Significance
Stromal-interacting molecule 1 (STIM1) proteins are essential for the function of store-operated Ca2+ entry (SOCE). Using transcriptomics, metabolomics, imaging, and inducible smooth muscle–specific STIM1 knockout mice expressing genetically encoded Ca2+ sensors, we reveal a crucial function of STIM1 in airway remodeling and airway hyperresponsiveness in asthma. STIM1-mediated Ca2+ oscillations in airway smooth muscle (ASM) cells are critical for ASM remodeling through metabolic and transcriptional reprogramming and cytokine secretion, including IL-6. These effects are driven by Ca2+-dependent activation of the transcription factor isoform NFAT4 specifically in ASM. Our data provide evidence that ASM STIM1 and SOCE are central triggers of asthma manifestations and advocate for the future use of STIM1 as a molecular target in asthma therapy.
Keywords: calcium signaling, asthma, smooth muscle remodeling, CRAC channels, metabolic reprogramming
Abstract
Airway remodeling and airway hyperresponsiveness are central drivers of asthma severity. Airway remodeling is a structural change involving the dedifferentiation of airway smooth muscle (ASM) cells from a quiescent to a proliferative and secretory phenotype. Here, we show up-regulation of the endoplasmic reticulum Ca2+ sensor stromal-interacting molecule 1 (STIM1) in ASM of asthmatic mice. STIM1 is required for metabolic and transcriptional reprogramming that supports airway remodeling, including ASM proliferation, migration, secretion of cytokines and extracellular matrix, enhanced mitochondrial mass, and increased oxidative phosphorylation and glycolytic flux. Mechanistically, STIM1-mediated Ca2+ influx is critical for the activation of nuclear factor of activated T cells 4 and subsequent interleukin-6 secretion and transcription of pro-remodeling transcription factors, growth factors, surface receptors, and asthma-associated proteins. STIM1 drives airway hyperresponsiveness in asthmatic mice through enhanced frequency and amplitude of ASM cytosolic Ca2+ oscillations. Our data advocates for ASM STIM1 as a target for asthma therapy.
Asthma is characterized as a chronic pulmonary disease with episodic or persistent airflow obstruction and affects ∼339 million people globally (1). In addition, it is estimated to cost the United States ∼81.9 billion dollars a year due to health care costs and productivity losses (2). Although many asthmatic patients have their symptoms under control with bronchodilators and corticosteroids, a significant subpopulation of these patients are refractory to these conventional treatments (3). Moreover, asthmatic patients have an accelerated decline in pulmonary function (4). Considering these high costs and morbidities, it is apparent that the current treatment modalities are inadequate, and additional studies are required to further understand the molecular mechanisms of asthma that could be targeted by novel therapies.
Although classically considered an inflammatory disease, increasing evidence points to the structural changes in the airways as key determinants in the pathogenesis of asthma (5). These structural changes, termed airway remodeling (AR), occur in both mild and severe asthmatics and tend to worsen with asthma severity (6, 7). AR includes goblet cell hyperplasia and metaplasia, fibrosis, thickening of the lamina reticularis, angiogenesis, and hyperplasia and hypertrophy of airway smooth muscle (ASM) (8). Specifically, ASM cells have been recognized as the effector cells of AR (5, 9, 10). During AR, ASM cells undergo a phenotypic switch from a quiescent and contractile phenotype into a synthetic phenotype. Synthetic ASM cells are highly proliferative, migratory, and secrete a plethora of cytokines and extracellular matrix molecules that worsen asthma outcome (11, 12).
Calcium (Ca2+) signaling is a key regulator of numerous short-term and long-term cellular functions including contraction and activation of transcriptional programs that control proliferation, migration, secretion, and metabolism (13). Smooth muscle phenotypic switch is characterized by extensive remodeling of the cellular ion channel repertoire with major changes in expression and regulation of Ca2+ channels and transporters (14–21).The store-operated Ca2+ entry (SOCE) pathway has emerged as particularly critical for vascular smooth muscle and cardiomyocyte remodeling during cardiovascular disease (22, 23). SOCE is activated by the binding of agonists, growth factors, and inflammatory mediators to receptors coupled to phospholipase-C (PLC) isoforms. PLC induces the cleavage of phosphatidylinositol 4,5-bisphosphate into the soluble inositol-1,4,5-trisphosphate (IP3) and the membrane-bound diacylglycerol. IP3 is then able to diffuse to the endoplasmic reticulum (ER) and stimulate IP3 receptors to deplete ER Ca2+ stores. ER Ca2+ store depletion causes Ca2+ to dissociate from the luminal EF-hand domains of the ER transmembrane protein, STIM1. This causes STIM1 to gain an extended conformation, oligomerize, and move into ER–plasma membrane junctions. At these junctions, STIM1 physically traps and activates the Ca2+ release–activated Ca2+ (CRAC) channels that are formed by hexamers of Orai proteins, leading to Ca2+ influx from the extracellular milieu into the cytosol (24, 25). Although mammals express another homolog of STIM1 called STIM2, quiescent smooth muscle cells primarily express STIM1 (24, 26, 27). When activated by physiological concentrations of agonists, SOCE-mediated Ca2+ signals take the form of regenerative Ca2+ oscillations (28, 29), which are crucial for the activation of isoforms of Ca2+-dependent transcription factors, including nuclear factor of activated T cells (NFAT) that drive transcriptional programs supporting proliferation, migration, metabolism, and cytokine secretion (30, 31).
By comparison to vascular smooth muscle and cardiomyocytes, very little is known about STIM1 and SOCE in ASM cells. Previous studies have shown that STIM1 is necessary for SOCE in cultured ASM cells (32, 33) and for ASM cell proliferation and migration induced by the chemoattractant, platelet-derived growth factor (PDGF)-BB (33, 34). Here, we hypothesize that STIM1 is a critical driver of ASM remodeling and airway hyperresponsiveness (AHR). We show that mice chronically challenged with the allergen house dust mite (HDM) have increased STIM1 protein expression in ASM. Using inducible smooth muscle–specific STIM1 knockout mice (STIM1smKO), we reveal that STIM1 is a core trigger of AR and AHR in asthmatic mice. ASM STIM1 is required for agonist-induced Ca2+ oscillations, activation of the NFAT4 isoform, interleukin-6 (IL-6) secretion, enhanced metabolic activity, and the reprogramming of ASM into a pro-remodeling transcriptional program. Intriguingly, transcriptional profiling showed that the enhanced metabolic activity of synthetic ASM is controlled by STIM1 and mediated by changes in both mitochondrial and glycolytic functions. We propose that because ASM STIM1 is required for both AR and AHR in asthma, STIM1 is a potential molecular target in asthma therapy.
Results
Chronic Asthmatic Mice Show Increased STIM1 Expression in ASM.
We generated STIM1smKO mice by crossing the current gold-standard tamoxifen-inducible smooth muscle Cre (Myh11 Cre) (35) with mice expressing STIM1 loxP-flanked alleles (STIM1fl/fl) (36). Two littermate controls were used for this study to control for both 1) an active Cre recombinase and 2) the presence of loxP sites: 1) Myh11 Cre mice intraperitonially injected with tamoxifen and 2) Myh11Cre STIM1fl/fl mice intraperitonially injected with vehicle. They will be henceforth referred to as 1) Myh11Cre and 2) STIM1fl/fl. Genotyping of the tracheas from STIM1smKO, Myh11Cre, and STIM1fl/fl confirms that all cohorts have the transgenic Myh11Cre, while the STIM1smKO and STIM1fl/fl mice have loxP-flanked alleles, and only the STIM1smKO mice have active Cre-Lox recombination. A STIM1fl/fl band can be seen in the STIM1 A reaction in the STIM1smKO cohort. It is likely that contaminating nonsmooth muscle tissue in these genotyping reactions from tracheas generate STIM1fl/fl PCR products in this cohort (SI Appendix, Fig. 1 A and B). Furthermore, Western blots of epithelium-denuded tracheas show that STIM1 protein expression is significantly reduced in the STIM1smKO mice compared to both the Myh11Cre and STIM1fl/fl littermates (Fig. 1 A and B). Interestingly, STIM2, another STIM homolog, was not detected in epithelium-denuded trachea from Myh11Cre mice. Splenic tissue, known for its high expression of STIM1 and STIM2 (36), was used as a positive control (SI Appendix, Fig. 1C).
Fig. 1.
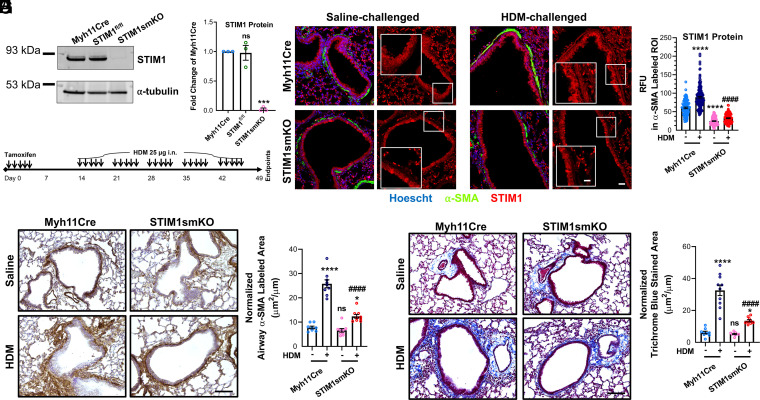
ASM STIM1 protein is augmented in asthmatic mice and is necessary for AR. (A) Representative Western blot showing STIM1 protein in the epithelial-denuded tracheas from Myh11Cre, STIM1fl/fl, and STIM1smKO mice. (B) Quantification of STIM1 protein from A in Myh11Cre (n = 3), STIM1fl/fl (n = 3), and STIM1smKO mice (n = 3) denuded tracheas using densitometry normalized to α-tubulin. (C) Scheme illustrating the time line of tamoxifen injections and intranasal HDM challenges. (D) Representative confocal images of lung slices from Myh11Cre and STIM1smKO mice challenged with either saline or HDM. Smooth muscle is labeled green with Cy3-congugated smooth muscle α-actin (α-SMA) antibody, STIM1 is labeled red with Alexa Flour 647 secondary and STIM1 antibody, and nuclei are counterstained blue with Hoescht. Images on the left are merged with all three colors, and images on the right only show STIM1. Zoomed insets focus on ASM. (Scale bars: 25 μm and 12.5 μm for inset.) (E) Quantification of relative fluorescence units (RFU; from many images like D) of STIM1 in regions of interest (ROIs) overlapping with α-SMA labeling in saline-challenged Myh11Cre (n = 98), HDM-challenged Myh11Cre (n = 185), saline-challenged STIM1smKO (n = 143), and HDM-challenged STIM1smKO mice (n = 122). (F) Representative immunohistochemistry images of lung slices from Myh11Cre and STIM1smKO mice challenged with either saline or HDM. Smooth muscle is labeled brown with α-SMA antibody and 3,3′-diaminobenzidine, and slides are counterstained with hematoxylin. (Scale bar: 100 μm.) (G) Quantification of α-SMA labeled area from A in saline-challenged Myh11Cre (n = 8), HDM-challenged Myh11Cre (n = 9), saline-challenged STIM1smKO (n = 9), and HDM-challenged STIM1smKO (n = 9) mice. (H) Representative images of lung slices stained with Masson’s trichrome where collagen is stained blue. (Scale bar: 100 μm.) (I) Quantification of blue stained area from C in saline-challenged Myh11Cre (n = 9), HDM-challenged Myh11Cre (n = 10), saline-challenged STIM1smKO (n = 9), and HDM-challenged STIM1smKO (n = 9) mice. ns, not significant, *P < 0.05, ***P < 0.001, ****P < 0.0001 when compared to saline-challenged Myh11Cre, ####P < 0.0001 when compared to HDM-challenged Myh11Cre (one-way ANOVA with Dunnett’s test for multiple comparisons).
