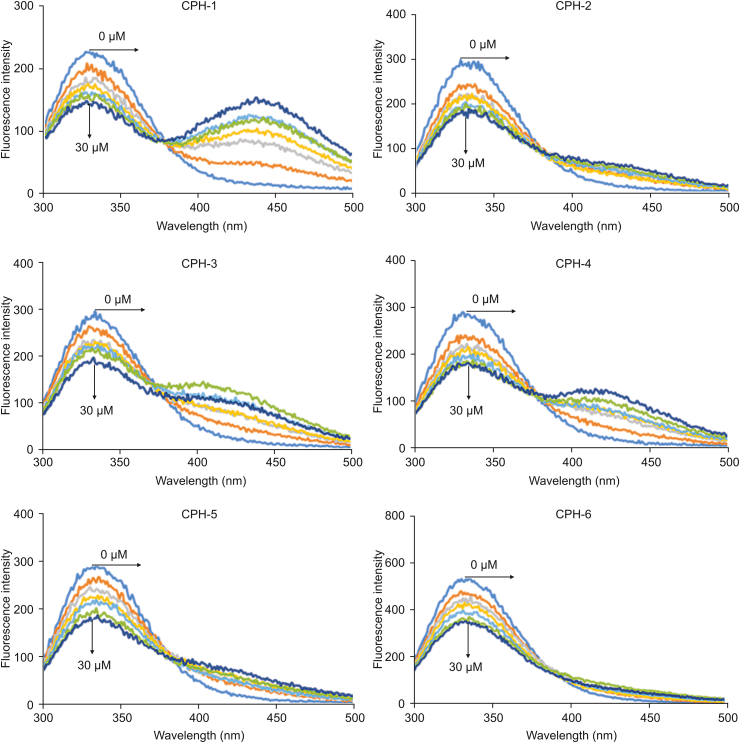
Intrinsic fluorescence of proteins is caused by three amino acids residues (tryptophan, tyrosine, and phenylalanine) containing aromatic groups. Among them, the tryptophan residues are believed to be main contributors [38]. A very significant aspect of proteins' intrinsic fluorescence is that its tryptophan residues have strong sensitivity to local environment [[39], [40], [41]]. The changes of fluorescence of tryptophan have been a useful tool to reveal the process of protein folding, aggregation and conformation transition [40]. As a result, the proteins' intrinsic fluorescence spectrum responding to small molecules can reflect valuable information about their interactions. In our study, the analysis of OleTSA fluorescence with increasing concentration of all derivatives (0, 5, 10, 15, 20, 25, and 30 μM, respectively) at 298 K was monitored. As shown in Fig. 3, decrease of fluorescence intensity was observed for all derivatives, indicating that the interaction between OleTSA and the CPHs can quench the enzyme's fluorescence signal. In addition, many CPHs in our research showed fluorescence signal at 400–500 nm with 280 nm excitation. In Fig. 3, with higher concentration of CPH-1, a stronger fluorescence signal was observed at 450 nm.
Fig. 3.
The changes in the intrinsic fluorescence intensity of OleTSA measured at the excitation wavelength (λem) of 280 nm in the presence of different concentrations of CPHs (0, 5, 10, 15, 20, 25, and 30 μM) at 298 K.
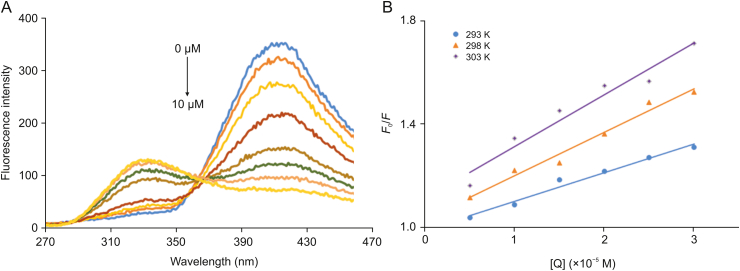
To gain insights into the interaction mechanism of CPHs with OleTSA, CPH-6 was chosen as a model for further investigation. The emission spectra of the compound CPH-6 with various concentrations of OleTSA at 298 K were collected (Fig. 4A). Under 250 nm excitation, CPH-6 emitted fluorescence signal in the range of 380–440 nm, which decreased with the increasing concentration of OleTSA. The result indicated that the fluorescence of the molecule could also be quenched by OleTSA, which could be attributed to the highly hydrophobic environment of the enzyme pocket.
