Abstract
Coagulase gene (coa) short sequence repeat region sequencing was used to measure relatedness among a collection of temporally and geographically diverse methicillin-resistant Staphylococcus aureus isolates. The results show that coa polymorphism is free of strong selective pressure and has a low index of variation that may be useful for long-term epidemiological investigations. coa typing is a useful addition to spa typing for analysis of S. aureus, including methicillin-resistant strains.
Recently, a method for sequencing and analyzing the polymorphic region of the protein A gene of Staphylococcus aureus (spa typing) was described for use in outbreak investigations (13). Although spa typing is appropriate for the study of short-term epidemiology, additional genetic markers with a slower rate of evolution are needed to help relate clonal groups of methicillin-resistant S. aureus (MRSA) isolates from temporally and geographically diverse locations (1, 12). Toward this end, we examined the utility of a second sequence target, the coagulase gene (coa) variable region, for use in conjunction with spa sequencing for the strain typing of MRSA.
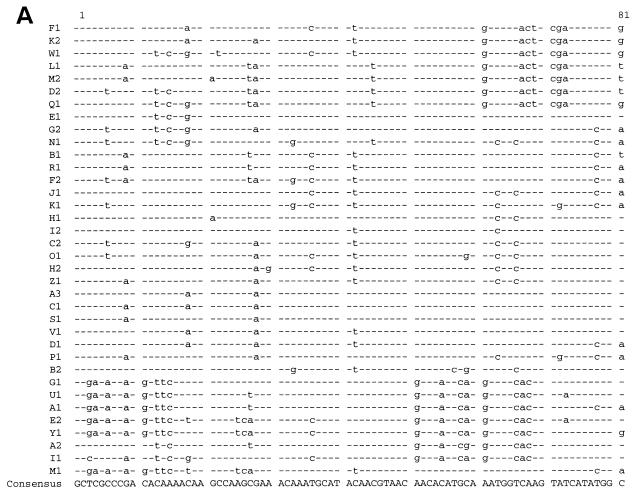
The coagulase protein is an important virulence factor of S. aureus. Like spa, coa has a polymorphic repeat region that can be used for differentiating S. aureus isolates (Fig. 1). The variable region of coa is comprised of 81-bp tandem short sequence repeats (SSRs) (16) that are variable in both number and sequence (2), as determined by restriction fragment length polymorphism analysis of PCR products (2–4).
FIG. 1.
DNA (A) and the deduced amino acid (B) sequences of coa repeats. Identical residues are identified by dashes.
To determine the suitability of coa variability for molecular typing of MRSA, the nucleotide sequences of the coa repeat regions of 54 archived MRSA isolates were determined. Eighteen MRSA isolates (group 1) were chosen for their geographic as well as temporal diversity (6). Another 36 isolates (group 2) represented the breadth of observed spa type diversity seen in MRSA isolates from New York City hospitals (Table 1) (13). Strain discrimination and the grouping of isolates determined by coa and spa typing were compared.
TABLE 1.
