Abstract
The severe acute respiratory syndrome (SARS) coronavirus encodes several RNA-processing enzymes that are unusual for RNA viruses, including Nsp15 (nonstructural protein 15), a hexameric endoribonuclease that preferentially cleaves 3′ of uridines. We solved the structure of a catalytically inactive mutant version of Nsp15, which was crystallized as a hexamer. The structure contains unreported flexibility in the active site of each subunit. Substitutions in the active site residues serine 293 and proline 343 allowed Nsp15 to cleave at cytidylate, whereas mutation of leucine 345 rendered Nsp15 able to cleave at purines as well as pyrimidines. Mutations that targeted the residues involved in subunit interactions generally resulted in the formation of catalytically inactive monomers. The RNA-binding residues were mapped by a method linking reversible cross-linking, RNA affinity purification, and peptide fingerprinting. Alanine substitution of several residues in the RNA-contacting portion of Nsp15 did not affect hexamer formation but decreased the affinity of RNA binding and reduced endonuclease activity. This suggests a model for Nsp15 hexamer interaction with RNA.
The Nidoviruses contain three families of viruses, including the Coronaviridae that cause numerous diseases in humans (1). Severe acute respiratory syndrome coronavirus (SARS-CoV)3 is a member of the Coronavirus genus (2, 3). It originated from animals but spread to humans, causing severe respiratory distress with a fatality rate of ∼10% (as shown by the World Health Organization, www.who.int/csr/sars/country/en/country2003_08_15.pdf). In addition to their medical importance, coronaviruses are of interest for their large ∼30-kb positive-strand genome and novel mechanisms that have evolved to replicate and transcribe this large RNA (5, 6). In keeping with the novel strategies used, coronaviruses encode several unusual RNA-processing enzymes, including an RNA endoribonuclease, an RNA methyltransferase, a second RNA-dependent RNA polymerase that generates primers for coronavirus replication (7), and a mechanism to decrease replication errors (8).
Coronavirus subgenomic RNAs are particularly interesting in that they all have the same 5′ leader sequence derived from the 5′ end of the genomic RNA. This organization requires recombination as part of transcription. Various mechanisms have been proposed for subgenomic RNA production, but a discontinuous transcription mechanism is increasingly favored (5). This model proposes that transcription regulatory sequences in the minus-strand RNA direct translocation of the ternary complex to the 5′ leader sequence, where transcription resumes. The minus-strand RNAs serves as the template for subgenomic RNA transcription. Ribonucleases that process the RNA intermediates for transcription have been proposed (9, 10, 11), but the mechanism for the process is still not completely understood.
Nsp15 (nonstructural protein 15) was predicted to be an RNA endoribonuclease as part of a bioinformatics analysis of the SARS-CoV genome (7). Substitutions of critical residues built into infectious clones of the human Coronavirus resulted in decreased infectivity and reduced genome RNA levels (12). A more thorough analysis of mutations in the Nsp15 ortholog of the related Arterivirus revealed that several residues, including those in the putative active site, reduced viral plaque size and decreased viral titers by up to five logs (13). Similar results were observed with Nsp15 of mouse hepatitis virus A59 (14). In both arterivirus and mouse hepatitis virus, reduced subgenomic RNA levels were associated with mutations in Nsp15. Other properties of virus infection were also affected. Therefore, it is likely that Nsp15 and its orthologs play multiple roles in viral infection (14).
To better understand how the activity of Nsp15 affects processing of viral RNAs, Bhardwaj et al. (15) and Ivanov et al. (12) independently produced recombinant Nsp15 of the SARS-CoV in Escherichia coli and demonstrated endoribonuclease activity that preferentially cleaves RNAs at uridylates. This activity was stimulated by Mn2+ but not by other divalent metals such as Mg2+ (15). More recently, Bhardwaj et al. (16) demonstrated that Nsp15 cleaves 3′ of uridylates through the formation of a 2′-3′ cyclic phosphodiester product.
Ribonucleases are generally grouped on the basis of whether they cleave 5′ or 3′ of the cognate phosphodiester (17). The former includes RNase H and RNase III, whereas the latter includes members of the RNase A family and RNase T1 (18). The two types of enzymes also differ in their metal ion dependence. The RNase H and III enzymes are metal-dependent, whereas RNase A and T1 are metal-independent (19). Thus, Nsp15 does not obviously belong to either group. Its cleavage is similar to RNase A, but its activity is enhanced by metal ion. This combination of properties is shared by XendoU, an endoribonuclease involved in small nucleolar RNA biosynthesis (19).
An unusual feature of Nsp15 is that it assembles into a hexamer in solution and that disruption of hexamer formation reduces endoribonuclease activity (20). Transmission electron microscopy analysis showed that Nsp15 consists of a dimer of trimers that interact in an end-to-end fashion (16). A low resolution co-crystal of Nsp15 and RNA suggested that the RNA binds to the outside of the hexamer and interacts with both trimers (16).
Two Nsp15 structures have been determined. Ricagno et al. (21) determined the x-ray structure of the SARS-CoV Nsp15 protein from crystals with one protomer in an asymmetric subunit. The crystal structure of mouse hepatitis virus Nsp15 was solved by Xu et al. (22), also with one subunit in the asymmetric unit. When arranged into a hexamer, the six active sites are exposed at the surface of the hexameric structure. Furthermore, the active site of Nsp15 can be modeled according to the residues of RNase A that share a common mechanism of catalysis (21, 22). In this structure, the C-terminal tail of Nsp15 folds back on the active site, suggesting that it may play a role in specific recognition of the cognate uridylate. These crystal structures, however, lack RNA, and the details of RNA interaction and many requirements of the subunit interactions remain unclear.
Using the coordinates of Ricagno et al. (21), we solved the structure of an active site mutant of the SARS-CoV Nsp15 by molecular replacement. Notably, this structure has six subunits within the asymmetric unit. Predictions of the structure were examined, leading to elucidation of the requirements for specificity in RNA cleavage, subunit interaction, and RNA binding.
