Abstract
Background
Most ovarian cancer patients are diagnosed at an advanced stage and have a high mortality rate. Current screening strategies fail to improve prognosis because markers that are sensitive for early stage disease are lacking. This medical need justifies the search for novel approaches using utero-tubal lavage as a proximal liquid biopsy.
Methods
In this study, we explore the extracellular transcriptome of utero-tubal lavage fluid obtained from 26 ovarian cancer patients and 48 controls using messenger RNA (mRNA) capture and small RNA sequencing.
Results
We observed an enrichment of ovarian and fallopian tube specific messenger RNAs in utero-tubal lavage fluid compared to other human biofluids. Over 300 mRNAs and 41 miRNAs were upregulated in ovarian cancer samples compared with controls. Upregulated genes were enriched for genes involved in cell cycle activation and proliferation, hinting at a tumor-derived signal.
Conclusion
This is a proof-of-principle that mRNA capture sequencing of utero-tubal lavage fluid is technically feasible, and that the extracellular transcriptome of utero-tubal lavage should be further explored in larger cohorts to assess the diagnostic value of the biomarkers identified in this study.
Impact
Proximal liquid biopsy from the gynecologic tract is a promising source for mRNA and miRNA biomarkers for diagnosis of early-stage ovarian cancer.
Keywords: Ovarian cancer, Utero-tubal lavage, Biomarker, Cell-free RNA, Liquid biopsy
Introduction
Ovarian cancer, the fifth leading cause of cancer-related mortality in women, a five-year survival rate below 45%, largely driven by late stage diagnoses [1]. In Europe, the incidence of ovarian cancer is 12.9 per 100,000 [2]. Ovarian cancer is often referred to as a ‘silent killer’ because local disease is usually asymptomatic and symptoms of advanced stage disease are nonspecific. More than 75% of affected women are diagnosed with metastatic disease that is rarely curable. Early detection of ovarian cancer is key as stage I disease has a 5-year survival rate of 93% [3]. Ovarian cancers are classified into histological subtypes, with various underlying transcriptional and mutational patterns. High-grade serous carcinoma is the most prevalent and most challenging subtype, which is common in genetically predisposed populations, such as germline BRCA1/2 mutation carriers, having an estimated lifetime risk of 54% and 23%, respectively. A risk-reducing bilateral salpingo-oophorectomy (RRBSO) around the age of 40, is the only effective approach to avoid ovarian cancer in these women, resulting in significant morbidity of early menopause [3,4]. Currently, no effective screening method for this cancer entity is available. Serum cancer antigen 125 (CA125), the most studied test for ovarian cancer screening, has important limitations [5,6]. Less than 50% of patients with early stage ovarian cancer have elevated CA125 levels, and elevated CA125 levels can also be observed in benign conditions, such as pelvic inflammatory disease, endometriosis, and ovarian cysts [7]. Human Epididymis Protein 4 (HE4) is another protein marker that has been evaluated as serum biomarker for ovarian cancer diagnosis and the test has received approval from the US Food and Drug Administration for use in women presenting with an ovarian mass in 2011 [8]. HE4 expression shows high specificity for ovarian cancer, however serum HE4 levels vary in smokers, in hormonal contraceptive users and the levels increase with aging [9]. Unfortunately, the use of CA125 and HE4 have not been effective in improving patient survival [10], [11], [12], [13]. Therefore, there is a clinical need for non-invasive, robust, and reliable diagnostics for ovarian cancer detection.
Extracellular RNAs (exRNAs) in blood and other biofluids have been identified as potential biomarkers for a wide range of diseases, including ovarian cancer [14], [15], [16], [17]. These so-called ‘liquid biopsies’ may offer a non-invasive alternative to tissue biopsies for diagnosis, prognosis and treatment response monitoring. The biomarker potential of extracellular RNAs in serum, plasma, urine and ascites has previously been investigated for the early diagnosis of ovarian cancer. These studies mainly focused on selected microRNAs using reverse transcription quantitative polymerase chain reaction (RT-qPCR) [14], while few applied RNA sequencing as a more unbiased and transcriptome wide approach [18], [19], [20].
There is increasing evidence that precursor lesions of high-grade serous carcinoma originate from the epithelium of the fallopian tube fimbriae rather than intraperitoneally. The fimbriae represent the distal end of the fallopian tube, adjacent to the ovaries [21], [22], [23]. The lag time from emergence of the first malignant cells to clinically overt high-grade ovarian cancer is approximately six years [21], and shedding of tumor cells from ovarian cancer and its precursor lesions into the gynecological tract has been reported [24]. Sampling the cells of the fimbriae or their secreted biological products, through proximal liquid aspirated from the gynecological tract, may thus reveal markers of the initial lesions. Utero-tubal lavage fluid can be obtained after flushing saline into the uterine cavity and fallopian tubes and holds promise for a minimally invasive liquid biopsy technique as this can be performed during a routine office-visit at the gynecology department [25,26]. Previous studies looking into the biomarker potential of utero-tubal lavage fluid primarily focused on circulating mutant p53 DNA [26] and on proteomic profiling of extracellular vesicles isolated from utero-tubal lavage samples [25]. So far, the RNA content of this fluid remains to be investigated.
The goal of this proof-of-concept study was to profile the extracellular transcriptome of utero-tubal lavage fluid using messenger RNA (mRNA) capture sequencing and small RNA sequencing to investigate the biomarker potential of extracellular mRNAs for ovarian cancer diagnosis.
