Abstract
Background
MicroRNAs (miRNAs) are good candidates as biomarkers for Lung cancer (LC). The aim of this article is to figure out the diagnostic value of both single and combined miRNAs in LC.
Methods
Normative meta-analysis was conducted based on PRISMA. We assessed the diagnostic value by calculating the combined sensitivity (Sen), specificity (Spe), positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) and the area under the curve (AUC) of single and combined miRNAs for LC and specific subgroups.
Results
A total of 80 qualified studies with a total of 8971 patients and 10758 controls were included. In non-small cell lung carcinoma (NSCLC), we involved 20 single-miRNAs and found their Sen, Spe and AUC ranged from 0.52-0.81, 0.66-0.88, and 0.68-0.90, respectively, specially, miR-19 with the maximum Sen, miR-20 and miR-10 with the highest Spe as well as miR-17 with the maximum AUC. Additionally, we detected miR-21 with the maximum Sen of 0.74 [95%CI: 0.62-0.83], miR-146 with the maximum Spe and AUC of 0.93 [95%CI: 0.79-0.98] and 0.89 [95%CI: 0.86-0.92] for early-stage NSCLC. We also identified the diagnostic power of available panel (miR-210, miR-31 and miR-21) for NSCLC with satisfying Sen, Spe and AUC of 0.82 [95%CI: 0.78-0.84], 0.87 [95%CI: 0.84-0.89] and 0.91 [95%CI: 0.88-0.93], and furtherly constructed 2 models for better diagnosis.
Conclusions
We identified several single miRNAs and combined groups with high diagnostic power for NSCLC through pooled quantitative analysis, which shows that specific miRNAs are good biomarker candidates for NSCLC and further researches needed.
Keywords: Lung cancer, microRNA, diagnostic biomarker, early diagnosis, NSCLC
1. Introduction
Lung cancer (LC) is a type of malignant neoplasm arising from bronchial mucosa or glands which accounts for the largest proportion of cancer globally in consideration of patient quantity as well as mortality [1, 2]. Histologically, LC is categorized as small cell lung carcinoma (SCLC) and non-small cell lung carcinoma (NSCLC) which is further classified as adenocarcinoma (AD), squamous cell carcinoma (SCC) and large cell carcinoma (LCC) [3]. NSCLC, accounting for 80–85% of LC, harbours specific molecular and genetic characteristics [4], indicating the likelihood to distinguish NSCLC from other subtypes of LC under the help of particular biomarkers. We have identified the association between interleukin polymorphisms and protein levels with lung cancer susceptibility as well as phenotypes in our previous study [5]. Current challenges lying on its early diagnosis: lung tissue biopsy, being regarded as “gold standard”, has to be done through invasive bronchoscopy or surgical excision [4]; CT [6] as well as PET-CT [7] are widely applied in the definition of TNM stage in NSCLC, which still lacks specific diagnostic directivity towards NSCLC. Novel diagnostic methods such as the detection of circulating tumour cells or other circulating biomarkers [8] need further confirmation [9]. Among all possible biomarkers under research, microRNAs (miRNAs) are considered to be one of the most promising objects in terms of early detection of NSCLC [10].
MiRNAs are non-coding RNA with the length varied from 18 to 25 nucleotides who involve in the regulation of gene expression through suppression of mRNA directly. We previously proved that miRNAs could serve as diagnostic biomarker in asthma [11]. The abnormal expression level of multiple miRNAs has been determined among diverse cancers [12,13]: oncogenic miRNAs are the ones overexpressed inside tumour cells which promote the development and proliferation of cancerous cells; tumour-suppressive miRNAs are the ones who are down-regulated during the process of tumorigenesis. MiRNAs could be expelled from a tumour or stromal cells to the body or secreted fluid in the form of exosomes [14], providing the possibility for detection of exosomal miRNAs to be novel but useful approach of a cancer diagnosis.
Till now, a group of miRNAs has been proved to participate in LC cancerization, proliferation, and metastasis and their target genes have been confirmed [15]: miR-21 serves as oncogene and participates in multiple pathways controlling NSCLC tumorigenesis such as proliferation and angiogenesis [16]; miR-148a suppresses invasion of NSCLC cells by affecting Wnt1 pathway [17]; exosomal miR-619-5p improves angiogenesis as well as metastasis in NSCLC by inhibiting RCAN1.4 [18]. Remarkably, specific miRNAs like miR-590-5p and miR-26b possess potential to be diagnostic or prognostic biomarkers in NSCLC [19,20]. Specific miRNA panels even show their potential on histological subcategorization of NSCLC [10]: the concentration of miR-181b-5p, miR-30a-3p, miR-30e-3p, and miR-361-5p suggests higher possibility for AD while the combination of miR-10b-5p, miR-15b-5p, and miR-320b points to SCC. However, whether certain miRNA or miRNA panel could serve as good biomarkers candidates for NSCLC diagnosis or detection of early-stage NSCLC remains unknown.
Therefore, we aimed to clarify whether miRNAs, single miRNAs and miRNA panels, can serve as a biomarker for NSCLC and other LC subtypes by performing a quantitative analysis of previously published miRNA expression profiling studies? Did the results show any difference among various sample sources and clinical stages?
2. Materials and methods
2.1. Search strategy
The Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines were followed [21]. We carefully search four databases (PubMed, Embase, Web of Science and Cochrane Library) by using keywords ((lung neoplasms) OR (lung tumour) OR (lung cancer) OR (lung carcinoma) OR (pulmonary neoplasms) OR (pulmonary cancer)) AND ((miRNA) OR (microRNA)) by Mar 31th, 2021. References and citing articles of the original articles were also searched artificially for further information extraction. All literatures were searched and screened by two independent staff because of objectivity principle. If there was a dispute, a third party was appreciated to make final decision.
2.2. Inclusion criteria based on PICOS
We strictly selected eligible studies by the principle of PICOS as follow:
Participants: patients attained the pathological diagnosis of LC who were further graded according to the 7th and 8th edition of lung cancer TNM grading by the International Association for the Study of Lung Cancer (including I, II, III, and IV) [22,23]. Available and detailed diagnosis of LC subtypes (including SCLC and NSCLC which further involved AD, SCC and LCC) defined by WHO classification in 2004 and 2015 [24,25] were appreciated.
Intervention: microarray or quantitative real-time polymerase chain reaction (qRT-PCR) for detection of the miRNAs’ expression levels in all participants;
Control: healthy people or cancer-free controls;
Outcomes: diagnostic data of individual miRNA or miRNA panels for lung cancer was provided, including true positive (TP), false positive (FP), false negative (FN), and true negative (TN);
Studies: case-control studies or cohort studies.
2.3. Exclusion criteria
We also carefully excluded articles with the following characteristics: (1) the literature not written in English; (2) researches not of the type as original articles, such as review, case report, letter, or conference summary; (3) the publication on the same topic from the same team which also shared overlapped participants;(4) deficiency of detailed data for combined analysis; (5) articles of each specific miRNA which owned less than 4 records; (6) articles of miRNA panels whose miRNA types were not all researched in our individual miRNA section.
2.4. Data extraction and quality assessment
We reviewed all the eligible publications and extracted the following information: the first author, year of publication, size of cases and controls, diagnosis of LC (including subtypes and staging), involved miRNAs and their expression outcome, as well as sources of the sample. We further normalized names of miRNAs from different studies basing on miRbase version 22 released in 2018 (http://www.mirbase.org/).
We used the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) [26], which consists of four key domains including patient selection, index test, reference standard, flow and timing, to evaluate the included papers by RevMan5.3 with the levels of “high”, “low” and “unclear”.
2.5. Statistical analysis
All the analyses were performed in STATA version 14 (Stata Corporation, College Station, TX, USA). The Spearman correlation coefficient was used to access the threshold effect, with r > 0.6 and p < .05, indicating a significant threshold effect between studies [27], which means effect size of included studies could be combined and further analyzed. The bivariate mixed‐effects mode [28] was used to calculate the indicators reflecting the diagnostic effect, such as sensitivity (Sen), specificity (Spe), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and corresponding 95% credible interval (CI). The summary receiver operator characteristic curve (SROC) which was plotted based on Sen and Spe and the area under the SROC curve (AUC) was also used to test the pooled diagnostic value of miRNAs.
Heterogeneity between studies was evaluated by the I2 test, with I2>25% indicating heterogeneity [29]. When the I2>50%, it suggested the heterogeneity between studies was high [30]. Then the subgroup and meta-analysis would be performed to find the sources of heterogeneity, which we performed from the aspect of sample sources (respiratory-based sample vs. blood-based sample) and control group (health and cancer-free).
We also performed the sensitivity analysis for further analysis of those studies that resulting in large heterogeneity [31]. By comparing the changes in effect size and 95% confidence intervals before and after the inclusion and exclusion of those studies, we accessed the impact of those studies on the process of pool analysis.
We used two methods, the Deek and the Funnel plot [32], to evaluate the publication bias based on the situation of a different number of studies in various miRNAs. When the number of studies more than 10, the Deek method was adopted, otherwise, the Funnel plot method was used. When the p > .05 in Deek or the Funnel plot is symmetrical, it suggests that there may be no obvious publication bias. Otherwise, the trim and fill method [33] will be used to review and identify the publication bias.