We used a clinically relevant asthmatic model that intranasally sensitizes and challenges mice to the allergen HDM. Chronic exposures of HDM have been shown to induce AHR, airway inflammation, and AR and fibrosis in mice (37–39). Myh11Cre and STIM1smKO mice were both challenged with HDM according to the protocol described in Fig. 1C. As a control, two additional groups from both mice genotypes were challenged with saline. At the end of this HDM-challenge protocol (at day 49; Fig. 1C), lung slices were isolated and colabeled with the following antibodies: anti-smooth muscle α-actin conjugated to Cy3 (α-SMA; green) and anti-STIM1 with a secondary antibody conjugated to Alexa Fluor 647 (red). STIM1 fluorescence (red) was then quantified specifically in ASM (green) from all mice cohorts. HDM-challenged Myh11Cre mice showed significantly more STIM1 protein expression in ASM compared to the saline-challenged Myh11Cre mice. Importantly, both cohorts of the STIM1smKO mice had significantly reduced STIM1 expression specifically in ASM with prominent STIM1 staining in adjacent airway epithelial cells and surrounding lung tissue (Fig. 1 D and E). The isotype controls had no fluorescent labeling for STIM1 (SI Appendix, Fig. 1D).
STIM1 Is Required for AR in Chronic Asthmatic Mice.
We and others have shown that STIM1 is necessary for remodeling of vascular smooth muscle during disease states, including chronic hypertension and restenosis (27, 40–43). Since AR shares morphological and mechanistic features with vascular remodeling (22), we reasoned that STIM1 might be necessary for AR in the chronic asthmatic HDM-challenge model. By labeling ASM in fixed lung slices with α-SMA and using immunohistochemistry (IHC), we observed that HDM-challenged Myh11Cre mice have increased ASM area as labeled with α-SMA (brown) compared to saline-challenged Myh11Cre mice, suggesting enhanced AR in agreement with previous findings (37–39). Remarkably, HDM-challenged STIM1smKO mice had significantly less ASM area compared to the HDM-challenged Myh11Cre mice. The saline-challenged STIM1smKO mice had similar ASM area to the saline-challenged Myh11Cre mice (Fig. 1 F and G). The isotype control antibody showed no IHC staining for α-SMA (SI Appendix, Fig. 1E). We also investigated fibrosis of these lung slices by using Masson’s Trichrome staining, which labels collagen and extracellular matrix fibers in blue. Consistently, HDM-challenged Myh11Cre mice had significantly more Trichrome blue–stained area than the saline-challenged Myh11Cre mice, and the HDM-challenged STIM1smKO mice had dramatically less Trichrome blue staining than the HDM-challenged Myh11Cre mice. Furthermore, the saline-challenged STIM1smKO mice had similar levels of Trichrome blue staining as the saline-challenged Myh11Cre mice (Fig. 1 H and I).
A prominent feature of asthma is leukocyte recruitment into the airways. Indeed, HDM-challenged Myh11Cre mice had significantly more leukocytes in their bronchoalveolar lavage (BAL) than saline-challenged Myh11Cre mice (SI Appendix, Fig. 1F). These include monocytes, lymphocytes, neutrophils, and eosinophils (SI Appendix, Fig. 1G). Interestingly, saline-challenged and HDM-challenged STIM1smKO mice had similar proportions of BAL leukocyte populations as the saline-challenged and HDM-challenged Myh11Cre mice, respectively (SI Appendix, Fig. 1 F and G). These results suggest that ASM STIM1 is critical for the development of AR in asthma, including smooth muscle thickening and fibrosis, and that this phenotype is independent of leukocyte recruitment to the airways.
STIM1 Is Crucial for ASM Proliferation and Migration.
To understand the molecular mechanisms by which STIM1 drives AR, we performed in vitro molecular studies on primary human ASM cells (HASMCs) (44). We have several cultures of HASMCs isolated from nonasthmatic and fatal asthmatic donors (SI Appendix, Fig. 2A). Cultured HASMCs express both STIM1 and STIM2, and we observed no difference in STIM1 and STIM2 protein expression between nonasthmatic and asthmatic donors using Western blotting (SI Appendix, Fig. 2 B–D). Furthermore, we measured SOCE among these donors using the ratiometric Ca2+ dye Fura2. HASMCs were stimulated first with 2 µM of the irreversible sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA) inhibitor thapsigargin in a nominally Ca2+-free bath solution to evaluate the extent of Ca2+ release from ER Ca2+ stores. Where indicated (SI Appendix, Fig. 2E), 2 mM Ca2+ was restored to the bath solution to measure the magnitude of SOCE. We found no difference in SOCE of HASMCs between nonasthmatic and asthmatic donors (SI Appendix, Fig. 2 E–G). Smooth muscle cells cultured in vitro dedifferentiate into a “synthetic” phenotype, remodel their Ca2+ signaling machinery, and up-regulate STIM proteins (11, 45, 46). Nevertheless, this in vitro dedifferentiation occurs equally in both nonasthmatic and asthmatic HASMCs, thus cancelling any in vivo differences between these two groups. This is consistent with results with vascular smooth muscle cells isolated from normotensive and hypertensive rats (41). Hence, for the remainder of our experiments, we used HASMCs from normal donors.
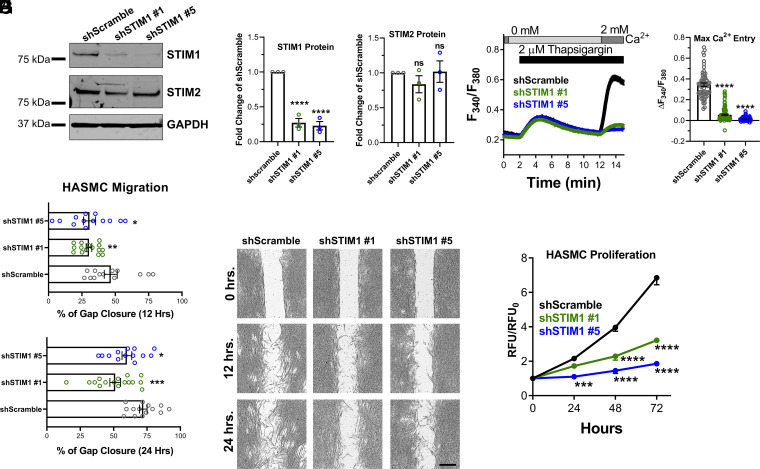
We generated stable STIM1 knockdown (KD) in HASMCs using two different small hairpin RNAs (shRNAs), and HASMCs stably expressing scrambled shRNA (shScramble) were used as a control. STIM1 protein expression was significantly diminished in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to shScramble HASMCs (Fig. 2 A and B). Furthermore, SOCE was nearly eliminated in both STIM1 KD HASMCs conditions compared to shScramble control (Fig. 2 D and E). There was no compensation in either STIM2 (Fig. 2 A and C), or Orai1protein expression (SI Appendix, Fig. 3 A–C); both Orai1-α and Orai1-β translational variants were analyzed after protein sample deglycosylation as described in Methods. Using RT-qPCR, Orai2 and Orai3 messenger RNA (mRNA) expression was also comparable between shScramble and shSTIM1 conditions (SI Appendix, Fig. 3 D and E). We then assessed the role of STIM1 in HASMC migration and proliferation. HASMC migration was measured using the gap closure assay. To eliminate contributions from HASMC proliferation, media was supplemented with 10 µg/mL mitomycin C. Migration was significantly impaired in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to shScramble HASMCs at 12 and 24 h (Fig. 2 F and G). Similarly, HASMC proliferation was measured over 72 h using the dye CyQUANT; shSTIM1#1 and shSTIM1#5 KD HASMCs had significant reduction in proliferation compared to shScramble HASMCs (Fig. 2H). We generated stable STIM1 KD HASMCs in another nonasthmatic donor (denoted as “donor 2”). STIM1 protein expression was also significantly reduced in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to shScramble HASMCs from donor 2 (SI Appendix, Fig. 3 F and G). The proliferation and migration of HASMCs from donor 2 were also significantly decreased in shSTIM1#1 and shSTIM1#5 KD conditions compared to shScramble control (SI Appendix, Fig. 3 H and I). We costained shScramble, shSTIM1#1, and shSTIM1#5 HASMCs with 7-aminoactinomycin D (7-AAD) and Annexin V to determine the extent of apoptosis in these cell groups. There was no statistically significant change between the shSTIM1 and shScramble HASMCs in the percentage of cells in early (positive for Annexin V), late (double positive for Annexin V and 7AAD), or total apoptosis (sum of early and late apoptosis) (SI Appendix, Fig. 4 A and B). Furthermore, cleaved caspase 3 protein expression, which is a marker of apoptosis (47), was similar between shScramble, shSTIM1#1, and shSTIM1#5 HASMCs (SI Appendix, Fig. 4 C–E).
Fig. 2.