Materials and methods
Donor material, collection and utero-tubal lavage preparation procedure
Sample collection was approved by the ethics committee of Chaim Sheba Medical Center, Rabin Medical Center and Meir Medical Center, Israel (ClinicalTrials.gov identifier: NCT03150121). Written informed consent was obtained from each participant in accordance with the Helsinki declaration. Recruited patients underwent gynecological surgical procedures under general anesthesia, including hysteroscopy, hysterectomy and/or RRBSO. Eligible indications included high-grade ovarian cancer (primary or interval debulking), suspicious ovarian mass, risk reduction, or various other benign gynecological disorders. Utero-tubal lavage samples were collected before surgery, after induction of anesthesia, by surgeons in the three participating centers. An intrauterine insemination catheter (InsemiTM-Cath, Cook Inc. Bloomington, USA) or rigid pipelle uterine sampler (Endosampler, MedGyn, Addison, USA) was inserted into the endometrial cavity through the cervical canal. Ten ml of saline were flushed into the uterine cavity and fallopian tubes and immediately retrieved (at an average volume of 4.6 ml per patient). The utero-tubal lavage samples were immediately centrifuged at 480xg for 15 min to eliminate cells and the supernatants were stored at -80 °C. RNA extraction, library preparation and sequencing methods are described in detail in the supplemental methods section. Raw data is presented in supplemental Tables 1–7.
Data analysis
Assessment of tissue and cell contribution to the extracellular transcriptome of utero-tubal lavage
Using total RNA-sequencing data from 27 normal human tissue types and 5 immune cell types from peripheral blood from the RNA Atlas [27], we created gene sets containing marker genes for each individual entity, as described in Hulstaert et al. [15]. We removed redundant tissues and cell types from the original RNA Atlas (e.g. granulocytes and monocytes were present twice; brain was kept and specific brain sub-regions such as cerebellum, frontal cortex, occipital cortex and parietal cortex were removed) and we used genes where at least one tissue or cell type had expression values greater or equal to 1 TPM normalized counts. A gene was considered to be a marker if its abundance was at least 5 times higher in the most abundant sample compared to the others. For the final analysis, only tissues and cell types with at least 3 markers were included, resulting in 26 tissues and 5 immune cell types. Gene abundance read counts from the biofluids in the discovery cohort of Hulstaert et al. and the gene abundance read counts of the utero-tubal lavage cohort from this study, were normalized using Sequin spikes as size factors in DESeq2 (v1.22.2) [15]. For all marker genes within each gene set, we computed the log2 fold changes between the median read count of a biofluid sample pair versus the median read count of all other biofluids.
Differential expression analysis with DeSEQ2
Further processing of the count tables was done with R (v3.5.1) making use of tidyverse (v1.2.1). Gene abundance expression read counts obtained were normalized using the sum of all reads mapping to Sequin spikes or RC spikes as size factors in DESeq2 (v1.20.0) [28]. To assess the biological signal in the case/control cohorts, we performed differential expression analysis between the patients and control groups using DESeq2 (v1.20.0). Only mRNAs present in at least 80% of each group (cancer or benign) with at least 7 read counts per sample were included in the analysis. Genes were considered differentially expressed when the absolute log2 fold change > 1 and at q < 0.05. Principle component analysis was performed and the first two principle components for the normalized sequencing data were plotted using the plotPCA function in R [29].
Differential exon usage with DEXSeq
To perform differential exon usage analysis the mapped sequencing data was preprocessed according to the two preparation Python scripts provided in the DEXSeq package (version 1.36.0, [30]). In first script a GTF file with gene models was transformed into a GFF file listing counting bins. In the second script such a GFF file and an alignment file in the BAM format were used to produce a list of exon counts. Next, the count tables consisting of exon counts were further processed with R (v3.5.1) making use of tidyverse (v1.2.1). Exon expression read counts were normalized using the sum of all reads mapping to Sequin spikes as size factors in DESeq2 (v1.20.0)(28). Differential expression analysis between the patients and control groups was performed using DESeq2 (v1.20.0). Exons were considered differentially expressed when the absolute log2 fold change > 1 and at q < 0.05. In order to identify genuine differentially abundant exons, only exons that were not part of differentially abundant genes were considered.
Gene set enrichment analysis
A pre-ranked gene set enrichment analysis was performed using the 50 hallmark gene sets (version 7.2.) available on the Molecular Signatures Database (1000 permutations, classic analysis) [30]. All mRNA lists were ordered based on the log-transformed fold change obtained after differential expression analysis with DeSEQ2 (ovarian cancer versus control). The R package Fast Gene Set Enrichment Analysis (fgsea, version 1.8.0) was used to determine normalized enrichment scores [31]. Significant enrichment was defined at false discovery rate < 0.05. A pre-ranked gene set enrichment analysis allows to select from an a priori defined list of gene sets those which have non-random behavior in a considered experiment.
Detection of fusion transcripts
Fusion transcript identification was performed using FusionCatcher (version 1.30) with default parameter settings [32]. Stringent filtering was applied to exclude potential false positive fusion transcripts. First, transcripts with a fusion description label indicative for a false positive result (i.e. the red annotations in supplemental Table 5) were excluded. Second, transcripts with reads mapping on both fusion partners were excluded. Third, transcripts with fusion partners less than 100 kb apart were also excluded. Only exon-exon fusions were included.
Classifier build using mRNA capture seq data, small RNA seq data and differential exon data
Pre-processing of the spike normalized data was performed as previously described [33]. Briefly, expression levels lower than a lower threshold of 10 were set to this lower threshold. Expression levels higher than an upper threshold of 30,000 were set to this upper threshold. Next, ratio filtering was applied, i.e. genes for which the ratio between the highest and lowest expression level was less than 5 were removed. Range filtering was applied, i.e. genes for which the difference between the highest and lowest expression level was less than 500 were removed. A base-2 logarithmic transformation was applied to the gene expression levels. The most significant genes between both groups (cancer and control) were defined using a pairwise t-test. Seven different classification methods were then applied to the data using the m most significant genes according to the preprocessing procedure, where n ranges over the values m = 5, 10, …, 500. The following classification methods with the publicly available R implementations, were used: [1] lasso[34] and elastic net logistic regression [35], computed using the glmnet package [2,36] adaptive [37] and relaxed lasso [38] for logistic regression, computed using the gcdnet and glmnet packages, respectively, [3] minimum concave penalized logistic regression [39], computed using the ncvreg package, [4] split-Lasso and split-EN logistic regression [40], computed using the SplitGLM package, [5] random forest [41], computed using the randomForest package, [6] random generalized linear model (GLM) [42], computed using the RGLM package and [7] extreme gradient boosting [43], computed using the xgboost package. Cross-validation was used to select the penalty parameters in methods 1–4 and the default settings were used for other tuning parameters.