3. Results
3.1. Characteristics of the included studies and quality assessment
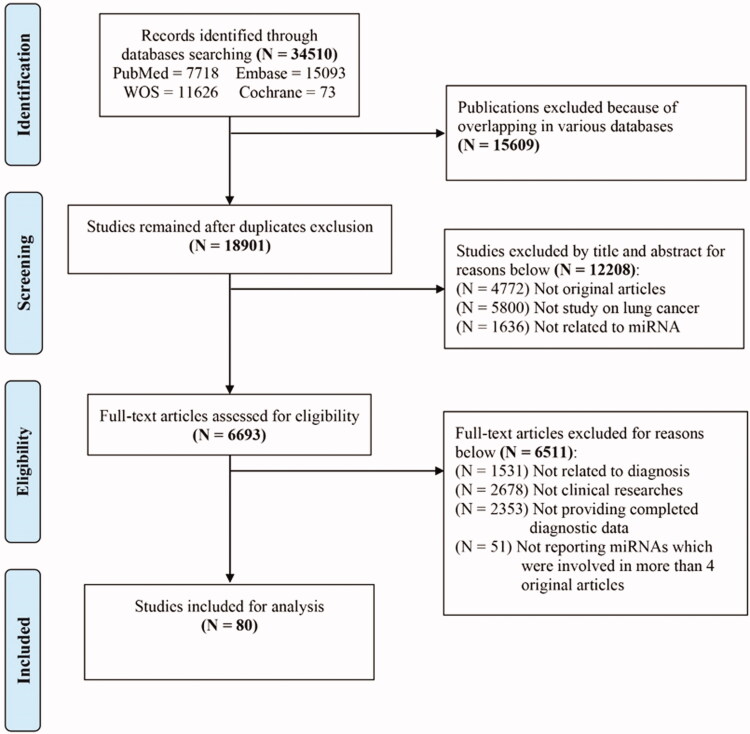
Based on the listed criteria of inclusion and exclusion in Method and PRISMA standard, a total of 80 studies with 8971 patients and 10758 controls were eventually included (Figure 1), among which, 24 single-miRNAs (miR-21, -210, -145, -155, -486, -17, -126, -223, -31, -20, -182, -146, -205, -19, -221, -200, Let-7, -125, -7, -10, -375, -150, -92, -25) were involved as reported in more than 4 articles in LC. Besides, a variety of panels with several miRNAs were also reported. Based on the QUADAS-2, these researches’ quality was at the middle and upper grades (Supplementary Appendix Figure 1). All involved original article harboured ethical approval.
Figure 1.
Flowchart of study selection based on the inclusion and exclusion criteria.
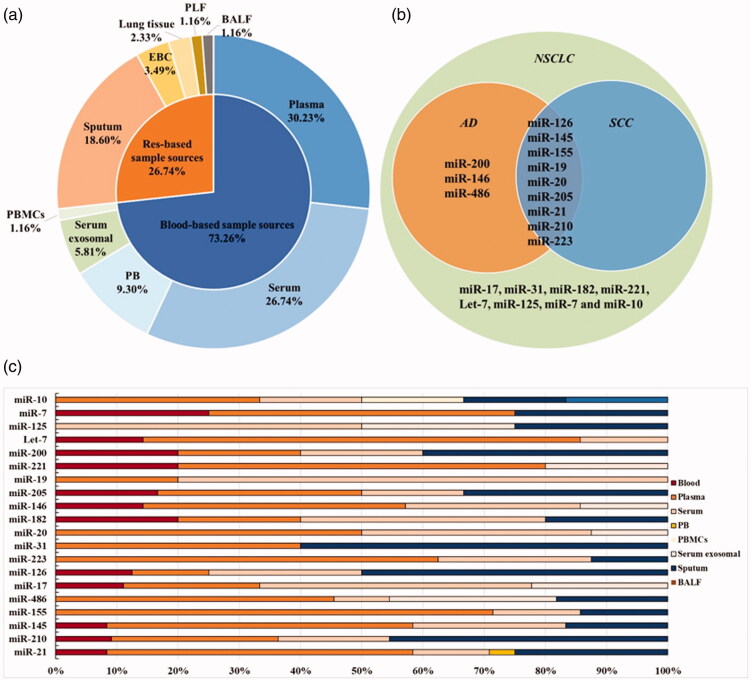
For all included researches (Table 1), the major LC subtype involved was NSCLC (69 studies, 86.3%), of which AD (13 studies) furtherly occupied the majority and SCC (10 studies) following after, while SCLC accounted for the least (1 study, 1.25%) and the rest (10 studies, 12.5%) lacking of clear description of LC subtypes. Additionally, except 15 articles (18.8%) without description of staging, we noticed that there were 44 studies (55%) stated I–IV stages, 17 studies (21.3%) focussed on early stage (I–II) and 4 studies (3.8%) concentrated on stage of III–IV. Furthermore, considering sample sources of researched miRNAs, blood derived samples sources (peripheral whole blood, serum, plasma or specific ingredients of peripheral blood mononuclear cell or serum exosomal) accounted for the main sources (73.26%) while respiratory system derived samples sources occupied the proportion of 26.74%, which included sputum, pleural lavage fluid, lung tissue, bronchoalveolar lavage fluid and exhaled breath condensate (Figure 2(a)). Therefore, we took these features, including LC subtypes, clinical staging and sample sources, into consideration in subgroup analysis.
Table 1.
The main features of eligible studies that related to the diagnosis of miRNA for LC.
| Study ID | Region | NO. of Case/Control | Tumor-type | Stage* | Sample sources | PMID |
|---|---|---|---|---|---|---|
| 2020.Zhang, Z. J. [34] | China | 330/312 | NSCLC | I-IV | Serum exosomal | 33178588 |
| 2020.Yang, C. [35] | China | 74/23 | NSCLC | III-IV | Serum | 32466856 |
| 2020.Wu, Q. [36] | USA | 48/48 | NSCLC | I-II | Serum exosomal | 32021461 |
| 2020.Wang, W. [37] | China | 54/28 | NSCLC | I | Blood | 32596148 |
| 2020.Wang, J. Y. [38] | China | 82/90 | AD | I-IV | Serum | 32388809 |
| 2020.Liu, X. [39] | China | 245/245 | NSCLC | I-IV | Serum | 31906699 |
| 2020.Liu, C. [40] | USA | 64/15 | NSCLC | I-II | Serum exosomal | 32265989 |
| 2020.Liao, J. [41] | USA | 132/127 | NSCLC | I-IV | Sputum, Plasma | 31994346 |
| 2020.Ghany, S. M. A. [42] | Egypt | 70/34 | NSCLC | NA | Plasma | NA |
| 2020.Fehlmann, T. [43] | USA | 606/2440 | LC | I-IV | Blood | 32134442 |
| 2020.Asakura, K. [44] | Japan | 1566/2178 | LC | I-IV | Serum | 32193503 |
| 2019.Zou, J. G. [45] | China | 50/30 | NSCLC | NA | Serum | 30779079 |
| 2019.Zhang, Y. [46] | China | 172/137 | NSCLC | I-III | Serum exosomal | 31146974 |
| 2019.Xi, K. [47] | China | 67/25 | NSCLC | I-II | Plasma | 31632908 |
| 2019.Wang, S. [48] | China | 50/24 | NSCLC | NA | Plasma | 31049003 |
| 2019.Szczyrek, M. [49] | Poland | 160/45 | NSCLC | I-IV | Plasma | 31115013 |
| 2019.Switlik, W. Z. [50] | Poland | 14/29 | AD | NA | Serum | 30950648 |
| 2019.Sui, A. [51] | China | 76/60 | NSCLC | I-IV | Serum | 30867756 |
| 2019.Sheervalilou, R. [52] | Iran | 47/41 | NSCLC | I-IV | Plasma | 32215263 |
| 2019.Roman-Canal, B. [53] | Spain | 14/21 | LC | NA | PLF | 31636323 |
| 2019.Li, J. [54] | China | 471/489 | AD, SCC | I-IV | Plasma | 31674214 |
| 2019.Hetta, H. F. [55] | USA | 40/20 | NSCLC | I-IV | Plasma | 31244320 |
| 2019.Abdollahi, A. [56] | Iran | 43/43 | NSCLC | I-IV | Blood | 31236600 |
| 2018.Yang, Y. L. [57] | China | 194/199 | NSCLC | III-IV | PBMCs | 30064233 |
| 2018.Yang, Y. [58] | China | 104/50 | NSCLC | I-IV | Serum | 29430184 |
| 2018.Xi, K. X. [59] | China | 42/15 | NSCLC | I-II | Plasma | 30174846 |
| 2018.Sun, Y. [60] | China | 28/28 | AD | I-IV | Plasma | 29103767 |
| 2018.Qiu, F. [61] | China | 58/42 | LC | NA | PB | 30556877 |
| 2018.Poroyko, V. [62] | USA | 20/10 | LC | NA | Serum exosomal | 29731983 |
| 2018.Mohamed, M. A. [63] | Egypt | 50/50 | LC | I-IV | Lung tissue | 29437031 |
| 2018.Leng, Q. [64] | USA | 56/28 | NSCLC, SCC, AD | I-IV | Plasma | 29783093 |
| 2018.Fan, L. H. [65] | China | 128/79 | NSCLC | I | Serum | NA |
| 2018.Bao, M. [66] | China | 80/75 | NSCLC | I-IV | Serum, Lung tissue |
31938164 |
| 2018.Bagheri, A. [67] | Iran | 30/30 | NSCLC, SCC, AD | I-IV | Sputum | 30485511 |
| 2018.Aiso, T. [68] | Japan | 56/26 | NSCLC | I-IV | Serum | 30405804 |
| 2018.Abu-Duhier, F.M. [69] | Saudi Arabia | 80/80 | AD, SCC | I-IV | Plasma | 30256067 |
| 2017.Zhang, H. [70] | China | 129/83 | NSCLC | I-II | Plasma | 28356944 |
| 2017.Yu, Y. [71] | USA | 50/30 | SCLC | I-IV | Plasma | 28106539 |
| 2017.Sheervalilou, R. [72] | Iran | 30/30 | NSCLC | I-III | BALF, Sputum | NA |
| 2017.Shang, A. Q. [73] | China | 127/112 | NSCLC | I-IV | Serum | 28253725 |
| 2017.Lv, S. [74] | China | 160/160 | AD | I-IV | Serum | 28123597 |
| 2017.Leng, Q. [75] | USA | 126/118 | NSCLC | I-IV | Plasma | 29340099 |
| 2017.Ibrahim, F. K. [76] | Egypt | 15/15 | LC | NA | EBC | NA |
| 2017.Bagheri, A. [77] | Iran | 17/17 | NSCLC | I-IV | Sputum, Lung tissue |
29090068 |
| 2016.Zhu, W. [78] | China | 112/104 | NSCLC | I-II | Serum | 27093275 |
| 2016.Zaporozhchenko, I.A. [79] | Russia | 75/50 | SCC, AD | II-IV | Plasma | 27768748 |
| 2016.Wang, X. [80] | China | 59/59 | NSCLC | I-IV | Plasma | 27499953 |
| 2016.Su, Yb. [81] | USA | 57/62 | NSCLC | I | Sputum | 27777637 |
| 2016.Su, Ya. [82] | USA | 117/174 | NSCLC | I | Sputum | 27176474 |
| 2016.Razzak, R. [83] | Canada | 43/10 | NSCLC | I-II | Sputum | 27122989 |
