Abstract
Background:
Prostate cancer is the second leading cause of cancer death in men, moreover when it develops metastasis. However, PSA detection in serum as current gold standard to measure disease progressivity had wide variability leading to confounding outcomes. MicroRNA-21 has diagnostic values for cancer over period of time researched, yet results are still inconclusive.
Objective:
The aim of the study was to conduct recent meta-analysis to assess reliability of miRNA-21 as diagnostic biomarker especially in progressivity of prostate cancer.
Methods:
Published papers from PubMed, Science Direct, and Embase” as of 1 July 2021 assessing circulating miRNA-21 in progressivity of prostate cancer patients were analyzed using Comprehensive Meta-Analysis tool. Pooled sensitivity, specificity, positive and negative likelihood ratio (LR) and SROC assessed with 95 % confidence intervals were estimated using fixed-effects or random-effects models.
Results:
In total, we included 6 papers total of 651 samples reporting miRNA-21 capability of detecting progressive prostate cancer. The pooled sensitivity and specificity showed 0.91 (95% CI 0.88-0.94, I2=0%) and 0.89 (95% CI 0.85-0.92, I2=44.8%), respectively. Positive and negative likelihood ratio showed 7.18 (95% CI 4.31-11.96, I2=56%) and 0.11 (95% CI 0.07-0.16, I2=11.8%). SROC were assessed and got Area Under Curve around 97.4%.
Conclusion:
miRNA-21 could serve as biomarkers of prostate cancer progressivity since remarkable diagnostic value of circulating miRNA-21 in prostate cancer metastasis process.
Keywords: miRNA-21, biomarker, prostate, cancer, progressivity, metastatic
1. BACKGROUND
As a complex disease caused by multiple environmental and genetic factors prostate cancer is the second leading cause of death from cancer in men, moreover when it develops metastasis (1). In Europe, it causes 92,300 deaths every year (2). As a routine laboratory test, in clinical practice prostate specific antigen (PSA) in serum is not specific for prostate cancer, and some PSA levels evaluated may cause false positives due to infection or hyperplasia. However, until nowadays PSA detection in serum still become current gold standard to measure disease progressivity, that had wide variability leading to confounding outcomes. Thus, it is necessary to find new and effective biomarkers in order to diagnose progressivity of prostate cancer (1).
MicroRNA is a family of non-coding small RNAs (19±22 nucleotides), stable and easy to accurately measure. MicroRNAs are differentially expressed in normal tissues and cancer, contributing to the development and progression of cancer (3). It is well-known that miRNA-21 is an anti-apoptotic agent that acts through the p53 network. It can target and inhibit the expression of the tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion (4, 5).
Regarding to the ability of miRNA-21 to detect progressivity of prostate cancer, a number of studies have been conducted. MicroRNA-21 has diagnostic values for cancer over period of time researched, yet results are still inconclusive. Porzycky et al found specificity of miRNA-21 to diagnose prostate cancer was 0.75, while Yang et al found it could reach 0.93 (6, 7). In order to accurately assess the diagnostic value of miRNA-21, we performed this meta-analysis.
2. OBJECTIVE
The aim of the study was to conduct recent meta-analysis to assess reliability of miRNA-21 as diagnostic biomarker especially in progressivity of prostate cancer.
3. METHODS
Study Design
A meta-analysis was undertaken to determine the sensitivity, specificity, positive and negative likelihood ratio and SROC of miRNA-21 to detect progressive prostate cancer. Published papers from PubMed, Science Direct, and Embase” as of 1 July 2021 assessing circulating miRNA-21 in prostate cancer patients were analyzed using Comprehensive Meta-Analysis tool and estimated using fixed-effects or random-effects models. The investigation was guided by Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) criteria (8).
Search strategy
A comprehensive searching up to 1 July 2021 through PubMed, Science Direct, and Embase was undertaken. Keyword terms combination from Medical Subjective Heading (MeSH) were utilized: [“microRNA-21” OR “miRNA-21” OR “miR-21”] AND [“prostate cancer” OR “prostate carcinoma”] AND [“diagnosis” OR “sensitivity” OR “specificity”]. English language restriction was applied. KPS and AFP independently searched the articles. In order to locate further publications, a manual reference search of pertinent research was done.
Eligibility criteria
The following studies met the inclusion criteria: a) retrospective, prospective, case-control or cross-sectional approaches, and published papers by manual search; b) assessing the diagnostic value of miRNA-21; and c) providing data for the calculation of diagnostic value and SROC with 95% Confidence Intervals. We rejected studies with irrelevant titles and/or abstracts, reviews, comments, dissertations that were not published into the journal, incomplete data, and inferior availability.