For each of the individual data layers (mRNA, miRNA, exon), and for all possible combinations of these data layers, the performance of the different classification methods was evaluated to select the best classifier for each of the data layers. To evaluate the performance of the classifiers, the data set was randomly split into training and test data. 75% of the samples (n = 55) were used to train the classification method and the remaining 25% (n = 19) was used as test data to evaluate its performance. This process of random splitting was repeated 100 times. The misclassification rate, the sensitivity, and the specificity, averaged over the 100 test sets is reported. The misclassification rate was used as criterion to select the best classifier.
Results
Patient population
Eighty-one utero-tubal lavage samples collected from 31 ovarian cancer patients and 50 patients with benign ovarian lesions were analyzed in this study. Upon RNA extraction and mRNA capture sequencing, 7 of the 81 samples (5 ovarian cancer and 2 control samples) were excluded for statistical analysis because of too low sequencing depth, resulting in a final cohort of 74 samples collected from 26 ovarian cancer patients with an average age of 61.5 years old and 48 patients with benign ovarian lesions with an average age of 63.8 years old. The demographic and clinical patient information is provided in Table 1. Details for all samples with the reason of exclusion for further analysis is provided in supplemental Table 2.
Table 1.
Patient characteristics for utero-tubal lavage samples included in the transcriptomic analysis.
| ovarian cancer (n = 26) | control (n = 48) | |
|---|---|---|
| age in years, mean (min-max) | 61.5 (46–78) | 63.8 (51–83) |
| stage | ||
| early stage (I-II) | 4 | - |
| late stage (III-IV) | 22 | - |
| BRCA status | ||
| germline mutation | 7 | 8 |
| no mutation | 17 | 0 |
| unknown | 2 | 40 |
| indication for surgery | ||
| high grade ovarian cancer | 26 | - |
| benign ovarian mass/cyst | - | 9 |
| menorrhagia | - | 3 |
| pelvic organ prolapse | - | 6 |
| leiomyomatous uterus | - | 2 |
| normal endometrium | - | 6 |
| RRBSO | - | 5 |
| mature teratoma | - | 2 |
| mucinous cystadenoma | - | 6 |
| other | - | 9 |
Messenger RNA profile of utero-tubal lavage fluid
Over all samples, the mapping rate to Ensembl genes was 90%, with a minimum of 20% and a maximum of 96% (Supplemental Fig. 1A). The total reads mapped to Ensembl genes varied from 1.2 million to 24.8 million per sample with a mean of 11.9 million reads per sample. Based on the coverage of artificial spike-in controls, the endogenous RNA mass per sample was calculated. The mean endogenous RNA mass detected per 1 mL fluid was 0.09 ng, with a minimum of 0.002 ng and a maximum of 0.72 ng. The endogenous RNA mass did not differ between the ovarian cancer group and the control group (Wilcoxon signed-rank test, two-sided, p = 0.196, Supplemental Fig. 1B). Despite the high variability in mapping rate across the samples, RNA complexity of the samples was very stable. The total number of unique mRNAs ranged from 11,887 to 17,850 with a mean of 15,451 mRNAs per sample (Supplemental Fig. 1A). In total, 8139 genes were detected in all samples.
Tissue contribution to the utero-tubal lavage fluid exRNAs
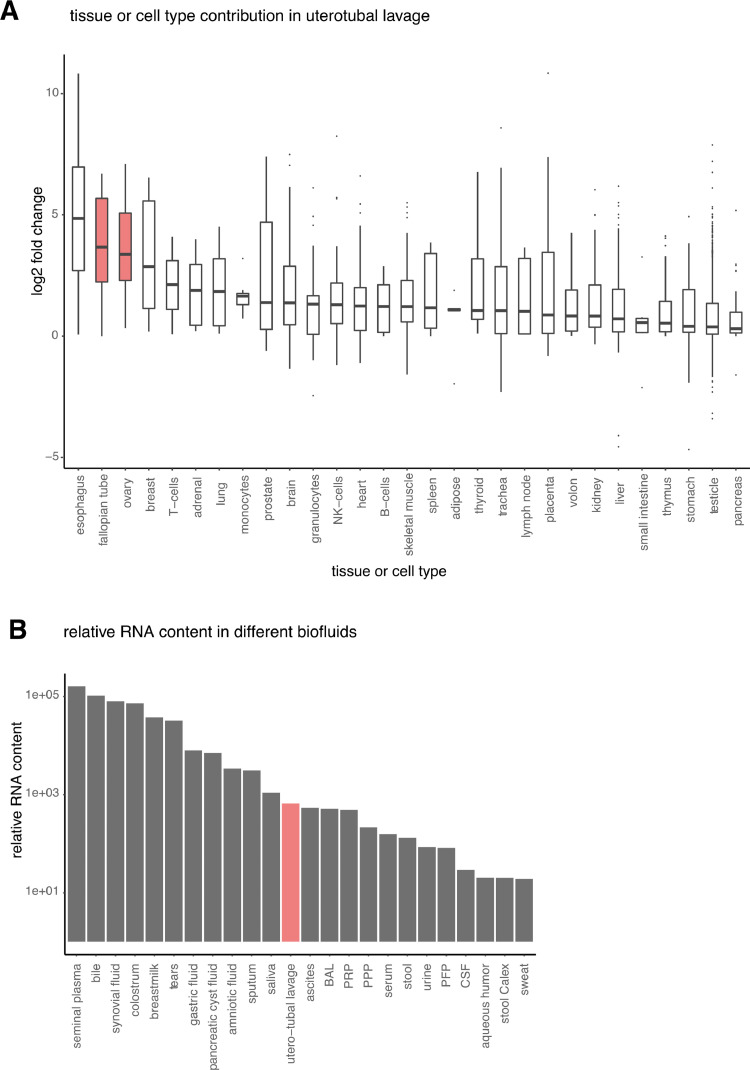
To assess which tissues or cell types contribute mRNA molecules to the utero-tubal lavage fluid RNA profile, we evaluated tissue- and cell-type-specific mRNA signatures. The boxplots in Fig. 1A highlight the relative contribution of tissues and cell types to utero-tubal lavage fluid compared to 23 human biofluids that were included in the Human Biofluid RNA Atlas [15]. Esophagus RNA markers were more abundant in utero-tubal lavage fluid than in the other biofluids, likely reflecting an epithelial RNA-signature shared between epithelial cells from esophagus and endometrium. Fallopian tube and ovary specific mRNAs were the second and third most enriched signatures, suggesting that the wash procedure enables detection of RNA originating from ovary and fallopian tube tissue. When comparing the relative RNA content of utero-tubal lavage to the 23 other biofluids, utero-tubal lavage fluid ranked as the twelfth highest concentrated fluid (Fig. 1B). The relative mRNA content of utero-tubal lavage was similar to that of ascites, broncho-alveolar lavage and platelet-rich-plasma. Utero-tubal lavage fluid contained 8-fold more RNA than platelet-free plasma, the most studied biofluid in the biomarker field.