| 2016.Jia, Y. C. [84] | China | 35/30 | NSCLC, SCLC | NA | Plasma | NA |
| 2016.Chen, J. L. [85] | China | 30/30 | NSCLC | I-II | EBC, Serum | NA |
| 2015.Zhao, W. [86] | China | 80/60 | NSCLC | NA | Serum | 26628958 |
| 2015.Yang, J. S. [87] | China | 152/300 | NSCLC | I-IV | Serum | 25501703 |
| 2015.Yan, H. J. [88] | China | 300/300 | NSCLC | I-IV | PB | 25765717 |
| 2015.Xing L. [89] | USA | 203/227 | LC | I-II | Sputum | 25593345 |
| 2015.Wang, R. J. [90] | China | 70/70 | NSCLC | NA | Serum | 25755772 |
| 2015.Wang, P. [91] | China | 142/111 | NSCLC | I-II | Serum | 25639977 |
| 2015.Su, J. [92] | USA | 56/73 | NSCLC | NA | Sputum | 26309391 |
| 2015.Li, W. [93] | China | 11/11 | NSCLC, SCC | NA | Plasma | 26237047 |
| 2015.Fan, L. [94] | China | 164/112 | NSCLC | I-III | Serum | 26695145 |
| 2015.Dou, H. [95] | China | 120/360 | NSCLC | I-IV | Plasma | 26309587 |
| 2014.Zhu, W. Y. [96] | China | 36/44 | NSCLC | I | Serum | 24945821 |
| 2014.Li, N. [97] | USA | 35/40 | NSCLC | NA | Sputum | 24281335 |
| 2014.Geng, Q. [98] | China | 151/85 | NSCLC, SCC, AD | I-II | Plasma | 25421010 |
| 2013.Zeng, X. L. [99] | China | 64/26 | NSCLC | I-IV | PBMCs | 24286416 |
| 2013.Tang, D. [100] | China | 96/92 | NSCLC, SCC, AD | I-III | Plasma | 23462458 |
| 2013.Mozzoni, P. [101] | Italy | 54/46 | NSCLC | I-III | Plasma, EBC | 24102090 |
| 2013.Anjuman, N. [102] | USA | 43/47 | NSCLC | I | Sputum | 24053570 |
| 2013.Abd El Fattah, A. A. [103] | Egypt | 65/37 | LC | NA | Serum | 23559272 |
| 2012.Wang, B. [104] | China | 31/39 | LC | I-IV | Serum | 22638884 |
| 2011.Wei, Jb. [105] | China | 77/36 | NSCLC | I-IV | Plasma | 21627863 |
| 2011.Wei, Ja. [106] | China | 63/30 | NSCLC | I-IV | Plasma | 23483517 |
| 2011.Shen Jb. [107] | USA | 58/29 | NSCLC, SCC, AD | III-IV | Sputum | 21116241 |
| 2011.Shen Ja. [108] | USA | 108/113 | NSCLC | I-IV | Sputum, Plasma | 21864403 |
| 2011.Li, Y. [109] | China | 20/10 | NSCLC | I-IV | PB | 22866162 |
| 2011.Jeong, H. C. [110] | Korea | 35/30 | NSCLC | I-IV | Blood | 21468581 |
| 2010.Yu, L. [111] | USA | 64/58 | NSCLC, AD | I-IV | Sputum | 21351266 |
| 2010.Xing, L. [112] | USA | 67/55 | SCC | III-IV | Sputum | 20526284 |
| 2010.Xie, Y. [113] | USA | 23/17 | NSCLC | I-IV | Sputum | 19446359 |
a,b: Different articles by the same author initials in the same year. *: The stage of clinical diagnosis in the CASE group, in which stage I-IV represents patients who are not classified as stage I-II or III-IV, including II-III, I-III, I-IV. NA: not available.
Tumor type: AD: Adenocarcinoma; SCC: Squamous cell carcinoma; SCLC: Small cell lung carcinoma; NSCLC: Non-Small Cell Lung Carcinoma; LC: lung cancer.
Sample sources: EBC: exhaled breath condensate; PB: Peripheral blood; PBMCs: Peripheral blood mononuclear cells; BALF: bronchoalveolar lavage; PLF: Pleural lavage fluid.
Figure 2.
The characteristics for included miRNAs of involved articles. (a) The proportion of sample sources for researched single-miRNAs and miRNA panels miRNAs in all included articles. (b)The relationship between total reported miRNAs and LC subtypes. (c) The proportion of specimen sources among 20 included single-miRNAs. Tumour type: AD: adenocarcinoma; SCC: squamous cell carcinoma; SCLC: small cell lung carcinoma; NSCLC: non-small cell lung carcinoma. Sample sources: EBC: exhaled breath condensate; PB: peripheral blood; PBMCs: peripheral blood mononuclear cells; BALF: bronchoalveolar lavage; PLF: pleural lavage fluid.
Considering that specified diagnosis of LC histologically usually provides clinical value for the selection of therapy, we intended to identify specific miRNAs connected with LC subtypes. Since NSCLC occupied the major histological subtype of LC, here we specially focus on the diagnostic potential of 20 miRNAs being involved in NSCLC (miR-21, -210, -145, -155, -486, -17, -126, -223, -31, -20, -182, -146, -205, -19, -221, -200, Let-7, -125, -7 and -10). When analyzing the associations between researched 20 miRNAs and NSCLC subtypes, we further found 9 miRNAs reported in both AD and SCC. Besides, 3 kinds miRNAs (miR-200, −146 and −486) were specifically reported in AD (Figure 2(b)). The proportion of specimen sources among theses 20 miRNAs were diagrammatically shown in Figure 2(c). In addition, due to data limitations, the diagnostic value of the other 4 miRNAs (miR-375, -150, -92 and -25) could only be explored in LC rather than NSCLC.
3.2. Single miRNA as a diagnostic biomarker in NSCLC
3.2.1. The diagnostic value of 20 miRNAs singlely in NSCLC
We analyzed 20 single-miRNAs mentioned above (Supplementary Appendix Table 1) and found there was no significant statistical threshold effect among involved miRNAs according to Spearman (Supplementary Appendix Table 2). Besides, slight heterogeneity as well as publication bias which could be corrected were observed, indicating the rationality of pooled analysis respectively (Supplementary Appendix Figure 2, Supplementary Appendix Table 3). As for the other 4 miRNAs (miR-375, -150, -92 and -25) in the unclassified LC, they are similar to the above 20 miRNAs and the diagnostic data and results are shown in the (Supplementary Appendix Table 4–6, Supplementary Appendix Figure 3–4).
Through pooled effect analysis on these 20 miRNAs, their Sen, Spe, and AUC varied from 0.52-0.81, 0.66-0.88 and 0.68-0.90, respectively. According to our results, miR-19 had the highest sensitivity of 0.81 [95%CI: 0.70-0.89], 2 miRNAs (miR-20 and miR-10) both shown the best specificity of 0.88 and miR-17 owned the highest AUC value of 0.90 [95%CI: 0.87-0.92]. Additionally, there were 12 miRNAs with AUC equal or higher than 0.8, 4 miRNAs with AUC equal or higher than 0.85, which suggested that these miRNAs were more worthy of attention in NSCLC diagnosis than other miRNAs. (Table 2, Supplementary Appendix Figure 3–4). In addition, we identified miR-21 could even be a potential satisfying biomarker for AD diagnosis considering its favoured sensitivity of 0.72 [95%CI: 0.57-0.83], specificity of 0.70 [95%CI: 0.46-0.87] and AUC of 0.76 [95%CI: 0.72-0.80].
Table 2.
Detailed assessment of overall diagnostic value of 20 single-miRNAs in NSCLC.