Fig. 4.
(A): Fluorescence spectra of CPH-6 measured at the excitation wavelength (λem) of 250 nm alongside with various concentrations of OleTSA (0, 0.2, 0.6, 2, 4, 6, 8, and 10 μM) at 298 K. (B): Stern-Volmer plot for binding OleTSA with CPH-6 at 293, 298, and 303 K, respectively.
The fluorescence data were further analyzed to reveal the origins of the quenching. It is known that dynamic quenching is the process of inactivation of the excited state of fluorophores after interaction with quenching molecules [42]. However, static quenching often generates nonfluorescent complex of the fluorophore and the quencher. The discrimination of dynamic and static quenching depends on the relationship of quenching constant and temperature [42]. The dynamic quenching constant increases with the rise of temperature while the other one shows the decline of quenching constant with the increase of temperature. The Stern-Volmer equation (Eq. (1)) can be applied to explain the quenching mechanism.
| (1) |
Here, F0 and F represent the fluorescence intensities of OleT without and with a quencher, [OleT] refers to the concentration of OleT, Kq means the biomolecular quenching rate constant, KSV is the Stern-Volmer constant, τ0 is the average lifetime of the molecule without quencher. The average fluorescence lifetime of the biomolecule is evaluated at about 10 ns [43,44], so the quenching constant Kq could be obtained according to the Eq. (2).
| (2) |
The plots of F0/F versus [OleT] are shown in Fig. 4B. The values of KSV and Kq were determined by the slope of the plots and Eq. (2). The results are listed in Table 1 together with the correlation coefficients. As shown in Table 1, a significant increase in Stern-Volmer constant values was observed with increasing temperatures, indicating that the dynamic quenching was responsible for the quenching process of OleTSA and CPH-6.
Table 1.
Stern-Volmer quenching constants of OleTSA and CPH-6 at 293 K, 298 K, and 303 K.
| System | Temp. (K) | Ksv (M−1) × 10−2 | Kq (M−1s−1) × 106 | Ra |
|---|---|---|---|---|
| OleTSA-CPH-6 | 293 | 11.1 | 11.1 | 0.9786 |
| OleTSA-CPH-6 | 298 | 16.9 | 16.9 | 0.9777 |
| OleTSA-CPH-6 | 303 | 20.1 | 20.1 | 0.9523 |
Correlation coefficient.
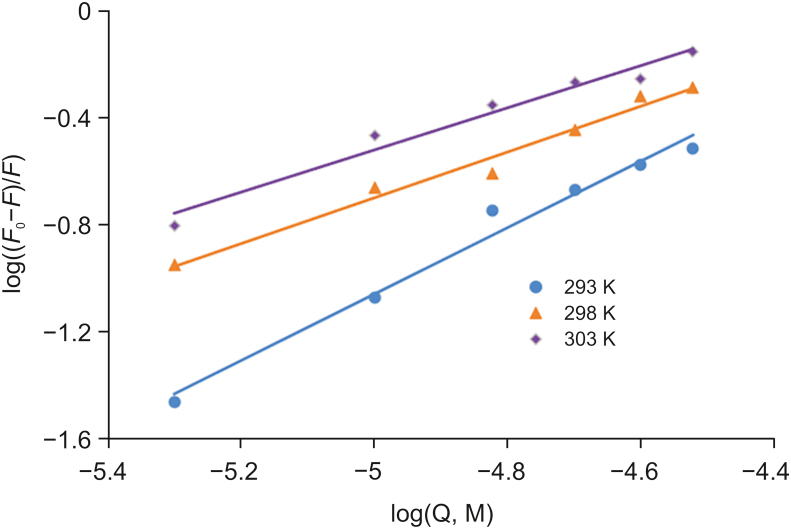
The binding constant (K) for OleT interaction with CPH-6 and the number of binding sites were described by double logarithmic plots as follows (Eq. (3)) [45,46].
| (3) |
Here, F0 and F represent the fluorescence intensities of OleT without and with a quencher, K denotes the binding constant of OleTSA with CPH-6, and n represents the number of binding sites. The characteristic binding parameters K and n were obtained from the plots of log ((F0–F)/F) versus log [OleT] (Fig. 5).