STIM1 is necessary for ASM cell proliferation and migration. (A) Representative Western blots showing STIM1 and STIM2 protein expression in HASMCs transfected with either shScramble or with STIM1 shRNA (shSTIM1#1 or shSTIM1#5). (B) Quantification of STIM1 and (C) STIM2 protein expression in shScramble (n = 3), shSTIM1#1 (n = 3), and shSTIM1#5 (n = 3) HASMCs from A using densitometry normalized to GAPDH. (D) Cytosolic Ca2+ measurements using the standard Ca2+ off/Ca2+ on protocol with 2 μM thapsigargin showing SOCE activity in shScramble (n = 61; black trace), shSTIM1#1 (n = 87; green trace), and shSTIM1#5 (n = 88; blue trace) HASMCs. (E) Quantification of maximal SOCE from D. (F) Quantification of HASMC migration at 12 and 24 h in shScramble (n = 13), shSTIM1#1 (n = 15), and shSTIM1#5 (n = 13) HASMCs. (G) Representative bright-field images from F. (Scale bar: 500 μm.) (H) Quantification in normalized relative fluorescent units (RFU) of proliferation of shScramble (n = 6; black trace), shSTIM1#1 (n = 6; green trace), and shSTIM1#5 (n = 6; blue trace) HASMCs over 72 h. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant (unpaired Student’s t test for two comparisons and ANOVA with Dunnett’s test for multiple comparisons).
STIM1 Mediates Pro-remodeling Transcriptional Reprogramming in ASM.
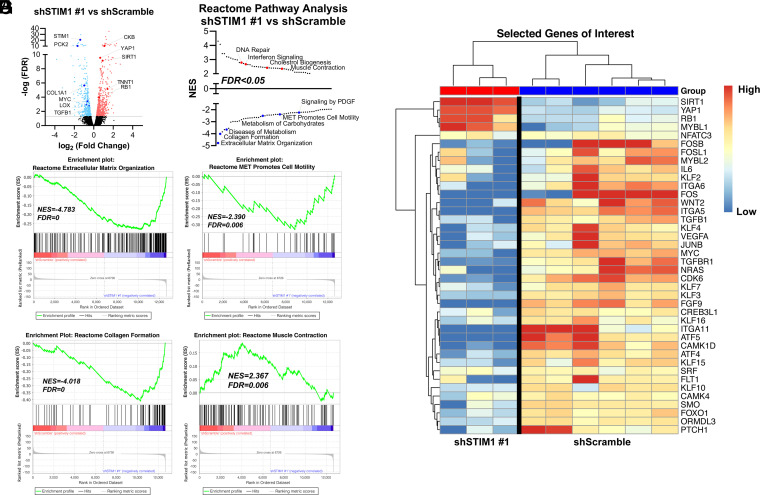
To identify downstream pathways regulated by STIM1 in ASM, we compared the transcriptional profiles of shScramble and shSTIM1 HASMCs using RNA sequencing (Dataset S3). Differential gene expression analysis revealed numerous genes significantly different between shScramble and shSTIM1#1 HASMCs (Fig. 3A) and between shScramble and shSTIM1#5 HASMCs (SI Appendix, Fig. 5A). Through gene set enrichment analysis (GSEA) using the reactome gene sets, numerous enriched pathways were revealed between shScramble and shSTIM1 HASMCs. Negatively enriched pathways found in shSTIM1 HASMCs include extracellular matrix organization, collagen formation, diseases of metabolism, metabolism of carbohydrates, signaling by PDGF, and mesenchymal to epithelial transition (MET) pathways that promote cell motility. Positively enriched pathways include DNA repair, interferon signaling, cholesterol biogenesis, and muscle contraction (Fig. 3 B–F and SI Appendix, Fig. 5 B and C). We validated several of these RNA-sequencing findings, especially targets involved in extracellular matrix production, using RT-qPCR. HASMCs transfected with shSTIM1#1 and shSTIM1#5 had significantly less COL5A2, LOX, and TGFβR1 mRNA than HASMCs transfected with shScramble (SI Appendix, Fig. 5 D–F). Expression profiling showed that STIM1 KD in HASMCs down-regulated the expression of pro-remodeling genes encoding transcription factors (MYC, FOXO1, KLF, ATF, FOS, and JUN), cytokines, agonists and receptors (IL-6, VEGF-A, TGFβ1/TGFβR1, Wnt2, Smoothened, and Patched1), signaling proteins (CaMK), and the orosomucoid-like 3 (ORMDL3) protein (48) that is specifically associated with human asthma (Fig. 3G). These transcriptional profiling results support a central function for STIM1 in promoting ASM dedifferentiation while suppressing the ASM quiescent phenotype.
Fig. 3.
STIM1 controls pro-remodeling transcriptional reprogramming. (A) Volcano plot comparing differentially expressed genes between shScramble and shSTIM1#1 HASMCs. Genes are plotted by log2 fold change and −log10 false discovery rate (FDR), with a threshold of an FDR <0.05 (gray line). In the shSTIM1#1 samples, significantly up-regulated and down-regulated genes are light red and light blue, respectively. (B) Pathway analysis using the reactome pathways gene set ranking significantly up-regulated (red) and down-regulated (blue) pathways based on normalized enrichment score (NES). (C–F) GSEA enrichment plots using the reactome pathways gene set comparing shSTIM1#1 and shScramble HASMCs showing a negative enrichment of extracellular matrix organization (C), MET promotes cell motility (D) and collagen formation genes (E), and a positive correlation in the enrichment of muscle contraction genes (F). For each GSEA enrichment plot, NES and FDR values are displayed. (G) Heatmap display of select genes, including major transcription factors, agonists, receptors, and effector proteins between HASMCs stably expressing shScramble and those expressing shSTIM1#1. Genes appear in rows, and samples appear in columns. Gene expression values were mean centered by gene.
STIM1 Is Required for NFAT4 Activation and Secretion of IL-6 by ASM Cells.
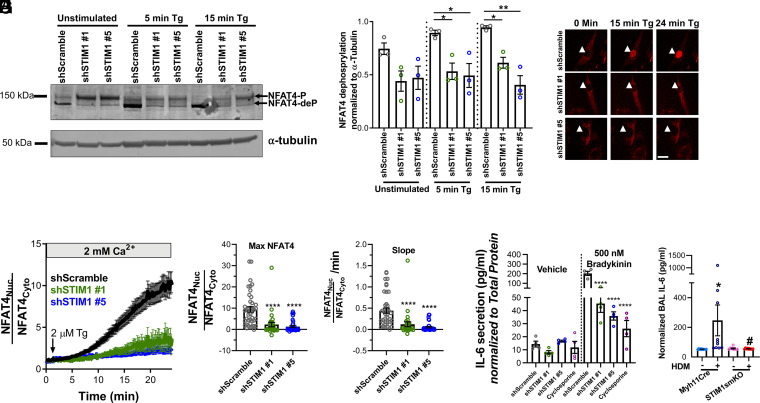
We next sought to identify the mechanisms by which STIM1-mediated Ca2+ signals transcriptionally reprogram ASM to be “synthetic.” SOCE has been well characterized in providing a specific spatiotemporal Ca2+ microdomain that stimulates the phosphatase calcineurin, which dephosphorylates numerous phosphorylation sites on the transcription factor NFAT, causing its translocation to the nucleus (28, 49, 50). Once in the nucleus, NFAT activates the transcription of other transcription factors, receptors, channels, and signaling pathways, including ones involved in proliferation, migration, and secretion (49). Furthermore, others have confirmed that NFAT4 (NFATC3) is the predominant NFAT isoform expressed in ASM cells (51–53). Following previously established techniques (54), we examined the ability of endogenous NFAT4 to be dephosphorylated in HASMCs upon maximal SOCE activation by 2 µM thapsigargin (SI Appendix, Fig. 5G). A total of 5 min after stimulation with thapsigargin, there was an increase in the proportion of dephosphorylated NFAT4 (NFAT4-deP) species relative to the phosphorylated species (NFAT4-P), as shown by shift in molecular weight on a Western blot. Importantly, both 1 µM cyclosporine (CsA), which inhibits calcineurin (55), and 5 µM gadolinium (Gd3+), which specifically inhibits SOCE at these low concentrations (56), blocked NFAT4 dephosphorylation (SI Appendix, Fig. 5 G and H). Interestingly, even under unstimulated conditions, there was significantly less dephosphorylated NFAT4 in shSTIM1#1 and shSTIM1#5 HASMCs compared to shScramble HASMCs (Fig. 4 A and B). After stimulation with thapsigargin (at 5 min and 15 min), NFAT4 dephosphorylation in shSTIM1#1- and shSTIM1#5-transfected HASMCs was significantly inhibited compared to shScramble HASMCs (Fig. 4 A and B). We also stably expressed NFAT4mCherry in shSTIM1#1, shSTIM1#5, and shScramble HASMCs and followed its nuclear translocation using time-lapse fluorescence microscopy upon stimulation with 2 µM thapsigargin (Fig. 4 C–F). NFAT4mCherry translocated slower and significantly less in shSTIM1#1- and shSTIM1#5-transfected HASMCs as compared to shScramble HASMCs (Fig. 4 C–F). These results strongly indicate that NFAT4 activation and subsequent NFAT4-dependent signaling in ASM cells requires STIM1.
Fig. 4.
STIM1 controls the activation of NFAT4. (A) Representative Western blot showing NFAT4-P and NFAT4-deP, respectively, in HASMCs transfected with shScramble, shSTIM1#1, or shSTIM1#5 and either unstimulated or stimulated for 5 or 15 min with 2 μM thapsigargin (Tg) in the presence of 2 mM Ca2+. (B) Quantification of percentage of NFAT4-deP to NFAT-P in shScramble (n = 3), shSTIM1#1 (n = 3), and shSTIM1#5 (n = 3) HASMCs from A using densitometry normalized to α-tubulin. (C) Representative images of shScramble, shSTIM1#1, and shSTIM1#5 HASMCs expressing NFAT4mCherry that are either unstimulated (time 0) or stimulated with 2 μM Tg in the presence of 2 mM Ca2+ for 15 min and 24 min. White arrowheads point to the cell nucleus. (Scale bar: 25 μm.) (D) Time-lapsed quantification of nuclear NFAT4mcherry translocation of shScramble (n = 41; black trace), shSTIM1#1 (n = 29; green trace), and shSTIM1#5 (n = 35; blue trace) HASMCs stimulated with 2 μM Tg in the presence of 2 mM Ca2+. Translocation is measured by dividing the fluorescence of a region of interest in the nucleus by that of a region of interest in the cytoplasm for each cell. Quantification of maximal (E) and slope (F) of NFAT4 translocation from C. (G) Using ELISA, IL-6 secretion is measured in shScramble (n = 4; gray circles), shSTIM1#1 (n = 4; green circles), shSTIM1#5 (n = 4; blue circles), and shScramble pretreated with 1 μM cyclosporine (n = 4; purple circles). HASMCs were stimulated overnight with either vehicle or 500 nM BK. For each condition, IL-6 secretion is normalized to total protein content in media. (H) Quantification of IL-6 in the BAL of saline-challenged Myh11Cre (n = 10), HDM-challenged Myh11Cre (n = 10), saline-challenged STIM1smKO (n = 9), and HDM-challenged STIM1smKO (n = 10) mice. *P < 0.05, **P < 0.01, ****P < 0.0001 when compared to shScramble or saline-challenged Myh11Cre, #P < 0.05 when compared to HDM-challenged Myh11Cre (one-way ANOVA with Dunnett’s test for multiple comparisons).