| miRNA-type | No. of research (Case/Control) | Sen [95% CI] | Spe [95% CI] | PLR [95% CI] | NLR [95% CI] | AUC [95% CI] | DOR [95% CI] |
|---|---|---|---|---|---|---|---|
| miR-21 | 23 (1641/1503) | 0.70 [0.64–0.76] | 0.77 [0.73–0.81] | 3.1 [2.6–3.6] | 0.39 [0.32–0.46] | 0.81 [0.77–0.84] | 8 [6–11] |
| miR-210 | 11 (670/645) | 0.71 [0.57–0.82] | 0.81 [0.70–0.88] | 3.7 [2.6–5.3] | 0.36 [0.26–0.51] | 0.83 [0.79–0.86] | 10 [7–15] |
| miR-145 | 11 (762/626) | 0.73 [0.57–0.84] | 0.75 [0.66–0.82] | 2.9 [2.1–4.0] | 0.36 [0.22–0.58] | 0.80 [0.76–0.83] | 8 [4–16] |
| miR-155 | 10 (697/555) | 0.77 [0.63–0.87] | 0.84 [0.72–0.91] | 4.8 [2.6–8.9] | 0.27 [0.16–0.46] | 0.88 [0.85–0.91] | 18 [7–47] |
| miR-486 | 10 (491/404) | 0.75 [0.71–0.78] | 0.76 [0.68–0.82] | 3.1 [2.3–4.2] | 0.33 [0.28–0.39] | 0.77 [0.73–0.80] | 9 [6–14] |
| miR-17 | 9 (1339/1170) | 0.78 [0.63–0.88] | 0.86 [0.77–0.92] | 5.7 [3.1–10.3] | 0.26 [0.15–0.46] | 0.90 [0.87–0.92] | 22 [8–60] |
| miR-126 | 8 (531/430) | 0.70 [0.51–0.84] | 0.85 [0.76–0.91] | 4.6 [2.8–7.6] | 0.36 [0.21–0.62] | 0.86 [0.83–0.89] | 13 [5–31] |
| miR-223 | 7 (605/419) | 0.78 [0.73–0.83] | 0.81 [0.72–0.87] | 4.0 [2.7–6.0] | 0.27 [0.21–0.35] | 0.85 [0.82–0.88] | 15 [8–27] |
| miR-31 | 7 (873/819) | 0.73 [0.63–0.81] | 0.78 [0.74–0.81] | 3.3 [2.8–3.8] | 0.35 [0.25–0.48] | 0.80 [0.77–0.84] | 9 [6–14] |
| miR-20 | 7 (329/239) | 0.68 [0.47–0.84] | 0.88 [0.72–0.95] | 5.6 [2.6–12.2] | 0.36 [0.21–0.62] | 0.86 [0.83–0.89] | 15 [6–37] |
| miR-182 | 6 (444/227) | 0.57 [0.43–0.70] | 0.79 [0.61–0.90] | 2.7 [1.5–4.9] | 0.54 [0.42–0.70] | 0.72 [0.68–0.76] | 5 [2–10] |
| miR-146 | 6 (306/247) | 0.58 [0.36–0.76] | 0.86 [0.62–0.96] | 4.0 [1.6–10.0] | 0.49 [0.33–0.74] | 0.78 [0.75–0.82] | 8 [3–22] |
| miR-205 | 6 (332/270) | 0.57 [0.43–0.69] | 0.78 [0.53–0.92] | 2.6 [1.2–5.7] | 0.55 [0.44–0.69] | 0.68 [0.64–0.72] | 5 [2–11] |
| miR-19 | 6 (470/363) | 0.81 [0.70–0.89] | 0.66 [0.53–0.77] | 2.4 [1.7–3.4] | 0.29 [0.18–0.45] | 0.81 [0.77–0.84] | 8 [4–16] |
| miR-221 | 6 (283/176) | 0.52 [0.28–0.74] | 0.75 [0.60–0.86] | 2.1 [1.2–3.8] | 0.64 [0.39–1.04] | 0.73 [0.68–0.76] | 3 [1–9] |
| miR-200 | 5 (200/154) | 0.58 [0.48–0.67] | 0.74 [0.60–0.84] | 2.2 [1.5–3.4] | 0.57 [0.46–0.70] | 0.69 [0.64–0.73] | 4 [2–7] |
| Let-7 | 5 (297/486) | 0.67 [0.57–0.75] | 0.74 [0.65–0.81] | 2.5 [1.7–3.9] | 0.45 [0.32–0.65] | 0.76 [0.72–0.80] | 6 [3–12] |
| miR-125 | 4 (466/456) | 0.67 [0.57–0.75] | 0.66 [0.57–0.73] | 1.9 [1.6–2.3] | 0.51 [0.41–0.62] | 0.71 [0.67–0.75] | 4 [3–5] |
| miR-7 | 4 (168/86) | 0.66 [0.44–0.83] | 0.86 [0.66–0.95] | 4.8 [2.1–10.6] | 0.39 [0.23–0.66] | 0.85 [0.81–0.88] | 12 [5–28] |
| miR-10 | 4 (351/345) | 0.79 [0.72–0.85] | 0.88 [0.73–0.96] | 6.8 [2.6–17.8] | 0.24 [0.16–0.34] | 0.85 [0.82–0.88] | 29 [8–103] |
Sen: sensitivity; Spe: specificity; PLR: positive likelihood ratio; NLR: negative likelihood ratio; AUC: area under curve; DOR: diagnostic odds ratio; CI: confidence interval.
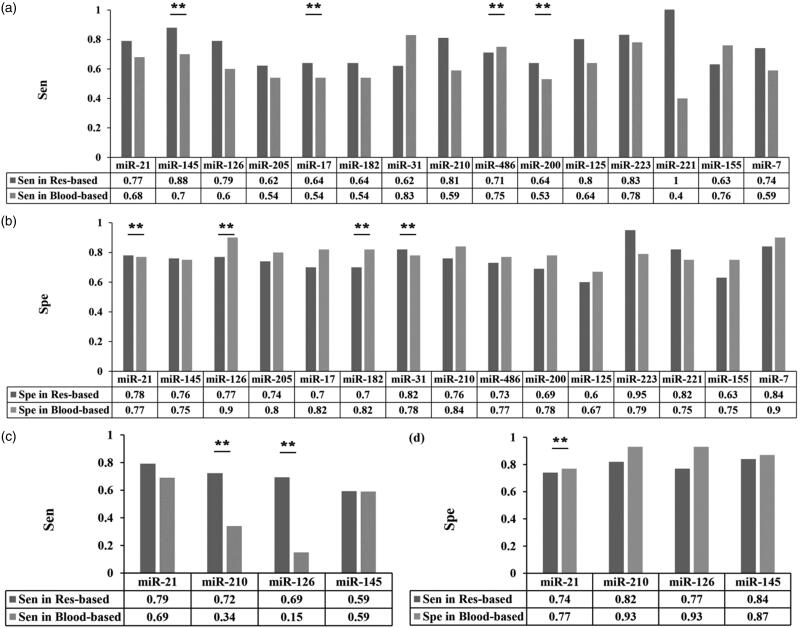
We further conducted subgroup analysis according to sample sources. We found that miR-145 had the most obvious difference in sensitivity (0.88 [95%CI: 0.68–1.00] vs. 0.70 [95%CI: 0.54–0.86]) (Figure 3(a)) and miR-126 showed the largest difference in specificity between the two kinds of samples (0.77 [95%CI: 0.44–0.82] vs. 0.90 [95%CI: 0.68–0.82]). (Figure 3(b)). These results with obvious otherness suggested different selection of miRNAs as biomarkers, depending on sample sources.
Figure 3.
The diagnostic value of several single-miRNAs in NSCLC between different sample sources. (a–d) The difference in diagnostic power of a certain miRNA between respiratory-based and blood-based samples for NSCLC (a–b) and early NSCLC (c–d) through comparison of sensitivity (a, c) and specificity (b, d). Colour in dark gray refers to respiratory-based samples while colour in light gray refers to blood-based samples.
3.2.2. The diagnostic value of 5 miRNAs singlely in early NSCLC
Since early diagnosing (stage I and II) of NSCLC help to reach better prognosis in NSCLC, we conducted analysis on assessment of diagnostic efficiency for miRNAs in early NSCLC. We further found 5 miRNAs (miR-21, -145, -126, -210, and -146) from above 20 miRNAs possessing diagnostic value for early lung cancer with Sen, Spe, and AUC varying from 0.49 to 0.74, 0.76 to 0.93, and 0.80 to 0.89, respectively. Noteworthily, miR-21 and miR-146, owned the highest value of Sen (0.74 [95%CI: 0.62–0.83]) and Spe (0.93 [95%CI: 0.79–0.98]) respectively. As for best performance assessing by AUC, miR-146 owned the highest value of 0.84, indicating them as candidate markers of early diagnosis of NSCLC (Table 3, Supplementary Appendix Figure 7–8).
Table 3.
The overall diagnostic value of single miRNAs in early NSCLC.
| miRNA-type | No. of research (Case/Control) | Sen [95% CI] | Spe [95% CI] | PLR [95% CI] | NLR [95% CI] | AUC [95% CI] | DOR [95% CI] |
|---|---|---|---|---|---|---|---|
| miR-21 | 10 (703/606) | 0.74 [0.62–0.83] | 0.76 [0.68–0.82] | 3.1 [2.4–3.9] | 0.35 [0.25–0.49] | 0.81 [0.78–0.84] | 9 [6–13] |
| miR-145 | 4 (342/260) | 0.59 [0.34–0.80] | 0.86 [0.68–0.95] | 4.2 [2.0–8.8] | 0.48 [0.28–0.82] | 0.83 [0.79–0.86] | 9 [3–23] |
| miR-126 | 4 (170/160) | 0.56 [0.30–0.79] | 0.81 [0.70–0.89] | 2.9 [1.9–4.5] | 0.55 [0.32–0.95] | 0.80 [0.77–0.84] | 5 [2–13] |
| miR-210 | 4 (251/298) | 0.64 [0.46–0.78] | 0.83 [0.76–0.88] | 3.8 [2.6–5.5] | 0.44 [0.28–0.68] | 0.84 [0.81–0.87] | 9 [4–18] |
| miR-146 | 4 (144/89) | 0.49 [0.25–0.75] | 0.93 [0.79–0.98] | 7.3 [2.6–20.2] | 0.54 [0.33–0.90] | 0.89 [0.86–0.92] | 13 [4–43] |
Sen: sensitivity; Spe: specificity; PLR: positive likelihood ratio; NLR: negative likelihood ratio; AUC: area under curve; DOR: diagnostic odds ratio; CI: confidence interval.
Considering the subtype-analysis of different sampling sources, we analyzed 4 of 5 single-miRNAs since miR-146 was merely available in blood-based sample. According to our results, miR-210 (0.72 [95%CI: 0.64-0.80] vs. 0.34 [95%CI: 0.14-0.55]) as well as miR-126 (0.69 [95%CI: 0.61-0.78] vs. 0.15 [95%CI: 0.01-0.30]) shown superiority in respiratory-based samples compared with blood-based samples in the aspect of sensitivity (Figure 3(c)). When it came to calculation of pooled specificity, only miR-21 occupied statistical difference. Still, their results for specificity appeared to be similar in the above two sampling sources (0.77 [95%CI: 0.68-0.87] vs. 0.74 [95%CI: 0.63-0.84], Figure 3(d)). Therefore, we supposed the diagnostic value on early NSCLC of miR-210 and miR-126 depended partly on their sample sources.