Quality assessment
The paper quality was assessed using the quality assessment of diagnostic accuracy studies-2 (QUADAS-2) tool.9 Two authors critically appraised the quality of the product (KPS and AFP). The QUADAS-2 was composed of patient selection, index test, reference standard, flow, and timing to measure risk of bias, and first three parameters were applied to assess applicability of the journal.
Data extraction
The following details were taken from each study: a) first author; b) publication year; c) country of center study; d) specimen type; e) source of control group; f) sample size; g) miRNA as control of normalization; h) miRNA as biomarker, i) diagnostic value including sensitivity, specificity, and likelihood ratio. Two separate authors (KPS and AFP) used a piloted data extraction form to obtain the data. A discussion was performed if there was a dispute between two writers.
Statistical analysis
The statistical analysis were using Comprehensive Meta-Analysis (CMA, New Jersey, USA) version 2.1 tool. The Z test was used to examine the significance of the pooled data (p <0.05 was considered statistically significant). Data were examined for potential publication bias first before significant risk variables were identified. A p-heterogeneity and I-square (I2) Q-analysis was used to measure the degree of heterogeneity. A random-effect model was used if there was heterogeneity (p <0.05) or I2>50%, and a fixed-effect model was used if there was not. The pooled sensitivity and specificity, positive and negative likelihood ratio, and area under curve (AUC) from summary receiver operating characteristics (SROC) curve with the corresponding 95% CI. Two authors did statistical analysis to avoid analytical inaccuracies (KPS and AFP).
4. RESULTS
Eligible studies
Initial searching found 625 papers, of which 542 articles were rejected due to primary screening unrelated titles and/or abstracts. There were a total of 81 papers selected for full-text evaluation and due to insufficient data provided and duplication, we got 6 papers to enter the next step. The other 70 studies were eliminated due to a lack of data that could be used to estimate the diagnostic value and 95 % confidence interval and 5 studies were eliminated by duplication cause. Our meta-analysis includes a total of 6 papers. The outcome of the literature search is displayed in a PRISMA flowchart (Figure 1). The baseline features of the research that we investigated are listed in Table 1.
Figure 1. A flowchart of article selection in our study.

Table 1. Baseline characteristics of studies included in our analysis.
| No. | Author, year | Country | Specimen | Control group | Prostate Cancer size | Control size | QUADAS-2 bias |
|---|---|---|---|---|---|---|---|
| 1 | Kotb et al 2014 | Egypt | Serum | Benign prostatic disease | 10 | 10 | Index test, reference standard, flow and timing were mentioned clearly, however patient selection were unclear |
| 2 | Huang et al 2015 | China | Mononuclear cell | Healthy patient | 75 | 75 | Patient selection, reference standard, flow and timing were mentioned clearly, however Index test were unclear |
| 3 | Gao et al 2016 | China | Plasma | Benign prostatic disease | 57 | 28 | Index test, reference standard, flow and timing were mentioned clearly, however patient selection were unclear |
| 4 | Yang et al 2016 | China | Mononuclear cell | Benign prostatic disease | 92 | 85 | Reference standard, flow and timing were mentioned clearly, however patient selection and index test were unclear |