Fig. 5.
Double logarithmic plots for OleTSA-CPH-6 complexes.
As shown in Table 2, the value of n at the tested temperatures was around equal to one, which demonstrated that there was a single set equivalent binding site in OleTSA with CPH-6. In addition, the value of K is estimated to be 15.2 × 104 M−1 at 293 K, suggesting a moderately binding affinity between these two substances. A decrease was obtained in the value of K with the increase of temperature from 293 K to 303 K, indicating that the protein can better accommodate the molecule at 293 K than the other two temperatures and the stability between them decreases with increased temperature.
Table 2.
Binding constants and the number of binding sites on OleTSA for CPH-6.
| System | Temp. (K) | K (M−1) × 104 | n | Ra |
|---|---|---|---|---|
| OleTSA-CPH-6 | 293 | 15.2 | 1.25 | 0.9805 |
| OleTSA-CPH-6 | 298 | 0.40 | 0.86 | 0.9788 |
| OleTSA-CPH-6 | 303 | 0.28 | 0.79 | 0.9693 |
Correlation coefficient.
3.2. Thermodynamic analysis
It is well studied that non-covalent interactive forces including hydrogen bonding, hydrophobic interactions, van der Waals force and electrostatic attraction contribute to the binding of small molecules with biomacromolecules [[47], [48], [49]]. Ross and Subramanian [47] reported the main binding forces based on the sign and magnitude of thermodynamic parameters such as enthalpy change (ΔH) and entropy change (ΔS). The principle includes: a) ΔH > 0 and ΔS > 0, hydrophobic forces; b) ΔH < 0 and ΔS < 0, van der Waals interactions and hydrogen bonds; c) ΔH < 0 and ΔS > 0, electrostatic interactions. The ΔH and ΔS are calculated from the liner Van't Hoff plot (Eq. (4)).
| (4) |
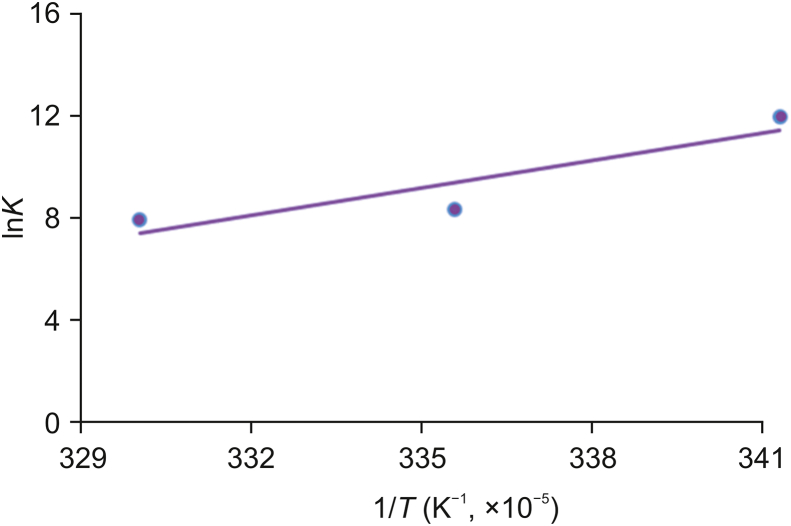
Here, K shows the associative binding constant at the specific temperature and R is the gas constant. The tested temperatures are 293, 298, and 303 K. ΔH and ΔS are obtained from the slope and intercept of the plot of lnK versus 1/T (Fig. 6). Free energy change (ΔG) is estimated from the following relationship (Eq. (5)).
| (5) |
Fig. 6.