EXPERIMENTAL PROCEDURES
Crystallization and Data Collection of Nsp15-H234AInitial crystallization trials were performed in sitting drops using 15 mg/ml of the Nsp15 mutant, H234A. The Hydra 96 robotic system (Robbins Scientific Co.) was used for Hampton and Wizard (Emerald Biostructures) crystallization screens. Three different conditions, all containing 2-methyl-2,4,-pentanediol as the precipitant, were judged promising for further optimization by the hanging drop vapor diffusion method of crystallization. Diffraction quality crystals were obtained at 10-15 mg/ml of H234A in 0.05-0.1 m MgCl2, 3-5% 2-methyl-2,4,-pentanediol, and 0.1 m tri-sodium citrate dehydrate (pH 5.6-5.9). The best crystal diffracted up to 2.64 Å at the synchrotron beam line 14-ID of Advance Photon Source, Chicago (Table 1 ). The crystal was mounted in cryoloop and flash-cooled in a liquid N2 stream after a brief soak in 30% ethylene glycol. The diffraction data set was indexed with unambiguous solution and processed using HKL2000 (23).
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Cell dimensions (Å) | a = b = 305.757 and c = 88.74 |
| Space group | R3 |
| Number of molecules per asymmetric unit (Z) | 6 |
| Resolution (Å) | 50-2.8 |
| Completeness (%)a | 92.0 (78.2) |
| Rsym (%)b | 6.5 (37.2) |
| I/sigma(I) | 17.5 (2.6) |
| Refinement statistics | |
| Resolution (Å) | 50-2.8 |
| Number of reflections in working data set | 66521 |
| Number of reflections in the test data set | 3507 |
| Rcryst (%)c | 19.7 |
| Rfree (%) | 25.3 |
| Ramachandran statistics | |
| Most favored (%) | 90 |
| Allowed (%) | 10 |
| Root mean square deviation | |
| From ideal bond lengths (Å) | 0.015 |
| From ideal bond angle (°) | 0.8 |
The values in parenthesis represent values for the highest resolution shell.
Rsym = ∑h∑i|Ih,i-<I>|/∑h∑iIh,i, where Ih,i is the ith observation of the reflection h2 whereas <Ih> is the means intensity of reflection h.
Rcryst = ∑|Fo|-|Fc|/|Fo|. Rfree was calculated with a fraction (5%) of randomly selected reflections excluded from refinement.
Structure Determination and RefinementThe crystal unit cell indicated the presence of six molecules in the R3 space group per asymmetric unit. The positions of each molecule were solved by molecular replacement using the program Phaser v.1.3 (24) with the previously published catalytically active structure of Nsp15 (Protein Data Bank code 2H85) (21) as the search model. Model building was accomplished using Xtalview (25) to examine the 2Fo - F c, F o - F c, and composite omit electron density maps and refinement using maximum likelihood restrained refinement method in CCP4-REFMAC (26, 27) were performed. After the model reached R factors of less than 30%, water molecules were added to the structure using automated water picking in Xtalview. Medium or tight noncrystallographic averaging increased the R factors, and therefore only loose noncrystallographic averaging restraints were applied during the REFMAC refinement. The final structure has an R factor of 19.7% and an R free value of 25.3% with good stereochemistry as analyzed by PROCHECK (28). The final refinement statistics are in Table 1. The figures were prepared using Spock (quorum.tamu.edu) and Chimera (30). The atomic coordinates of the final model have been deposited in the Protein Data Bank with the code 2RHB.
Molecular DockingDock 6.0 and its accessory programs (31) were used to carry out the flexible docking of 3′-UMP or CMP (from Protein Data Bank code 4RSK-RNASE A:3′UMP complex structure) into the Nsp15 crystal structure (Protein Data Bank code 2H85), which was kept rigid. DMS (32), a software that computes molecular surface, was used to create the molecular surface of the receptor. The negative image of the binding site was defined using Sphgen (31) within the 10 Å radius of the catalytic site residues His234, His249, Ser293, Lys289, and Tyr342 to adopt the sphere-matching algorithm of Dock 6.0. An incremental construction (Anchor-and-Grow method) was used to allow for flexibility of the ligand. Automatic matching mode was used with the 20 configurations/ligand building cycle. Interaction between the ligand and the catalytic site was evaluated by the grid score (33, 34) followed by visual inspection.
Constructing the Nsp15-S293A ModelThe atomic coordinates of Nsp15 crystal structure (Protein Data Bank code 2H85) were used to construct a model of Nsp15-S293A using Modeler (Insight II, www.accelrys.com). Residues within a 10 Å radius of A293 were subjected to a medium level simulated annealing optimization with respect to the variable target function of Modeler.
Site-directed Mutagenesis and Protein PurificationAmino acid substitutions were introduced in the Nsp15 expression plasmid using the QuikChange site-directed mutagenesis protocol as recommended by Stratagene. The entire open reading frame was sequenced to confirm the presence of directed mutation and absence of unintended mutations. WT and mutant proteins with a His tag at their N termini were expressed in E. coli and purified by metal ion affinity chromatography, followed by Mono Q ion exchange chromatography as described previously (15). The purified proteins were stored in 50 mm Tris (pH 7.9), 300 mm NaCl, 1 mm dithiothreitol, 50% (v/v) glycerol at -20 °C. The protein concentrations were determined by absorbance at 280 nm.
Size Exclusion ChromatographyWT or mutant protein samples (∼100 μg) were filtered through a Superdex 200 column using a Pharmacia fast protein liquid chromatography system calibrated with the following molecular mass standards (kDa in parentheses): blue dextran (2000); thyroglobulin (660); ferritin (440); catalase (232); aldolase (140); bovine serum albumin (67); and chymotrypsinogen (25) according to the protocol described in Guarino et al. (20).
Reversible Protein-RNA Cross-linking and Peptide MappingH249A (10 μm) was incubated with or without 20 μm biotinylated RNA, Bio-rUC10 in 20 mm Hepes (pH 7.5), 5 mm MnCl2, and 1 mm dithiothreitol at room temperature for 5 min followed by the addition of formaldehyde to a final concentration of 0.1% in a 25-μl reaction. The cross-linking reaction was stopped by the addition of glycine at 0.2 m final concentration. After 5 min, sequencing grade trypsin (Trypsin Gold; Promega, Madison, WI) was added at a protease:substrate ratio 1:50 (w/w) in 100 mm NH4HCO3 (pH 7.8) at 37 °C and allowed to digest overnight. Streptavidin magnetic beads (New England Biolab, Beverly, MA) were used to capture biotinylated RNA and RNA-peptide conjugates as described previously by Kim et al. (4). RNA-peptide conjugates were reversed by incubating the samples at 70 °C for 1 h. The samples were centrifuged at 3000 × g for 5 min, and the supernatants containing the peptides were desalted using a Ziptip (Millipore, Bedford, MA). The bound peptides were eluted in 2.5 μl of 70% acetonitrile and 0.1% trifluoroacetic acid. The eluted samples were analyzed by MALDI-ToF.