| 5 | Yang et al 2016 | China | Mononuclear cell | Healthy patient | 92 | 97 | All parameters (patient selection, index test, reference standard, flow and timing) were mentioned clearly |
| 6 | Porzycki et al 2018 | Poland | Serum | Healthy patient | 20 | 10 | All parameters (patient selection, index test, reference standard, flow and timing) were mentioned clearly |
Data synthesis
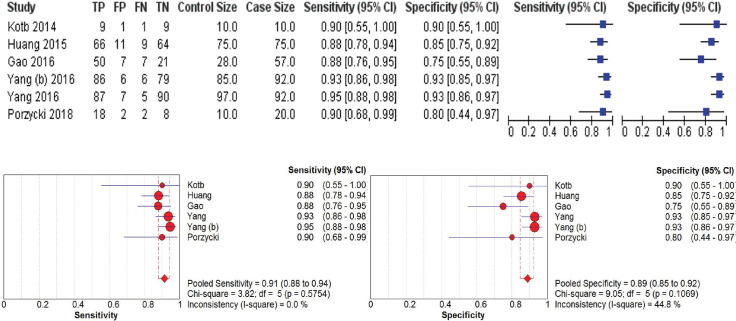
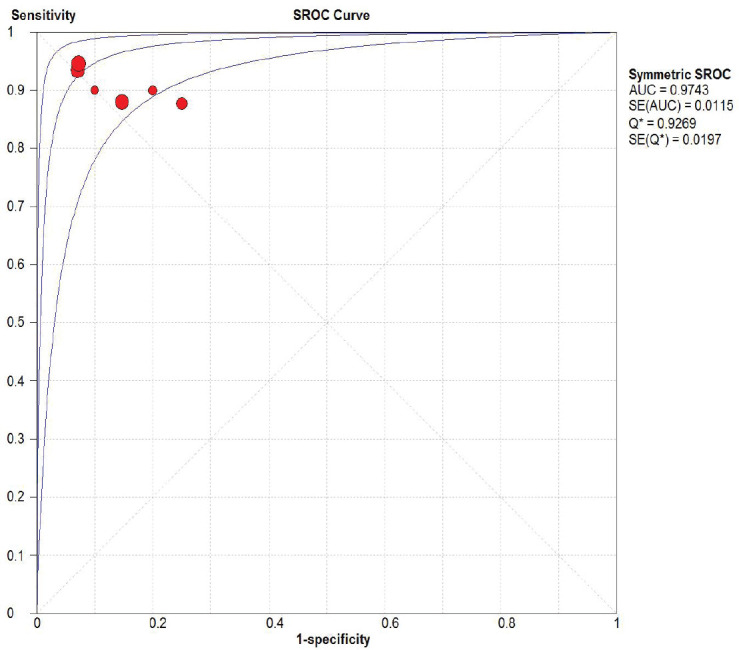
A total of 651 samples reporting miRNA-21 in prostate cancer patients from 6 papers was synthesized. The pooled sensitivity and specificity showed high value around 0.91 or 91% (95% CI 0.88-0.94, I2=0%) and 0.89 or 89% (95% CI 0.85-0.92, I2=44.8%), respectively as shown in Figure 2. Positive likelihood ratio showed 7.18 (95% CI 4.31-11.96, I2=56%) which is also high for biomarker candidate to propose. Negative likelihood ratio was also supportive toward the result was around 0.11 (95% CI 0.07-0.16, I2=11.8%). Likelihood ratios were showed in Figure 3. SROC were assessed and got broad range of Area Under Curve assessing miRNA-21 for progressive prostate cancer or castrate resistant prostate cancer around 97.4% as shown in Figure 4.
Figure 2. Forest Plot of Sensitivity and Specificity of miRNA-21 in Prostate Cancer Detection.
Figure 3. Forest Plot of Positive and Negative Likelihood Ratio.
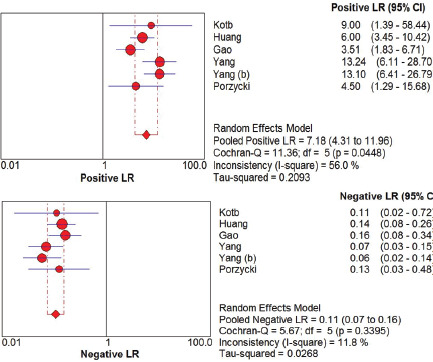
Figure 4. Pooled SROC analysis of miRNA-21 Capability in Detecting Prostate Cancer.
Potential publication bias
The publication bias assessment among articles was conducted using QUADAS-2 assessment tool. The reporting bias was summarized in Table 1.
5. DISCUSSION
The recurrence of progressive prostate cancer or castrate resistant prostate cancer (CRPC) is usually related to the excessive activation of androgen receptor. Recent studies have shown that several oncogenic miRNAs are associated with abnormal androgen receptor (AR) activation. In particular, miRNA-21 is an AR-regulated miRNA, and its expression level continues to increase from androgen-dependent prostate cancer to CRPC (10). Overexpression of miRNA-21 can support xenograft tumor growth and induce castration resistance phenotype (11). In addition to the androgen response element (ARE), other cis-elements such as AP-1 and STAT-3 have been found in the promoter region of miRNA-21 (12, 13). AP-1 activity is closely related to CRPC recurrence and STAT-3 has also been shown to be involved in prostate cancer metastasis (14, 15). In general, the high level of miRNA-21 may be attributed to the abnormal expression of transcriptional activators (such as AR and AP-1). The subsequent effects of miRNA-21 overexpression in turn promoted the occurrence of prostate tumors. Several target genes of miRNA-21 have been shown to inhibit tumor progression by inhibiting invasion, promoting apoptosis and cell cycle arrest. For example, the myristoylated alanine-rich protein kinase c substrate (MARCKS) is a direct target of miRNA-21 which plays a key role in mitogenesis, cell motility, and membrane transport. Therefore, miRNA-21 promotes cell motility, apoptosis resistance, and invasiveness of PC3 and DU-145 cells in part by targeting MARCKS (16). At the same time, a recent study showed that the reversal induction of cysteine-rich protein with Kazal motif (RECK) is another new target of miRNA-21; RECK and miRNA-21 have been shown inverse correlation from different stages of prostate cancer (17). The previous meta-analysis provided mean and SD or miRNA-21 expression levels fold change in prostate cancer patients versus control but did not provide any diagnostic information of it (18).