The Van't Hoff plot for the binding of OleTSA with CPH-6.
As shown in Table 3, all the thermodynamic parameters ΔG, ΔH and ΔS were negative at the tested temperatures. Negative values of free energy (ΔG) revealed that the binding of OleTSA with CPH-6 was spontaneous. Both the values of ΔH and ΔS were negative, indicating that the main forces in the OleTSA-CPH-6 complex binding were hydrogen bond and van der Waals force.
Table 3.
Thermodynamic parameters for the interaction of OleTSA with CPH-6 at tested temperature.
| System | Temp. (K) | ΔH (kJ/mol) | ΔS (kJ/mol∙K) | ΔG (kJ/mol) |
|---|---|---|---|---|
| OleTSA-CPH-6 | 293 | −296.1 | −915.7 | −29.1 |
| OleTSA-CPH-6 | 298 | −296.1 | −915.7 | −20.6 |
| OleTSA-CPH-6 | 303 | −296.1 | −915.7 | −20.0 |
3.3. UV–vis spectrum analysis
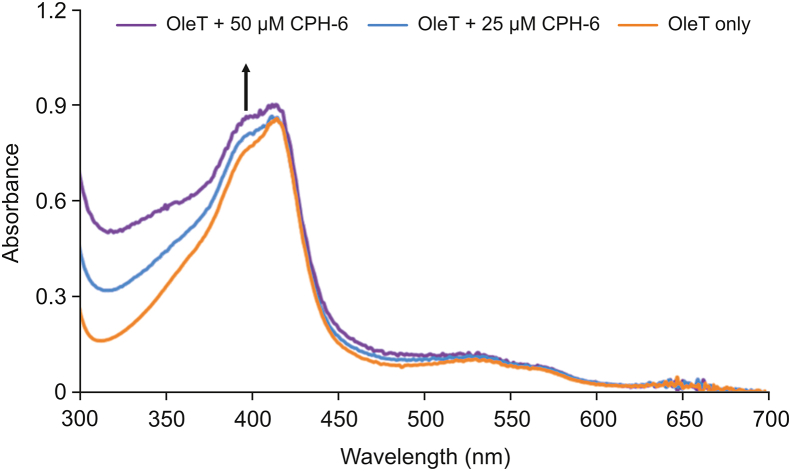
It has been well established that UV–vis spectra of P450s are sensitive to the redox state of the heme as well as its local environment [50]. To further study the binding effect of CPH-6 to OleTSA, the spectrum of OleTSA was monitored by UV–vis with or without of CPH-6. As shown in Fig. 7, the fluorescence intensity of OleTSA with 25 μM or 50 μM CPH-6 was adjusted by the extraction of the background fluorescence of corresponding concentration of substrate to eliminate the interference. It was found that the substrate-free OleTSA was in the mixed state at the beginning and the addition of 25 μM CPH-6 led to a slight increase of high spin. With a higher concentration (50 μM), this increment went further, indicating that CPH-6 can alter the microenvironment of heme iron of OleTSA.
Fig. 7.
UV–Vis binding titrations of CPH-6 with OleTSA. 10 μM OleTSA was used.