Affinity Chromatography10 μg of WT or mutant proteins were incubated on ice with ∼2 mg polyU-agarose in buffer A (50 mm Tris, pH 7.5, 5% glycerol, 1 mm β-mercaptoethanol, 10 mm MnCl2) containing 30 mm NaCl. After 90 min, the agarose beads were washed by centrifugation at 6,000 rpm for 10 min to remove unbound protein. Bound protein was eluted with buffer A containing increasing concentrations of 50, 100, 200, 300, 400, and 500 mm NaCl. The fractions were analyzed by SDS-polyacrylamide gel electrophoresis followed by staining with Coomassie Brilliant Blue. Protein was quantitated using the Coomassie Plus Protein Assay kit from Pierce.
Endoribonuclease AssaysThe real time endoribonuclease assay used a substrate purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). The substrate has a 5′ carboxyl fluorescein fluorophore at the 5′ end and tetramethylrhodamine at the 3′ end, which quenches 5′ carboxyl fluorescein fluorescence. Of the four nucleotides, only the cognate cleavage nucleotide is a ribonucleotide, whereas the others are deoxyribonucleotides. Protein and RNA substrate were incubated in 20 mm Hepes (pH 7.5), 5 mm MnCl2, 1 mm dithiothreitol, as indicated. Substrate cleavage was monitored with an excitation wavelength of 492 nm and an emission wavelength of 518 nm (16). Changes in fluorescence were measured over time using an LS55 spectrometer (PerkinElmer Life Sciences).
RESULTS AND DISCUSSION
The Crystal Structure of Nsp15 Mutant H234AA mutant version of Nsp15 protein with an alanine substitution for one of the active site histidines, H234A, was deemed suitable for crystallization because it could be expressed and purified at a level more than 10-fold higher levels than the WT Nsp15. Furthermore, it, like WT Nsp15, purified as a hexamer (20) and was competent for RNA binding (16).
A single crystal of H234A protein, hereafter named Nsp15H, diffracted up to 2.64 Å with unit cell parameters a = b = 305.757 and c = 88.740 in the R3 space group. The asymmetric unit contained six molecules, and the positions of each subunit of Nsp15H were solved by molecular replacement using the structure of the catalytically active Nsp15 V291G-D300G double mutant (Protein Data Bank code 2H85) (21) as the search model. The Ricagno et al. (21) model, hereafter named Nsp15VD, has only one molecule/asymmetric unit. The asymmetric unit of Nsp15H provides additional insight into the heterogeneity among individual subunits of the hexamer and allows comparison with the WT active site in Nsp15VD.
The overall structure of Nsp15H is very similar to that of Nsp15VD, with a root mean square deviation difference of 0.53-0.73 Å (344Cα atoms of each subunit of Nsp15 H were super-imposed on Nsp15VD structure using the LSQKAB routine of CCP4 (26)). Both Nsp15H and Nsp15VD consist of a dimer of trimers, which is consistent with the solution state and with single Nsp15 particles analyzed by transmission electron microscopy and cryoelectron microscopy (16).
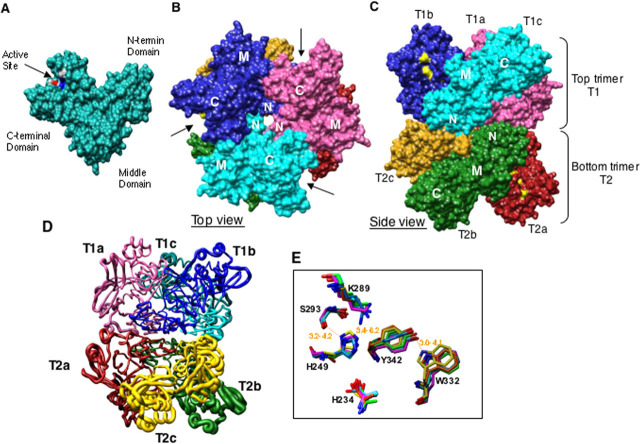
Each subunit contains nine α-helices and twenty-one β-strands. A subunit is further organized into three domains: an N-terminal domain (residues 1-61), a middle domain (residues 62-181), and a C-terminal domain (residues 182-345) (Fig. 1A ). Alanine substitutions of highly conserved residues have demonstrated that the C-terminal domain contains the active site (20), which faces away from the center of the hexamer and contains the extreme C-terminal residues, consistent with the observations of Ricagno et al. (21). When viewed from the top of the hexamer, a pore through the trimer is evident, with the N-terminal domains of the trimer lining the bottom of the pore (Fig. 1B). This pore has an inner diameter of ∼12 Å and does not interact with the substrate RNA according to the cryoelectron microscopy structure (16). The middle and the N-terminal domains have extensive contacts with the other subunits of the hexamer. These interactions are evident in the side view of the Nsp15 hexamer (Fig. 1C).
FIGURE 1.
Structure of SARS-CoV Nsp15.A, surface representation of a subunit showing three domains. Active site residues are colored by element and indicated by an arrow. B, arrangement of six subunits from top view of hexamer. N-terminal, middle, and C-terminal domains are indicated as N, M, and C, respectively. The arrows indicate the positions of catalytic sites in top trimer. C, side view of hexamer showing arrangement and N- to N-terminal interaction of the top trimer (T1) with bottom trimer (T2). Six subunits are labeled as a-c and are colored as follows: T1a, pink; T1b, blue; T1c, cyan; T2a, red; T2b, green; T2c, golden. This color scheme is used throughout the figures. Catalytic residues are colored yellow. D, worm diagram drawn based on b-factor. Worm thickness is directly proportional to flexibility, i.e. the thickest region indicates most flexible. E, overlap of the catalytic residues within the active sites of the six subunits. The structures are anchored by the backbond of His249.