6. CONCLUSION
Our current study has identified that miRNA-21 could serve as biomarkers of prostate cancer detection since remarkable diagnostic value of circulating miRNA-21 in prostate cancer progressivity detection.
Author‘s contribution:
The investigation was arranged by KPS, BBP, HS, HK, and AFP who also performed research, provided research materials, and collated and processed data. KPS and AFP were responsible for data analysis and interpretation. KPS, BBP, HS, HK, and AFP contributed with the initial and final versions of the article as well as practical assistance. All authors were in control of the manuscript‘s substance after critically reviewing and approving the final text.
Conflicts of interest:
There are no conflicts of interest.
Financial support and sponsorship:
None.
REFERENCES
- 1.Collak FK, Demir U, Ozkanli S, Kurum E, Zerk PE. Increased expression of YAP1 in prostate cancer correlates with extraprostatic extension. Cancer Biol Med. 2017;14(4):405–413. doi: 10.20892/j.issn.2095-3941.2017.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson HC, Shah SI, Abel PD, et al. Contemporary hormone therapy with LHRH agonists for prostate cancer: avoiding osteoporosis and fracture. Cent European J Urol. 2015;68(2):165–168. doi: 10.5173/ceju.2015.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W, Zhang E, Lin C. MicroRNAs in tumor angiogenesis. Life Sci. 2015;136:28–35. doi: 10.1016/j.lfs.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 4.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Guo JX, Shao ZQ. miR-21 targets and inhibits tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion: an experimental study. Asian Pac J Trop Med. 2017;10(1):87–91. doi: 10.1016/j.apjtm.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018;50(9):1619–1626. doi: 10.1007/s11255-018-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B, Liu Z, Ning H, et al. MicroRNA-21 in peripheral blood mononuclear cells as a novel biomarker in the diagnosis and prognosis of prostate cancer. CBM. 2016;17(2):223–230. doi: 10.3233/CBM-160634. [DOI] [PubMed] [Google Scholar]
- 8.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 10.Ribas J, Ni X, Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormonedependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9:923–929. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita S, Ito T, Mizutani T, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Iliopoulos D, Jaeger SA, Hirsch HA, et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajanne R, Miettinen P, Tenhunen M, et al. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int J Oncol. 2009;35:1175–1182. doi: 10.3892/ijo_00000434. [DOI] [PubMed] [Google Scholar]
- 15.Abdulghani J, Gu L, Dagvadorj A, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172:1717–1728. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Li D, Sha J, et al. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 17.Reis ST, Pontes-Junior J, Antunes AA, et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012;12:14. doi: 10.1186/1471-2490-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: a meta-analysis of related studies. J Cell Biochem. 2018;119(3):2763–2786. doi: 10.1002/jcb.26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y, Guo Y, Wang Z, et al. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma. 2016;63(4):623–628. doi: 10.4149/neo_2016_417. [DOI] [PubMed] [Google Scholar]
- 20.Huang W, Kang XL, Cen S, Wang Y, Chen X. High-level expression of microRNA-21 in peripheral blood mononuclear cells is a diagnostic and prognostic marker in prostate cancer. Genet Test Mol Biomarkers. 2015;19(9):469–475. doi: 10.1089/gtmb.2015.0088. [DOI] [PubMed] [Google Scholar]
- 21.Kotb S, Mosharafa A, Essawi M, Hassan H, Meshref A, Morsy A. Circulating miRNAs 21 and 221 as biomarkers for early diagnosis of prostate cancer. Tumor Biol. 2014;35(12):12613–12617. doi: 10.1007/s13277-014-2584-7. [DOI] [PubMed] [Google Scholar]