A significant property of ASM cells in asthma is their function as immune effectors that secrete inflammatory mediators, which further aggravate AR including smooth muscle proliferation, migration, and deposition of extracellular matrix (57–59). One such inflammatory mediator that requires SOCE and NFAT activation is IL-6 (60–63). Our RNA-sequencing findings showed that IL-6 expression in ASM cells is strongly down-regulated upon STIM1 KD (Fig. 3G). Thus, we sought to determine whether STIM1 is specifically required for the secretion of IL-6 in HASMCs. Using an enzyme-linked immunosorbent assay (ELISA), we measured IL-6 secretion in the media from serum-starved HASMCs stimulated with various asthma-related agonists (e.g., 500 nM Bradykinin [BK] and 100 µM methacholine [MeCh]), growth factors (20 ng/mL PDGF), and allergens (10 µg/mL HDM) in serum-free media for 18 h. All stimuli caused a significant increase in IL-6 secretion from HASMCs (Fig. 4G and SI Appendix, Fig. 5I). Interestingly, shSTIM1#1 and shSTIM1#5 KD HASMCs had significantly less IL-6 secretion than shScramble HASMCs in response to all stimuli. Furthermore, shScramble HASMCs treated with 1 µM CsA showed similar levels of stimulated IL-6 secretion to those of STIM1 KD HASMCs (Fig. 4G and SI Appendix, Fig. 5I), suggesting that IL-6 secretion requires calcineurin/NFAT signaling downstream of STIM1. Similar results were observed when we studied a different batch of HASMCs originating from another donor (donor 2; SI Appendix, Fig. 5J). Furthermore, we validated these findings with the chronic asthmatic mouse model (Fig. 1C). We show that IL-6 secretion in the BAL of asthmatic mice challenged with HDM critically depends on ASM STIM1 (Fig. 4H). Using ELISA, HDM-challenged Myh11Cre mice had significantly more BAL IL-6 compared to saline-challenged Myh11Cre mice. Strikingly, BAL IL-6 was significantly reduced in the HDM-challenged STIM1smKO mice but was similar between the saline-challenged cohorts (Fig. 4H). As a control, we measured BAL Immunoglobulin E (IgE), which is predominantly secreted by plasma cells. Bal IgE was significantly increased in HDM-challenged mice compared to saline-challenged mice. Importantly, the BAL IgE levels of HDM-challenged STIM1smKO mice were statistically similar to those of HDM-challenged Myh11Cre mice. However, there was a trend for decreased BAL IgE in the HDM-challenged STIM1smKO mice, which may suggest a possible interaction of smooth muscle and plasma cells that is mediated by smooth muscle STIM1 (SI Appendix, Fig. 5K). These data strongly argue that the up-regulation of STIM1 specifically in ASM cells of asthmatic mice mediates IL-6 secretion through activation of NFAT4.
STIM1 Regulates ASM Proliferation and Migration through NFAT4.
To establish that STIM1 regulates ASM proliferation and migration specifically through NFAT4 activation, we generated a constitutively active mutant of NFAT4 (CA-NFAT4) based on constructs previously reported (64, 65). Following the homology of a previously published constitutively active mutant of NFAT1 (66), we generated the CA-NFAT4 construct by introducing 25 alanine substitutions at serine phosphorylation sites in the SRR1, SP-1, SP-2, and SP-3 regions (SI Appendix, Fig. 6A). We added a carboxyl-terminal mCherry tag to this CA-NFAT4 construct and cloned it into a lentiviral vector to allow stable expression in HASMCs. Wild-type NFAT4mCherry and mCherry alone were also independently cloned into the same lentiviral vector. Unstimulated HASMCs expressed NFAT4mCherry primarily in the cytosol, and following thapsigargin stimulation, the construct translocated into the nucleus. The CA-NFAT4mCherry primarily localized to the nucleus in unstimulated HASMCs, suggesting constitutive activation of this construct (SI Appendix, Fig. 6B). We then expressed CA-NFAT4mCherry, NFAT4mCherry, and mCherry into stable shScramble, shSTIM1#1, and shSTIM1#5 HASMCs and measured proliferation and migration in complete culture media. Results from shSTIM1#1 and shSTIM1#5 were normalized to the respective shScramble. We found that CA-NFAT4 fully rescues proliferation and migration in shSTIM1#1 and shSTIM1#5 HASMCs (SI Appendix, Fig. 6 C and D). Interestingly, wild-type NFAT4 also rescued proliferation and migration in shSTIM1#1 and shSTIM1#5 HASMCs, although to a lesser extent than the constitutively active mutant. These results suggest that wild-type NFAT4 overexpression in STIM1 KD cells likely leads to a significantly higher portion of NFAT4 migrating to the nucleus compared to cells expressing endogenous NFAT4 levels. These results show that STIM1 promotes ASM cell proliferation and migration through NFAT4.
Loss of STIM1 Quells ASM Cell Metabolism.
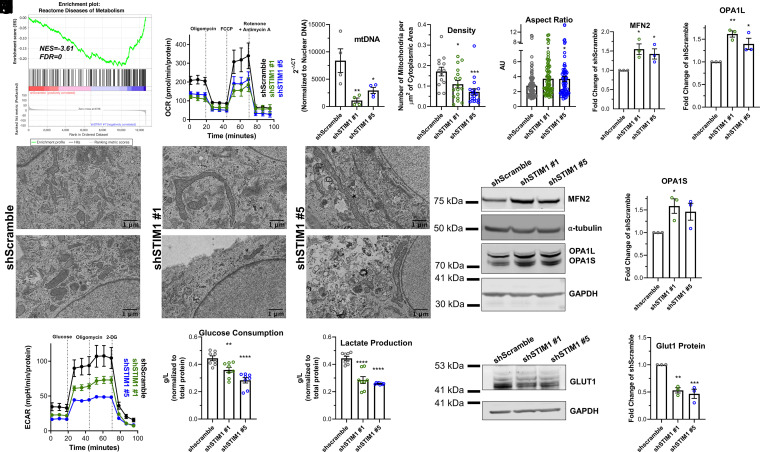
An interesting finding that emerged from our transcriptional profiling and GSEA (Fig. 3 A and B) was that the “Diseases of Metabolism” pathway was significantly down-regulated in shSTIM1 HASMCs compared to shScramble HASMCs (Fig. 5A and SI Appendix, Fig. 7A). Recent studies suggest that ASM cells from asthmatic bronchi have enhanced respiration and mitochondrial mass and hypothesized that this augmented metabolism likely fuels AR (67, 68). Interestingly, recent evidence from T cells suggested that through activation of NFAT, SOCE is a major regulator of metabolic gene programs that support both oxidative phosphorylation and glycolysis (69). Thus, we evaluated mitochondrial respiration in HASMCs using the Seahorse mitochondrial stress test. Basal and maximal respiration were significantly reduced in shSTIM1#1 and shSTIM1#5 HASMCs compared to shScramble HASMCs (Fig. 5B and SI Appendix, Fig. 7 B and C). Nonmitochondrial respiration was unchanged (SI Appendix, Fig. 7D). Similar results were found with HASMCs obtained from another donor (donor 2; SI Appendix, Fig. 7I). Using qPCR, we found that mitochondrial DNA was significantly decreased in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to shScramble HASMCs (Fig. 5C). Using transmission electron microscopy (TEM), we revealed that mitochondrial density was also considerably reduced in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to shScramble HASMCs (Fig. 5 D and E and SI Appendix, Fig. 8A). Similarly, using flow cytometry, shSTIM1#1 and shSTIM1#5 KD HASMCs showed decreased Mitotracker staining compared to shScramble HASMCs (SI Appendix, Fig. 8B). Interestingly, TEM images showed that mitochondria were considerably longer in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to mitochondria of shScramble HASMCs (Fig. 5 D and F). This includes a larger aspect ratio (Fig. 5F) and decreased circularity, roundness, and solidity in shSTIM1#1 and shSTIM1#5 KD HASMCs (SI Appendix, Fig. 8 C–E). In further support of this finding, the protein expression of mitofusin 2 (MFN2) and OPA1, which are two GTPases important for outer and inner mitochondrial membrane fusion, respectively (70), were significantly increased in shSTIM1#1 and shSTIM1#5 KD HASMCs compared to shScramble HASMCs (Fig. 5 G–J). However, we did not observe any changes in the expression of proteins of the electron transport chain (ATP5A, UQCRC2, MTCO1, SDHB, and NDUFB8) between shScramble, shSTIM1#1, and shSTIM1#5 HASMCs (SI Appendix, Fig. 8 F and G). Similarly, there was no change in protein expression of the mitochondrial biogenesis transcription regulators: peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC1-α) and peroxisome proliferator-activated receptor-γ (PPAR-γ) between shScramble, shSTIM1#1, and shSTIM1#5 HASMCs (SI Appendix, Fig. 8 H–K). Proteins were quantified with the bicinchoninic acid (BCA) assay, equal amounts of total proteins were loaded for each condition, and GAPDH was used as a loading control. These results suggest that STIM1 regulates mitochondrial respiration, density, and fusion independently of the mitochondrial biogenesis pathways governed by PGC1-α and PPAR-γ.
Fig. 5.