Heterogeneity in the Six Subunits in Nsp15There are significant differences among the subunits of Nsp15H that are not observed in Nsp15VD. To better describe these differences, the three subunits of the upper trimer, as oriented in Fig. 1 (B-D), are labeled T1a, T1b, and T1c, and the lower trimer subunits are labeled T2a, T2b, and T2c. The T2 subunits have B factors of 49.3, whereas the T1 subunits have B factors of 69.7 Å2, respectively. The increased flexibility is shown by the worm diagram in Fig. 1D. In addition, flexibility is significantly higher in the C-terminal domains of each subunit than for the rest of that subunit (Fig. 1D). Within subunits T1 and T2, the root mean square deviation difference in the N-terminal domain is 0.27-0.34 Å, whereas the C-terminal domain had a root mean square deviation difference of 0.51-0.57 Å. The crystal packing may have some effect on the observed variations. This is insufficient, however, to affect the entire domain of each subunit. Moreover, the trimers showed significant differences between each other despite being in a similar environment.
Heterogeneity is most obvious in the subunit catalytic pockets formed by residues His234, His249, Trp332, Tyr342, and Lys289. Superposition of the active site residues showed the degree of conformational flexibility between the subunits (Fig. 1E). Although the positions of Lys289 and His234 are highly similar in each of the subunits, Tyr342 is especially flexible and adopted different rotomer conformations in the different subunits (Fig. 1E). In addition, Trp332 is disordered in subunit T2c and not visible in the electron density map. The distance between the side chain oxygen atom of Tyr342 to ϵ1-nitrogen atom of Trp332 varied between 3.0 and 4.1 Å in Nsp15H.
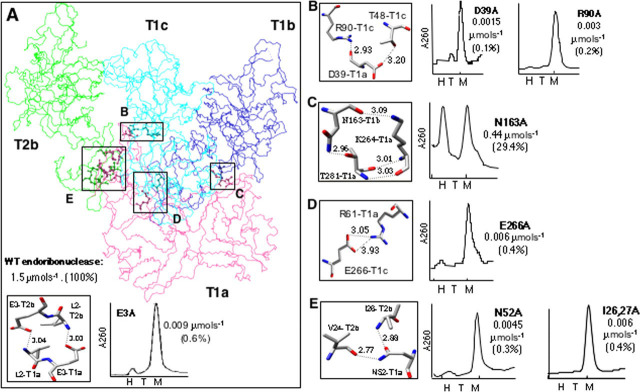
Subunit InteractionsBased on the examination of a panel of mutants, Guarino et al. (16, 20) proposed that hexamer formation is a prerequisite for enzymatic activity and RNA binding. A key mutant in this analysis was E3A, located in the N-terminal domain of Nsp15. It was inactive at lower concentrations but recovered some catalytic function at higher concentrations that should favor oligomerization (20). Structural data suggests that E3 and the adjacent L2 residue of one subunit are involved in trimer-trimer interactions with L2 and E3 of a subunit in a different trimer (Fig. 2A , inset). This interlocking structure thus provides a reasonable explanation for the effects of the E3 mutation on Nsp15 oligomerization (20). Furthermore, because oligomerization is required for activity, it is important to examine oligomerization in greater detail.
FIGURE 2.
Subunit interaction.A, wire diagram of subunit T1a and its interaction with other subunits. Contact regions are boxed and labeled (boxes B-E). The inset shows molecular arrangement and atomic distances between E3 and L2 (left) and the gel filtration profile of the E3A mutant of Nsp15 (20). H, T, and M denote positions corresponding to elution volume of a hexamer, trimer, and monomer of Nsp15, respectively. B-E, contacting residues and the calculated distances between them (left). Atomic distances are calculated using Chimera. Gel filtration chromatography elution profiles of the indicated mutant proteins (right). The values below the mutant name refer to the cleavage rate relative to WT in parentheses.
The crystal structure predicts multiple interactions that should help stabilize the Nsp15 hexamer (Fig. 2A). We examined four of the contacting surfaces, named B through E, by making mutations and examining their effects on Nsp15 structure and function. In Fig. 2A the bottom side of the T1 trimer (T1a, T1b, and T1c) is shown interacting with only one subunit of the T2 trimer (T2b). Contacting surfaces B, C, and D make contacts within a trimer, whereas E contains interactions between trimers.supplemental Table S1 provides the distances involved in these subunit contacts. The interactions are predicted to be similar for both Nsp15VD and Nsp15H. However, Arg61 of Nsp15H salt bridges with Glu266 (contact point D;supplemental Table S1). Side chain atoms of all of these residues are well ordered.
In interaction surface B, the backbone oxygen atom of residue Asp39 of subunit T1a forms an ionic bond with the H1-nitrogen atom of Arg90 in T1c (Fig. 2B). In addition, the δ2-oxygen atom of Asp39 forms a H-bond with the γ1-oxygen atom of Thr48 in T1c. To test the functional relevance of these interactions, we analyzed hexamer formation and enzymatic activity of the mutant versions D39A and R90A. Oligomerization state was analyzed using a rapid and reproducible gel filtration assay. The validity of this assay has been previously confirmed by analytical centrifugation assays and also a native gel analysis (20). Both D39A and R90A eluted in a gel filtration column as monomers (Fig. 2B), confirming the importance of these residues in hexamer formation.
A real time fluorescent assay previously described in Bhardwaj et al. (16) was used to monitor endoribonuclease activity. The substrate contains three deoxyribonucleotides, one cleavable ribonucleotide (rU), a 5′ fluorophore, and a 3′ quencher. Cleavage of the substrate results in an increase in fluorescence, which can be measured in real time. Functional analysis of D39A and R90A, using the real time endoribonuclease assay, revealed that the activity of these mutants was less than 1% of the WT protein.
In surface C, the backbone oxygen atom of N163 in T1b interacts with the ζ-nitrogen atom of K264 in T1a. In addition, the δ2-nitrogen atom of N163 in T1b bonds the γ2-oxygen atom of T281 in T1a (Fig. 2C). Alanine substitution of Asn163 should disrupt the latter interaction and resulted in a nearly equal distribution between the monomer and the hexamer, suggesting that hexamer formation was affected but not abolished. In the real time endoribonuclease activity assay, N163A retained 29% of the activity of the WT protein.