Characteristics of group 1 and 2 isolates
| Group | City (borough) | State or provincea | Country | Yr(s) of isolation |
spab
|
coab
|
spa and coa groupc | ||
|---|---|---|---|---|---|---|---|---|---|
| Type | Repeats | Type | Repeats | ||||||
| II | New York | NY | United States | 1996 | 20 | YHB2CMBQBLO | 11 | MNPQ | C |
| I | San Antonio | TX | United States | 1986–1987 | 7 | YHGCMBQBLO | 11 | MNPQ | C |
| II | New York (Queens) | NY | United States | 1996 | 7 | YHGCMBQBLO | 11 | MNPQ | C |
| II | New York (Brooklyn) | NY | United States | 1996 | 7 | YHGCMBQBLO | 11 | MNPQ | C |
| II | New York | NY | United States | 1996 | 7 | YHGCMBQBLO | 11 | MNPQ | C |
| I | London | Ontario | Canada | 1997 | 40 | YFGFMBQBLO | 3 | MNOPQ | C |
| I | London | England | 1989 | 55 | YGFMBQBLQBLPO | 3 | MNOPQ | C | |
| I | Denmark | 1960s | 4 | YHFGFMBQBLO | 3 | MNOPQ | C | ||
| I | Toronto | Ontario | Canada | 1986 | 4 | YHFGFMBQBLO | 3 | MNOPQ | C |
| II | New York | NY | United States | 1996 | 4 | YHFGFMBQBLO | 3 | MNOPQ | C |
| I | London | England | 1960s | 1 | YHGFMBQBLO | 3 | MNOPQ | C | |
| I | New York | NY | United States | 1986–1987 | 1 | YHGFMBQBLO | 3 | MNOPQ | C |
| II | New York | NY | United States | 1996 | 1 | YHGFMBQBLO | 3 | MNOPQ | C |
| I | New York | NY | United States | 1990 | 48 | YHGFMBQBLO | 3 | MNOPQ | C |
| I | Denmark | 1960s | 59 | YHGFMBQBLO | 3 | MNOPQ | C | ||
| I | Dublin | Ireland | 1991 | 46 | YMBQBLO | 3 | MNOPQ | C | |
| I | New York | NY | United States | 1994 | 4 | YHFGFMBQBLO | 3 | MNOPQ | C |
| I | Dublin | Ireland | 1991 | 1 | YHGFMBQBLO | 3 | MNOPQ | C | |
| I | Geneva | Switzerland | 1962 | 1 | YHGFMBQBLO | 3 | MNOPQ | C | |
| I | Cairo | Egypt | 1961 | 1 | YHGFMBQBLO | 3 | MNOPQ | C | |
| I | Uganda | 1966 | 1 | YHGFMBQBLO | 3 | MNOPQ | C | ||
| I | New York (Brooklyn) | NY | United States | 1990 | 12 | TJMGMK | 3 | MNOPQ | C |
| I | Iowa City | IA | United States | 1986–1987 | 8 | UJ | 8 | USVDW | |
| I | San Francisco | CA | United States | 1986–1987 | 9 | UGFMEEBBPB | 12 | E2F2G2H212J2K2 | |
| I | Toronto | Ontario | Canada | 1996 | 30 | XKAKAOM | 14 | GHIJKM2 | B |
| II | New York (Staten Island) | NY | United States | 1996 | 19 | XKAKAOMQ | 14 | GHIJKM2 | B |
| I | Japan | 1981 | 43 | WGKAKAOMQ | 2 | GHIJKL | B | ||
| I | New York (Brooklyn) | NY | United States | 1990 | 3 | WGKAOMQ | 2 | GHIJKL | B |
| I | Davisd | CA | United States | 1978 | 3 | WGKAOMQ | 2 | GHIJKL | B |
| I | London | England | 1989 | 3 | WGKAOMQ | 2 | GHIJKL | B | |
| II | New York (Brooklyn) | NY | United States | 1996 | 3 | WGKAOMQ | 2 | GHIJKL | B |
| I | Montreal | Quebec | Canada | 1989 | 44 | WGMQ | 2 | GHIJKL | B |
| I | New Orleans | LA | United States | 1986–1987 | 6 | A2AKBEKBKB | 10 | YZA2B2C2D2 | D |
| I | Toronto | Ontario | Canada | 1996 | 31 | XKAKBBEBKB | 10 | YZA2B2C2D2 | D |
| I | Toronto | Ontario | Canada | 1996 | 15 | A2AKEEMBMK | 18 | YZD2 | D |
| I | London | Ontario | Canada | 1997 | 15 | A2AKEEMBMK | 18 | YZD2 | D |
| I | Memphis | TN | United States | 1986–1987 | 10 | TIMBMDMGMK | 4 | AREDEF | A |
| I | Dublin | Ireland | 1990 | 47 | TMDMGMK | 21 | ARCDEF | A | |
| I | Winnepeg | Manitoba | Canada | 1989 | 56 | SMBDMGMK | 6 | ABSDEF | A |
| I | Edmonton | Alberta | Canada | 1989 | 45 | TJMBDMGMK | 6 | ABSDEF | A |