3.3. Combining miRNAs into panels as diagnostic biomarker in NSCLC
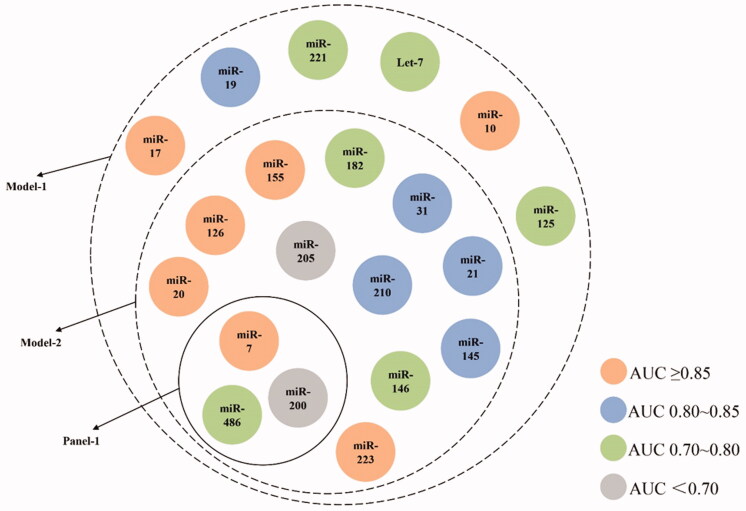
According to our results above, the performance of a certain miRNA seemed to reach satisfaction considering either sensitivity or specificity, making it difficult to choose one particularly for NSCLC diagnosis. For investigation of NSCLC diagnostic value of combining miRNAs, we firstly conducted pooled effect size analysis on available miRNA-panels (involving multiple miRNAs researched at the same time in participants) which were reported in no less than 4 articles and uniquely contained miRNAs as mentioned in above section (Supplementary Appendix Table 7). To sum up, there were only 1 panel (Panel-1) fulfilling the above criteria, containing miR-210, miR-31 and miR-21 for NSCLC (Figure 4). Still, it performed well according to AUC (0.91 [95%CI: 0.88-0.93]) and even better than any single miRNAs in both Sen and Spe of 0.82 [95%CI: 0.78-0.84] and 0.87 [95%CI: 0.84-0.89] respectively (>0.8), suggesting enhanced diagnostic value in NSCLC of combining miRNAs as panels.
Figure 4.
Containment relationship of analyzed single-miRNAs as different combined groups for NSCLC. The colour of each miRNA depends on their AUC value shown in Table 2. Panel-1contains three miRNAs (miR-21, -31, -210), Model-1 contains 20 miRNAs and Model-2 contains 14 miRNAs.
Additionally, there were also other miRNA-panels only containing above 20 miRNAs but reported in less than 4 articles (Supplementary Appendix Table 7).For further verification of these favourable miRNAs, we conducted effect size analysis by constructing models consisting of numerous reported panels. During this process, there was no requirement of the publication count for each panel except that the components of miRNA types are among the 20 miRNAs mentioned in previous section. The positive outcome of these models was defined as either of the involved panels owned positive finding. We firstly constructed Model-1 that finely retained these 20 miRNAs. As for total NSCLC, Model-1, the combination of 20 miRNAs, illustrated much better efficiency compared to any single miRNA with Sen of 0.77 [95%CI: 0.74-0.80], Spe of 0.84 [95%CI: 0.82-0.86] and AUC of 0.88 [95%CI: 0.85-0.91] since no miRNA could achieve such satisfying sensitivity and specificity at the same time. (Figure 4, Supplementary Appendix Figure 9, 10). Still, considering the large quantity of involved miRNAs, we furtherly set up Model-2 by limiting AUC cut-off value of each involved panel as no less than 0.80 to optimise the best combination of miRNA for construction of diagnostic panels with less miRNAs while owning better diagnostic value (with the sensitivity of 0.79 [95%CI: 0.75-0.82], specificity of 0.86 [95%CI: 0.83–0.88] and AUC value of 0.90 [95%CI: 0.87-0.92]). All the results above suggested the diagnostic potential of Model-1 and -2 in clinical practice.
4. Discussion
In this pooled study, we firstly quantificationally assessed the diagnostic value of 20 single-miRNAs for NSCLC and furtherly conducted subgroup analysis on sample sources. We found that, according to the cut-off of AUC > 0.85, there were 4 single-miRNAs ranked top, containing miR-155, miR-17, miR-126 and miR-20 for NSCLC. Additionally, miR-21 and miR-146 also shown promising diagnostic value for early-stage NSCLC. Besides, we also found that miRNA panel which contains miR-210, miR-31 and miR-21 illustrated much better efficiency compared to any single-miRNA and constructed 2 models for optimization. All our results suggested that single-miRNAs and miRNA panels show high diagnostic values and are good diagnosis biomarker for NSCLC.
Although there are plenty of original articles on the diagnostic function of miRNAs, meta-analysis of single-miRNA and their potential for LC diagnosis is limited in quantity. A current published systemic review [114] discussed the diagnostic and prognostic potential of miRNAs in LC. Still, they focussed on the changing trend of miRNA expression and carried out a criterion to grade miRNAs for screening of candidate in LC diagnosis while we directly obtained potential biomarkers including single-miRNAs and miRNA panels through quantitative analysis. On the other hand, they found 7 miRNAs (miR-21, miR-25, miR-27b, miR-19b, miR-125b, miR-146a, and miR-210) with talent as latent therapic targets for LC, 5 of which were also included in our analysis (miR-21, miR-19, miR-125, miR-146 and miR-210). Furtherly, we identified miR-19 with maximum sensitivity in LC, miR-146 with maximum specificity in LC, miR-21 with maximum sensitivity in early-stage LC and miR-210 with maximum AUC value in early-stage LC. Therefore, our study not only serve as quantitative evidence supporting the former review, but also intuitively illustrated specific diagnostic power of each single-miRNAs, miRNA panels and novel models, which provides more convincible testimony for clinical practice of miRNAs as NSCLC biomarkers.
There are many miRNAs, e.g. miR-17 [115,116] and miR-223 [117], with good diagnostic performance also shown related biological functions. However, some of the miRNAs who also contribute to LC neoplasia fail to serve as candidate for LC diagnosis according to our results. Members in miR-200 family participate in a series of LC-related function such as migration, invasion and mesenchymal-to-epithelial transition [118]. Still, their diagnostic power was less than satisfactory. MiR-375 was considered to associated with NSCLC development and Cladin-1 was proved to be its target [119], while whether miR-375 could be suitable diagnostic biomarker of LC is lacking of evidence. This situation suggested that there may be candidate miRNAs did not included in our current research limited by our strict inclusion criteria, and high quality study screened for a wider group of miRNAs are need.
A hallmark of early-stage LC is speculated for a long time in practice. Since extracellular secretion of miRNAs were observed during the process of cancerization in LC [120], the expression levels of some miRNAs might be lower at the beginning of LC tumorigenesis in samples sourced from body fluid (like serum) or secreta (like sputum). In this article, we defined early-stage LC as stage I or II after considering former related articles [121] and in view of different treatment strategies recommended for each stage [122]. According to our data, we found the expression level of miR-145 and miR-486 vary during different stages of LC regardless of specific LC subtypes: the expression of miR-145 [37,51,59,67,68,70,90,98,100,111] and miR-486 [36,40,63,80,84,89,101,107] was upregulated in LC at stage I–II while that was downregulated after the studied stages covered from I to IV. Considering the anti-tumour effect of miR-145 and miR-486 [123–126], we supposed that the tumour itself might tend to reduce intracellular miR-145 and miR-486 level for its good growth. Still, the underlying specific mechanism of this interesting finding needs further study because we did not conduct a subgroup analysis of confounding factors like LC subtypes or sample sources considering limited information provided by original articles.
According to our study, blood is a good sample source in the case of detecting miRNAs as diagnostic marker for LC. Since LC rooted in the normal respiratory tract, we supposed that samples from the respiratory system might be more powerful subjects than blood-derived samples. Detection of LC by sputum cytology was not satisfying according to earlier studies [127,128]. What’s more, articles using other respiratory samples have limitations not only on quantity but also on biomarkers researchers chose to detect (most of them focus on DNA, cytokines, and other proteins) [129–132]. According to our included articles, detection of miRNA from respiratory system only account for 20%. More original studies using respiratory samples are needed for further study of their diagnosis value.
Still, we found 3 miRNAs including miR-21, miR-486, and miR-31 whose statistical difference between sample sources was significant. Interestingly, miR-31 had higher Sen in haematic samples while its specificity was higher in respiratory samples. The possible mechanism is still waited to be determined. According to the results of other statistically positive miRNAs (miR-210, miR-126, miR-182, and miR-200b), haematic samples owned higher Spe than respiratory ones. Here we have to admit that blood is a good sample source in the detection of miRNAs not only for it is highly accessible regardless of hospital-level but also because of the anti-RNase characteristic of miRNAs themselves [133], which might cause the current condition that researchers prefer to choose blood as sample source in the detection of miRNAs. Besides, whether the examination for sampling was invasive or not should be taken into consideration when it came to clinical practice.
Last but not less, the combination of miRNAs with other biomarkers as novel panels might serve as powerful tool for LC diagnosis. Currently, miRNA panels are widely used in the diagnosis of various cancers, such as pancreatic cancer [134,135], gastric cancer [136], bladder cancer [137] and glioma [138]. For example, the Dartmouth-Hitchcock Medical Centre demonstrated a qRT-PCR assay containing 5 miRNAs to diagnose pancreatic cancer [135] while Qian even conducted a meta-analysis in Glioma and found that panels of multiple miRNAs enhanced the diagnostic sensitivity [138]. We also conduct analysis on the diagnostic potential of miRNA panels in this article. However, our panels are not satisfying enough since we could not verify its power and the involved miRNAs were numerous. There are also other biomarker panels of LC, which contains peptides or proteins [139] (such as ProGRP, NSE, CEA, CYFRA21-1, HE4), genes [140] (such as TP53, STK11, RTK) and lncRNA [141].