STIM1 is crucial for metabolic activity of ASM cells. (A) GSEA enrichment plots using the reactome pathways gene set comparing shSTIM1#1 and shScramble HASMCs showing a negative correlation in the enrichment of diseases of metabolism. (B) Oxygen consumption rate (OCR) in shScramble (n = 7; black trace), shSTIM1#1 (n = 9; green trace), and shSTIM1#5 (n = 10; blue trace) HASMCs consecutively stimulated with 0.5 μM oligomycin, 2 μM Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin A. Measurements are normalized to total proteins in each sample. (C) Quantification of mitochondrial DNA (mtDNA) in shScramble (n = 4), shSTIM1#1 (n = 4), and shSTIM1#5 (n = 4) HASMCs using RT-qPCR. mtDNA cycle threshold (CT) values are normalized to CT values of genomic DNA products. (D) Two representative TEM images of shScramble, shSTIM1#1, and shSTIM1#5 HASMCs. (Scale bar: 1 μm.) (E) Quantification of the number of mitochondria per square micrometer of cytoplasmic area in micrographs from shScramble (n = 13), shSTIM1#1 (n = 17), and shSTIM1#5 (n = 19) HASMCs. (F) Quantification of the aspect ratio of mitochondria from shScramble (n = 99), shSTIM1#1 (n = 72), and shSTIM1#5 (n = 70) HASMCs. (G) Western blot showing MFN2, OPA1L, and OPA1S proteins in shScramble, shSTIM1#1, and shSTIM1#5 HASMCs. Quantification of MFN2 (H), OPA1L (I), and OPA1S (J) proteins from G in shScramble (n = 3), shSTIM1#1 (n = 3), and shSTIM1#5 (n = 3) HASMCs using densitometry normalized to α-tubulin and GAPDH. (K) Extracellular acidification rate (ECAR) in shScramble (n = 5; black trace), shSTIM1#1 (n = 7; green trace), and shSTIM1#5 (n = 6; blue trace) HASMCs consecutively stimulated with 10 mM glucose, 1 μM oligomycin, and 50 mM 2-deoxy-glucose (2-DG). Measurements are normalized to total proteins in each sample. Quantification of glucose consumption (L) and lactate production (M) in the media of shScramble (n = 8), shSTIM1#1 (n = 8), and shSTIM1#5 (n = 8) HASMCs using a bioanalyzer. (N) Representative Western blot showing GLUT1 protein in shScramble, shSTIM1#1, and shSTIM1#5 HASMCs. (O) Quantification of GLUT1 protein expression from N in shScramble (n = 3), shSTIM1#1 (n = 3), and shSTIM1#5 (n = 3) HASMCs using densitometry normalized to GAPDH. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (ANOVA with Dunnett’s test for multiple comparisons).
Furthermore, using the Seahorse glycolytic stress test, we found that basal acidification, glycolysis, glycolytic capacity, and nonglycolytic acidification are decreased in shSTIM1#1- and shSTIM1#5-transfected HASMCs compared to shScramble HASMCs (Fig. 5K and SI Appendix, Fig. 7 E–H). Similar results were obtained with HASMCs originating from another donor (donor 2; SI Appendix, Fig. 7J). We also measured glucose consumption and lactate production in HASMCs using a YSI bioanalyzer. Compared to shScramble HASMCs, shSTIM1#1 and shSTIM1#5 HASMCs had significantly reduced glucose consumption and lactate production (Fig. 5 L and M). The protein expression of the major glucose uniporter in ASM cells, GLUT1 was also significantly decreased in shSTIM1#1 and shSTIM1#5 HASMCs compared to shScramble HASMCs (Fig. 5 N and O). Using liquid chromatography and mass spectrometry (Dataset S4), we also discovered that many amino acids are increased in shSTIM1#1 and shSTIM1#5 HASMCs compared to shScramble HASMCs (SI Appendix, Fig. 9 A–C), suggesting that preventing up-regulation of STIM1 leads ASM cells to accumulate amino acids that are otherwise used to support the proliferative smooth muscle phenotype.
STIM1smKO HDM-Challenged Mice Show Decreased ASM Mitochondrial Density.
Both the Myh11Cre and STIM1fl/fl littermate mice were challenged with either saline or HDM in the chronic asthmatic mouse model (Fig. 1C). At day 49, fixed airways were embedded in TEM grids. ASM can be identified in the medial layers of airways (SI Appendix, Fig. 10A; red arrows). Consistent with previous findings (67, 68), mitochondrial density was significantly augmented in ASM of HDM-challenged Myh11Cre mice compared to saline-challenged Myh11Cre mice. Interestingly, and consistent with our in vitro results utilizing synthetic shSTIM1 KD HASMCs, HDM-challenged STIM1smKO mice had considerably less mitochondrial density in their ASM than HDM-challenged Myh11Cre mice. ASM cells from saline-challenged group mice had similar mitochondrial density (SI Appendix, Fig. 10 A and B). Consistent with the in vitro results utilizing synthetic shSTIM1 KD HASMCs, ASM cells from saline-challenged STIM1smKO mice had elongated mitochondria with larger aspect ratio compared to ASM cells from saline-challenged Myh11Cre mice, although this elongated mitochondrial phenotype was more drastic in cultured HASMCs (SI Appendix, Fig. 10C).
ASM STIM1 Promotes AHR and ASM Ca2+ Oscillations in Asthmatic Mice.
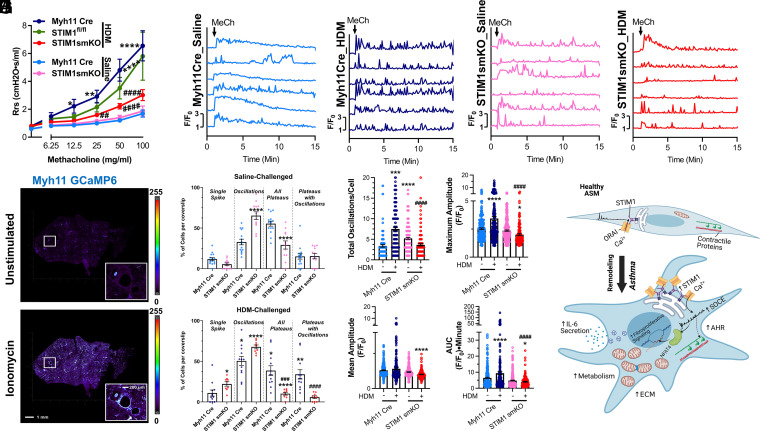
Our mechanistic insights thus far show that inhibition of STIM1 expression in ASM cells leads to impaired NFAT4 activation, transcriptional reprogramming, and quelled metabolism. These data led us to hypothesize that HDM-challenged STIM1smKO mice might have mitigated asthma manifestations. A cardinal feature of asthma is AHR, or the tendency of asthmatic airways to excessively contract in response to stimuli (71). Using the small animal ventilator, Flexivent, we measured AHR in living mice to the muscarinic agonist MeCh. HDM-challenged Myh11Cre and STIM1fl/fl mice had enhanced AHR compared to saline-challenged Myh11Cre mice (Fig. 6A). Astoundingly, HDM-challenged STIM1smKO had significantly less AHR compared to HDM-challenged Myh11Cre and STIM1fl/fl mice, while AHR was similar between the saline-challenged cohorts (Fig. 6A). These results strongly argue that ASM STIM1 is required for AHR in asthma.
Fig. 6.
ASM STIM1 is necessary for cytosolic Ca2+ oscillations and AHR. (A) Airway resistance (Rrs) in response to increasing concentrations of MeCh from saline-challenged Myh11Cre (n = 12; light blue trace), HDM-challenged Myh11Cre (n = 13; dark blue trace), HDM- challenged STIM1fl/fl mice (n = 5; green trace), saline-challenged STIM1smKO (n = 12; pink trace), and HDM-challenged STIM1smKO (n = 14; red trace) mice. (B) Representative confocal images of lung slices from Myh11Cre mice expressing GCaMP6. The top image is unstimulated, and the bottom image is stimulated with 10 µM ionomycin in the presence of 2 mM Ca2+. Inset displays an airway. (Scale bar: 1 mm and 200 µm for inset.) (C) Representative traces of Ca2+ oscillations in the ASM of lung slices from Myh11Cre and STIM1smKO mice expressing GCaMP6 and challenged with either saline or HDM. Lung slices were stimulated with 6.25 mg/mL MeCh in the presence of 2 mM Ca2+ at 1 min. Ca2+ oscillations are measured by the GCaMP6 fluorescence at each time point normalized to the GCaMP6 fluorescence at 0 min (F/F0). (D and E) Quantification of the percentage of ASM cells responding with 1) a single Ca2+ spike, 2) regenerative Ca2+ oscillations returning to baseline, 3) all varieties of Ca2+ plateaus, and 4) a subset of Ca2+ plateaus from 3) 3 superimposed with Ca2+ oscillations in lung slices from (D) saline-challenged Myh11Cre (n = 15, light blue circles) and saline-challenged STIM1smKO (n = 12, pink circles) mice, and (E) HDM-challenged Myh11Cre (n = 12, dark blue circles), and HDM-challenged STIM1smKO (n = 9, red circles) mice. Representative movies of these four experimental conditions are SI movies S6 through S9, respectively. “n” represents the number of lung slices. Quantification in ASM cells of (F) total Ca2+ oscillations per 14 min, (G) mean amplitude, (H) maximal amplitude, (I) AUC of the Ca2+ signal for entire trace from saline-challenged Myh11Cre (n = 108), HDM-challenged Myh11Cre (n = 205), saline-challenged STIM1smKO (n = 194), and HDM-challenged STIM1smKO mice (n = 127). “n” represents number of ASM cells. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 when compared to saline- challenged Myh11 Cre; ##P < 0.01, ###P < 0.001, ####P < 0.0001 when compared to HDM-challenged Myh11 Cre (one-way ANOVA with Dunnett’s test for multiple comparisons). (J) Summary of the findings from this study (created with BioRender.com).
To elucidate how ASM STIM1 regulates AHR, we considered the possibility that STIM1 in ASM cells may directly be modulating the Ca2+ signal generated by each dose of muscarinic agonist. Indeed, previous studies showed that agonist-induced rapid cytosolic Ca2+ oscillations are critical for ASM contraction (20, 72). SOCE, but not voltage-gated Ca2+ channels, has been shown as essential for maintaining Ca2+ oscillations in lung slices (73). To study agonist-induced Ca2+ oscillations in ASM from chronic asthmatic mice (Fig. 1C), we crossed STIM1smKO and Myh11Cre with GCaMP6fl/fl mice (74), and genotyping confirmed the insertion of the GCaMP6 cassette (SI Appendix, Fig. 11 A and B). This model allowed us to measure cytosolic Ca2+ oscillations specifically in ASM in live freshly isolated lung slices ex vivo. GCaMP6 fluorescence offered a large dynamic range as determined by comparing unstimulated lung slices and maximally stimulated slices with 10 μM of the ionophore ionomycin (Fig. 6B). We then stimulated lung slices from saline-challenged and HDM-challenged Myh11Cre and STIM1smKO mice with the lowest concentration of MeCh (6.25 mg/mL) used in the AHR experiments. Six representative cells are shown in Fig. 6C for each of the four experimental conditions. We noted that ASM cells from the four different experimental conditions showed different proportions of four distinct patterns of Ca2+ signals in response to MeCh. These four patterns of Ca2+ signals are 1) cells that responded with a single transient that eventually receded to baseline (single spike); 2) cells that responded with repetitive Ca2+ oscillations; 3) cells that responded with a Ca2+ plateau for the duration of the recording; and 4) a subgroup of cells among the latter group (i.e., group 3) that responded with Ca2+ plateaus with regenerative Ca2+ oscillations on top of these plateaus (see SI Appendix, Fig. 12 for representative traces for each group of cells). We defined a Ca2+ oscillation as a spike that had an amplitude of at least 1.25-fold of the basal fluorescence value and returned to baseline. Plateaus were defined as sustained Ca2+ signals for at least 5 min after stimulation that receded to ≥25% of the initial peak. The proportion of cells with plateaus was calculated for each coverslip. For the cells without plateaus, the number of oscillations for that entire 14 min were reported.