| II | New York (Bronx) | NY | United States | 1996 | 29 | TJMBMDMGGMK | 1 | ABCDEF | A |
| II | New York | NY | United States | 1996 | 23 | TJMBMDMGK | 1 | ABCDEF | A |
| I | New York | NY | United States | 1990 | 2 | TJMBMDMGMK | 1 | ABCDEF | A |
| I | Davis | CA | United States | 1976 | 2 | TJMBMDMGMK | 1 | ABCDEF | A |
| II | New York | NY | United States | 1996 | 2 | TJMBMDMGMK | 1 | ABCDEF | A |
| II | New York | NY | United States | 1996 | 2 | TJMBMDMGMK | 1 | ABCDEF | A |
| II | New York (Bronx) | NY | United States | 1996 | 2 | TJMBMDMGMK | 1 | ABCDEF | A |
| II | New York | NY | United States | 1996 | 2 | TJMBMDMGMK | 1 | ABCDEF | A |
| I | New York (Brooklyn) | NY | United States | 1991 | 14 | TJMBMDMGMKK | 1 | ABCDEF | A |
| I | Memphis | TN | United States | 1986–1987 | 11 | TJMBMDMMK | 1 | ABCDEF | A |
| II | New York (Brooklyn) | NY | United States | 1996 | 26 | TJMBMGMK | 1 | ABCDEF | A |
| II | Stony Brook | NY | United States | 1996 | 25 | TJMDMGMK | 1 | ABCDEF | A |
| II | New York | NY | United States | 1996 | 28 | TKJMBMDMGMKK | 1 | ABCDEF | A |
| II | Westchester | NY | United States | 1996 | 27 | TMDGMMK | 15 | UA3DW | A |
NY, New York; CA, California; TN, Tennessee; LA, Louisiana; IA, Iowa.
spa and coa types are arbitrarily assigned for each unique sequence identified. Repeat codes are derived from organization of individual repeats (random alphabetical code). To make the notation shorter, letter codes ending with the digit “1” have been simplified by removing the number. It should be noted that two isolates were assigned separate spa types (48 and 59), despite an identical repeat pattern. They contained a sequence polymorphism in a flanking region that is normally conserved (not shown).
spa and coa groups are organized based on sequence alignment and congruence of the spa and coa repeat similarity. The assignment of a particular spa or coa group (A, B, C, and D) was based on the reported spa repeat similarity groupings of New York City MRSA (Shopsin et al. [13]).
Davis, University of California at Davis.
coa and spa sequencing and analysis were performed as described previously (13). The forward and reverse primers used for amplification and direct sequencing of the coa variable-region were coaF (5′-TGCTGGTACAGGTATCCGTGAAT-3′) and coaR (5′-AGAAGCACATAGAATGCATGA-3′), respectively. Primers were designed from available sequences (5) (GenBank accession no. X167457). coa (and spa) repeats were analyzed using the program FINDPATTERNS from Genetics Computer Group Wisconsin Package 9.1. The following ambiguous search sequence identified all unique coagulase variable region repeats in the isolates analyzed: GCN{4}CCN{8}ANAANNCN{5}ANANNAANGCNNA NAANGNNACNACNNAN{5}ANGGN{11}ANNGN. A repeat type, designated by an alphabetical code, was assigned to each of the identified repeats (e.g., repeat A1, B1, C2…). coa strain types were then defined by the string of repeat types (from the 5′ to the 3′ ends of the repeat region) that represented the organization of the coa SSR region of each of the strains [e.g., coa repeat types A(1)+B(1)+C2+C2+E3 = coa strain type (#)-ABC2C2E3; to make the notation shorter, letter codes ending in 1 have been simplified by removing the number]. All unique coa strain types were assigned a numerical code.