Fig. 1.
Assessment of the tissues contributing RNA molecules to utero-tubal lavage fluid. (A) Boxplots showing the log2 fold change for a gene set with markers specific for a certain tissue or cell type. The log2 fold change is calculated between the median read count of all utero-tubal lavage samples and the median read count of all other biofluids. The tissues or cell types for which markers were selected based on the RNA Atlas Project are shown on the x-axis. (B) Barplot showing the relative mRNA content in 24 human biofluids. BAL, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; PFP, platelet-free plasma; PPP, platelet-poor plasma; PRP, platelet-rich plasma.
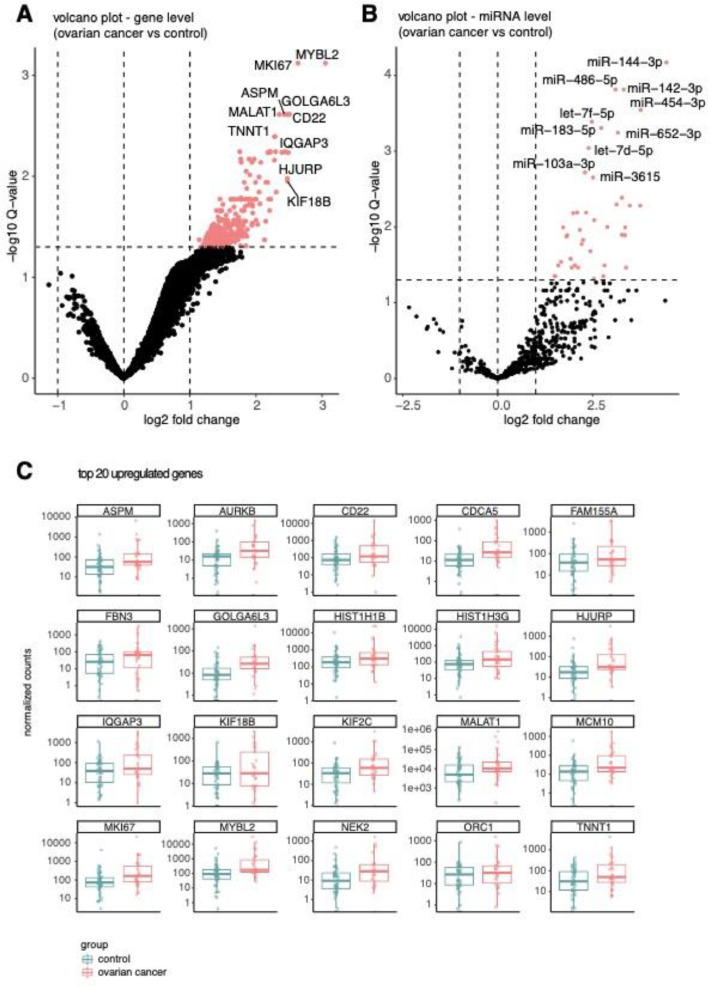
Differential abundance analysis revealed upregulation of mRNAs and miRNAs in cancer samples
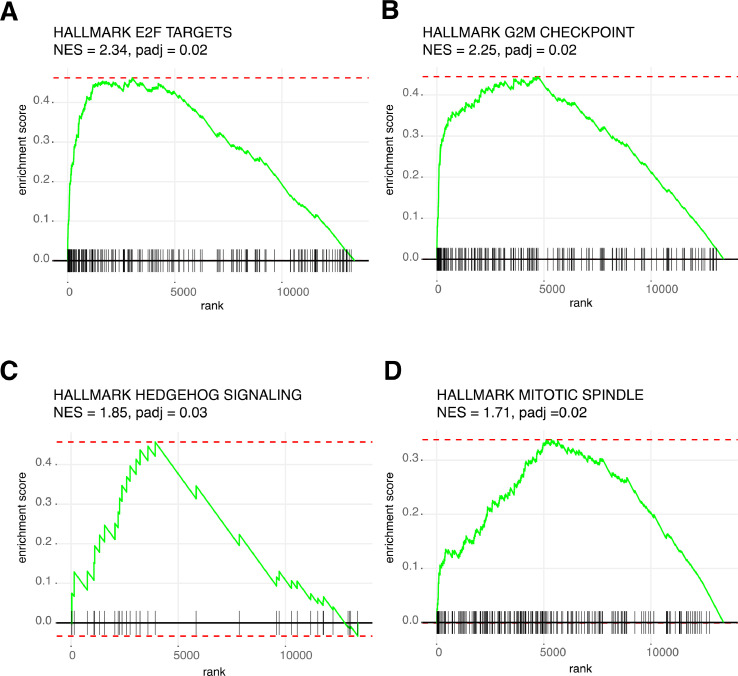
Differential abundance analysis revealed 330 mRNAs that were significantly more abundant in utero-tubal lavage fluid from ovarian cancer patients compared to that from controls (Fig. 2A). Amongst the 330 mRNAs are bona-fide proliferation markers, such as Ki-67 and aurora kinase B (AURKB). A list with the results of the differential abundance analysis can be found in Supplemental Table 4. The normalized abundance of the 20 most differentially abundant mRNAs is shown in Fig. 2B. Of note, principal component analysis of all expressed genes did not reveal clustering based on the clinical diagnosis of the donor (Supplemental Fig. 2). Gene set enrichment analysis of Hallmark gene sets demonstrated a statistically significant enrichment of four gene sets representing cell cycle deregulation: genes encoding cell cycle related targets of E2F transcription factors (normalized enrichment score (NES) = 2.34, padj = 0.02), genes involved in the G2/M checkpoint, as in progression through the cell division cycle (NES = 2.25, padj = 0.02), genes up-regulated by activation of hedgehog signaling (NES = 1.85, padj = 0.03) and genes that are important for mitotic spindle assembly (NES = 1.71, padj = 0.02; Fig. 3). PAX8, CA125 (MUC16) and HE4 (WFDC2), known lineage markers, were not differentially abundant between ovarian cancer patients and control samples (Supplemental Fig. 3). Differential abundance analysis on miRNA level revealed that 41 miRNAs were more abundant in the ovarian cancer group compared to the control group (Fig. 2B, Supplemental Table 4). Five of these miRNAs (let-7d-5p, miR-203a, miR-200b, miR-200c, miR-191) have previously been linked to the pathogenesis of ovarian cancer and were more abundant in plasma, serum or ascites of ovarian cancer patients compared to healthy controls [14].