Most ASM cells from control Myh11Cre mice treated with saline responded with a Ca2+ plateau (55.4 ± 3.06%; Fig. 6D), with the remaining cells responding with a single spike (11.77 ± 1.8%) or with Ca2+ oscillations (32.82 ± 3.45%) as defined in Methods and in SI Appendix, Fig. 12. However, most ASM cells from STIM1smKO mice treated with saline responded with Ca2+ oscillations (65.3 ± 4.65%; Fig. 6D) at the expense of Ca2+ plateaus (29.21 ± 5.0%) with 5.48 ± 1.63% of cells responding with a single spike, consistent with the requirement of STIM1 in supporting Ca2+ plateaus (29). Strikingly, in the HDM-challenged Myh11Cre mice, the proportion of ASM cells that responded with Ca2+ oscillations increased significantly, (to 50.46 ± 5.56%) and among the cells that responded with plateaus (38.59 ± 5.95%), the majority (33.95 ± 5.7%) also displayed Ca2+ oscillations on top of these plateaus (Fig. 6E). ASM cells from HDM-challenged STIM1smKO mice mostly responded with Ca2+ oscillations, with fewer cells displaying oscillations on top of plateaus (5.79 ± 1.65%; Fig. 6E). Importantly, although the vast majority of ASM cells from HDM-challenged STIM1smKO mice responded with Ca2+ oscillations, they had significantly reduced Ca2+ oscillatory frequency (3.64 ± 0.27 oscillations/14 min compared to 7.5 ± 0.4 oscillations/14 min in the HDM-challenged Myh11Cre group), with most cells showing a rundown of Ca2+ oscillations with time. Interestingly, the HDM-challenged STIM1smKO ASM trended to have less Ca2+ oscillations than the saline-challenged STIM1smKO ASM (Fig. 6 C and F). The reason for this difference requires further investigations, but it is possible that HDM causes changes in other proteins regulating the activities of either SOCE, IP3 receptor–dependent Ca2+ release, or membrane potential.
When we considered the Ca2+ signals as a whole without distinction between the different patterns of Ca2+ signals, the mean of the amplitude, the maximum amplitude, and the area under the curve (AUC) of these Ca2+ signals were significantly larger in ASM cells from the HDM-challenged Myh11Cre group compared to the other groups and were significantly blunted in ASM cells from the HDM-challenged STIM1smKO mice (Fig. 6 G–I). These results suggest that during chronic asthma, most ASM cells respond with a high frequency of repetitive Ca2+ oscillations in response to agonists and that even cells responding with Ca2+ plateaus display oscillations on top of these plateaus. STIM1 is crucial for sustaining the high frequency and amplitude of these Ca2+ oscillations.
Discussion
It is clearly established that AR and, in particular, the increase of ASM mass are pathological hallmarks of asthma that contribute to disease severity (5, 8–10, 75–78). During ASM remodeling, ASM cells undergo a phenotypic switch from a quiescent to a synthetic phenotype (11, 12). The molecular mechanisms of this phenotypic switch remain largely unclear. Evidence points to an important association between ASM cell remodeling and the molecular reprogramming of surface receptors, ion channels, and Ca2+ signaling pathways. Specifically, STIM1, which is a critical component of SOCE in most cell types, is required for vascular smooth muscle remodeling in diseases such as chronic hypertension and restenosis (26, 27, 41, 43). We previously showed that STIM1 protein is necessary for ASM cell proliferation and migration and that STIM1 expression is increased in ASM cells isolated from ovalbumin-challenged asthmatic mice (33). Others have shown that the pharmacological inhibitors of SOCE, 3,5-bis(trifluoromethyl) pyrazole and RP3128 mitigate airway inflammation and AHR in asthmatic rodent models (79–81). However, the specific role of ASM STIM1 in AR, AHR, and asthma has remained unknown. Although these SOCE inhibitors have questionable specificity, our work suggests that SOCE inhibition specifically in ASM through STIM1 deletion is sufficient to mitigate AR and AHR in asthma.
Here, we show that ASM STIM1 protein expression is increased in the chronic HDM-challenged asthmatic mice and that STIM1 is necessary for AR through activation of NFAT4. We show that HDM-challenged STIM1smKO mice have less airway fibrosis and reduced secretion of IL-6, a cytokine known to induce airway fibrosis (82). As the mechanisms of airway fibrosis are rather undefined (83), ASM cells have emerged as significant immunomodulatory cells capable of secreting cytokines and extracellular matrix (57). IL-6, which is also secreted by activated airway epithelial cells (62, 63), has been shown to stimulate smooth muscle cell proliferation and migration (60, 84). HDM has been demonstrated to induce SOCE-mediated production of IL-6 in airway epithelial cells (85). Hence, STIM1-mediated SOCE by HDM and agonists in airway cells and subsequent IL-6 secretion likely form a positive feedback loop that maintains AR. Our results show that IL-6 production requires activation of NFAT4, which agrees with previous studies (60, 86). STIM1 likely regulates the secretion of other profibrotic and inflammatory mediators by ASM. Indeed, our RNA sequencing showed that KD of STIM1 in HASMCs reduces the expression of transforming growth factor-β (TGF-β) and its receptor. Previous reports described a link between SOCE and fibrosis, presumably through SOCE-dependent up-regulation of TGF-β signaling (87, 88).
A striking finding from our transcriptional profiling experiments was that the asthma-associated gene ORMDL3 was down-regulated while muscle contractile pathways were positively enriched in our STIM1 KD HASMCs, suggesting that STIM1 KD in synthetic HASMCs induces cell differentiation back into the quiescent/contractile phenotype of healthy airways. Although further studies are required to understand the molecular mechanisms involved, STIM1-dependent signaling may serve as a molecular checkpoint in the dedifferentiation process of ASM remodeling. Previous studies showed that mitochondrial mass and metabolism are increased during AR in asthma (67, 68). A key discovery from our study is that STIM1 is a central driver of metabolic reprogramming that supports AR. Recent findings showed that SOCE is required for proliferation, secretion of cytokines, and clonal expansion of T cells through metabolic reprogramming and up-regulation of both oxidative phosphorylation and glycolytic genes (69). Interestingly, we found that STIM1 KD enhances MFN2 and OPA1 protein expression and ASM mitochondrial fusion. These data suggest that during AR, STIM1 is required for mitochondrial fission and enhanced mitochondrial function to bolster AR independently of mitochondrial biogenesis. Indeed, previous studies have shown that mitochondrial fission reflects a proliferative phenotype (89), while others reported that MFN2 expression is reduced in ASM of patients with asthma (90) and that cytokines induce mitochondrial fission in ASM (91). Future studies scrutinizing these proteins in isolated mitochondria would likely shed light on the specific pathways by which STIM1 controls mitochondrial density and dynamics.
Although AHR is a useful tool in the diagnosis and characterization of asthma (92), the molecular mechanisms of AHR are rather dubious (71). The contribution of AR to AHR is especially controversial. Some animal studies showed that overexpression of structural genes linked to asthma leads to AR and AHR in the absence of airway inflammation (93, 94), suggesting that AR contributes to AHR presumably because more and bigger ASM cells would support enhanced contractility (95). However, other studies have found no association between AR and AHR (96, 97). While our data showed that both AHR and AR are reduced in HDM-challenged STIM1smKO mice, STIM1 KD promotes the up-regulation of smooth muscle contractile genes. This suggests that AHR is likely mediated by receptor-dependent Ca2+ signaling pathways reminiscent of nonexcitable cells (e.g., SOCE) instead of voltage-dependent Ca2+ signaling (e.g., CaV1.2) commonly associated with muscle contractility. Clearly, additional studies are needed to determine any causal link between AR and AHR and the molecular pathways involved. Future studies with simultaneous contractility and Ca2+ imaging in isolated bronchial rings from the four different cohorts of mice studied herein would further elucidate the role of STIM1 and AR in airway contraction.
The enhanced frequency of Ca2+ oscillations has been recognized as an important determinant of AHR (73, 98, 99). In particular, it has been shown that the frequency of Ca2+ oscillations is directly related to the degree of sustained ASM contraction, which is important for AHR (99, 100). Frequent and repetitive Ca2+ oscillations are likely more adept at inducing sustained ASM contraction than monophasic Ca2+ spikes or plateaus that recede over time to near baseline. In addition, these signals are most likely driving the activation on long-term remodeling pathways such as ASM proliferation, migration, fibrosis, and IL-6 secretion. These Ca2+ oscillations are a product of agonist-induced ER Ca2+ release from the IP3R (and, possibly, contributions from ryanodine receptors) and reuptake into the ER by SERCA (101). Our results demonstrate that ASM cells from HDM-challenged mice have increased Ca2+ oscillation frequency and amplitude compared to ASM cells from saline-challenged mice and present with a high proportion of ASM cells that respond with Ca2+ oscillations on top of sustained plateaus, which provides an explanation as to why asthmatic ASM exhibit AHR. Importantly, we show that ASM cells from HDM-challenged STIM1smKO mice have dramatically fewer Ca2+ oscillations than ASM cells from HDM-challenged Myh11Cre mice. Our results are consistent with previous pharmacological studies showing that SOCE, but not voltage-gated CaV1.2 Ca2+ channels, is important for maintaining Ca2+ oscillations in lung slices (73). Since STIM1 mediates significant molecular rewiring of ASM Ca2+ channels, receptors, and effectors during asthma, it is reasonable to propose that up-regulation of STIM1-mediated Ca2+ signals in asthmatic ASM cells mediates both short-term contractility that promotes AHR through receptor-evoked Ca2+ oscillations and long-term signaling that drives AR through NFAT4-dependent transcriptional and metabolic reprogramming. Future studies are needed to elucidate how these Ca2+ oscillations modulate smooth muscle contraction and how agonists and HDM itself, which has been shown to stimulate the protease-activated receptor expressed, drive AHR in asthma.
In summary, ASM STIM1 is up-regulated in asthma and is critical for both AR and AHR by mediating receptor-evoked Ca2+ signaling through SOCE, activation of Ca2+ oscillations in ASM, NFAT4, and downstream pro-remodeling transcriptional programs (Fig. 6J). Although, historically, Ca2+ chelators and L-type Ca2+ channel blockers showed disappointing efficacy in the treatment of asthma (102, 103), these treatments have likely failed to specifically target the unique spatiotemporal Ca2+ signals mediated by STIM1 that are necessary for AR and AHR. We propose that drugs that interfere with STIM1 function and block SOCE would likely be efficacious in alleviating AHR and AR in asthmatic patients. Since STIM1 and SOCE are also critical for immune and epithelial cell function and these cell types are key components of airway inflammation in asthma (24), STIM1 and SOCE may represent the ideal target for asthma therapy.