In surface D, the H1-nitrogen atom of Arg61 in subunit T1a interacts with the ϵ1-oxygen atom of Glu266 in T1c. Gel filtration analysis of the mutant protein E266A showed that it eluted primarily as a monomer. Furthermore its endoribonuclease activity was less than 0.4% of WT (Fig. 2D). With the three sets of mutants that affected intratrimer interactions, it is interesting to note that only mutant N163A retained some ability to form hexamers and also retained some endoribonuclease activity.
Interactions within surface E highlight the importance of intertrimer interaction. One such interaction is between the δ1-oxygen and δ1-nitrogen atoms of Asn52 in T1a with backbone nitrogen atom of Ile26 and oxygen atom of Val24, respectively, in T2b. When mutant N52A or I26A,I27A were tested, both proteins existed primarily as monomers. Again, both mutants were severely debilitated for endoribonuclease activity, with cleavage rate of rU being less than 1% of WT (Fig. 2E).
Interestingly, mutants E3A, N52A, and I26,27A, which were predicted to form trimers because these residues are involved in trimer-trimer interactions, all eluted as monomers and not trimers as we expected (Fig. 2, A, inset, and E). These results suggest that a trimer is not a stable intermediate in the formation of hexamers and that the equilibrium favors monomers and hexamers. Some trimer (<10%) was seen previously in gel filtration columns (20). It is possible that these reflect a dissociation event.
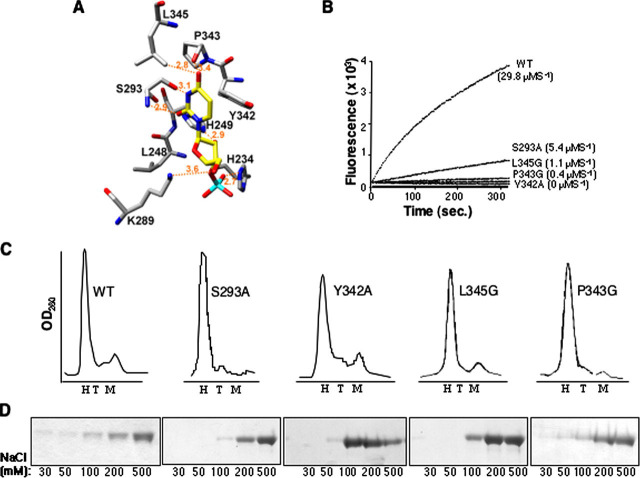
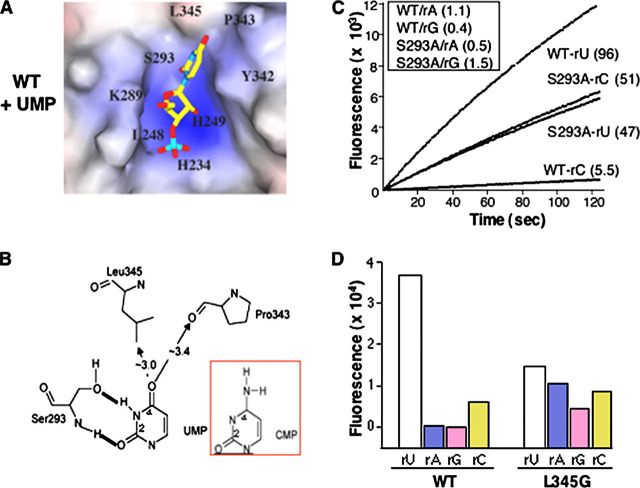
Functional Analysis of Active Site ResiduesAn interesting aspect of the Nsp15 active site is that the C-terminal tail contributes to its formation. To obtain an initial model for substrate recognition at the Nsp15 active site, we performed a molecular docking analysis with UMP. The results suggested that some of the active site residues might contribute to specific interactions with U and help discriminate against a C (Fig. 3A). First, Ser293 could potentially H-bond with the N3 imino hydrogen and the C2 carbonyl oxygen of the uracil. This information is consistent with the proposal of Ricagno et al. (21), who postulated that Ser293 was comparable in spatial location to Thr45, the residue in RNase A that hydrogen bonds to the base and specifies cleavage. Second, the phenyl side chain of Tyr342 could stack against the uracil, resulting in Pro343 and Leu345 occupying the space above the C4 carbonyl of uracil. In addition, three basic residues, Lys289, His234, and His249 that have been previously identified by bioinformatics analysis to participate in catalysis (7) could form an ionic surface that interacts with the ribose portion of the uridylate and the adjacent phosphate. Consistent with their proposed roles in catalysis, these three residues are positioned to act on the ribose 2′ OH and the adjacent phosphodiester group (Fig. 3A). It is known that mutations in any of these three residues reduced RNA cleavage to background (20).
FIGURE 3.
Analysis of the Nsp15 catalytic site.A, molecular docking of UMP into the Nsp15 active site. The backbone of active site residues is shown in gray, and that of UTP is in yellow; nitrogen and oxygen atoms are blue and red, respectively. The phosphorus is cyan. Atomic interaction is denoted by broken lines, and the calculated distances are indicated. B, results of cleavage of substrate rU with WT and mutant Nsp15, using real time fluorescent endoribonuclease assay. The identity of protein and rate of reaction is indicated to the right of the graph. The endoribonuclease activity was tested at 0.05 μm of WT or mutant proteins. C, Gel filtration of Nsp15. Elution profiles for WT Nsp15 and mutant versions as indicated next to the curve. The expected positions for hexamer (H), trimer (T), and monomer (M) are shown in each panel. D, SDS-PAGE analysis of polyU-agarose affinity chromatography fractions. WT or mutant Nsp15 proteins were incubated with polyU-agarose for 90 min at 30 mm NaCl. Unbound fraction was removed by a low speed centrifugation, and the slurry was washed sequentially with increasing NaCl concentration. Samples of each NaCl concentration wash were analyzed by SDS-PAGE and Coomassie Blue staining.
To examine whether and how residues in the catalytic site, including the C-terminal tail, affect endoribonuclease activity, we made mutant versions of Nsp15 that changed Ser293 and Tyr342 to alanines to disrupt H-bond or stacking interactions, which these residues might have with substrate. Pro343 and Leu345 were mutated to glycines to widen the catalytic pocket. Endoribonuclease analyses with the rU substrate revealed that the WT protein had a cleavage rate of ∼1.5 μmol-1. The rate for mutant S293A was ∼0.25 μmol-1, P343G and L345G cleaved the substrate at 0.02 and 0.055 μmol-1 respectively, whereas the activity of Tyr342 was undetectable (Fig. 3B). These results confirm that the residues identified in the catalytic pocket are involved in catalysis.
Based on the structure, we did not expect oligomerization to be affected by the changes in proteins S293A, Y342A, P343G, and L345G. To test this directly, however, we examined the oligomerization state of these proteins by gel filtration. WT Nsp15 and all four of these mutants eluted in a position corresponding to hexamers (∼240 kDa) (Fig. 3C), confirming that the C-terminal tail does not affect oligomerization but is required for endoribonuclease activity.
To determine whether these catalytic site mutants were affected for interactions with the RNA substrate, we performed affinity chromatography using polyU-agarose resin. To prevent RNA cleavage, the chromatography was performed with a specific active site inhibitor that decreased Nsp15 endoribonuclease activity to background (to be published). Fig. 3D shows SDS-polyacrylamide gel electrophoretic analysis of protein fractions eluting at different salt concentrations. WT Nsp15 eluted from polyU agarose at 200 and 500 mm NaCl (Fig. 3D). The elution profiles of mutants S293A and P343G were similar to WT, whereas mutants Y342A and L345G eluted at lower salt (Fig. 3D). These results show that RNA binding was affected by alterations in some of the active residues. Among this panel of mutants, Tyr342 showed the most dramatic effect on binding and was also the most affected with respect to activity. This does not always correlate, however, because H249A is catalytically inactive and demonstrates WT binding to polyU-agarose (data not shown).
Specificity for UridylateMolecular docking of UMP into the Nsp15 active site (Fig. 4A) suggests that N3 and O2 of the uridylate could form H-bonds with γ-oxygen atom and backbone nitrogen atoms, respectively, of Ser293, whereas O4 could interact with either Leu345 or Pro343 via H-bonds (Fig. 4B). This possibility is supported by our earlier results showing that modification of either N3 or O4 of uridylate affected cleavage by Nsp15 (16). Cytosine should not form equally strong interactions with the active site residues. Although the O2 of cytosine could interact with backbone nitrogen atom of Ser293, the γ-oxygen atom of Ser293 would need to reorient to form a second H-bond with N3 of cytosine. Additionally, the bulkier NH2 group at C4 could sterically hinder the fit of cytosine in the pocket (Fig. 4B, inset).
FIGURE 4.
Examination of substrate specificity.A, UMP docked into catalytic pocket of WT Nsp15. B, schematic to denote uridine recognition by S293 and the approximate distances between UMP and nearby catalytic site residues. C, results of real time fluorescent endoribonuclease assay. 0.25 μm of WT or mutant S293A proteins were used in these assays, and cleavage of substrates rU, rC, rA, and rG was measured in real time. The results of rU and rC cleavage are shown in real time. The slopes of each curve represent the rate of reaction. The identities of protein and substrate along with rate of reaction are indicated to the right of the curves. Cleavage rates of rA and rG are shown in the inset. D, cleavage of fluorescent substrates by WT or L345G proteins. Each protein (0.25 μm) was incubated with one of the four fluorogenic substrates (rU, rC, rA, or rG) for 20 min at room temperature and measured the fluorescence for each treatment. The increased cleavage of the rA substrate by L345G was confirmed in kinetic assay in two other independent experiments (data not shown).
Molecular modeling and energy minimization of mutant S293A with CMP or UMP indicate that in absence of Ser293, Pro343 could interact with either O4 of uridylate or the C4 amino group of cytosine to stabilize binding (supplemental Fig. S1 A). In addition, the side chains of Lys289, Leu248, and Leu345 would adjust to better complement the electronics of a cytosine in the active site (supplemental Fig. S1 B). The S293A mutation should also result in a decreased anionic nature of the active site cleft and a narrowing of the active site cavity, which could accommodate a uridylate or a cytidylate equally well, although with a lower affinity as compared with the affinity of WT protein to a uridylate. Taken together, our observations from the models is that uridylate should be a preferred substrate over purines or a cytidylate because it can form stronger specific interactions and has a better steric fit in the catalytic pocket.
To test these predictions, first we analyzed the effect of mutations that would potentially alter the interaction between uridylate and the catalytic residue Ser293. In addition to the already characterized S293A, we changed Ser293 to asparagine (S293N) or threonine (S293T). All three mutant proteins eluted in a gel filtration column as hexamers (data not shown). To examine the specificity for RNA cleavage with greater sensitivity, we used the fluorescent real time assay and with 0.25 μm of S293A or WT Nsp15, a concentration that is 5-fold higher than our typical assay. We also compared the normal rU substrate and a similar substrate having cytidylate as the only ribonucleotide (named rC), as well as substrates with guanosine (rG) and adenosine (rA). WT reproducibly cleaved rU with a rate of 4.8 μmol-1 and rC with a rate of 0.285 μmol-1 (Fig. 4C). The ratio of rU to rC cleavage, or the specificity factor, is 16.8 for WT. There was no cleavage by the WT protein with rG or rA (Fig. 4D and data not shown).
Mutant S293N was severely decreased in activity, with rates of cleavage for rU and rC at less than 0.017 μmol-1. S293T cleaved rU and rC with rates of 4.6 and 0.28 μmol-1, respectively, thus generating a specificity factor of 16.4, which is comparable with that of WT (supplemental Fig. S2). The fact that S293T is not affected in endoribonuclease activity is not surprising given that the comparable residue in the mouse hepatitis virus Nsp15 is a threonine, and the infectivity of EAV was not significantly affected either residue at the comparable position in its endoribonuclease ortholog (13).
S293A had a more interesting activity profile. It had reduced cleavage rate, with rU being cleaved at 2.35 μmol-1 (Fig. 4C). Unlike WT enzyme, however, S293A cleaved rU and rC with comparable rates (2.55 and 2.35 μmol-1), resulting in a specificity factor of 1.1. Furthermore, S293A has comparable Km values for rU (26.7 + 8.5 μm) and rC (28 + 2.6 μm), values similar to that of WT Nsp15 for rU (30.3 ± 9.5 μm). Like WT, S293A did not cleave rA or rG, thus discrimination against purine substrates was not altered. These results support our molecular docking predictions and the idea proposed by Ricagno et al., (21). These modeling results suggest that Ser293 participates in specific recognition of the substrate by Nsp15 and further extends the similarities in the cleavage mechanism for Nsp15 and RNase A.
Next, we analyzed the mutants P343G and L345G. We predicted that these substitutions could potentially widen the pocket and/or increase flexibility in the active site. P343G was reduced in catalytic activity to 1.3% of WT and had a specificity factor of 10.4 ± 2 for rU/rC. The more interesting result was with L345G. Leucine at this position should sterically block the binding of a purine in the WT protein (Fig. 4A). Substitution of glycine at this position decreased overall cleavage activity and resulted in a lower rU/rC specificity factor of 2.3 ± 0.2 relative to WT. Interestingly, L345G cleaved purine substrates at a significantly higher rate than WT or other catalytic mutants (Fig. 4D). Because leucine 345 is the penultimate residue, these results suggest that the C terminus of Nsp15 contributes to the specific recognition of uridylates in part by preventing the access of other nucleosides into the active site.
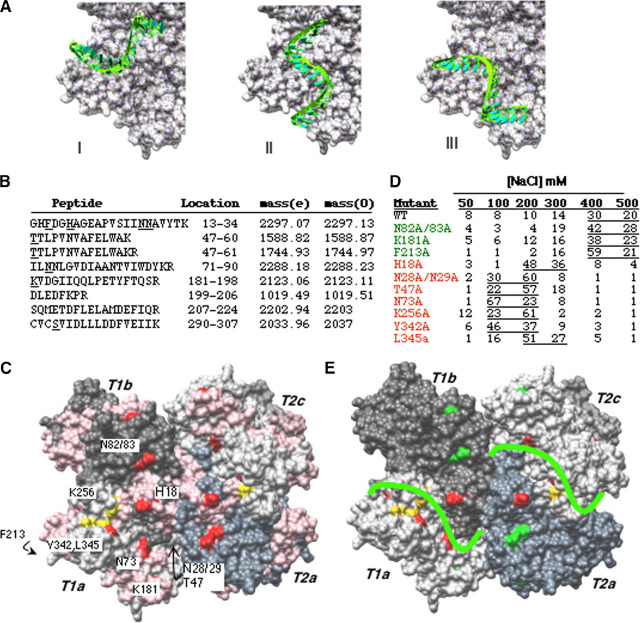
Nsp15-RNA InteractionsImages of Nsp15 in 2D crystals in complex with RNA indicates that an RNA of 16 nucleotides binds across the interface between two trimers (16). Furthermore, only hexamers bind RNA in solution, indicating that RNA binding follows oligomerization (16). The catalytic pockets are surface-exposed in both Nsp15VD and Nsp15H crystal structures. These pockets, however, are too narrow in both structures to accommodate an RNA molecule, and a conformational change is necessary for RNA binding. The flexibility observed in this region may be a part of the mechanism to allow RNA docking into the catalytic pocket.
Three different RNA access paths for the catalytic site of Nsp15 hexamer were identified based on surface complementarity and were finalized by manual docking of a single-stranded RNA of sixteen uridylates (built using the biopolymer module of Insight II (Accelrys Inc.)). These dockings were done with some steric clashes that needed to be resolved by slight conformational changes in the protein. The three potential paths for RNA binding are shown in Fig. 5A . To examine whether any or all of these potential paths contact RNA, we used a method that does not contain built-in bias in selecting RNA-contacting residues, a reversible cross-linking peptide fingerprinting method that was previously used to identify the template-binding portions of the hepatitis C virus RNA-dependent RNA polymerase (4). Briefly, a biotinylated RNA is cross-linked to a protein by use of formaldehyde. The reaction is then treated with trypsin to digest the protein after lysines and arginines, and the RNA and RNA-peptide complexes are enriched by streptavidin affinity chromatography. After reversing the cross-links, the peptides are purified from the RNA and subjected to MALDI-ToF spectroscopy. The peptides are identified from peptides generated by a virtual cleavage.
FIGURE 5.
A Model for a complex between Nsp15 and oligonucleotide RNA.A, putative RNA binding path of sNsp15 hexamer. For clarity, only subunit T1A is emphasized. A U16 A-RNA oligonucleotide is manually docked through the narrow catalytic site of the enzyme. B, summary of Nsp15 peptides obtained by biotin-streptavidin affinity chromatography and reversible cross-linking and trypsin digestion. The peptides were purified using Zip-tip as recommended by the manufacturer and subjected to MALDI-ToF. The expected masses obtained by virtual tryptic digestion of Nsp15 are denoted by (e) and the masses of observed peptide peaks from MALDI-ToF analysis are denoted by (0). The peptide position on the protein sequence is indicated by protein sequence as well as residue numbers. C, location of peptides identified by reversible RNA cross-linking and MALDI-ToF analysis. Identified peptides are colored pink. Potential RNA binding residues, along trajectories selected for mutational analysis are colored red. Active site residues are colored by element. D, affinity chromatography analysis of Nsp15 mutant and wild type proteins. All of the proteins were allowed to bind to polyU-agarose at 30 mm sodium chloride concentration followed by washing the slurry with sodium chloride concentrations indicated on top. Protein in each fraction was quantified by the Coomassie Plus protein assay kit from Pierce and is indicated as a percentage of the amount loaded. E, favored RNA binding path on Nsp15. Color scheme is as follows: protein subunits, shades of gray; active site residues, yellow; residues that affected affinity of protein for RNA, red; residues that did not affect RNA binding, green. A green line indicating RNA path is drawn on protein surface.
The biotinylated RNA used in the reversible cross-linking peptide fingerprinting contained multiple uridylates to facilitate binding by Nsp15 (16) and several cytidylates that contain amines that can be cross-linked with formaldehyde. The catalytically inactive mutant H249A was used in these experiments to maximize the production of RNA-protein complexes by avoiding catalytic cleavage of the substrate. A typical mass spectrum for the H249A-RNA cross-linked sample is shown in the lower panel of supplemental Fig. S3, whereas the result from a control reaction performed in the absence of RNA is in the upper panel. After subtracting the peaks corresponding to streptavidin and/or trypsin, eight ions that could be attributed to H249A were identified (Fig. 5B). Peptides containing over-lapping sequences to these eight were also observed when the samples were treated with chymotrypsin (data not shown), providing a confirmation for their assignment.
When the peptides were mapped back to the Nsp15 structure, the pattern shown in Fig. 5C was observed. Notably, the affinity-purified peptides border the predicted paths identified by bioinformatics analysis. In addition, peptides corresponding to the active site were found. The location of the peptides, however, did not allow us to identify a single path, suggesting either that all three are used at some level or that this analysis is insufficient to discriminate among the possibilities.
To obtain a better resolution of the RNA paths, we used the locations of the RNA-interacting peptides in the crystal structure and sequence conservation to guide the design of additional amino acid substitutions. This analysis required assumptions of the more likely residues based on the probability of particular side chain atoms to participate in formaldehyde cross-linking as well as orientation of the side chains within the Nsp15 hexamer structure. Formaldehyde preferentially cross-links residues that have basic, aromatic, or sulfur-containing side chains (29). Based on the orientation of the side chains, alanine substitutions of residues His18, Asn73, Asn82, Asn83, Arg181, Phe213, and Lys256 were constructed. In addition, based on their conservation and presence in the affinity purified tryptic peptides, alanine substitutions at Asn28, Asn29, and Thr47 were also included in the mutational analysis. S293A, P343G, Y342A, and L345G were previously made in our analysis of the active site, and they are known to have effects on RNA binding, respectively (Fig. 3D). Because of their proximity, Asn28 and Asn29 were mutated together, as were Asn82 and Asn83. Because hexamer formation is a prerequisite for RNA binding, all of the mutants were examined for RNA binding and were found to be hexameric (data not shown).
Next, we used polyU affinity resin to compare the stability of binding of mutant Nsp15 proteins with that of WT. Mutants K181A, F213A, and N82A,N83A bound to polyU resin at levels comparable with WT, with the peak of the proteins eluting at 400 mm NaCl (Fig. 5D). Mutants H18A, T47A, N28/29A, N73A, and K256A were significantly decreased for RNA binding, with the peak of the proteins eluting at 200 mm NaCl or lower, suggesting that these residues directly or indirectly contribute to RNA binding. When located onto the Nsp15 structure, the effects of the mutants indicate that Path III is most likely involved in stable binding to RNA (Fig. 5, A and E).
Final CommentsSARS-CoV Nsp15 displays a mechanism of endoribonuclease action that is identical to that of RNase A and likely uses a general acid-base mechanism for catalysis. RNase A is known to employ two histidines and a lysine for catalysis. Functional studies have demonstrated that two histidines (His234 and His249) and one lysine (Lys289) are important catalytic residues for Nsp15 as well. Our structure and molecular docking with UMP confirms the previous thesis of Ricagno et al. (21) that the catalytic pocket of Nsp15 bears a striking resemblance to that RNase A. Furthermore, we have functionally elucidated the relevance of a number of residues in the Nsp15 hexamer that are important for specific recognition of uridine and discrimination against purines and cytidine. We also identified several residues that stabilize interaction with the substrate. Although the catalytic center is formed by the C terminus, residues from N as well as C termini were found to help stabilize protein-substrate interactions. Identified RNA-binding residues that are located on the “side” of the Nsp15 hexamer supports our earlier proposal based on cryoelectron microscopy analyses and further identifies a path that feeds the RNA into active site. The information now available on the Nsp15 active site, the RNA binding path, and the contact sites required for oligomerization should be of interest in the design of Nsp15-specific inhibitors.
Footnotes
The atomic coordinates and structure factors (code 2RHB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SARS
- severe acute respiratory syndrome
- CoV
- coronavirus
- WT
- wild type
- MALDI-ToF
- matrix-assisted laser desorption ionization time-of-flight.
Supplementary Material
References
- 1.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai M.M., Cavanagh D. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spaan W., Cavanagh D., Horzinek M.C. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y.C., Russell W.K., Ranjith-Kumar C.T., Thomson M., Russell D.H., Kao C.C. J. Biol. Chem. 2005;280:38011–38019. doi: 10.1074/jbc.M508145200. [DOI] [PubMed] [Google Scholar]
- 5.Sawicki S.G., Sawicki D.L. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 6.Lai M.M. Annu. Rev. Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 7.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An S., Maeda A., Makino S. J. Virol. 1998;72:8517–8524. doi: 10.1128/jvi.72.11.8517-8524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasternak A.O., Spaan W.J., Snijder E.J. J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 11.Brian D.A., Baric R.S. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posthuma C.C., Nedialkova D.D., Zevenhoven-Dobbe J.C., Blokhuis J.H., Gorbalenya A.E., Snijder E.J. J. Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J.L., Kao C.C. J. Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhardwaj K., Guarino L., Kao C.C. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. J. Mol. Biol. 2006;361:243–2563. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saida F., Uzan M., Bontems F. Nucleic Acids Res. 2003;31:2751–2758. doi: 10.1093/nar/gkg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deshpande R.A., Shankar V. Crit. Rev. Microbiol. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- 19.Gioia U., Laneve P., Dlakic M., Arceci M., Bozzoni I., Caffarelli E. J. Biol. Chem. 2005;280:18996–190026. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- 20.Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. J. Mol. Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. J. Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Storoni L.C., McCoy A.J., Read R.J. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 25.McRee D.E. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 26.The CCP4 Suite (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50, 760-763 [DOI] [PubMed]
- 27.Murshudov G.N., Vagin A.A., Dodson E.D. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R.A., McArthur M.W., Moss D.S., Thornton J.M. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 29.Metz B., Kersten G.F., Hoogerhout P., Brugghe H.F., Timmermans H.A., de Jong A., Meiring H., ten Hove J., Hennink W.E., Crommelin D.J., Jiskoot W. J. Biol. Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. J. Compu. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Kuntz I.D., Blaney J.M., Oatley S.J., Langridge R., Ferrin T.E. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 32.Ferrin T.E., Huang C.C., Jarvis L.E., Langridge R. J. Mol. Graph. 1988;6:13–27. [Google Scholar]
- 33.Shoichet B.K., Bodian D.L., Kuntz I.D. J. Comp. Chem. 1992;13:380–397. [Google Scholar]
- 34.Meng E.C., Shoichet B.K., Kuntz I.D. J. Comp. Chem. 1992;13:505–524. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.