There are still some limitations to this analysis. Due to insufficiency of available data, we did not conducted analysis on ethnic or NSCLC subtypes’ subgroups. Besides, the clinical relevance of miRNAs involved in the miRNA panel and models we constructed in this article was still lacking and the further optimization of this panel was waited to conduct. We also had to admit that the score of our patients’ quality according to QUADAS-2 was dissatisfactory because all of our included articles were case-control study. Even so we still brought those articles into analysis not only because their total score of quality were favoured, but also for the lacking of high-quality original articles.
5. Conclusion
In summary, we identified several single miRNAs as potential diagnostic biomarkers of NSCLC as well as sub-analysis of early-stage NSCLC through pooled calculation of their sensitivity, specificity and AUC value. In addition, we found 6 miRNAs with statistical difference between blood-based and respiratory-based sample sources. Innovatively, we discovered 1 miRNA panel (miR-210, miR-31 and miR-21) with great diagnostic power among available panels and furtherly defined 2 models of miRNA groups with satisfying value on NSCLC diagnosing. All our results suggested that single miRNA and miRNA panel could serve as a diagnosis biomarker for NSCLC, which also need to be further verified in independence clinical samples.
Supplementary Material
Acknowledgement
We also acknowledge the contributions of all participants and researchers for all included papers related to this topic.
Funding Statement
This research was supported by the National Natural Science Foundation of China [No. 82001357, No. 31500999], the Hunan Provincial Natural Science Foundation of China [No. 2020JJ5951, No. 2021JJ80079], the Changsha Municipal Natural Science Foundation [No. kq2014123], the Scientific Research Project of Hunan Provincial Health Commission [No. 202103021406], the Youth Science Foundation of Xiangya Hospital [No. 2019Q17], the Degree & Postgraduate Education Reform Project of Central South University [No. 2020JGB125, No. 2021YJSKSA10] and the Undergraduate Education Reform Project of Central South University [No. 2020jy146, No.2020kcsz032].
Author contributions
Minhan Yi: conception and design, administrative support, provision of study materials or patients, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. Zexi Liao: collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. Langmei Deng: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. Li Xu: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. Yun Tan: collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. Kun Liu: collection and assembly of data, manuscript writing, final approval of manuscript. Ziliang Chen: collection and assembly of data, manuscript writing, final approval of manuscript. Yuan Zhang: conception and design, administrative support, provision of study materials or patients, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.The Lancet. Lung cancer: some progress, but still a lot more to do. The Lancet. 2019;394(10212):1880 doi: 10.1016/S0140-6736(19)32795-3. [DOI] [PubMed] [Google Scholar]
- 3.Inamura K. Lung cancer: understanding its molecular pathology and the 2015 WHO classification. Front Oncol. 2017;7:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen JE, Minna JD.. Molecular biology of lung cancer: clinical implications. Clin Chest Med. 2011;32(4):703–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding K, Yi M, Li L, et al. Interleukin polymorphisms and protein levels associated with lung cancer susceptibility and phenotypes. Expert Rev Clin Immunol. 2021;17(9):1029–1040. [DOI] [PubMed] [Google Scholar]
- 6.Saettele TM, Ost DE.. Multimodality systematic approach to mediastinal lymph node staging in non-small cell lung cancer. Respirology. 2014;19(6):800–808. [DOI] [PubMed] [Google Scholar]
- 7.Fischer BM, Mortensen J, Hansen H, et al. Multimodality approach to mediastinal staging in non-small cell lung cancer. Faults and benefits of PET-CT: a randomised trial. Thorax. 2011;66(4):294–300. [DOI] [PubMed] [Google Scholar]
- 8.Knight SB, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7(9):170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X, Wang L, Wang X, et al. Evaluation of cell surface vimentin positive circulating tumor cells as a diagnostic biomarker for lung cancer. Front Oncol. 2021;11:672687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–5319. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Yi M, Tan Y, et al. A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma. Ther Adv Respir Dis. 2020;14:1753466620981863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svoronos AA, Engelman DM, Slack FJ.. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76(13):3666–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thind A, Wilson C.. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inamura K, Ishikawa Y.. MicroRNA in lung cancer: Novel biomarkers and potential tools for treatment. JCM. 2016;5(3):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bica-Pop C, Cojocneanu-Petric R, Magdo L, et al. Berindan-Neagoe I. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci. 2018;75(19):3539–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Min L, Ren C, et al. miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLOS One. 2017;12(2):e0171751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DH, Park S, Kim H, et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 2020;475:2–13. [DOI] [PubMed] [Google Scholar]
- 19.Jiang L-P, Zhu Z-T, He C-Y.. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future Oncol. 2016;12(9):1105–1115. [DOI] [PubMed] [Google Scholar]
- 20.Khandelwal A, Seam RK, Gupta M, et al. Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020;111(3):826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groome PA, Bolejack V, Crowley JJ, Participating Institutions, et al. The IASLC lung cancer staging project: validation of the proposals for revision of the T, N, and M descriptors and consequent stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol. 2007;2(8):694–705. [DOI] [PubMed] [Google Scholar]
- 23.Detterbeck FC, Chansky K, Groome P, IASLC Staging and Prognostic Factors Committee, Advisory Boards, and Participating Institutions, et al. The IASLC lung cancer staging project: Methodology and validation used in the development of proposals for revision of the stage classification of NSCLC in the forthcoming (eighth) edition of the TNM classification of lung cancer. J Thorac Oncol. 2016;11(9):1433–1446. [DOI] [PubMed] [Google Scholar]
- 24.Travis WD, Brambilla E, Nicholson AG, WHO Panel, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. [DOI] [PubMed] [Google Scholar]
- 25.Travis WD, Brambilla E. Muller-Hermelink Hk. Pathology and genetics: tumours of the lung, pleura, thymus and heart: World health organization. Classification of tumours. Lyon:IARC Press. 2004. [DOI] [PubMed] [Google Scholar]
- 26.Whiting PF, Rutjes AW, Westwood ME, QUADAS-2 Group, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. [DOI] [PubMed] [Google Scholar]
- 27.Spearman's rank correlation coefficient. BMJ. 2018;362:k4131. [DOI] [PubMed] [Google Scholar]
- 28.Reitsma JB, Glas AS, Rutjes AWS, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982–990. [DOI] [PubMed] [Google Scholar]
- 29.Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11(2):193–206. [DOI] [PubMed] [Google Scholar]
- 30.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patsopoulos NA, Evangelou E, Ioannidis JP.. Sensitivity of between-study heterogeneity in meta-analysis: proposed metrics and empirical evaluation. Int J Epidemiol. 2008;37(5):1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biljana M, Jelena M, Branislav J, et al. Bias in meta-analysis and funnel plot asymmetry. Stud Health Technol Inform. 1999;68:323–328. [PubMed] [Google Scholar]
- 33.Jennions MD, Møller AP.. Publication bias in ecology and evolution: an empirical assessment using the 'trim and fill' method. Biol Rev. 2002;77(2):211–222. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Tang Y, Song X, et al. . Tumor-derived exosomal mirnas as diagnostic biomarkers in non-small cell lung cancer. Front Oncol. 2020;10. doi: 10.3389/fonc.2020.560025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C, Jia X, Zhou J, et al. . The mir-17-92 gene cluster is a blood-based marker for cancer detection in non-small-cell lung cancer. Am J Med Sci. 2020;360(3):248–260. doi: 10.1016/j.amjms.2020.05.004. 32466856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Q, Yu L, Lin X, et al. Combination of Serum miRNAs with Serum Exosomal miRNAs in Early Diagnosis for Non-Small-Cell Lung Cancer. CMAR. 2020;ume 12:485–495. doi: 10.2147/CMAR.S232383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Chen D, Chen W, et al. Early detection of Non-Small cell lung cancer by using a 12-microRNA panel and a nomogram for assistant diagnosis. Front Oncol. 2020;10:855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Zhang C, Peng X, et al. . A combination of four serum mirnas for screening of lung adenocarcinoma. Hum Cell. 2020;33(3):830–838. doi: 10.1007/s13577-020-00346-6. 32388809 [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Tang C, Song X, et al. . Clinical value of CTLA4-associated micrornas combined with inflammatory factors in the diagnosis of non-small cell lung cancer. Ann Clin Biochem. 2020;57(2):151–161. doi: 10.1177/0004563220901564. 31906699 [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Kannisto E, Yu G, et al. Non-invasive detection of exosomal MicroRNAs via tethered cationic lipoplex nanoparticles (tCLN) biochip for lung cancer early detection. Front Genet. 2020;11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao J, Shen J, Leng Q, et al. . MicroRNA-based biomarkers for diagnosis of non-small cell lung cancer (NSCLC). Thorac Cancer. 2020;11(3):762–768. doi: 10.1111/1759-7714.13337. 31994346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghany S, Ali E, Ahmed AE, et al. . Circulating mirna-30a and mirna-221 AS novel biomarkers for the early detection of non-small-cell lung cancer. Middle East Journal of Cancer. 2020;11(1):50–58. [Google Scholar]