Fig. 2.
Differentially abundant mRNAs and miRNAs. (A) Volcano plot of differentially abundant mRNAs in ovarian cancer (n = 26) versus controls (n = 48). Upregulated genes with an adjusted p-value of less than 0.05 are shown in pink. No downregulated genes are detected. (B) Volcano plot of differentially abundant miRNAs in ovarian cancer (n = 26) versus controls (n = 48). Upregulated miRNAs with an adjusted p-value of less than 0.05 are shown in pink. No downregulated miRNAs are detected. (C) Boxplots comparing the Sequin spike normalized read counts per group for the top 20 most differentially abundant genes. The normalized read count per sample is shown as a dot. Samples obtained from ovarian cancer patients are pink, samples obtained from controls are blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 3.
Enrichment plots of the four gene sets of the hallmark pathways that are enriched in ovarian cancer versus controls (adjusted p-value < 0.05). (A) Enrichment plot for genes encoding cell cycle related targets of E2F transcription factors (NES = 2.34, padj = 0.02). (B) Enrichment plot for genes involved in the G2/M checkpoint, as in progression through the cell division cycle (NES = 2.25, padj = 0.02). (C) Enrichment plot for genes upregulated by activation of hedgehog signaling (NES = 1.85, padj = 0.03). (D) Enrichment plot for genes important for mitotic spindle assembly (NES = 1.71, padj = 0.02). NES, normalized enrichment score; padj, adjusted p-value.
Differential exon usage analysis identified exons that are more abundant in cancer
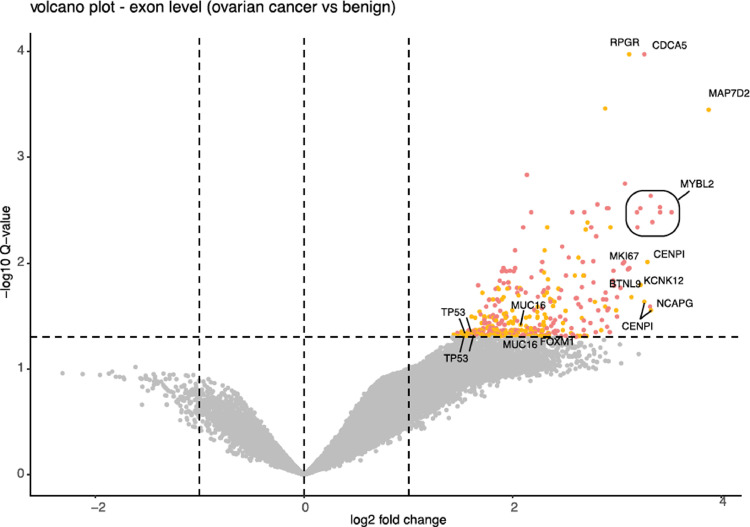
Altered gene expression levels represent only a part of the complex transcriptional program in cancer cells. Alternative splicing, the differential inclusion and exclusion of exonic sequences in mRNA, is an additional mechanism that impacts the transcriptome. In a complementary analysis, we profiled differential exon usage in utero-tubal lavage fluid from cancer and control samples. Differential exon usage analysis revealed 407 exons that were significantly more abundant in utero-tubal lavage fluid from ovarian cancer patients compared to that from controls (Fig. 4). A list with the results of the differential abundance analysis can be found in Supplemental Table 4. Of interest, 203 out of the 407 differential exons did not overlap with differentially abundant genes that were previously identified. Among these differentially abundant exons were exonic sequences belonging to TP53. TP53 is a tumor suppressor gene mutated in over 95% of all ovarian cancer cases, leading to either complete or partial loss of function. The exon segments that are differentially abundant in our cohort match with exon 5 of the main TP53 isoform [44], which encodes for the highly conserved DNA-binding domain of the p53 protein and which contains the majority of the somatic mutations detected in ovarian cancer [45]. Also for MUC16, encoding the CA125 protein, and the oncogene Forkhead box M1 (FOXM1), increased abundance of selected exons was identified in samples from cancer patients.
Fig. 4.
Differentially abundant exons. (A) Volcano plot of differentially abundant exonic parts in ovarian cancer (n = 26) versus controls (n = 48). Upregulated exons with an adjusted p-value of less than 0.05 are shown in pink and yellow. Exonic sequences that belong to genes that are differentially abundant at gene level are shown in pink. Exonic sequences that do not belong to differentially abundant genes are shown in yellow. No downregulated genes are detected. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Utero-tubal lavage fluid does not contain bona-fide ovarian cancer fusion transcripts
Fusion gene analysis of all 74 transcriptomes revealed a total of 414 raw fusion predictions in 26 ovarian cancer samples and 816 raw fusion predictions in 48 control samples. After stringent filtering, 64 high confidence fusion transcripts remained in the cancer group and 173 high confidence fusion transcripts were detected in the control group. A detailed list of the high confidence fusion transcripts is provided in Supplemental Table 5. The median number of high confidence fusion transcripts per control sample was 3 (min 0, max 11) and the median number of high confidence fusion transcripts per cancer sample was 2 (min 0, max 7). No significant difference in the number of fusion transcripts between both groups was detected (Mann-Whitney-U test, two-sided, p-value = 0.11). There was no overlap between the transcripts detected in our cohort and reported fusion transcripts in ovarian cancer [46], [47], [48].