3.4. 31P NMR study
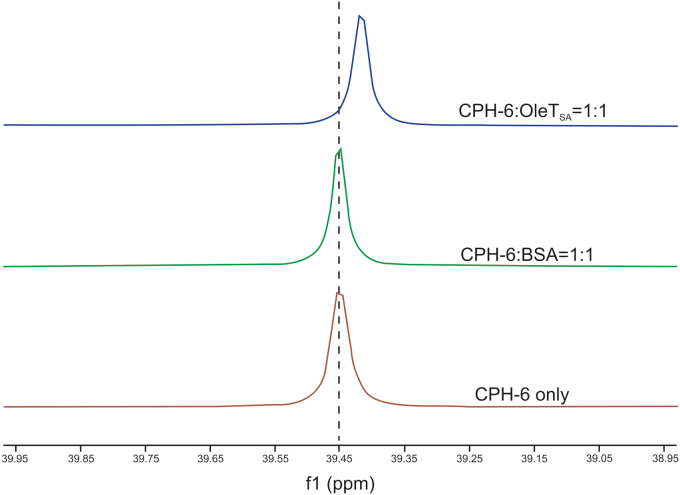
The interactions described above between CPH-6 and OleTSA were confirmed by 31P NMR analysis. As shown in Fig. 8, for the OleTSA-CPH-6 complex with 1:1 M ratio, 31P chemical shift value was moved to high field compared to the pure molecule CPH-6, suggesting that the environment of CPH-6 changed after adding into OleTSA. As a control, the 31P chemical shift of CPH-6 did not change after incubation with BSA in the same condition. With increased OleTSA concentration (i.e., OleTSA-CPH-6 complex with 2:1 M ratio), further shift was observed (Fig. S2). These results confirmed the interaction between OleTSA and CPH-6.
Fig. 8.
31P NMR spectra of CPH-6-OleTSA, CPH-6-BSA, and CPH-6 only. 50 μM of OleTSA or BSA, and 50 μM of CPH-6 were used.
3.5. OleTSA enzymatic activity assay
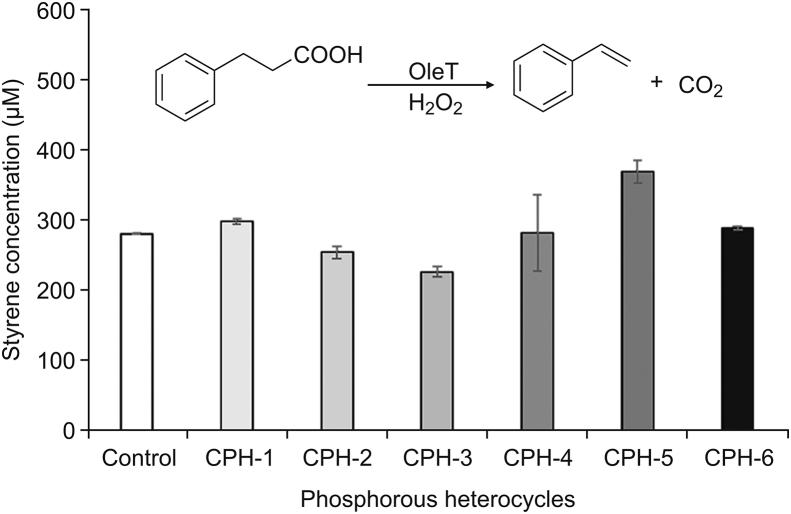
OleTSA catalyzes the decarboxylation of hydrocinnamic acid to form styrene [51], which can be easily detected by LC. The effect of phosphorous heterocycles on OleTSA activity was characterized by using hydrocinnamic acid as substrate (Fig. 9). Interestingly, the CPHs with different side groups showed different effects on OleTSA activity. For example, OleTSA showed around 30% enhanced activity with treatment by CPH-5 (cyclohexyl phosphorous heterocycle with fluoro group) while around 20% activity was inhibited by CPH-3 (cyclopentyl phosphorous heterocycle). Further characterization of enzyme-CPHs complex structure will provide useful information to understand these effects. Nevertheless, the different effects resulting from the fine-tuned small molecule structure represent a viable way to leverage enzyme activity. Such information can be also used for P450 enzyme effector design.
Fig. 9.
Different CPHs showed different effects on OleTSA activity (expressed as amount of styrene produced). Reaction condition: 5 μM OleTSA, 2 mM hydrocinamic acid, 2 mM H2O2, 200 μM CPHs, room temperature for 30 min. Error bars represent standard deviations of duplicate experiments (n=2).