- 43.Fehlmann T, Kahraman M, Ludwig N, et al. . Evaluating the use of circulating microrna profiles for lung cancer detection in symptomatic patients. JAMA Oncol. 2020;6(5):714–723. doi: 10.1001/jamaoncol.2020.0001. 32134442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asakura K, Kadota T, Matsuzaki J, et al. . A mirna-based diagnostic model predicts resectable lung cancer in humans with high accuracy. Commun Biol. 2020;3(1):134 doi: 10.1038/s42003-020-0863-y. PMC: 32193503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou J-G, Ma L-F, Li X, et al. . Circulating microrna array (miR-182, 200b and 205) FOR THE early diagnosis and poor prognosis predictor of non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(3):1108–1115. doi: 10.26355/eurrev_201902_17001. 30779079 [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Zhang Y, Yin Y, et al. . Detection of circulating exosomal mir-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathology- Research and Practice. 2019;215(8):152466 doi: 10.1016/j.prp.2019.152466. [DOI] [PubMed] [Google Scholar]
- 47.Xi K, Wang W, Wen Y, et al. . Combining plasma mirnas and computed tomography features to differentiate the nature of pulmonary nodules. Front Oncol. 2019;9:975 doi: 10.3389/fonc.2019.00975. PMC: 31632908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Wang Z, Wang Q, et al. . Clinical significance of the expression of mirna-21, mirna-31 AND mirna-let7 in patients with lung cancer. Saudi J Biol Sci. 2019;26(4):777–781. doi: 10.1016/j.sjbs.2018.12.009. 31049003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szczyrek M, Kuźnar-Kamińska B, Grenda A, et al. . Diagnostic value of plasma expression of micrornas complementary to drosha and dicer in lung cancer patients. Eur Rev Med Pharmacol Sci. 2019;23(9):3857–3866. doi: 10.26355/eurrev_201905_17813. 31115013 [DOI] [PubMed] [Google Scholar]
- 50.Świtlik WZ, Karbownik MS, Suwalski M, et al. . Serum mir-210-3p as a potential noninvasive biomarker of lung adenocarcinoma: A preliminary study. Genet Test Mol Biomarkers. 2019;23(5):353–358. doi: 10.1089/gtmb.2018.0275. 30950648 [DOI] [PubMed] [Google Scholar]
- 51.Sui A, Zhang X, Zhu Q.. Diagnostic value of serum miR197 and miR145 in non-small cell lung cancer. Oncol Lett. 2019;doi: 10.3892/ol.2019.9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheervalilou R, Lotfi H, Shirvaliloo M, et al. . Circulating mir-10b, mir-1 AND mir-30a expression profiles in lung cancer: Possible correlation with clinico-pathologic characteristics and lung cancer detection. Int J Mol Cell Med. 2019;8(2):118–129. doi: 10.22088/IJMCM.BUMS.8.2.118. 32215263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roman-Canal B, Moiola CP, Gatius S, et al. EV-associated miRNAs from pleural lavage as potential diagnostic biomarkers in lung cancer. Sci Rep. 2019;9(1):15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Fang H, Jiang F, et al. . External validation of a panel of plasma microrna biomarkers for lung cancer. Biomark Med. 2019;13(18):1557–1564. doi: 10.2217/bmm-2019-0213. 31674214 [DOI] [PubMed] [Google Scholar]
- 55.Hetta HF, Zahran AM, El-Mahdy RI, et al. . Assessment of circulating mirna-17 AND mirna-222 expression profiles as non-invasive biomarkers in egyptian patients with non-small-cell lung cancer. Asian Pac J Cancer Prev. 2019;20(6):1927–1933. doi: 10.31557/APJCP.2019.20.6.1927. 31244320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdollahi A, Rahmati S, Ghaderi B, et al. . A combined panel of circulating microrna as a diagnostic tool for detection of the non-small cell lung cancer. QJM. 2019;112(10):779–785. doi: 10.1093/qjmed/hcz158. 31236600 [DOI] [PubMed] [Google Scholar]
- 57.Yang YL, Wang W, Xu LP.. Predictive value of microrna-10b expression in peripheral blood mononuclear cells in evaluating short- AND long-term efficacy of chemotherapy for patients with advanced non-small-cell lung cancer. Neoplasma. 2018;65(4):610–619. doi: 10.4149/neo_2018_170110N20. 30064233 [DOI] [PubMed] [Google Scholar]
- 58.Yang Y, Chen K, Zhou Y, et al. . Application of serum microrna-9-5p, 21-5p, and 223-3p combined with tumor markers in the diagnosis of non-small-cell lung cancer in Yunnan in southwestern China. Onco Targets Ther. 2018;11:587–597. doi: 10.2147/OTT.S152957. 29430184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xi KX, Zhang XW, Yu XY, et al. The role of plasma miRNAs in the diagnosis of pulmonary nodules. J Thorac Dis. 2018;10(7):4032–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Y, Mei H, Xu C, et al. . Circulating microrna-339-5p and-21 in plasma as an early detection predictors of lung adenocarcinoma. Pathol Res Pract. 2018;214(1):119–125. doi: 10.1016/j.prp.2017.10.011. 29103767 [DOI] [PubMed] [Google Scholar]
- 61.Qiu F, Gu WG, Li C, et al. . Analysis on expression level and diagnostic value of mir-19 AND mir-21 in peripheral blood of patients with undifferentiated lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8367–8373. [DOI] [PubMed] [Google Scholar]
- 62.Poroyko V, Mirzapoiazova T, Nam A, et al. . Exosomal mirnas species in the blood of small cell and non-small cell lung cancer patients. Oncotarget. 2018;9(28):19793–19806. doi: 10.18632/oncotarget.24857. 29731983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohamed MA, Mohamed EI, El-Kaream SAA, et al. . Underexpression of mir-486-5p but not overexpression of mir-155 is associated with lung cancer stages. MIRNA. 7(2):120–127. doi: 10.2174/2211536607666180212124532. [DOI] [PubMed] [Google Scholar]
- 64.Leng Q, Wang Y, Jiang F.. A direct plasma mirna assay for early detection and histological classification of lung cancer. Transl Oncol. 2018;11(4):883–889. doi: 10.1016/j.tranon.2018.05.001. 29783093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan LH, Chen H, Teng JL, et al. Evaluation of Serum-Paired miRNA ratios for early diagnosis of Non-Small cell lung cancer using quantum Dot-Based suspension array. J Nanomater. 2018;2018:1–9. [Google Scholar]
- 66.Bao M, Pan S, Yang W, et al. . Serum mir-10a-5p and mir-196a-5p as non-invasive biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2018;11(2):773–780. [PMC free article] [PubMed] [Google Scholar]
- 67.Bagheri A, Khorshid HRK, Tavallaie M, et al. . A panel of noncoding rnas in non-small-cell lung cancer. J Cell Biochem. 2018;doi: 10.1002/jcb.28111. 30485511 [DOI] [PubMed] [Google Scholar]
- 68.Aiso T, Ohtsuka K, Ueda M, et al. Serum levels of candidate microRNA diagnostic markers differ among the stages of non-small-cell lung cancer. Oncol Lett. 2018;16(5):6643–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abu-Duhier FM, Javid J, Sughayer MA, et al. . Clinical significance of circulatory mirna-21 AS AN efficient non-invasive biomarker for the screening of lung cancer patients. Asian Pacific Journal of Cancer Prevention : APJCP. 2018;19(9):2607–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Mao F, Shen T, et al. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. 2017;13(2):669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y, Zuo J, Tan Q, et al. . Plasma mir-92a-2 AS A biomarker for small cell lung cancer. CBM. 18(3):319–327. doi: 10.3233/CBM-160254. [DOI] [PubMed] [Google Scholar]
- 72.Sheervalilou R, Khamaneh AM, Sharifi A, et al. Using miR-10b, miR-1 and miR-30a expression profiles of bronchoalveolar lavage and sputum for early detection of non-small cell lung cancer. Biomedicine and Pharmacotherapy. 2017;88:1173–1182. [Google Scholar]
- 73.Shang AQ, Xie YN, Wang J, et al. . Predicative values of serum microrna-22 AND microrna-126 levels for non-small cell lung cancer development and metastasis: A case-control study. Neoplasma. 2017;64(3):453–459. doi: 10.4149/neo_2017_317. 28253725 [DOI] [PubMed] [Google Scholar]
- 74.Lv S, Xue J, Wu C, et al. . Identification of a panel of serum micrornas as biomarkers for early detection of lung adenocarcinoma. J Cancer. 2017;8(1):48–56. doi: 10.7150/jca.16644. 28123597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leng Q, Lin Y, Jiang F, et al. . A plasma mirna signature for lung cancer early detection. Oncotarget. 2017;8(67):111902–111911. doi: 10.18632/oncotarget.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ibrahim FK, Ali-Labib R, Galal IH, et al. . MicroRNA-155 expression in exhaled breath condensate of patients with lung cancer. Egyptian Journal of Chest Diseases and Tuberculosis. 2017;66(4):687–691. [Google Scholar]
- 77.Bagheri A, Khorram Khorshid HR, Mowla SJ, et al. . Altered mir-223 expression in sputum for diagnosis of non-small cell lung cancer. Avicenna J Med Biotechnol. 2017;9(4):189–195. [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu W, Zhou K, Zha Y, et al. . Diagnostic value of serum mir-182, mir-183, mir-210, and mir-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11(4):e0153046 doi: 10.1371/journal.pone.0153046. PMC: 27093275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaporozhchenko IA, Morozkin ES, Skvortsova TE, et al. . Plasma mir-19b and mir-183 AS potential biomarkers of lung cancer. PLoS One. 2016;11(10):e0165261 doi: 10.1371/journal.pone.0165261. PMC: 27768748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, Zhi X, Zhang Y, et al. Role of plasma MicroRNAs in the early diagnosis of non-small-cell lung cancers: a case-control study. J Thorac Dis. 2016;8(7):1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Su Y, Fang H, Jiang F. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin Epigenetics. 2016;8:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su Y, Fang H, Jiang F.. Integrating DNA methylation and microrna biomarkers in sputum for lung cancer detection. Clin Epigenet. 2016;8(1)doi: 10.1186/s13148-016-0275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Razzak R, Bédard ELR, Kim JO, et al. . MicroRNA expression profiling of sputum for the detection of early and locally advanced non-small-cell lung cancer: A prospective case-control study. Curr Oncol. 2016;23(2):e86–e94. doi: 10.3747/co.23.2830. 27122989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jia Y, Zang A, Feng Y, et al. . miRNA-486 AND mirna-499 in human plasma evaluate the clinical stages of lung cancer and play a role as a tumor suppressor in lung tumorigeneisis not pathogenesis. Bangladesh J Pharmacol. 2016;11(1):264. doi: 10.3329/bjp.v11i1.25318. [DOI] [Google Scholar]
- 85.Chen JL, Chen JR, Han HN, Zhou F, Lv XD, Ma H. Clinical significance of miRNA21 in exhaled breath condensate of non-small-cell lung cancer. International J Clin Exp. 2016;9(9):17232–8. [Google Scholar]