Methods
HASMCs isolated from healthy and asthmatic donors were maintained in culture under standard conditions and used to perform biochemical assays, mitochondrial fuel flux assays, glucose and lactate measurements, and analysis of migration and proliferation. These cells were also transfected with shRNA against STIM1 and used in Ca2+ measurements using Fura2 and fluorescence microscopy. Ca2+ measurements on live lung slices from control and asthmatic mice were achieved through smooth muscle–specific expression of GCaMP6f in mice. Mitochondrial structure in HAMSCs and in lung slices from mice were studied using TEM. Lung function in anesthetized control and asthmatic mice was measured using the Flexivent system in which respiratory mechanics were evaluated in response to increasing doses of MeCh as described in detail in SI Appendix. All details of cell-culture conditions, complementary DNA construct cloning, stable expressions, Western blotting, RT-qPCR, Ca2+ imaging, proliferation and migration assays, flow cytometry, mitochondrial fuel flux assays, glucose and lactate measurements, transcriptome sequencing and analysis, TEM imaging, metabolomics, mouse models, AHR measurements, GCaMP6f measurements, IHC, IF (immunofluorescence), and BAL cell counts, and ELISA measurements are provided in SI Appendix. Details are also provided of sources and reference numbers for reagents, recombinant DNA, and oligonucleotides necessary for replication in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Yongsoo Kim and Steffy Manjila for using their vibratome and for help optimizing the conditions for GCaMP experiments, Dr. Han Chen from The Pennsylvania State University College of Medicine Electron Microscopy (EM) facility for assistance with TEM imaging, and Dr. Drew Jones and Mr. Leonard Ash from New York University Langone Health Metabolomics Core Resource Laboratory for assistance with metabolomics experiments. We are grateful to the Flow Cytometry and Informatic and Data Analysis Core facilities from The Pennsylvania State University College of Medicine. This work was supported by NIH/National Heart, Lung, and Blood Institute Grants R35-HL150778 (to M.T.), F30-HL147489-01A1 (to M.T.J.), TL1TR002016-04 (to M.T.J.), and P01-HL114471 (to R.A.P).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114557118/-/DCSupplemental.
Data Availability
All materials and experimental protocols, the sources and catalog numbers of reagents, sequences of oligonucleotides, sources of recombinant DNA, and genotypes and sources of animals necessary for replication of the study are included in SI Appendix. The source data has not been deposited in a publicly accessible database. All raw unprocessed data from the study are included in Dataset S1. Unprocessed gels and Western blots are included in Dataset S2. Raw RNA-sequencing data are included in Dataset S3, and raw metabolomics data are included in Dataset S4. All data are available upon request.
References
- 1.GBD 2016 Disease and Injury Incidence and Prevalence Collaborators, Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nurmagambetov T., Kuwahara R., Garbe P., The economic burden of asthma in the United States, 2008-2013. Ann. Am. Thorac. Soc. 15, 348–356 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Chung K. F., et al. , International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur. Respir. J. 43, 343–373 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Lange P., Parner J., Vestbo J., Schnohr P., Jensen G., A 15-year follow-up study of ventilatory function in adults with asthma. N. Engl. J. Med. 339, 1194–1200 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Hough K. P., et al. , Airway remodeling in asthma. Front. Med. (Lausanne) 7, 191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson S. J., et al. , The relationship between eosinophilia and airway remodelling in mild asthma. Clin. Exp. Allergy 43, 1342–1350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourdin A., et al. , Specificity of basement membrane thickening in severe asthma. J. Allergy Clin. Immunol. 119, 1367–1374 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Wadsworth S., Sin D., Dorscheid D., Clinical update on the use of biomarkers of airway inflammation in the management of asthma. J. Asthma Allergy 4, 77–86 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepe C., et al. , Differences in airway remodeling between subjects with severe and moderate asthma. J. Allergy Clin. Immunol. 116, 544–549 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Rosethorne E. M., Charlton S. J., Airway remodeling disease: Primary human structural cells and phenotypic and pathway assays to identify targets with potential to prevent or reverse remodeling. J. Exp. Pharmacol. 10, 75–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright D. B., et al. , Phenotype modulation of airway smooth muscle in asthma. Pulm. Pharmacol. Ther. 26, 42–49 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Zuyderduyn S., Sukkar M. B., Fust A., Dhaliwal S., Burgess J. K., Treating asthma means treating airway smooth muscle cells. Eur. Respir. J. 32, 265–274 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Hill-Eubanks D. C., Werner M. E., Heppner T. J., Nelson M. T., Calcium signaling in smooth muscle. Cold Spring Harb. Perspect. Biol. 3, a004549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.House S. J., Potier M., Bisaillon J., Singer H. A., Trebak M., The non-excitable smooth muscle: Calcium signaling and phenotypic switching during vascular disease. Pflugers Arch. 456, 769–785 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahn K., et al. , Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc. Natl. Acad. Sci. U.S.A. 106, 10775–10780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepherd M. C., et al. , KCa3.1 Ca2+ activated K+ channels regulate human airway smooth muscle proliferation. Am. J. Respir. Cell Mol. Biol. 37, 525–531 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Snetkov V. A., Hirst S. J., Ward J. P., Ion channels in freshly isolated and cultured human bronchial smooth muscle cells. Exp. Physiol. 81, 791–804 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. X., Wang L., Zheng Y. M., Canonical transient potential receptor-3 channels in normal and diseased airway smooth muscle cells. Adv. Exp. Med. Biol. 1131, 471–487 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Thompson M. A., Prakash Y. S., Pabelick C. M., Arachidonate-regulated Ca(2+) influx in human airway smooth muscle. Am. J. Respir. Cell Mol. Biol. 51, 68–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jude J. A., Wylam M. E., Walseth T. F., Kannan M. S., Calcium signaling in airway smooth muscle. Proc. Am. Thorac. Soc. 5, 15–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao C., et al. , Microdomain elements of airway smooth muscle in calcium regulation and cell proliferation. J. Physiol. Pharmacol., 10.26402/jpp.2018.2.01 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Johnson M., Trebak M., ORAI channels in cellular remodeling of cardiorespiratory disease. Cell Calcium 79, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emrich S. M., Yoast R. E., Trebak M., Physiological functions of CRAC channels. Annu. Rev. Physiol., 10.1146/annurev-physiol-052521-013426 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Prakriya M., Lewis R. S., Store-operated calcium channels. Physiol. Rev. 95, 1383–1436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trebak M., Putney J. W. Jr., ORAI calcium channels. Physiology (Bethesda) 32, 332–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potier M., et al. , Evidence for STIM1- and Orai1-dependent store-operated calcium influx through ICRAC in vascular smooth muscle cells: Role in proliferation and migration. FASEB J. 23, 2425–2437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W., et al. , Orai1-mediated I (CRAC) is essential for neointima formation after vascular injury. Circ. Res. 109, 534–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoast R. E., et al. , The native ORAI channel trio underlies the diversity of Ca2+ signaling events. Nat. Commun. 11, 2444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emrich S. M., et al. , Omnitemporal choreographies of all five STIM/Orai and IP3Rs underlie the complexity of mammalian Ca2+ signaling. Cell Rep. 34, 108760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gwack Y., Feske S., Srikanth S., Hogan P. G., Rao A., Signalling to transcription: Store-operated Ca2+ entry and NFAT activation in lymphocytes. Cell Calcium 42, 145–156 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kudryavtseva O., Aalkjaer C., Matchkov V. V., Vascular smooth muscle cell phenotype is defined by Ca2+-dependent transcription factors. FEBS J. 280, 5488–5499 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Peel S. E., Liu B., Hall I. P., ORAI and store-operated calcium influx in human airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 38, 744–749 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spinelli A. M., et al. , Airway smooth muscle STIM1 and Orai1 are upregulated in asthmatic mice and mediate PDGF-activated SOCE, CRAC currents, proliferation, and migration. Pflugers Arch. 464, 481–492 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suganuma N., et al. , STIM1 regulates platelet-derived growth factor-induced migration and Ca2+ influx in human airway smooth muscle cells. PLoS One 7, e45056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirth A., et al. , G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 14, 64–68 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Oh-Hora M., et al. , Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo L. N., et al. , A 4-week model of house dust mite (HDM) induced allergic airways inflammation with airway remodeling. Sci. Rep. 8, 6925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z. G., et al. , Neutralization of TSLP inhibits airway remodeling in a murine model of allergic asthma induced by chronic exposure to house dust mite. PLoS One 8, e51268 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piñeiro-Hermida S., et al. , Attenuated airway hyperresponsiveness and mucus secretion in HDM-exposed Igf1r-deficient mice. Allergy 72, 1317–1326 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Bisaillon J. M., et al. , Essential role for STIM1/Orai1-mediated calcium influx in PDGF-induced smooth muscle migration. Am. J. Physiol. Cell Physiol. 298, C993–C1005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson M. T., et al. , L-type Ca2+ channel blockers promote vascular remodeling through activation of STIM proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 17369–17380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aubart F. C., et al. , RNA interference targeting STIM1 suppresses vascular smooth muscle cell proliferation and neointima formation in the rat. Mol. Ther. 17, 455–462 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassan M., et al. , Essential role of smooth muscle STIM1 in hypertension and cardiovascular dysfunction. Arterioscler. Thromb. Vasc. Biol. 36, 1900–1909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panettieri R. A., Murray R. K., DePalo L. R., Yadvish P. A., Kotlikoff M. I., A human airway smooth muscle cell line that retains physiological responsiveness. Am. J. Physiol. 256, C329–C335 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Trebak M., STIM/Orai signalling complexes in vascular smooth muscle. J. Physiol. 590, 4201–4208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berra-Romani R., Mazzocco-Spezzia A., Pulina M. V., Golovina V. A., Ca2+ handling is altered when arterial myocytes progress from a contractile to a proliferative phenotype in culture. Am. J. Physiol. Cell Physiol. 