In the coa variable regions of group 1 and 2 isolates, sequence determination and analysis identified 36 different coa repeat types, representing 18 amino acid sequence combinations (Fig. 1). spa SSRs showed less diversity, with 24 unique repeat types identified (spa repeat sequences not shown).
Since the coa gene product is a phenotypic trait that may have a role in virulence, selective forces could affect the rate of coa repeat evolution, uncoupling it from that of the overall S. aureus genome and obscuring genetic relationships based on coa variation. To evaluate the selective forces acting on repeat units, synonymous and nonsynonymous substitution rates were determined using the GCG program DIVERGE, which analyzes pairwise, codon-by-codon comparisons of protein coding sequences based on the method by Li et al. (7). Substitution rates were calculated from aligned repeat sequences (Fig. 1), which were created using the GCG program PILEUP. coa repeats had a higher number of synonymous substitutions per synonymous site (ds = silent) relative to nonsynonymous substitutions per nonsynonymous site (dn = amino acid altering), indicating an average ds/dn ratio of 6.3 for coa. A ds/dn ratio near 1 (the neutral expectation) suggests that coa repeats are subject to weak conservation. In agreement with this hypothesis, amino acid replacements maintained the same hydrophobicity, indicating that polymorphisms may be selectively neutral. Thus, as with spa (13), coa repeats do not evolve from positive selection, i.e., variability is likely to be uncoupled from environmental selection.
As mentioned above, the organization of coa and spa repeats in the SSR regions of each isolate was used to determine a spa or coa strain type. A total of 13 unique coa and 33 spa types were identified, and coa polymorphic regions varied in size from three to seven repeats (Table 1). Together, spa and coa typing resulted in 33 strain types (the same as spa alone). Thus, coa typing did not distinguish isolates with the same spa type. Even though spa has fewer repeat motifs (or greater repeat conservation), its variable region is more polymorphic and therefore more discriminatory than that of coa, apparently due to a greater rate of repeat recombination. Each spa type was associated with only one coa type, which, given the temporal and geographic diversity of the isolates, suggests that in MRSA, primary differentiation of coa is for the most part followed by polymorphism of spa.
Distinguishing MRSA isolates for outbreak investigation requires the analysis of rapidly evolving markers, which can obscure relationships among clones that have had more time to diverge (15). The slower rate of change (clock speed) associated with coa typing (it has less discrimination than spa typing [13]) is more appropriate for answering questions of global epidemiology. Thus, coa typing can be used to enhance the value of spa typing by providing more supported inferences on strain lineage and clonality among isolates with similar or identical spa repeat organization, since congruence between different markers is an indication of linkage disequilibrium or clonality (coa and spa are estimated to be approximately 100 kb apart and are therefore unlikely to be coinherited in a single transfer event (Steve Gill [TIGR] personal communication). The use of more than one genetic marker for relating strains is desirable (14) and likely to become increasingly important because recombination will eventually diversify MRSA to the extent that clonal types within a given region can no longer be distinguished by a single locus.
To determine congruence between coa and spa typing results among the population sampled, isolates were organized based on the similarity of their coa and spa repeat regions (Table 1), which placed 53 of 54 isolates into four major groups (labeled groups A, B, C, and D). This association was based on the assumption that accumulated point mutations are consistent between strains and thus may be an indication of more recent common ancestry (13). Because of the high sequence identity among coa repeat regions in our study isolates, the same major groupings may be obtained by dendrograms constructed from progressive, pairwise alignments of coa sequences (not shown). However, since more extensive duplication or deletion of repetitive units obviates comparison using algorithms based on alignments, visual analysis of repeat organization is a more simple and reliable means of relating strains.