4. Conclusions
In conclusion, this research was to study the interactions between CPHs and OleTSA by fluorescence spectra, UV–vis and 31P NMR spectroscopies. Dynamic quenching mechanism was identified. The binding constant value of 15.2 × 104 M−1 at 293 K indicated a moderately binding affinity between CPHs and OleTSA. Hydrogen bonding and van der Waals force were identified as main forces involved in the interactions on the basis of thermodynamic parameters analysis. UV–vis spectroscopies suggested that the presence of CPHs could affect the enzyme's interior environment. The 31P NMR studies revealed that binding of CPHs to OleTSA had an influence on the chemical shift of 31P NMR of the molecule. Moreover, the effect of such molecules on OleTSA enzymatic activity indicated potential functions of these molecules and provided useful information for enzyme stimulator design.
Declaration of competing interest
The authors declare that there have no conflicts of interest.
Acknowledgments
This work was supported by the China Scholarship Council and Fujian University-Industry Research Cooperation Project (Project No.: 2018N5013). The authors also wish to thank Thomas M. Makris for providing the OleT plasmid, thank Olivia Manley and Suman Das for their help with protein purification and thank Fiaz Ahmed for offering the help with fluorescence temperature-controlled experiment from Department of Chemistry and Biochemistry of University of South Carolina.
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.12.004.
Contributor Information
Yingwu Yin, Email: ywyin@xmu.edu.cn.
Yuxing Gao, Email: gaoxingchem@xmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Westheimer F.H. Why nature chose phosphates. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 2.Seto H., Kuzuyama T. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 1999;16:589–596. doi: 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- 3.Zon G. Cyclophosphamide analogues. Prog. Med. Chem. 1982;19:205–246. doi: 10.1016/s0079-6468(08)70330-8. [DOI] [PubMed] [Google Scholar]
- 4.Stec W.J. Organophosphorus Chemistry, Cambridge: Royal Society of Chemistry; 1982. Cyclophosphamide and its congeners; pp. 145–174. [Google Scholar]
- 5.Kafarski P., Lejczak B. Biological activity of aminophosphonic acids. Phosphorus Sulfur Silicon Relat. Elem. 1991;63:193–215. [Google Scholar]
- 6.Colvin O.M. An overview of cyclophosphamide development and clinical applications. Curr. Pharm. Des. 1999;5:555–560. [PubMed] [Google Scholar]
- 7.Kafarski P., Lejczak B. Aminophosphonic acids of potential medical importance. Curr. Med. Chem. Anticancer Agents. 2001;1:301–312. doi: 10.2174/1568011013354543. [DOI] [PubMed] [Google Scholar]
- 8.Demkowicz S., Rachon J., Daśko M., et al. Selected organophosphorus compounds with biological activity. Applications in medicine. RSC Adv. 2016;6:7101–7112. [Google Scholar]
- 9.Rodriguez J.B., Gallo-Rodriguez C. The role of the phosphorus atom in drug design. ChemMedChem. 2019;14:190–216. doi: 10.1002/cmdc.201800693. [DOI] [PubMed] [Google Scholar]
- 10.Richardson R.J. Assessment of the neurotoxic potential of chlorpyrifos relative to other organophosphorus compounds: a critical review of the literature. J. Toxicol. Environ. Health. 1995;44:135–165. doi: 10.1080/15287399509531952. [DOI] [PubMed] [Google Scholar]
- 11.Pope C.N., Brimijoin S. Cholinesterases and the fine line between poison and remedy. Biochem. Pharmacol. 2018;153:205–216. doi: 10.1016/j.bcp.2018.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilard V., Martino R., Malet-Martino M., et al. Chemical stability and fate of the cytostatic drug ifosfamide and its N-dechloroethylated metabolites in acidic aqueous solutions. J. Med. Chem. 1999;42:2542–2560. doi: 10.1021/jm980587g. [DOI] [PubMed] [Google Scholar]
