Significance
Many effectors of the Coxiella burnetii Dot/Icm transporter are important for its virulence, but the lack of understanding of their biochemical activity prevents the use of them as potential therapeutic targets. Here, we found that the effector Cbu0513 (CinF) is a protein phosphatase that attacks IκBα, a key regulatory protein in NF-κB signaling. Unlike its homologs such as ST0318 from the archaeon Sulfolobus tokodaii, CinF has lost the enzymatic activity as a fructose-1,6-bisphosphate aldolase/phosphatase. Instead, it specifically targets IκBα to make it resistant to proteasome-mediated degradation in cells stimulated with NF-κB agonists. Our finding has expanded the strategy used by bacterial pathogens to inhibit host immunity, which may provide leads for the development of immune modulators for disease treatment.
Keywords: NF-κB, protein phosphatase, type IV secretion, effectors
Abstract
Coxiella burnetii is a bacterial pathogen that replicates within host cells by establishing a membrane-bound niche called the Coxiella-containing vacuole. Biogenesis of this compartment requires effectors of its Dot/Icm type IV secretion system. A large cohort of such effectors has been identified, but the function of most of them remain elusive. Here, by a cell-based functional screening, we identified the effector Cbu0513 (designated as CinF) as an inhibitor of NF-κB signaling. CinF is highly similar to a fructose-1,6-bisphosphate (FBP) aldolase/phosphatase present in diverse bacteria. Further study reveals that unlike its ortholog from Sulfolobus tokodaii, CinF does not exhibit FBP phosphatase activity. Instead, it functions as a protein phosphatase that specifically dephosphorylates and stabilizes IκBα. The IκBα phosphatase activity is essential for the role of CinF in C. burnetii virulence. Our results establish that C. burnetii utilizes a protein adapted from sugar metabolism to subvert host immunity.
Immune cells are equipped with various receptors to sense signals that belong to either pathogen-associated molecular patterns or damage-associated molecular patterns. The detection of these signals often activates specific and sometimes interwinding signaling cascades, leading to pathogen clearance by the production of cytokines, antimicrobial peptides, or elimination of the infected cells via various forms of cell death (1).
Coxiella burnetii, the causative agent of Q fever, is a highly adaptive pathogen that has evolved a number of strategies to effectively evade immune detection. This pathogen is highly infectious, and fewer than 10 bacteria have been shown to cause diseases in animal models (2); it also exhibits remarkable resistance to physical stresses such as desiccation, high temperature, osmotic pressure, and ultraviolet light, which facilitates its long-time persistence and long distance spread in the environment. Because of these features, C. burnetii had been listed as a category B biothreat agent (3).
Infections by C. burnetii often lead to an acute self-limiting illness characterized by headache and fever or chronic diseases such as endocarditis and hepatitis (4). A major pathogenic determinant of C. burnetii is lipopolysaccharide (LPS). Virulent wild-type strains (phase I) of C. burnetii elaborate LPS molecules that contain a tetra-acylated lipid A with long fatty acid chains, which plays an important role in immune evasion by mediating resistance to complement (5). This smooth LPS also prevents the activation of dendritic cells (6). The loss of a set of genes involved in the addition of terminal O antigen sugars to the LPS gave rise to phase II strains with marked reduction in virulence on immune competent animals (7, 8). Phase II strains are still comparably competent in intracellular replication in cultured cells or isolated primary macrophages (9) and thus are extremely useful in the study of C. burnetii pathogenesis owning to the requirement of less stringent containment measures.
The second major virulence factor is the Dot/Icm type IV secretion system originally identified in Legionella pneumophila (10), which, like C. burnetii, belongs to the Legionellales order. