Abstract
About 50 y ago, Crow and Kimura [An Introduction to Population Genetics Theory (1970)] and Ohta and Kimura [Genet. Res. 22, 201–204 (1973)] laid the foundations of conservation genetics by predicting the relationship between population size and genetic marker diversity. This work sparked an enormous research effort investigating the importance of population dynamics, in particular small population size, for population mean performance, population viability, and evolutionary potential. In light of a recent perspective [J. C. Teixeira, C. D. Huber, Proc. Natl. Acad. Sci. U.S.A. 118, 10 (2021)] that challenges some fundamental assumptions in conservation genetics, it is timely to summarize what the field has achieved, what robust patterns have emerged, and worthwhile future research directions. We consider theory and methodological breakthroughs that have helped management, and we outline some fundamental and applied challenges for conservation genetics.
Keywords: conservation, genetic variation, population size, threatened species, adaptation
A Brief History of Conservation Genetics and Conservation Biology
Conservation Genetics (CG) and the larger discipline of Conservation Biology (CB) were founded at about the same time, during the second half of the 20th century. This was when awareness grew of the negative impact of human activity on the abundance of many wild species (e.g., ref. 1). CB was then charged with tasks, such as enumerating the causes of species’ declines and identifying counteracting measures, recognizing that populations remain vulnerable when they persist at a low density or small size (2). Threats associated with low density and small population size include stochastic events, positive density-dependent population growth, and negative feedback by genetic and evolutionary mechanisms (3, 4). CG has mostly focused on the latter, as well as providing tools for understanding demographically driven processes in wild and captive populations (5, 6).
Sewall Wright’s theory on the role of genetic drift in small populations laid the foundations for CG in the first half of the 20th century. Wright (7) linked small population census size with random changes in allele frequencies (genetic drift) and their population genetic consequences. He outlined how census size directly translates into drift-effective population size (Ne), and how small Ne is linked with the loss of heterozygosity and fixation or loss of alleles. However, drift-effective population size is difficult to study in natural populations as high-quality demographic data are required, depicting numbers of reproducing male and female individuals, variation in reproductive success, and variation in these parameters over time (8). As a surrogate, various polymorphic, biparentally, and codominantly inherited genetic markers came into use: allozymes first (9), then microsatellites (10), and more recently single nucleotide polymorphisms (SNPs, see below). Allozymes and microsatellites allow for many allelic states at a locus, whereas for SNPs there are generally only two alleles that are considered. Evolutionary models linking marker variation with Ne were produced by Crow and Kimura (11) and Ohta and Kimura (12). These models stimulated empirical efforts to estimate Ne of wild populations based on heterozygosity at marker loci. However, Ne was often not deduced explicitly because knowledge on migration or gene flow was lacking. Instead, genetic marker variation of populations served as the sole estimate to deduce population demographics.
Another main task of CG has been to assess the threat to small populations by genetic and evolutionary processes. An immediate genetic threat that may reduce population size is when inbreeding occurs because of reproduction between closely related individuals; this results in decreased fitness arising from homozygosity of recessive deleterious alleles, termed “inbreeding depression” (13). Further genetic threats are linked to the reduced potential to adapt due to genetic drift. Two bodies of theory have focused on the relationship between Ne and genetic variation for polygenic traits, and between Ne and the response to selection. They generally predict a decrease in additive genetic variance with decreasing Ne (e.g., ref. 14), and a decrease in the selection response with decreasing Ne (e.g., refs. 15 and 16)). Later theory considered the role of small population size in triggering the increase in frequency of deleterious mutations, the accumulation of genetic load, the types of genetic load (e.g., segregating load, fixed load), and how load can result in population extinction over short to intermediate time scales (e.g., ref. 17). These processes can be affected by connectedness between populations, with migration introducing new genetic variants that can alleviate the immediate negative effects of inbreeding, increase Ne, and increase genetic variation.
CG has its theoretical roots in population genetics, and a clear focus on both theory and application has made the field strong. The strongest inferences in CG apply to populations and make sense in a comparative context, often with a focus on recent changes in genetic diversity over time, such as since the beginning of land-use change, or over heterogeneous landscapes affected by human activity. CG focuses on management actions when a loss of genetic diversity is linked to widespread negative population trends in recent times (e.g., refs. 18 and 19), where the subsequent evolutionary and genetic consequences—reviewed in this article—can be detrimental for the persistence of populations (and an entire species if all remaining populations are affected).
Applied Conservation Genetics
When assessing whether species are threatened, the main considerations taken into account by the International Union for Conservation of Nature (IUCN) are the number of mature individuals, the extent to which a species occupies its former range, and the rate at which it is declining (or recovering) with respect to this range. Genetic diversity is acknowledged as one of the three levels of biodiversity, but genetic factors are rarely included in IUCN assessments, highlighted as a shortcoming in the IUCN Red List process (20). Similarly, the IUCN threats classification scheme, which is currently being updated (https://www.iucnredlist.org/resources/threat-classification-scheme), does not directly recognize the loss of genetic variation within a species, but instead focuses on external threats related to loss of suitable habitat or exploitation of target species.
Nevertheless, genetic information has been integral to the development of management plans for threatened populations and species (for a recent example, see Box 1). Molecular genetic tools have been used to estimate population-level parameters, such as longer-term Ne, evidence for recent bottlenecks, and inbreeding in threatened species (examples are given in ref. 5). They have helped in understanding patterns of gene flow, movement across landscapes, and processes, such as hybridization and contemporary versus historic genetic isolation. Genetic tools have also been useful in gaining knowledge of reproductive patterns, social structure, and life cycles of threatened species, and in estimating the accumulation of deleterious mutations. These parameters are important in deciding what management actions are necessary for threatened populations and species, in an effort to increase demographic numbers and genetic variation, and alleviate the expression of genetic load while preserving genetic uniqueness. Finally, monitoring levels of genetic variation in threatened and restored populations of animals and plants has been used to track the long-term viability of populations (21).
Box 1.
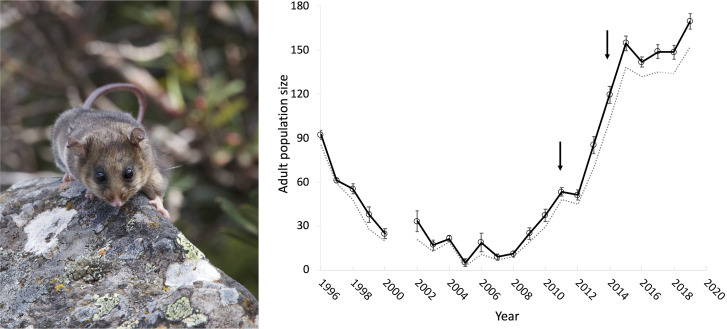
The impact of CG-based decisions on threatened species recovery is particularly well illustrated by examples of genetic rescue where the positive consequences of introducing genetic variation have been evident. The endangered mountain pygmy-possum is one such example, where Mitrovski et al. (19) documented a rapid decline in genetic diversity measured through microsatellite markers reflecting a bottleneck that paralleled a population collapse over at least a 10-y period (from 1996) at Mount Buller in Victoria, Australia. Threats were mitigated through habitat reconstruction and an invasive predator control program, but the population size did not increase, and genetic data showed this was due to inbreeding, an exceedingly low Ne to near 10 individuals and the loss of over 80% of the population’s genetic diversity (19, 66). A genetic rescue strategy was developed that included moving six genetically healthy males from the closest known population at Mount Higginbotham (Victoria). These two populations have likely been separated for more than 20,000 y. Despite the high levels of genetic differentiation between these populations, Weeks et al. (23) showed that this was likely due to genetic drift, reducing concerns around outbreeding depression (69). The strategy was implemented in 2011 and rapid population growth followed the introduction (66). In 2014 an additional six males were moved from another highly genetically diverse population (Timms Spur, Victoria) to the Mount Buller population to further increase genetic diversity. By 2019, the population had grown to the highest level ever recorded on the mountain and is now one of the largest populations across their range in Australia (Fig. 1) (updated from ref. 66).
Fig. 1.
Adult population size for Burramys parvus at Mount Buller, Victoria, Australia. Estimates are based on capture–recapture data and a robust design model with means across years (bars are SEs) connected by the solid line. Dashed line represents yearly unique captures. Arrows indicate the years in which six males were introduced from the Mount Higginbotham and Timms Spur populations to the Mount Buller population.
Furthermore, CG has helped threatened species management with the identification of populations and species that should be the focus of management actions. The identification of evolutionary significant units (ESUs) has been used to determine which populations should be conserved separately. ESUs are often recognized because they are highly genetically differentiated (particularly for mtDNA), presumably because they are likely to be on different evolutionary (and potentially adaptive) trajectories to each other (22). The level of genetic divergence appropriate for defining ESUs is not necessarily straightforward. The main goal is to favor genetic distinctness, without promoting the effects of genetic drift on generating genetic divergence (23). Regardless of this issue, the ESU concept can lead to practical outcomes such as defining putative cryptic species adapted to different environmental conditions (24).
Despite the common and successful application of genetic tools in CG, genetic factors are still too often ignored in species recovery plans, particularly outside of the United States (25). One reason is, as indicated above, that genetic status is not directly considered in the IUCN listing process. A recent study suggested that its consideration would make little sense as genetic variation across species was unrelated to current IUCN status (26). However, in CG estimates of genetic diversity at the level of a species is not normally used to inform about demographic issues. Here we reemphasize that CG typically focuses on patterns of variation within and across populations of a species, but mostly avoids comparisons among species, because many parameters—including mutation rate, long-term demographic changes, and life history attributes—will contribute to genetic diversity of a species, and these parameters are likely to vary among species (27). However, low genetic diversity at the population level and in a comparative context over time and space will typically be a good indicator of conservation status. Indeed, low genetic variation within vertebrate populations as evaluated by microsatellite markers was positively associated with the IUCN status of species in one comparison (28).
Another reason for underappreciating the potential of CG in conservation management is a lack of government policies on how to adopt CG and evolutionary processes in planning (29). Here, a combined effort by researchers and policy makers is needed to decide on how genetic data could be systematically implemented in the phases of risk assessment, formulation of goals, planning of actions, and monitoring of success.
Learnings from the Last Decades
There are clear foundations for implementing CG-based strategies in conservation management (for another recent review, see ref. 30). Here we briefly cover five key findings from the last few decades and then explore future directions.
Small Census Size and Severe Demographic Bottlenecks Are Associated with Reduced Population Genetic Variation.
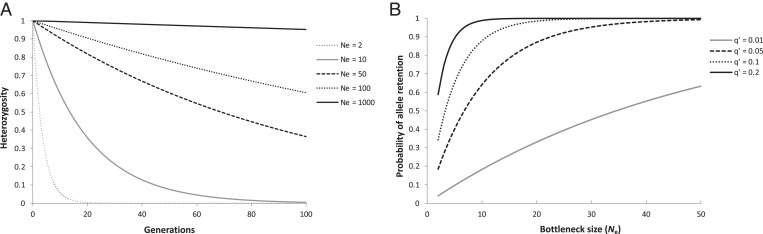
Crow and Kimura (11) formulated how loss of heterozygosity and additive genetic variance (VA) of quantitative traits in neutral parts of the genome depends on Ne, and that populations of small size harbor less genetic variation and lose genetic variation faster than larger ones. The work of Crow and Kimura further predicts that loss of genetic variation is an exponential process across generations. For example, random genetic drift decreases heterozygosity at the rate of 1/[2Ne] per generation (Fig. 2A). Even if changes in population size are intermittent, they will have a substantial impact on the frequency spectrum of alleles in a population. In particular, there will be a loss of alleles following a population bottleneck, particularly for rare alleles (Fig. 2B) (31).
Fig. 2.
(A) Theoretically expected effects of random genetic drift on heterozygosity through time (generations) for varying effective population (Ne) sizes and (B) the effects of the size of a single generation bottleneck on the retention of rare alleles at a single locus with initially two alleles (11).
There is very strong support for an association between small census size estimated in natural populations and low genetic marker variation. A metalevel analysis including animals and plants on the relationship between (log-transformed) population census size and genetic variation reported overall average correlation coefficients between 0.46 and 0.54, depending on the diversity estimate considered (32). A formal meta-analysis on 41 plant species revealed a correlation of 0.41 across studies (33). Several other meta-analyses also confirmed a strong negative link between habitat loss and fragmentation or overharvesting and genetic marker diversity, indicating reductions in population size across different and large taxonomic groups (34–36). Furthermore, studies have highlighted factors strengthening this negative relationship, such as body size in animals (35).
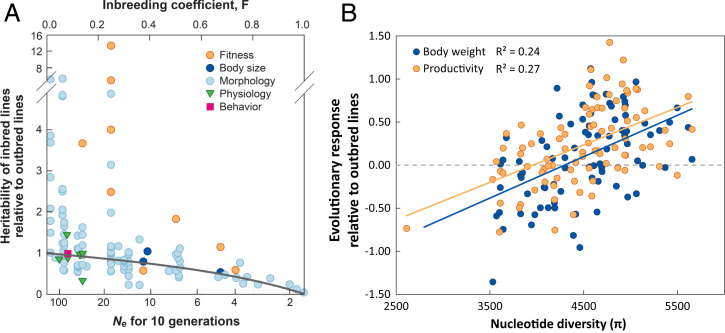
Evidence for a link between small population size and genetic variance for quantitative traits is mixed. A decline in heritability, which is genetic variance standardized by phenotypic variance, has been well documented in experimental studies focusing on traits typically associated with higher additive than nonadditive effects (Fig. 3A), but not in traits with stronger nonadditive effects, such as life-history traits (37). A large meta-analysis on heritability estimated in natural populations revealed no support for a positive relationship with population size (38). Several reasons for nonsupportive findings are possible, including the effects of selection on VA, changes in gene effects due to genetic drift, and population isolation (39). Wood et al. (38) also pointed to the need to improve estimates of census size and to provide more estimates for rare and specialist species.
Fig. 3.
(A) Association between effective population size or inbreeding and adaptive genetic variation for different trait classes (from ref. 39, copied with permission). The panel presents the change in heritability in inbred populations relative to outbred controls, measured in 21 experimental studies on animals and plants. The horizontal axis is expressed as either inbreeding coefficient (upper axis) or the value of Ne that would generate the same level of inbreeding over 10 generations. (B) Association between neutral molecular diversity and responses to selection, measured as slopes of changes in phenotypes across generations of experimental evolution in laboratory populations of D. melanogaster; from Ørsted et al. (44).
Small Census Size Is Associated with Inbreeding Depression and Low Population Mean Performance.
Small population size increases the likelihood of matings between related individuals even if mating is random. The increase in inbreeding (per generation) is predicted to be proportional to Ne (ΔF = 1/2Ne) (11). Thus, a population experiencing reductions in population size (population bottlenecks) are expected to have a higher degree of homozygosity due to inbreeding compared to the prebottleneck population, and this makes the expression of recessive deleterious mutations segregating in the population more likely (13, 40). These recessive deleterious alleles are typically at low frequency, and their summed effects, called segregating or inbreeding load, may not be different between historically large and recently small populations. However, this load poses a threat in small populations over time because its expression is a function of the inbreeding coefficient, which is higher in small populations. The reduced fitness of inbred individuals can also arise from reduced expression of overdominance due to a reduction in heterozygosity in an inbred population.
Reduced fitness because of inbreeding is well-documented (5, 13). In many wild populations of small size, inbreeding depression can cause severe fitness declines [e.g., 50% (41)]. The degree of inbreeding depression for a particular trait differs among individuals and populations, even when these have similar levels of inbreeding. Inbreeding depression is expected to be higher for traits with relatively high dominance variance, which is typically the case for traits closely associated with fitness (42). Also, for traits associated with fitness but with other nonadditive gene-action (i.e., partial dominance or overdominance), performance typically declines with increasing inbreeding (5, 43). Data obtained using experimentally bottlenecked populations and molecular genomic tools have supported the notion that inbreeding depression contributes to extinction (e.g., ref. 44), which is also supported by a meta-analysis including wild populations (45).
In studies on inbreeding effects, only some components of fitness are usually assessed and the consequences of inbreeding are therefore likely to be underestimated. Nevertheless, highly inbred populations sometimes do not to suffer from inbreeding, such as in the cases of the northern elephant seal, European beaver, and island fox of California (46–48). Such examples have led some authors to conclude that genetic problems are not important relative to demographic and environmental risks (26), but this is countered by the overwhelming empirical evidence that inbreeding depression is often substantial in natural populations. Reasons why some highly inbred individuals and populations seem unaffected by inbreeding include stochastically low genetic load segregating in populations (49) and purging of deleterious mutations (50). However, purging under drift with random mating may only work against highly recessive and strongly deleterious mutations (51), and empirical studies indicate that significant purging is not common [e.g., in 12% of 119 studied zoo populations (52)]. An added level of complexity arises from the environmental dependence of both inbreeding depression and purging. Favorable environmental conditions can moderate the expression of inbreeding depression (53), with the flip side that inbreeding effects are typically more severe in harsh environments, to which natural populations are increasingly being exposed (40). Furthermore, the genetic architecture of many traits is context-dependent (54, 55), such that unpurged alleles may be neutral or beneficial in one environment but contribute to genetic load in another (56). Thus, if a few small and inbred populations do well despite high inbreeding levels, there is no guarantee that this will continue to be the case as environments change.
Small Populations or Populations with Low Marker Variation Experience more Drift Load.
Small populations suffer from reduced mean performance due to the accumulation of deleterious mutations. When a population becomes small initially, it is expected to mainly express segregating or inbreeding load because of increased inbreeding. Some of that load might be purged, although inbreeding by small population size may only purge the most deleterious mutations (see discussion above). However, when small size persists and gene flow is rare, segregating and new deleterious alleles are expected to increase in frequency as genetic drift overwhelms purifying selection, and some of these mutations will eventually become fixed, and this is known as drift load (17). This accumulated drift load is predicted to decrease the performance of small populations, leading to the mutational meltdown that should markedly constrain longer-term population persistence (17).
A meta-analysis mainly based on plant studies found that both census size and genetic diversity were indeed positively correlated with population mean performance across studies, with overall correlation coefficients of 0.44 and 0.30, respectively (33). Many of the individual studies that contributed to the meta-analysis had been performed in the field, and therefore some of the effect may not be due to the expression of load but due to marginal conditions where populations of smaller size occur. However, studies have also estimated drift load by comparing the fitness of offspring in crosses between versus within populations; a performance gain is interpreted as the magnitude of drift load (and may include inbreeding depression). A meta-analysis provided strong support for the notion that small and inbred populations experience a dramatic increase in fitness measures due to outbreeding, with a median increase of 48% when the environment is benign and 114% under more stressful conditions (57); this provides strong evidence that genetic load is a substantial problem in persistently small populations.
From a conservation perspective, it may seem irrelevant whether performance declines are due to inbreeding depression or due to drift load, as both lower population mean performance. However, management actions may differ in dealing with these situations. If declines in mean performance are due to recent inbreeding and the expression of inbreeding load, management actions may need to focus on increasing census size and improving connectivity between habitat patches (and thereby increasing Ne). When drift load has accumulated and many deleterious mutations are at high frequency or fixed, management options include genetic rescue strategies by assisted gene flow. Conversely, populations that have undergone a slow decline in population size have a greater opportunity to purge deleterious mutations and management actions should initially target habitat improvement, although increasing genetic variation through assisted gene flow is also likely to be important for long-term resilience to environmental change.
Small Populations or Populations with Low Marker Variation Show a Reduced Response to Selection.
There are several ways in which small population size can negatively affect a selection response, and these are likely to act in combination. First, as standing genetic variation is predicted to be lower in small populations (e.g., ref. 14), the response to selection should be lower. Second, new mutations that selection might favor are less likely to emerge in small populations (e.g., ref. 15). Third, if selection is weak, it may be overridden by genetic drift, which should also reduce a selection response (e.g., ref. 16). Fourth, increased linkage associated with small population size is predicted to lead to Hill–Robertson interference, where linkage between alleles reduces the effectiveness of selection and results in a lower selection response (58).
Despite the mixed empirical evidence for reduced VA or heritability in small populations, evidence for reduced selection response is rather consistent. A meta-analysis on studies of local adaptation in herbaceous plants showed that populations of smaller census size were less adapted to local conditions, as revealed in transplant experiments (59). The result could be due to any of the four mechanisms mentioned above. Furthermore, experimental studies showed that smaller populations had reduced responses to selection: for example, 57% over 55 generations in Drosophila melanogaster populations with Ne ∼ 8 compared with Ne ∼ 200 (reviewed in ref. 39). The association between genetic marker diversity and response to selection has also received strong experimental support. For example, a recent highly replicated experimental evolution study on D. melanogaster provided evidence for a strong positive association between genome-wide heterozygosity and evolutionary response (Fig. 3B) (44). Evidence for the importance of Hill–Robertson interference at linked selected sites includes sequence analyses of protein-coding genes in Drosophila, showing that it diminished the rate of adaptive evolution by about 27% (60). However, the problem may be even more important when linkage involves deleterious mutations (43).
Nevertheless, strong responses to selection can sometimes be observed in small populations, a phenomenon well known from animal breeding where significant genetic responses in populations with small effective population sizes (typically less than 100) can occur (61). Because population growth and variation in relative fitness is typically negatively associated (62), selection may act more effectively in populations declining in size, producing a heightened selection response in the short term (63). However, these selection responses will not be maintained in the long term. And while additive genetic variance might also increase in populations going through bottlenecks when there are dominance interactions between alleles within loci or when there is epistasis (64), such increases come with an increased frequency of deleterious alleles and do not translate into increased responses to directional selection (65).
Restored Gene Flow and Genetic Mixing Can Overcome Low Population Performance.
Gene flow is an evolutionary force that may both aid and limit adaptation. On the one hand, gene flow can help small and isolated populations in the short term by introducing alleles that mask the expression of deleterious recessive ones, and in the longer term by introducing new genetic variants that increase rates of adaptive evolution. On the other hand, gene flow can lead to outbreeding depression when genetic incompatibility arises from combining distantly related genomes, or it can lead to an influx of poorly adapted alleles into a population from an area where different conditions predominate.
The immediate benefit of gene flow to small, isolated populations has been well documented in the meta-analysis on outbreeding in natural populations (57). Consequently, genetic rescue—the deliberate introduction of new genes into small populations suffering from the expression of deleterious mutations—can be a powerful means of managing small, declining populations (e.g., ref. 66; also see Box 1). However, as noted under the third point above, understanding population history can be important for predicting the benefits of genetic rescue and for identifying source populations that can be used in a genetic rescue attempt (67; but see also ref. 68). Ideally, rescue attempts should be combined with other programs, such as habitat restoration or predator/competitor control, that allow for population expansion (66).
However, risks of outbreeding depression have often been overstated. Furthermore, they can be appropriately managed (66, 67, 69) and need to be balanced against the benefits of genetic rescue. Increases in genetic diversity measured using neutral genetic markers following genetic rescue have been explicitly linked to population recovery and growth following steep declines and improvements in fitness of hybrid individuals (66). Despite some genetic homogenization through genetic introductions, alleles associated with local adaptation may not be swamped by gene flow (70). Advances in genomics provide insights into the consequences of genetic rescue on adaptive genetic diversity (70).
Additional Insights
Three key aspects are important take home messages from the learnings briefly outlined above. First, in CG and CB more generally, overall relationships as emphasized above should be the focus, rather than the exceptions. There will always be some variation around the main axes of relationships, such as between population size and genetic diversity, and part of that is due to genetic drift, which is a stochastic force that leads to marked variation in allele frequencies between generations and small populations. Second, neutral evolution and neutral genetic diversity as measured by CG studies remain directly relevant to adaptive evolution. Neutral markers provide an estimate of genetic drift, which lowers genetic variation and counters the response to directional selection. Lower population mean performance due to increased drift load in small populations means that, at some point, the population cannot persist given the selective deaths that come with an adaptive response. Species that have naturally fragmented distributions, for which drift load is particularly important, may be limited in their capacity to persist in the long term, especially in the context of rapid environmental change, where drift load will combine with low overall genetic diversity to limit persistence of small populations (71). Third, the loss of genetic variation due to drift raises the issue of how high Ne of an isolated population must be to ensure that sufficient genetic variation is maintained for long-term persistence under environmental changes. Originally, Franklin (72) proposed that an Ne of 500 is needed for an isolated population to retain evolutionary potential in the long run. This number was based on the equilibrium between new genetic variation arising from mutations and the loss of genetic variation from random genetic drift (Fig. 2), for traits that are relatively unaffected by selection or are subject to stabilizing selection. Revised recommendations (by considering differences in gene effects) suggest that an Ne above 1,000 is more realistic to sustain long-term viable populations (73). As Ne is typically an order-of-magnitude lower than the census size, numbers may need to be above 10,000. Unfortunately, studies validating these numbers are scarce, and they are challenging to meet, for instance, for many species with extensive space requirements. CG principles should therefore guide management decisions within the reality that optimal conditions might never be reached, and with an awareness that some connectivity among populations can greatly contribute to assuring evolutionary potential.
Recent Developments
Although there is a solid backbone of theory and experimental data behind CG practices, many emerging tools particularly linked to next-generation sequencing (NGS) are set to provide fine-scale and new insights in the next few years. NGS techniques, such as reduced-representation sequencing (e.g., RADseq) and whole-genome sequencing (WGS) of population-pooled samples or individuals, are now commonly applied in CG [e.g., RADseq on grasshoppers (74); individual WGS of an endangered pheasant species (75)]. The massive amount of data generated by NGS allows for testing complex demographic scenarios, linking changes in Ne back to the more distant past, and examining the impact of migration and inbreeding across the genome. The estimation of genetic load has recently been addressed in several contributions to CG (e.g., ref. 76). Eventually, diversity estimates at loci related to expressed traits (quantitative-trait loci, QTLs) may help predict potentially adaptive genetic variation and evolutionary potential (77).
Here we summarize how the newest technologies and analytical methods could provide unprecedented insights into the importance of genetic diversity in conservation, particularly in the face of environmental change (e.g., eQTLs) (see ref. 78 for a detailed review of approaches of conservation genomics). We stress that good practice involves carefully planned studies with precise goals, using appropriate analytical methods to meet these goals, which often dictates the sampling effort, wet-laboratory protocols, sequencing strategy, and depth of sequencing required. In particular, studies should equalize sample sizes for all populations when using NGS data, ensure that sequencing depth is adequate, and that SNP-based heterozygosity estimates are not biased by population structure (79).
Genomic Inference on Population Demography.
RADseq and WGS produce thousands to millions of SNPs, such that the amount of data can often compensate for fewer samples. For some species and analyses, higher sample numbers per population (e.g., ref. 50) combined with RADseq seems better; for others, two to three individuals per population combined with WGS at good depth or pooling (e.g., 50 samples per population combined with WGS) should be considered. Also, some downstream analyses on the detailed demography need more data resources than others. Some require a reference genome from the target species or a closely related species. The de novo assembly of reference genomes for nonmodel organisms has become an option because of a drop in cost combined with improved sequencing technology: for example, the sequencing of long DNA fragments with high accuracy (e.g., PacBio SMRT sequencing, Sequel II). An important step for demographic inference is to filter for variants that are not strongly affected by selection (80).
A pragmatic approach is to first screen data for population structure to verify whether the a priori definition of a population used for sampling passes genetic testing. Evidence for structure and little gene flow may lead to a focus on each population separately, followed by comparative analyses. In this case, a simple way of assessing recent change in population size is the discrepancy between diversity estimates, such as those based on heterozygosity and on allelic richness [e.g., Tajima’s D, for an example of an application (75)]. More complex approaches consider SNP diversity across DNA sequences and allow the estimation of changes in Ne over time. There has been a recent increase in the use of coalescent-based reconstruction of Ne by changes in the local density of heterozygous sites, but resolution for the most recent period is typically weak [e.g., the software PMSC (81); for an application see ref. 82]. A method that depicts recent changes in Ne (>5 generations ago) is derived from haplotype-based estimation of Ne and uses the decay of linkage disequilibrium [e.g., the software LinkNe (83), as applied in ref. 18]. This approach requires a reference genome and sampling should include about 50 diploid individuals per population. When there is evidence of considerable migration, the statistical comparison of alternative demographic models seems more appropriate. With SNP data summarized as folded or unfolded site frequency spectra, the latter polarized to an outgroup, most likely demographic parameters can be inferred [e.g., software fastsimcoal2 (84), as applied by ref. 85]. This allows the estimation of split times of populations, past and more recent migration, and changes in population size. The study design ideally considers a few populations, and importantly some that serve as references with little change in size and migration.
Another strong approach for deducing changes in demography involves longitudinal sampling of populations and the tracking of genetic variation over time. Such studies should benefit from larger sampling efforts within populations, as might be achievable via pool-sequencing. In the face of a worldwide biodiversity crisis, it will be important to monitor genetic diversity over time, to systematically resample sites and relate changes in genetic diversity with continuous human impacts on wild populations, using unbiased estimates of heterozygosity that include monomorphic as well as polymorphic sequenced sites (79) so that comparisons can be made across datasets and studies.
Screening for the Abundance of Deleterious Mutations: Assessing the Magnitude of Genetic Load.
High-quality marker and genomic tools are available to assess the threat of inbreeding depression via estimating the level of inbreeding in otherwise strongly outcrossing species (e.g., ref. 86). For the estimation of genetic load, it is imperative to have an annotated reference genome from one or several closely related outgroup species. Derived SNPs of coding regions are filtered for and then annotated functionally for their likely impact [e.g., SnpEff (87)] or by assessing each SNP site for conservation across homologous sites of a large database; SNPs at highly conserved sites are assumed to be more deleterious [e.g., SIFT algorithm (88)]. Under the assumption that deleterious mutations are mostly recessive, it is not the counts per se that are relevant in depicting genetic load, but homozygosity for derived variants (89).
An area that requires further exploration is the analysis and interpretation of genomic data within the context of fitness. For example, it is not yet clear how distributions of dominance coefficients link with the magnitude of mutational effects. For knockout mutants in yeast, complete recessivity is approached asymptotically as mutations become increasingly deleterious (90), supporting the expectation that lethals and sublethals are fully recessive and other deleterious mutations mostly partially recessive. We also lack knowledge on whether the intensity of selection acting on polymorphisms are represented by measures of deleteriousness that have come into use in genomic analysis. Therefore, it remains hard to judge if reduced genetic load estimated from genomic data in species of conservation concern (91) translates into fitness effects. In the meantime, it seems reasonable to work with estimates depicting load that consider full or high recessivity of deleterious mutations and to be cautious with the interpretation of results.
Screening for Adaptive Genetic Variation.
Although we have here emphasized the continued importance of assessing overall levels of genetic variation, including neutral variation, we agree with Teixeira and Huber (26) on the point that adaptive genes should be investigated in quantitative genetic studies. This can be based on a priori knowledge or an association study, and is particularly relevant when there are a limited number of loci involved in an adaptive response. Even for some polygenic traits, a few QTL loci may explain considerable phenotypic trait variation. Variation at these QTLs may have a high predictive power. For example, in the simulation study of Caballero and García-Dorado (77), diversity at those loci could predict the response to short- and long-term selection very well (often r > 0.5) under different demographic scenarios; in contrast, additive genetic variance (VA) was the best predictor for a short-term selection response. This offers some optimism that evolutionary potential might eventually be predicted once adequate genomic resources become available, including more annotated reference genomes.
Genome-wide association studies provide an approach for identifying genomic regions contributing to evolutionary responses (e.g., ref. 92). The approach can be used flexibly and applied to environmental instead of trait data, and by performing analyses at the level of populations instead of individuals (e.g., ref. 93). To avoid circularity when assessing genetic variation at QTLs, the samples used for finding them and for estimating evolutionary potential should not overlap, but they should also not be completely unrelated in case the genetic basis of adaptive evolution varies among populations. With most populations and species facing climate warming, it may be of interest to determine whether and where relevant genetic variation within species can be found (e.g., ref. 92); approaches are reviewed in Capblancq et al. (94). Screening for adaptive variation may make sense in the context of environmental change or targeted gene flow (e.g., ref. 82). However, also general genetic diversity—unrelated to QTLs—will remain a good general predictor of selection response, as demonstrated by Ørsted et al. (44) (Fig. 3B).
Conclusions
Conservation genetics remains a rapidly developing discipline highly relevant to the management of the increasing number of threatened populations and species. The early paradigms emphasizing the important methods of maintaining genetic variation within populations and minimizing inbreeding are critical to the long-term success of threatened populations. The ongoing effects of climate change, habitat loss, and fragmentation mean that these concerns are increasingly critical as more and more natural populations become threatened due to rapid decreases in their species range and in abundance. Overall, genetic marker variation is an important parameter to predict the evolvability of populations when there is limited information on the genetic basis of traits and where the highly polygenic nature of selection responses makes it difficult to predict genetic changes.
Study designs in CG can be developed that take advantage of emerging technologies and tools. An a priori research question is key, ideally including hypotheses about causes of decline, and rigorous spatial or temporal sampling of populations over a spatial scale that ideally includes a mixture of intact populations and affected populations. Several good examples exist in the literature, including studies on the decline of salmon and trout populations and its causes (18, 95). The use of high-density genomic SNP-based markers should follow good practice in data preparation and filtering, and in standardized output of variables, such as autosomal heterozygosity (79). An initial analysis should always test whether populations are distinct entities and whether there have been changes in size over recent times. As detailed analyses emerge, it should be possible to refine knowledge around genetic and evolutionary threats to small populations. With this added knowledge, more information should be extractable from low-resource genetic analysis applied to populations and species of conservation concern.
While genetic diversity is per se not a good indicator of conservation status of a taxon, a relatively recent decline in genetic diversity across populations can be used as an indicator that should be considered with other CB threats. Here, an ideal approach is to sample populations over time and analyze changes in genetic variation (e.g., refs. 18 and 95). In principle, stored samples (of herbaria or museum collections) can be used for comparison, but sample sizes are typically low, DNA degradation is common, and estimates are associated with high variances (96). An interesting area for the future is to examine changes in genetic parameters across interacting sets of organisms, given the cascade of effects that can develop as key functional groups of organisms are lost (97). Mutation accumulation has so far been underappreciated in CG. Performance declines in natural populations associated with mutation accumulation have been estimated to be 50% in Arabidopsis lyrata (98) and more data on this issue is needed.
CG offers important tools to be used in monitoring and managing populations. It is unfortunate that these tools are so infrequently applied in the direct management of threatened species and that they are often only given cursory consideration in management and recovery plans. Management strategies, like genetic mixing and the creation of corridors, will become increasingly important in attempts to minimize biodiversity loss in the face of environmental deterioration, and CG tools provide key inputs in applying these options. CG will be a critical component of our urgent need to be managing species for the future and not just the present.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
There are no data underlying this work.
References
- 1.Sprunt I. V. A., et al. , Comparative productivity of six bald eagle populations. Trans. N. Am. Wildl. Nat. Resourc. Conf. 38, 96–106 (1973). [Google Scholar]
- 2.Soulé M. E., What is Conservation Biology? A new synthetic discipline addresses the dynamics and problems of perturbed species, communities, and ecosystems. Bioscience 35, 727–734 (1985). [Google Scholar]
- 3.Courchamp F., Clutton-Brock T., Grenfell B., Inverse density dependence and the Allee effect. Trends Ecol. Evol. 14, 405–410 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Lande R., Genetics and demography in biological conservation. Science 241, 1455–1460 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Frankham R., Ballou J. D., Briscoe D. A., Introduction to Conservation Genetics (Cambridge University Press, 2002). [Google Scholar]
- 6.Hedrick P. W., Miller P. S., Conservation genetics, techniques and fundamentals. Ecol. Appl. 2, 30–46 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Wright S., Evolution in Mendelian populations. Genetics 16, 97–159 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caballero A., Developments in the prediction of effective population size. Heredity 73, 657–679 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Hubby J. L., Lewontin R. C., A molecular approach to the study of genic heterozygosity in natural populations. I. The number of alleles at different loci in Drosophila pseudoobscura. Genetics 54, 577–594 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litt M., Luty J. A., A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 44, 397–401 (1989). [PMC free article] [PubMed] [Google Scholar]
- 11.Crow J. F., Kimura M., An Introduction to Population Genetics Theory (Harper & Row, N.Y., 1970). [Google Scholar]
- 12.Ohta T., Kimura M., A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 89, 367–370 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Keller L. F., Waller D. M., Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (2002). [Google Scholar]
- 14.Houle D., The maintenance of polygenic variation in finite populations. Evolution 43, 1767–1780 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Hill W. G., Predictions of response to artificial selection from new mutations. Genet. Res. 40, 255–278 (1982). [DOI] [PubMed] [Google Scholar]
- 16.Robertson A., A theory of limits in artificial selection. Proc. R. Soc. Lond. B Biol. Sci. 153, 234–249 (1960). [Google Scholar]
- 17.Lynch M., Conery J., Bürger R., Mutational meltdowns in sexual populations. Evolution 49, 1067–1080 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Lehnert S. J., et al. , Genomic signatures and correlates of widespread population declines in salmon. Nat. Commun. 10, 2996 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitrovski P., Hoffmann A. A., Heinze D. A., Weeks A. R., Rapid loss of genetic variation in an endangered possum. Biol. Lett. 4, 134–138 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner B. A., Hoban S., Luikart G., IUCN Red List and the value of integrating genetics. Conserv. Genet. 21, 795–801 (2020). [Google Scholar]
- 21.Schwartz M. K., Luikart G., Waples R. S., Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22, 25–33 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Moritz C., Conservation units and translocations, strategies for conserving evolutionary processes. Hereditas 130, 217–228 (1999). [Google Scholar]
- 23.Weeks A. R., Stoklosa J., Hoffmann A. A., Conservation of genetic uniqueness of populations may increase extinction likelihood of endangered species: The case of Australian mammals. Front. Zool. 13, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman R. A., Weeks A. R., Hoffmann A. A., Balancing genetic uniqueness and genetic variation in determining conservation and translocation strategies: A comprehensive case study of threatened dwarf galaxias, Galaxiella pusilla (Mack) (Pisces: Galaxiidae). Mol. Ecol. 22, 1820–1835 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Pierson J. C., et al. , Genetic factors in threatened species recovery plans on three continents. Front. Ecol. Environ. 14, 433–440 (2016). [Google Scholar]
- 26.Teixeira J. C., Huber C. D., The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. U.S.A. 118, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romiguier J., et al. , Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515, 261–263 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Willoughby J. R., et al. , The reduction of genetic diversity in threatened vertebrates and new recommendations regarding IUCN conservation rankings. Biol. Conserv. 191, 495–503 (2015). [Google Scholar]
- 29.Cook C. N., Sgrò C. M., Aligning science and policy to achieve evolutionarily enlightened conservation. Conserv. Biol. 31, 501–512 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Kardos M., et al. , The crucial role of genome-wide genetic variation in conservation. bioRxiv [Preprint] (2021) 10.1101/2021.07.05.451163. Accessed 6 July 2021. [DOI] [PMC free article] [PubMed]
- 31.Nei M., Maruyama T., Chakraborty R., The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975). [DOI] [PubMed] [Google Scholar]
- 32.Frankham R., Relationship of genetic variation to population size in wildlife. Conserv. Biol. 10, 1500–1508 (1996). [Google Scholar]
- 33.Leimu R., Mutikainen P., Koricheva J., Fischer M., How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 94, 942–952 (2006). [Google Scholar]
- 34.Pinsky M. L., Palumbi S. R., Meta-analysis reveals lower genetic diversity in overfished populations. Mol. Ecol. 23, 29–39 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Rivera-Ortiz F. A., Aguilar R., Arizmendi M. D. C., Quesada M., Oyama K., Habitat fragmentation and genetic variability of tetrapod populations. Anim. Conserv. 18, 249–258 (2015). [Google Scholar]
- 36.González A. V., Gómez-Silva V., Ramírez M. J., Fontúrbel F. E., Meta-analysis of the differential effects of habitat fragmentation and degradation on plant genetic diversity. Conserv. Biol. 34, 711–720 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Van Buskirk J., Willi Y., The change in quantitative genetic variation with inbreeding. Evolution 60, 2428–2434 (2006). [PubMed] [Google Scholar]
- 38.Wood J. L., Yates M. C., Fraser D. J., Are heritability and selection related to population size in nature? Meta-analysis and conservation implications. Evol. Appl. 9, 640–657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willi Y., Van Buskirk J., Hoffmann A. A., Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433–458 (2006). [Google Scholar]
- 40.Hedrick P. W., Kalinowski S. T., Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31, 139–162 (2000). [Google Scholar]
- 41.Keller L. F., Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia). Evolution 52, 240–250 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Wright L. I., Tregenza T., Hosken D. J., Inbreeding, inbreeding depression and extinction. Conserv. Genet. 9, 833–843 (2007). [Google Scholar]
- 43.Hoffmann A. A., Sgrò C. M., Kristensen T. N., Revisiting adaptive potential, population size, and conservation. Trends Ecol. Evol. 32, 506–517 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Ørsted M., Hoffmann A. A., Sverrisdóttir E., Nielsen K. L., Kristensen T. N., Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLoS Genet. 15, e1008205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spielman D., Brook B. W., Frankham R., Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U.S.A. 101, 15261–15264 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellegren H., Hartman G., Johansson M., Andersson L., Major histocompatibility complex monomorphism and low levels of DNA fingerprinting variability in a reintroduced and rapidly expanding population of beavers. Proc. Natl. Acad. Sci. U.S.A. 90, 8150–8153 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abadía-Cardoso A., Freimer N. B., Deiner K., Garza J. C., Molecular population genetics of the northern elephant seal Mirounga angustirostris. J. Hered. 108, 618–627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson J. A., Brown C., Kim B. Y., Lohmueller K. E., Wayne R. K., Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr. Biol. 28, 3487–3494.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laws R. J., Jamieson I. G., Is lack of evidence of inbreeding depression in a threatened New Zealand robin indicative of reduced genetic load? Anim. Conserv. 14, 47–55 (2011). [Google Scholar]
- 50.Kimura M., Maruyama T., Crow J. F., The mutation load in small populations. Genetics 48, 1303–1312 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glémin S., How are deleterious mutations purged? Drift versus nonrandom mating. Evolution 57, 2678–2687 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Boakes E. H., Wang J., Amos W., An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity 98, 172–182 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Reed D. H., Fox C. W., Enders L. S., Kristensen T. N., Inbreeding-stress interactions: Evolutionary and conservation consequences. Ann. N. Y. Acad. Sci. 1256, 33–48 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Ørsted M., Hoffmann A. A., Rohde P. D., Sørensen P., Kristensen T. N., Strong impact of thermal environment on the quantitative genetic basis of a key stress tolerance trait. Heredity 122, 315–325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang W., et al. , Context-dependent genetic architecture of Drosophila life span. PLoS Biol. 18, e3000645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bijlsma R., Bundgaard J., Van Putten W. F., Environmental dependence of inbreeding depression and purging in Drosophila melanogaster. J. Evol. Biol. 12, 1125–1137 (1999). [Google Scholar]