A multi-omics classifier outperforms diagnostic classifiers based on single layer RNA sequencing data
Prior to the pre-processing step, the mRNA data contained 13,386 predictors, the miRNA data contained 1527 predictors and the exon data contained 509 predictors. The misclassification rate, sensitivity, and specificity for the individual data layers and for all possible combinations of the data layers are summarized in supplementary Table 6. The best classifiers were obtained with elastic net logistic regression when the 15 most significant markers were retained from each of the individual data layers after the pre-processing step. The multi-omics classifier based on combined mRNA, miRNA and exon data yielded the best performance, achieving an overall misclassification rate of 21%, a sensitivity of 66% and a specificity of 88%. A receiver operating characteristic (ROC) curve was built by repeating the analysis on 100 random splits of the data into training and test sets with a varying threshold ranging from 0 to 1 (Supplemental Fig. 4). The lowest misclassification rate was achieved with a threshold of 0.5 and resulted in an area-under-the-curve of 0.86. An overview of the most important predictors for the multi-omics classifier is provided in Supplemental Table 7.
Discussion
Utero-tubal liquid biopsy can be collected in a minimally invasive way and is an intriguing fluid to study in the context of ovarian cancer diagnosis, due to its contact with the epithelium of the fallopian tube, where these tumors arise. Here, we provide proof-of-principle that isolating RNA from utero-tubal lavage fluid is technically feasible. The mRNA capture sequencing data that was generated from utero-tubal lavage contains mRNA signatures specific for ovary and fallopian tube and can thus be used to explore liquid biopsy applications for ovarian cancer diagnosis.
Bulk RNA sequencing allows to inspect RNA derived from the tumor as well as RNA representing the complex tumor-microenvironment. Our study revealed an upregulation of mRNAs involved in cell cycle regulation and proliferation in utero-tubal lavage fluid from ovarian cancer patients compared to control samples. Over 300 mRNAs were upregulated in ovarian cancer compared to control samples. V-Myb avian myeloblastosis viral oncogene homolog-like 2 (MYBL2), the most differentially abundant mRNA, showed an 8-fold upregulation in ovarian cancer patients compared with healthy donors. MYBL2 is a physiological regulator of cell proliferation, cell survival and cell differentiation, but it is frequently deregulated in ovarian cancer, contributing to tumorigenesis and progression [49,50]. The marker of proliferation Ki-67 is also among the upregulated genes, probably reflecting the persistent cell proliferation of ovarian cancer cells [51]. Two members of the E2F family of transcription factors (E2F1 and E2F8) are more abundant in ovarian cancer samples compared to controls. Deregulation of E2F transcription factors has been reported as a crucial player in ovarian cancer pathogenesis [51], [52], [53] and E2F1 has been suggested as therapeutic target. At exon level, centromere protein I (CENPI) is more abundant in the cancer group compared to the control group. CENPI is a known target gene of E2F1 that promotes chromosome instability in cancer [54]. Cell division cycle associated gene 5 (CDCA5) and aurora kinase B (AURKB) are upregulated in both utero-tubal lavage of ovarian cancer and in tumor tissue of ovarian cancer patients relative to benign ovarian tissue [55]. Both at gene and at exon level, no downregulated RNA markers were detected, which is in line with the hypothesis that ovarian cancer samples contain tumor derived RNA that is absent in the control samples. Beside identification of differentially abundant genes and exons, we also interrogated the presence of fusion genes in our dataset. In our cohort, high confidence fusion transcripts were detected in both cancer and control samples. Fusion gene analysis in ovarian cancer tissue and in ascites from relapsed patients has been reported in only a few studies and the contribution of fusions in this cancer entity remains unclear [46], [47], [48].
To our knowledge, this is the first time that RNA sequencing has been successfully applied to utero-tubal lavage samples of ovarian cancer patients to profile the extracellular RNA content. Barnabas et al. explored the proteomic profile of extracellular vesicles isolated from utero-tubal lavage fluid and constructed a 9-protein classifier for ovarian cancer diagnosis with 70% sensitivity and 76.2% specificity [25]. None of the 9 proteins that were included in the classifier, showed corresponding upregulation of mRNA in our cohort. It is known that the correlation between mRNA transcripts and generated protein expressions can be low due to differences in half lives and the post transcription machinery. Based on the available mRNA data, miRNA data and exon data, a multi-omics classifier was built to predict ovarian cancer. Combining the three different data layers resulted in the best classifier, with a sensitivity of 66% and specificity of 88%, indicating the added value of combining complementary data layers. A limitation of our study is that we did not have large enough cohort to segregate germline BRCA mutations carriers from BRCA-WT cases and controls, which could have highlighted a more robust classifier. This approach should be taken in future studies, since accumulating data hints that the expressional profile of BRCA-mutated müllerian epithelium significantly differs from the WT pattern.
Another caveat to our study is that CA125 serum levels were lacking. As a result, the performance of our classifier could not be compared with the current gold-standard. A joint analysis of the transcriptomic and proteomic data could thus reveal useful insights that may not be deciphered from the separate analysis of mRNA or protein expressions. Biomarker development efforts to date clearly indicate that no individual biomarker, can provide sufficient sensitivity at high specificity for the early detection of ovarian cancer. In order to identify a robust multi-marker algorithm, it is necessary to explore alternative biofluids, such as utero-tubal lavage, and to combine different -omics approaches for biomarker discovery.