The apparent nonrandom association between genetic markers (linkage disequilibrium) among isolates from diverse temporal and geographic locations suggests only four predominant lineages among MRSA strains. This striking observation may be explained by at least three hypotheses. First, there may simply be four primary lineages of the coa gene and/or S. aureus. Second, the predominance of particular clones may result from a longer association of the mec resistance element with the chromosomes of particular isolates (9) and the possibility of a low-rate of horizontal transfer of mec DNA among S. aureus lineages. A third but not mutually exclusive hypothesis is that not all MRSA clones are equally fit.
In addition, it appears that local (New York City) MRSA populations can contain the full extent of genetic variation that exists worldwide. The global recovery of the same genotypes is an indication of restricted recombination, possibly related to the lack of natural transformability of S. aureus (10, 11). Recovery of the breadth of global clonal types, however, may only be common in urban regions. Elsewhere, geographic isolation probably creates even more homogenous hospital populations, which are difficult to discriminate. Thus, the effectiveness of typing for determining the likelihood of strain transmission will have strong regional variation.
In summary, current methods for tracking the spread of MRSA over large geographic areas commonly require the use of multiple techniques that lack many of the inherent advantages in terms of interpretation, databasing, and portability of results afforded by sequence-based methods. The multilocus sequence typing technique combines sequence information from several housekeeping genes (approximately seven to eight genes) in a manner similar to multilocus enzyme electrophoresis (8). However, because of the polymorphism associated with repeat regions, SSR sequencing can discriminate isolates with a minimum of loci (as well as nucleotides), and strain types can be described using a simple numerical format and alphabetical repeat designation that lends itself to use in a wide range of laboratories (13). coa typing now expands the role of SSR sequence typing from infection control and outbreak investigation (via the spa locus) by providing the clinical microbiologist and researcher with the opportunity to use a simple, rapid, and practical method to monitor variation in MRSA populations and relate strains from divergent regions and time periods.
Acknowledgments
We thank Karl Drlica and J. P. Bifani for their careful reading and critical comments on the manuscript and Steve Gill of The Institute for Genomic Research (TIGR) for his genomic mapping data.
REFERENCES
- 1.Ayliffe G A. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24(Suppl. 1):S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 2.Goh S H, Byrne S K, Zhang J L, Chow A W. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hookey J V, Edwards V, Cookson B D, Richardson J F. PCR-RFLP analysis of the coagulase gene of Staphylococcus aureus: application to the differentiation of epidemic and sporadic methicillin-resistant strains. J Hosp Infect. 1999;42:205–212. doi: 10.1053/jhin.1999.0595. [DOI] [PubMed] [Google Scholar]
- 4.Hookey J V, Richardson J F, Cookson B D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaida S, Miyata T, Yoshizawa Y, Igarashi H, Iwanaga S. Nucleotide and deduced amino acid sequences of staphylocoagulase gene from Staphylococcus aureus strain 213. Nucleic Acids Res. 1989;17:8871. doi: 10.1093/nar/17.21.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 7.Li W H, Wu C I, Luo C C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 8.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musser J, Selander R. Genetic analysis of natural populations of Staphylococcus aureus. New York, N.Y: VCH; 1990. [Google Scholar]
- 10.Musser J M, Kroll J S, Moxon E R, Selander R K. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci USA. 1988;85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Rourke M, Stevens E. Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J Gen Microbiol. 1993;139(Pt. 11):2603–2611. doi: 10.1099/00221287-139-11-2603. [DOI] [PubMed] [Google Scholar]
- 12.Roberts R B, Tennenberg A M, Eisner W, Hargrave J, Drusin L M, Yurt R, Kreiswirth B N. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb Drug Resist. 1998;4:175–183. doi: 10.1089/mdr.1998.4.175. [DOI] [PubMed] [Google Scholar]
- 13.Shopsin B, Gomez M, Montgomery S O, Smith D H, Waddington M, Dodge D E, Bost D A, Riehman M, Naidich S, Kreiswirth B N. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R, et al. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]