- 86.Zhao W, Zhao JJ, Zhang L, et al. . Serum mir-21 level: A potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med. 2015;8(9):14759–14763. [PMC free article] [PubMed] [Google Scholar]
- 87.Yang J-S, Li B-J, Lu H-W, et al. . Serum mir-152, mir-148a, mir-148b, and mir-21 AS novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36(4):3035–3042. doi: 10.1007/s13277-014-2938-1. 25501703 [DOI] [PubMed] [Google Scholar]
- 88.Yan HJ, Ma JY, Wang L, Gu W. Expression and significance of circulating microRNA-31 in lung cancer patients. Med Sci Monit. 2015;21:722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xing L, Su J, Guarnera MA, et al. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin Cancer Res. 2015;21(2):484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang RJ, Zheng YH, Wang P, Zhang JZ. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(1):765–71. [PMC free article] [PubMed] [Google Scholar]
- 91.Wang P, Yang D, Zhang H, et al. . Early detection of lung cancer in serum by a panel of microrna biomarkers. Clin Lung Cancer. 2015;16(4):313–319.e1. doi: 10.1016/j.cllc.2014.12.006. 25639977 [DOI] [PubMed] [Google Scholar]
- 92.Su J, Anjuman N, Guarnera MA, et al. . Analysis of lung flute-collected sputum for lung cancer diagnosis. Biomark Insights. 2015;10:55–61. doi: 10.4137/BMI.S26883. 26309391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W, Wang Y, Zhang Q, et al. . MicroRNA-486 AS A biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS One. 2015;10(8):e0134220 doi: 10.1371/journal.pone.0134220. PMC: 26237047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fan L, Qi H, Teng J, et al. . Identification of serum mirnas by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumor Biol. 2016;37(6):7777–7784. doi: 10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 95.Dou H, Wang Y, Su G, et al. . Decreased plasma let-7c and mir-152 AS noninvasive biomarker for non-small-cell lung cancer. Int J Clin Exp Med. 2015;8(6):9291–9298. [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu W-Y, Luo B, An J-Y, et al. . Differential expression of mir-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest. 2014;32(8):394–401. doi: 10.3109/07357907.2014.922569. 24945821 [DOI] [PubMed] [Google Scholar]
- 97.Li N, Ma J, Guarnera MA, et al. . Digital PCR quantification of mirnas in sputum for diagnosis of lung cancer. J Cancer Res Clin Oncol. 2014;140(1):145–150. doi: 10.1007/s00432-013-1555-5. 24281335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geng Q, Fan T, Zhang B, et al. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeng XL, Zhang SY, Zheng JF, et al. . Altered mir-143 AND mir-150 expressions in peripheral blood mononuclear cells for diagnosis of non-small cell lung cancer. Chin Med J (Engl). 2013;126(23):4510–4516. [PubMed] [Google Scholar]
- 100.Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22(6):540–548. [DOI] [PubMed] [Google Scholar]
- 101.Mozzoni P, Banda I, Goldoni M, et al. Plasma and EBC microRNAs as early biomarkers of non-small-cell lung cancer. Biomarkers. 2013;18(8):679–686. [DOI] [PubMed] [Google Scholar]
- 102.Anjuman N, Li N, Guarnera M, et al. . Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. Clinical and Translational Medicine. 2013;2(1). doi: 10.1186/2001-1326-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abd-El-Fattah AA, Sadik NAH, Shaker OG, et al. . Differential micrornas expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67(3):875–884. doi: 10.1007/s12013-013-9575-y. 23559272 [DOI] [PubMed] [Google Scholar]
- 104.Wang B, Zhang Q.. The expression and clinical significance of circulating microrna-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138(10):1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PubMed] [Google Scholar]
- 105.Wei J, Liu L-K, Gao W, et al. . Reduction of plasma microrna-21 is associated with chemotherapeutic response in patients with non-small cell lung cancer. Chin J Cancer Res. 2011;23(2):123–128. doi: 10.1007/s11670-011-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei J, Gao W, Zhu C-J, et al. . Identification of plasma microrna-21 AS A biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. 2011;30(6):407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91(4):579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen J, Liu Z, Todd NW, et al. . Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microrna biomarkers. BMC Cancer. 2011;11:374 doi: 10.1186/1471-2407-11-374. PMC: 21864403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Y, Li W, Ouyang Q, et al. . Detection of lung cancer with blood microrna-21 expression levels in chinese population. Oncol Lett. 2011;2(5):991–994. doi: 10.3892/ol.2011.351. 22866162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jeong HC, Kim EK, Lee JH, et al. . Aberrant expression of let-7a mirna in the blood of non-small cell lung cancer patients. Mol Med Rep. 2011;4(2):383–387. doi: 10.3892/mmr.2011.430. 21468581 [DOI] [PubMed] [Google Scholar]
- 111.Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127(12):2870–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xing L, Todd NW, Yu L, et al. . Early detection of squamous cell lung cancer in sputum by a panel of microrna markers. Mod Pathol. 2010;23(8):1157–1164. doi: 10.1038/modpathol.2010.111. 20526284 [DOI] [PubMed] [Google Scholar]
- 113.Xie Y, Todd NW, Liu Z, et al. . Altered mirna expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhong S, Golpon H, Zardo P, et al. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl Res. 2021;230:164–196. [DOI] [PubMed] [Google Scholar]
- 115.Borzi C, Ganzinelli M, Caiola E, et al. LKB1 down-modulation by miR-17 identifies NSCLC patients with worse prognosis eligible for energy-stress based treatments. J Thor Oncol. 2021;16(8):1298–1311. [DOI] [PubMed] [Google Scholar]
- 116.Zhao J, Fu W, Liao H, Dai L, Jiang Z, Pan Y, et al. The regulatory and predictive functions of miR-17 and miR-92 families on cisplatin resistance of non-small cell lung cancer. Bmc Cancer. 2015;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang H, Chen J, Zhang S, et al. MiR-223 regulates autophagy associated with cisplatin resistance by targeting FBXW7 in human non-small cell lung cancer. Cancer Cell Int. 2020;20(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahn Y-H, Ko YH.. Diagnostic and therapeutic implications of microRNAs in non-small cell lung cancer. IJMS. 2020;21(22):8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu H, Jiang L, Sun C, et al. Decreased circulating miR-375: a potential biomarker for patients with non-small-cell lung cancer. Gene. 2014;534(1):60–65. [PubMed] [Google Scholar]
- 120.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108(9):3713–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403):eaan2415. doi: 10.1126/scitranslmed.aan2415. 28814544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. [DOI] [PubMed] [Google Scholar]
- 123.Lu Q, Shan S, Li Y, et al. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. Faseb J. 2018;32(7):3957–3967. [DOI] [PubMed] [Google Scholar]
- 124.Mataki H, Seki N, Mizuno K, et al. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget. 2016;7(44):72084–72098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang J, Tian X, Han R, et al. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33(9):1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Borzi C, Calzolari L, Centonze G, et al. Mir-660-p53-mir-486 network: a new key regulatory pathway in lung tumorigenesis. IJMS. 2017;18(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Laurie W, Szaloky LG.. Sputum cytology in the diagnosis of bronchial carcinoma. Med J Aust. 1971;1(5):247–251. [DOI] [PubMed] [Google Scholar]
- 128.Oswald NC, Hinson KF, Canti G, et al. The diagnosis of primary lung cancer with special reference to sputum cytology. Thorax. 1971;26(6):623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Arai T, Yasuda Y, Takaya T, et al. Application of telomerase activity for screening of primary lung cancer in broncho-alveolar lavage fluid. Oncol Rep. 1998;5(2):405–408. [DOI] [PubMed] [Google Scholar]
- 130.Chen W, Zhao W, Zhang L, et al. MALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via WNT signaling pathway. Oncotarget. 2017;8(55):94317–94329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen Z, Xu Z, Sun S, et al. TGF-beta 1, IL-6, and TNF-alpha in bronchoalveolar lavage fluid: useful markers for lung cancer? Sci Rep. 2014;4:5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chan HP, Lewis C, Thomas PS.. Exhaled breath analysis: novel approach for early detection of lung cancer. Lung Cancer. 2009;63(2):164–168. [DOI] [PubMed] [Google Scholar]
- 133.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. [DOI] [PubMed] [Google Scholar]
- 134.Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136(11):2616–2627. [DOI] [PubMed] [Google Scholar]
- 135.de Abreu FB, Liu X, Tsongalis GJ.. miRNA analysis in pancreatic cancer: the dartmouth experience. Clin Chem Lab Med. 2017;55(5):755–762. [DOI] [PubMed] [Google Scholar]
- 136.Huang Z, Zhu D, Wu L, et al. Six Serum-Based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. 2017;26(2):188–196. [DOI] [PubMed] [Google Scholar]
- 137.Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110(1):408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou Q, Liu J, Quan J, et al. MicroRNAs as potential biomarkers for the diagnosis of glioma: a systematic review and meta-analysis. Cancer Sci. 2018;109(9):2651–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Y, Li M, Zhang Y, et al. Age-stratified and gender-specific reference intervals of six tumor markers panel of lung cancer: a geographic-based multicenter study in China. J Clin Lab Anal. 2021;35(6):e23816–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Passaro A, Attili I, Rappa A, et al. Genomic characterization of concurrent alterations in non-small cell lung cancer (NSCLC) harboring actionable mutations. Cancers. 2021;13(9):2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shao J, Lyu W, Zhou J, et al. A panel of five-lncRNA signature as a potential biomarker for predicting survival in gastric and thoracic cancers. Front Genet. 2021;12:666155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.