Financial support
E.H. is funded by the Fund for Scientific Research Flanders (FWO; 1133120N). Annelien Morlion is supported by the Special Research Fund (BOF) scholarship of Ghent University (BOF.DOC.2019.0047.01). This work is supported by Kom Op Tegen Kanker (Stand up to Cancer), the Flemish cancer society. K.L. is supported by the Israel Science Foundation (1104/17), the Israel Cancer Research Fund, Israel Cancer Association Early Detection grant, and the SPARK-Tel Aviv University grant.
Data availibility
The RNA-sequencing dataset generated by the authors in the study is available at the European Genome-phenome Archive (EGA) under accession number EGAS00001005498.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgments
Acknowledgments
Prof. Tamar Geiger, Prof. Noam Shomron, Dr. Guy Shapira and Mrs. Georgina Barnabas (Tel Aviv University, Israel) for review of the results and correlation with proteomic profiling of the samples.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.12.008.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M., et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Arts-de Jong M., Harmsen M.G., Hoogerbrugge N., Massuger L.F., Hermens R.P., de Hullu J.A. Risk-reducing salpingectomy with delayed oophorectomy in BRCA1/2 mutation carriers: patients’ and professionals’ perspectives. Gynecol Oncol. 2015;136:305–310. doi: 10.1016/j.ygyno.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Harmsen M.G., Arts-de Jong M., Hoogerbrugge N., Maas A., Prins J.B., Bulten J., et al. Early salpingectomy (TUbectomy) with delayed oophorectomy to improve quality of life as alternative for risk-reducing salpingo-oophorectomy in BRCA1/2 mutation carriers (TUBA study): a prospective non-randomised multicentre study. BMC Cancer. 2015;15:593–601. doi: 10.1186/s12885-015-1597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buys S.S., Partridge E., Black A., Johnson C.C., Lamerato L., Isaacs C., et al. Effect of screening on ovarian cancer mortality: the prostate, lung, colorectal and ovarian (PLCO) cancer screening randomized controlled trial. JAMA. 2011;305:2295–2303. doi: 10.1001/jama.2011.766. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs I.J., Menon U., Ryan A., Gentry-Maharaj A., Burnell M., Kalsi J.K., et al. Ovarian cancer screening and mortality in the UK collaborative trial of ovarian cancer screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945–956. doi: 10.1016/S0140-6736(15)01224-6. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meden H., Fattahi-Meibodi A. CA 125 in benign gynecological conditions. Int J Biol Mark. 1998;13:231–237. doi: 10.1177/172460089801300411. [DOI] [PubMed] [Google Scholar]
- 8.Moore R.G., Miller M.C., Disilvestro P., Landrum L.M., Gajewski W., Ball J.J., et al. Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet Gynecol. 2011;118:280–288. doi: 10.1097/AOG.0b013e318224fce2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dochez V., Caillon H., Vaucel E., Dimet J., Winer N., Ducarme G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J Ovarian Res. 2019:12. doi: 10.1186/s13048-019-0503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu K.H., Skates S., Hernandez M.A., Bedi D., Bevers T., Leeds L., et al. A 2-stage ovarian cancer screening strategy using the Risk of Ovarian Cancer Algorithm (ROCA) identifies early-stage incident cancers and demonstrates high positive predictive value. Cancer. 2013;119:3454–3461. doi: 10.1002/cncr.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore R.G., McMeekin D.S., Brown A.K., DiSilvestro P., Miller M.C., Allard W.J., et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2009;112:40–46. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlan B.Y., Thorpe J., Watabayashi K., Drescher C.W., Palomares M., Daly M.B., et al. Use of CA125 and HE4 serum markers to predict ovarian cancer in elevated-risk women. Cancer Epidemiol Biomark Prev. 2014;23:1383–1393. doi: 10.1158/1055-9965.EPI-13-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sölétormos G., Duffy M.J., Othman Abu Hassan S., Verheijen R.H.M., Tholander B., Bast R.C., et al. Clinical use of cancer biomarkers in epithelial ovarian cancer: updated guidelines from the european group on tumor markers. Int J Gynecol Cancer Off J Int Gynecol Cancer Soc. 2016;26:43–51. doi: 10.1097/IGC.0000000000000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulstaert E., Morlion A., Levanon K., Vandesompele J., Mestdagh P. Candidate RNA biomarkers in biofluids for early diagnosis of ovarian cancer: a systematic review. Gynecol Oncol. 2020 doi: 10.1016/j.ygyno.2020.11.018. Academic Press Inc. [DOI] [PubMed] [Google Scholar]
- 15.Hulstaert E., Morlion A., Avila Cobos F., Verniers K., Nuytens J., Vanden Eynde E., et al. Charting extracellular transcriptomes in the human biofluid RNA atlas. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108552. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura K., Sawada K., Yoshimura A., Kinose Y., Nakatsuka E., Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016:48. doi: 10.1186/s12943-016-0536-0. BioMed Central Ltd.Jun 24page. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiland M., Gao X.H., Zhou L., Mi Q.S., M W., XH G., et al. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol Taylor and Francis Inc. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., Xu S., Liu X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high‑throughput sequencing. Oncol Lett. 2019 doi: 10.3892/ol.2019.10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias K.M., Fendler W., Stawiski K., Fiascone S.J., Vitonis A.F., Berkowitz R.S., et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. Elife. 2017;6:e28932. doi: 10.7554/eLife.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji T., Zheng Z.G., Wang F.M., Xu L.J., Li L.F., Cheng Q.H., et al. Differential microRNA expression by Solexa sequencing in the sera of ovarian cancer patients. Asian Pacific J Cancer Prev. 2014;15:1739–1743. doi: 10.7314/apjcp.2014.15.4.1739. [DOI] [PubMed] [Google Scholar]