- 13.Budzisz E., Brzezinska E., Krajewska U., et al. Cytotoxic effects, alkylating properties and molecular modelling of coumarin derivatives and their phosphonic analogues. Eur. J. Med. Chem. 2003;38:597–603. doi: 10.1016/s0223-5234(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 14.Sørensen M.D., Blæhr L.K.A., Christensen M.K., et al. Cyclic phosphinamides and phosphonamides, novel series of potent matrix metalloproteinase inhibitors with antitumour activity. Bioorg. Med. Chem. 2003;11:5461–5484. doi: 10.1016/j.bmc.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Patil S.L., Bhalgat C.M., Burli S., et al. Synthesis, antibacterial and antioxidant properties of newer 3-(1-benzofuran-2-yl)-5-substituted aryl-1, 2-oxazole. Int. J. Chem. Sci. Appl. 2010;1:42–49. [Google Scholar]
- 16.Clarion L., Jacquard C., Sainte-Catherine O., et al. Oxaphosphinanes: new therapeutic perspectives for glioblastoma. J. Med. Chem. 2012;55:2196–2211. doi: 10.1021/jm201428a. [DOI] [PubMed] [Google Scholar]
- 17.Roy S., Nandi R.K., Ganai S., et al. Binding interaction of phosphorus heterocycles with bovine serum albumin: a biochemical study. J. Pharm. Anal. 2017;7:19–26. doi: 10.1016/j.jpha.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S., Saxena S.K., Mishra S., et al. Spectroscopic evidence of phosphorous heterocycle–DNA interaction and its verification by docking approach. J. Fluoresc. 2018;28:373–380. doi: 10.1007/s10895-017-2199-7. [DOI] [PubMed] [Google Scholar]
- 19.Denisov I.G., Makris T.M., Sligar S.G., et al. Structure and chemistry of cytochrome P450. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 20.Guengerich F.P. Characterization of human cytochrome P450 enzyme. Faseb. J. 1992;6:745–748. doi: 10.1096/fasebj.6.2.1537465. [DOI] [PubMed] [Google Scholar]
- 21.Anzenbacher P., Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics, Cell. Mol. Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamb D.C., Waterman M.R., Kelly S.L., et al. Cytochromes P450 and drug discovery. Curr. Opin. Biotechnol. 2007;18:504–512. doi: 10.1016/j.copbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Veith A., Moorthy B. Role of cytochrome P450s in the generation and metabolism of reactive oxygen species, Cytochromes P450 and drug discovery. Curr. Opin. Toxicol. 2018;7:44–51. doi: 10.1016/j.cotox.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foroozesh M., Sridhar J., Goyal N., et al. Coumarins and P450s, studies reported to-date. Molecules. 2019;24:1620. doi: 10.3390/molecules24081620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stjernschantz E., Vermeulen N.P.E., Oostenbrink C. Computational prediction of drug binding and rationalisation of selectivity towards cytochromes P450. Expert. Opin. Drug Metab. Toxicol. 2008;4:513–527. doi: 10.1517/17425255.4.5.513. [DOI] [PubMed] [Google Scholar]
- 26.Ogu C.C., Maxa J.L. Drug interactions due to cytochrome P450. Proc (Bayl Univ Med Cent) 2000;13:421–423. doi: 10.1080/08998280.2000.11927719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad S., Mazumdar S., Mitra S. Binding of camphor to Pseudomonas putida cytochrome P450cam: steady-state and picosecond time-resolved fluorescence studies. FEBS Lett. 2000;477:157–160. doi: 10.1016/s0014-5793(00)01745-2. [DOI] [PubMed] [Google Scholar]
- 28.Shumyantseva V.V., Bulko T.V., Petushkova N.A., et al. Fluorescent assay for riboflavin binding to cytochrome P450 2B4. J. Inorg. Biochem. 2004;98:365–370. doi: 10.1016/j.jinorgbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Shao J., Chen J., Li T., et al. Spectroscopic and molecular docking studies of the in vitro interaction between puerarin and cytochrome P450. Molecules. 2014;19:4760–4769. doi: 10.3390/molecules19044760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsch G.A., Carlson B.T., Guengerich F.P. 7,8-benzoflavone binding to human cytochrome P450 3A4 reveals complex fluorescence quenching, suggesting binding at multiple protein sites. J. Biomal. Struct. Dyn. 2018;36:841–860. doi: 10.1080/07391102.2017.1301270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma D., Pan J., Yin L., et al. Copper-catalyzed direct oxidative C−H functionalization of unactivated cycloalkanes into cycloalkyl benzo[b]phosphole oxides. Org. Lett. 2018;20:3455–3459. doi: 10.1021/acs.orglett.8b01108. [DOI] [PubMed] [Google Scholar]