295, C779–C790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porter A. G., Jänicke R. U., Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 6, 99–104 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Pham A. K., et al. , ORMDL3 expression in ASM regulates hypertrophy, hyperplasia via TPM1 and TPM4, and contractility. JCI Insight 6, 136911 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crabtree G. R., Olson E. N., NFAT signaling: Choreographing the social lives of cells. Cell 109 (suppl.), S67–S79 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Zhang X., et al. , A calcium/cAMP signaling loop at the ORAI1 mouth drives channel inactivation to shape NFAT induction. Nat. Commun. 10, 1971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevenson A. S., Gomez M. F., Hill-Eubanks D. C., Nelson M. T., NFAT4 movement in native smooth muscle. A role for differential Ca(2+) signaling. J. Biol. Chem. 276, 15018–15024 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Zhao L., Sullivan M. N., Chase M., Gonzales A. L., Earley S., Calcineurin/nuclear factor of activated T cells-coupled vanilliod transient receptor potential channel 4 ca2+ sparklets stimulate airway smooth muscle cell proliferation. Am. J. Respir. Cell Mol. Biol. 50, 1064–1075 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mancarella S., et al. , Targeted STIM deletion impairs calcium homeostasis, NFAT activation, and growth of smooth muscle. FASEB J. 27, 893–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feske S., Draeger R., Peter H. H., Eichmann K., Rao A., The duration of nuclear residence of NFAT determines the pattern of cytokine expression in human SCID T cells. J. Immunol. 165, 297–305 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Matsuda S., Koyasu S., Mechanisms of action of cyclosporine. Immunopharmacology 47, 119–125 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Trebak M., Bird G. S., McKay R. R., Putney J. W. Jr., Comparison of human TRPC3 channels in receptor-activated and store-operated modes. Differential sensitivity to channel blockers suggests fundamental differences in channel composition. J. Biol. Chem. 277, 21617–21623 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Damera G., Tliba O., Panettieri R. A. Jr., Airway smooth muscle as an immunomodulatory cell. Pulm. Pharmacol. Ther. 22, 353–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ammit A. J., et al. , Tumor necrosis factor-alpha-induced secretion of RANTES and interleukin-6 from human airway smooth muscle cells: Modulation by glucocorticoids and beta-agonists. Am. J. Respir. Cell Mol. Biol. 26, 465–474 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Huang C. D., Tliba O., Panettieri R. A. Jr., Amrani Y., Bradykinin induces interleukin-6 production in human airway smooth muscle cells: Modulation by Th2 cytokines and dexamethasone. Am. J. Respir. Cell Mol. Biol. 28, 330–338 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Nilsson L. M., et al. , Novel blocker of NFAT activation inhibits IL-6 production in human myometrial arteries and reduces vascular smooth muscle cell proliferation. Am. J. Physiol. Cell Physiol. 292, C1167–C1178 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Nilsson-Berglund L. M., et al. , Nuclear factor of activated T cells regulates osteopontin expression in arterial smooth muscle in response to diabetes-induced hyperglycemia. Arterioscler. Thromb. Vasc. Biol. 30, 218–224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jairaman A., Yamashita M., Schleimer R. P., Prakriya M., Store-operated Ca2+ release-activated Ca2+ channels regulate PAR2-activated Ca2+ signaling and cytokine production in airway epithelial cells. J. Immunol. 195, 2122–2133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kountz T. S., et al. , Differential regulation of ATP- and UTP-evoked prostaglandin E2 and IL-6 production from human airway epithelial cells. J. Immunol. 207, 1275–1287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urso K., et al. , NFATc3 controls tumour growth by regulating proliferation and migration of human astroglioma cells. Sci. Rep. 9, 9361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minami T., et al. , Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J. Biol. Chem. 279, 50537–50554 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Okamura H., et al. , Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 6, 539–550 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Girodet P. O., et al. , Bronchial smooth muscle remodeling in nonsevere asthma. Am. J. Respir. Crit. Care Med. 193, 627–633 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Trian T., et al. , Bronchial smooth muscle remodeling involves calcium-dependent enhanced mitochondrial biogenesis in asthma. J. Exp. Med. 204, 3173–3181 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaeth M., et al. , Store-operated Ca2+ entry controls clonal expansion of T cells through metabolic reprogramming. Immunity 47, 664–679.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westermann B., Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Chapman D. G., Irvin C. G., Mechanisms of airway hyper-responsiveness in asthma: The past, present and yet to come. Clin. Exp. Allergy 45, 706–719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanderson M. J., Delmotte P., Bai Y., Perez-Zogbhi J. F., Regulation of airway smooth muscle cell contractility by Ca2+ signaling and sensitivity. Proc. Am. Thorac. Soc. 5, 23–31 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Boie S., Chen J., Sanderson M. J., Sneyd J., The relative contributions of store-operated and voltage-gated Ca2+ channels to the control of Ca2+ oscillations in airway smooth muscle. J. Physiol. 595, 3129–3141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madisen L., et al. , Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bai T. R., Evidence for airway remodeling in chronic asthma. Curr. Opin. Allergy Clin. Immunol. 10, 82–86 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Bentley J. K., Hershenson M. B., Airway smooth muscle growth in asthma: Proliferation, hypertrophy, and migration. Proc. Am. Thorac. Soc. 5, 89–96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ebina M., Takahashi T., Chiba T., Motomiya M., Cellular hypertrophy and hyperplasia of airway smooth muscles underlying bronchial asthma. A 3-D morphometric study. Am. Rev. Respir. Dis. 148, 720–726 (1993). [DOI] [PubMed] [Google Scholar]
- 78.Ebina M., et al. , Hyperreactive site in the airway tree of asthmatic patients revealed by thickening of bronchial muscles. A morphometric study. Am. Rev. Respir. Dis. 141, 1327–1332 (1990). [DOI] [PubMed] [Google Scholar]
- 79.Ohga K., et al. , The suppressive effects of YM-58483/BTP-2, a store-operated Ca2+ entry blocker, on inflammatory mediator release in vitro and airway responses in vivo. Pulm. Pharmacol. Ther. 21, 360–369 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Sutovska M., Kocmalova M., Franova S., Vakkalanka S., Viswanadha S., Pharmacodynamic evaluation of RP3128, a novel and potent CRAC channel inhibitor in guinea pig models of allergic asthma. Eur. J. Pharmacol. 772, 62–70 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Yoshino T., et al. , YM-58483, a selective CRAC channel inhibitor, prevents antigen-induced airway eosinophilia and late phase asthmatic responses via Th2 cytokine inhibition in animal models. Eur. J. Pharmacol. 560, 225–233 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Rincon M., Irvin C. G., Role of IL-6 in asthma and other inflammatory pulmonary diseases. Int. J. Biol. Sci. 8, 1281–1290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Royce S. G., Cheng V., Samuel C. S., Tang M. L., The regulation of fibrosis in airway remodeling in asthma. Mol. Cell. Endocrinol. 351, 167–175 (2012). [DOI] [PubMed] [Google Scholar]
- 84.Wang Z., Newman W. H., Smooth muscle cell migration stimulated by interleukin 6 is associated with cytoskeletal reorganization. J. Surg. Res. 111, 261–266 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Jairaman A., Maguire C. H., Schleimer R. P., Prakriya M., Allergens stimulate store-operated calcium entry and cytokine production in airway epithelial cells. Sci. Rep. 6, 32311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tripathi P., et al. , Activation of NFAT signaling establishes a tumorigenic microenvironment through cell autonomous and non-cell autonomous mechanisms. Oncogene 33, 1840–1849 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mai X., et al. , Blockade of orai1 store-operated calcium entry protects against renal fibrosis. J. Am. Soc. Nephrol. 27, 3063–3078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang B., et al. , Store-operated Ca2+ entry (SOCE) contributes to angiotensin II-induced cardiac fibrosis in cardiac fibroblasts. J. Pharmacol. Sci. 132, 171–180 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Liesa M., Palacín M., Zorzano A., Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89, 799–845 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Aravamudan B., et al. , Cigarette smoke-induced mitochondrial fragmentation and dysfunction in human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 306, L840–L854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delmotte P., Sieck G. C., Interaction between endoplasmic/sarcoplasmic reticulum stress (ER/SR stress), mitochondrial signaling and Ca(2+) regulation in airway smooth muscle (ASM). Can. J. Physiol. Pharmacol. 93, 97–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fowler S. J., Dempsey O. J., Sims E. J., Lipworth B. J., Screening for bronchial hyperresponsiveness using methacholine and adenosine monophosphate. Relationship to asthma severity and beta(2)-receptor genotype. Am. J. Respir. Crit. Care Med. 162, 1318–1322 (2000). [DOI] [PubMed] [Google Scholar]
- 93.Das S., et al. , GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc. Natl. Acad. Sci. U.S.A. 113, 13132–13137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miller M., et al. , ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J. Immunol. 192, 3475–3487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perez-Zoghbi J. F., et al. , Ion channel regulation of intracellular calcium and airway smooth muscle function. Pulm. Pharmacol. Ther. 22, 388–397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kermode J. A., et al. , The effect of airway remodelling on airway hyper-responsiveness in asthma. Respir. Med. 105, 1798–1804 (2011). [DOI] [PubMed] [Google Scholar]
- 97.McParland B. E., Macklem P. T., Pare P. D., Airway wall remodeling: Friend or foe? J. Appl. Physiol. 95, 426–434 (2003). [DOI] [PubMed] [Google Scholar]
- 98.Parameswaran K., Janssen L. J., O’Byrne P. M., Airway hyperresponsiveness and calcium handling by smooth muscle: A “deeper look”. Chest 121, 621–624 (2002). [DOI] [PubMed] [Google Scholar]
- 99.Perez J. F., Sanderson M. J., The frequency of calcium oscillations induced by 5-HT, ACH, and KCl determine the contraction of smooth muscle cells of intrapulmonary bronchioles. J. Gen. Physiol. 125, 535–553 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bai Y., Sanderson M. J., Airway smooth muscle relaxation results from a reduction in the frequency of Ca2+ oscillations induced by a cAMP-mediated inhibition of the IP3 receptor. Respir. Res. 7, 34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berridge M. J., Bootman M. D., Roderick H. L., Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 (2003). [DOI] [PubMed] [Google Scholar]
- 102.Downes H., Hirshman C. A., Calcium chelators increase airway responsiveness. J. Appl. Physiol. (1985) 59, 92–95 (1985). [DOI] [PubMed] [Google Scholar]
- 103.Ann Twiss M., Harman E., Chesrown S., Hendeles L., Efficacy of calcium channel blockers as maintenance therapy for asthma. Br. J. Clin. Pharmacol. 53, 243–249 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials and experimental protocols, the sources and catalog numbers of reagents, sequences of oligonucleotides, sources of recombinant DNA, and genotypes and sources of animals necessary for replication of the study are included in SI Appendix. The source data has not been deposited in a publicly accessible database. All raw unprocessed data from the study are included in Dataset S1. Unprocessed gels and Western blots are included in Dataset S2. Raw RNA-sequencing data are included in Dataset S3, and raw metabolomics data are included in Dataset S4. All data are available upon request.