- 21.Labidi-Galy S.I., Papp E., Hallberg D., Niknafs N., Adleff V., Noe M., et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. 2017;8:1093. doi: 10.1038/s41467-017-00962-1. Nature Publishing Group; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levanon K., Crum C., Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol. 2008;26:5284–5293. doi: 10.1200/JCO.2008.18.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perets R., Drapkin R. It's totally tubular....riding the new wave of ovarian cancer research. Cancer Res. 2016;76:10–17. doi: 10.1158/0008-5472.CAN-15-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanciu P.I., Ind T.E.J., Barton D.P.J., Butler J.B., Vroobel K.M., Attygalle A.D., et al. Development of peritoneal carcinoma in women diagnosed with serous tubal intraepithelial carcinoma (STIC) following risk-reducing Salpingo-oophorectomy (RRSO) J Ovarian Res. 2019:12. doi: 10.1186/s13048-019-0525-1. J Ovarian Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnabas G.D., Bahar-Shany K., Sapoznik S., Helpman L., Kadan Y., Beiner M., et al. Microvesicle proteomic profiling of uterine liquid biopsy for ovarian cancer early detection. Mol Cell Proteom. 2019;18 doi: 10.1074/mcp.RA119.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maritschnegg E., Wang Y., Pecha N., Horvat R., Van Nieuwenhuysen E., Vergote I., et al. Lavage of the uterine cavity for molecular detection of Müllerian duct carcinomas: a proof-of-concept study. J Clin Oncol. 2015;33:4293–4300. doi: 10.1200/JCO.2015.61.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenzi L., Chiu H.S., Avila Cobos F., Gross S., Volders P.J., Cannoodt R., et al. The RNA atlas expands the catalog of human non-coding RNAs. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00936-1. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016:374. doi: 10.1098/rsta.2015.0202. Philos Trans A Math Phys Eng Sci; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. Genome Res; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M., Sergushichev A. Fast gene set enrichment analysis. Bioinformatics. 2016 [Google Scholar]
- 32.Nicorici D., Satalan M., Edgren H., Kangaspeska S., Murumagi A., Kallioniemi O., et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014:011650. [Google Scholar]
- 33.Dudoit S., Fridlyand J., Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97:77–86. [Google Scholar]
- 34.Regression shrinkage and selection via the Lasso on JSTOR [Internet]. [cited 2021 Oct 20]. Available from: https://www.jstor.org/stable/2346178
- 35.Zou H., Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B Stat Methodol. 2005;67:301–320. [Google Scholar]
- 36.Simon N., Friedman J., Hastie T., Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. American Statistical Association; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou H. The adaptive lasso and its oracle properties. J Am Stat Assoc. 2006;101:1418–1429. Taylor & Francis; [Google Scholar]
- 38.Meinshausen N. Relaxed lasso. Comput Stat Data Anal. 2007;52:374–393. North-Holland; [Google Scholar]
- 39.Zhang CH. Nearly unbiased variable selection under minimax concave penalty. Ann Stat. 2010;38:894–942. [Google Scholar]
- 40.Christidis A.A., Van A.S, Zamar R. Split modeling for high-dimensional logistic regression. 2021;
- 41.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 42.Song L., Langfelder P., Horvath S. Random generalized linear model: a highly accurate and interpretable ensemble predictor. BMC Bioinform. 2013;14:5. doi: 10.1186/1471-2105-14-5. BMC Bioinformatics; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T., International CG-P of the 22nd ACM SIGKDD, 2016 undefined. Xgboost: a scalable tree boosting system. dl.acm.org. Association for Computing Machinery; 2016;13-17-Augu:785–94.
- 44.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. Hum Mutat; [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Cao L., Nguyen D., Lu H. TP53 mutations in epithelial ovarian cancer. Transl Cancer Res. 2016;5:650–663. doi: 10.21037/tcr.2016.08.40. Transl Cancer Res; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christie E., Pattnaik S., Beach J., Copeland A., Rashoo N., Fereday S., et al. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09312-9. Nat Commun; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Earp M.A., Raghavan R., Li Q., Dai J., Winham S.J., Cunningham J.M., et al. Characterization of fusion genes in common and rare epithelial ovarian cancer histologic subtypes. Oncotarget. 2017;8:46891–46899. doi: 10.18632/oncotarget.16781. Impact Journals LLC; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzyzanowski P.M., Sircoulomb F., Yousif F., Normand J., La Rose J., E Francis K., et al. Regional perturbation of gene transcription is associated with intrachromosomal rearrangements and gene fusion transcripts in high grade ovarian cancer. Sci Rep. 2019;9 doi: 10.1038/s41598-019-39878-9. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musa J., Aynaud M.M., Mirabeau O., Delattre O., Grünewald T.G.P. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017;8:e2895. doi: 10.1038/cddis.2017.244. Cell Death Dis;–e2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanner M.M., Grenman S., Koul A., Johannsson O., Meltzer P., Pejovic T., et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res An Off J Am Assoc Cancer Res Nat Biotechnol. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 51.López-Reig R., López-Guerrero J.A. The hallmarks of ovarian cancer: proliferation and cell growth. Eur J Cancer Suppl. 2020;15:27–37. doi: 10.1016/j.ejcsup.2019.12.001. EJC Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhan L., Zhang Y., Wang W., Song E., Fan Y., Wei B., et al. E2F1: a promising regulator in ovarian carcinoma. Tumor Biol. 2016;37:2823–2831. doi: 10.1007/s13277-015-4770-7. Tumour Biol; [DOI] [PubMed] [Google Scholar]
- 53.Eoh K.J., Kim H., Lee J.W., Kim L.K., Park S.A., Kim H., et al. E2F8 induces cell proliferation and invasion through the epithelial–Mesenchymal transition and notch signaling pathways in ovarian cancer. Int J Mol Sci. 2020;21:5813. doi: 10.3390/ijms21165813. Int J Mol Sci; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thangavelu P., Lin C., Vaidyanathan S., Nguyen T., Dray E., Duijf P. Overexpression of the E2F target gene CENPI promotes chromosome instability and predicts poor prognosis in estrogen receptor-positive breast cancer. Oncotarget. 2017;8:62167–62182. doi: 10.18632/oncotarget.19131. Oncotarget; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C., Chen S., Luo M., Yan H., Pang L., Zhu C., et al. The role of the CDCA gene family in ovarian cancer. Ann Transl Med. 2020;8:190. doi: 10.21037/atm.2020.01.99. Ann Transl Med; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.