- 32.Rude M.A., Baron T.S., Brubaker S., et al. Terminal olefin (1-Alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 2011;77:1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh C.H., Huang X., Amaya J.A., et al. The enigmatic P450 decarboxylase OleT is Capable of, but evolved to frustrate, oxygen rebound chemistry. Biochemistry. 2017;56:3347–3357. doi: 10.1021/acs.biochem.7b00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C., Shen F., Wang S., et al. An engineered self-sufficient biocatalyst enables scalable production of linear alpha olefins from carboxylic acids. ACS Catal. 2018;8:5794–5798. [Google Scholar]
- 35.Wise C.E., Hsieh C.H., Poplin N.L., et al. Dioxygen activation by the biofuel-generating cytochrome P450 OleT. ACS Catal. 2018;8:9342–9352. [Google Scholar]
- 36.Amaya J.A., Rutland C.D., Makris T.M. Mixed regiospecificity compromises alkene synthesis by a cytochrome P450 peroxygenase from Methylobacterium populi. J. Inorg. Biochem. 2016;158:11–16. doi: 10.1016/j.jinorgbio.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Amaya J.A. University of South Carolina; Los Angeles, USA: 2018. Mechanisms of Decarboxylation in the CYP152 Family of Cytochrome P450s (Dissertation) [Google Scholar]
- 38.Chen Y., Barkley M.D. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 39.Sharma A., Schulman S.G. Wiley Press; New York: 1999. Introduction to Fluorescence Spectroscopy. [Google Scholar]
- 40.Pontremoli C., Barbero N., Viscardi G., et al. Insight into the interaction of inhaled corticosteroids with human serum albumin: a spectroscopic-based study. J. Pharm. Anal. 2018;8:37–44. doi: 10.1016/j.jpha.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sindrewicz P., Li X., Yates E.A., et al. Intrinsic tryptophan fluorescence spectroscopy reliably determines galectin-ligand interactions. Sci. Rep. 2019;9:11851. doi: 10.1038/s41598-019-47658-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakowicz J.R. Springer; New York: 1999. Principle of Fluorescence Spectroscopy. [Google Scholar]
- 43.Lakowic J.R., Weber G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buddanavar A.T., Nandibewoor S.T. Multi-spectroscopic characterization of bovine serum albumin upon interaction with atomoxetine. J. Pharm. Anal. 2017;7:148–155. doi: 10.1016/j.jpha.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anbazhagan V., Renganathan R. Study on the binding of 2,3-diazabicyclo[2.2.2]oct-2-ene with bovine serum albumin by fluorescence spectroscopy. J. Lumin. 2008;128:1454–1458. [Google Scholar]
- 46.Min J., Meng-Xia X., Dong Z., et al. Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin. J. Mol. Struct. 2004;692:71–80. [Google Scholar]
- 47.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 48.Némethy G., Scheraga H.A. Structure of water and hydrophobic bonding in proteins. I. A model for the thermodynamic properties of liquid water. J. Chem. Phys. 1962;36:3382–3400. [Google Scholar]
- 49.Shahabadi N., Fatahi A. Multispectroscopic DNA-binding studies of a tris-chelate nickel(II) complex containing 4,7-diphenyl 1,10-phenanthroline ligands. J. Mol. Struct. 2010;970:90–95. [Google Scholar]
- 50.Luthra A., Denisov I.G., Sligar S.G. Spectroscopic features of cytochrome P450 reaction intermediates. Arch. Biochem. Biophys. 2011;507:26–35. doi: 10.1016/j.abb.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munro A.W., McLean K.J., Grant J.L., et al. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem. Soc. Trans. 2018;46:183–196. doi: 10.1042/BST20170218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.