- 57.Frankham R., Genetic rescue of small inbred populations: Meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 24, 2610–2618 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Hill W. G., Robertson A., The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (1966). [PubMed] [Google Scholar]
- 59.Leimu R., Fischer M., A meta-analysis of local adaptation in plants. PLoS One 3, e4010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castellano D., Coronado-Zamora M., Campos J. L., Barbadilla A., Eyre-Walker A., Adaptive evolution is substantially impeded by Hill-Robertson interference in Drosophila. Mol. Biol. Evol. 33, 442–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill W. G., Kirkpatrick M., What animal breeding has taught us about evolution. Annu. Rev. Ecol. Evol. Syst. 41, 1–19 (2010). [Google Scholar]
- 62.Moorad J. A., A demographic transition altered the strength of selection for fitness and age-specific survival and fertility in a 19th century American population. Evolution 67, 1622–1634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moorad J. A., Wade M. J., Selection gradients, the opportunity for selection, and the coefficient of determination. Am. Nat. 181, 291–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barton N. H., Turelli M., Effects of genetic drift on variance components under a general model of epistasis. Evolution 58, 2111–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Wang J., Caballero A., Keightley P. D., Hill W. G., Bottleneck effect on genetic variance. A theoretical investigation of the role of dominance. Genetics 150, 435–447 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weeks A. R., et al. , Genetic rescue increases fitness and aids rapid recovery of an endangered marsupial population. Nat. Commun. 8, 1071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kyriazis C. C., Wayne R. K., Lohmueller K. E., Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett. 5, 33–47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffmann A. A., Miller A. D., Weeks A. R., Genetic mixing for population management: From genetic rescue to provenancing. Evol. Appl. 14, 634–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frankham R., et al. , Predicting the probability of outbreeding depression. Conserv. Biol. 25, 465–475 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Fitzpatrick S. W., et al. , Genomic and fitness consequences of genetic rescue in wild populations. Curr. Biol. 30, 517–522.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Willi Y., Hoffmann A. A., Demographic factors and genetic variation influence population persistence under environmental change. J. Evol. Biol. 22, 124–133 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Franklin I. R., “Evolutionary change in small populations” in Conservation Biology: An Evolutionary-Ecological Perspective, Soule M. E., Wilcox B. A., Eds. (Sinauer Associates Inc., 1980), pp. 135–149. [Google Scholar]
- 73.Frankham R., Bradshaw C. J. A., Brook B. W., Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 170, 56–63 (2014). [Google Scholar]
- 74.Hoffmann A. A., et al. , An endangered flightless grasshopper with strong genetic structure maintains population genetic variation despite extensive habitat loss. Ecol. Evol. 11, 5364–5380 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang P., et al. , Genomic consequences of long-term population decline in brown eared pheasant. Mol. Biol. Evol. 38, 263–273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu L., et al. , Genetic consequences of long-term small effective population size in the critically endangered pygmy hog. Evol. Appl. 14, 710–720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caballero A., García-Dorado A., Allelic diversity and its implications for the rate of adaptation. Genetics 195, 1373–1384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hohenlohe P. A., Funk W. C., Rajora O. P., Population genomics for wildlife conservation and management. Mol. Ecol. 30, 62–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt T. L., Jasper M., Weeks A. R., Hoffmann A. A., Unbiased population heterozygosity estimates from genome-wide sequence data. Methods Ecol. Evol. 12, 1888–1898 (2021). [Google Scholar]
- 80.Pouyet F., Aeschbacher S., Thiéry A., Excoffier L., Background selection and biased gene conversion affect more than 95% of the human genome and bias demographic inferences. eLife 7, e36317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oh K. P., Aldridge C. L., Forbey J. S., Dadabay C. Y., Oyler-McCance S. J., Conservation genomics in the Sagebrush Sea: Population divergence, demographic history, and local adaptation in sage-grouse (Centrocercus spp.). Genome Biol. Evol. 11, 2023–2034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollenbeck C. M., Portnoy D. S., Gold J. R., A method for detecting recent changes in contemporary effective population size from linkage disequilibrium at linked and unlinked loci. Heredity 117, 207–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V. C., Foll M., Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X., et al. , Out of Tibet: Genomic perspectives on the evolutionary history of extant pikas. Mol. Biol. Evol. 37, 1577–1592 (2020). [DOI] [PubMed] [Google Scholar]
- 86.Grossen C., Guillaume F., Keller L. F., Croll D., Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nat. Commun. 11, 1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar P., Henikoff S., Ng P. C., Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Henn B. M., et al. , Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. U.S.A. 113, E440–E449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Agrawal A. F., Whitlock M. C., Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187, 553–566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van der Valk T., de Manuel M., Marques-Bonet T., Guschanski K., Estimates of genetic load suggest frequent purging of deleterious alleles in small populations. bioRxiv [Preprint] (2021). 10.1101/696831. Accessed 22 April 2021. [DOI]
- 92.Exposito-Alonso M., Burbano H. A., Bossdorf O., Nielsen R., Weigel D.; 500 Genomes Field Experiment Team, Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 573, 126–129 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Walden N., Lucek K., Willi Y., Lineage-specific adaptation to climate involves flowering time in North American Arabidopsis lyrata. Mol. Ecol. 29, 1436–1451 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Capblancq T., Fitzpatrick M. C., Bay R. A., Exposito-Alonso M., Keller S. R., Genomic prediction of (mal)adaptation across current and future climatic landscapes. Annu. Rev. Ecol. Evol. Syst. 51, 245–269 (2020). [Google Scholar]
- 95.Hansen M. M., Fraser D. J., Meier K., Mensberg K.-L. D., Sixty years of anthropogenic pressure: A spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Mol. Ecol. 18, 2549–2562 (2009). [DOI] [PubMed] [Google Scholar]
- 96.Wandeler P., Hoeck P. E., Keller L. F., Back to the future: Museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Zipkin E. F., DiRenzo G. V., Ray J. M., Rossman S., Lips K. R., Tropical snake diversity collapses after widespread amphibian loss. Science 367, 814–816 (2020). [DOI] [PubMed] [Google Scholar]
- 98.Perrier A., Sánchez-Castro D., Willi Y., Expressed mutational load increases toward the edge of a species’ geographic range. Evolution 74, 1711–1723 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.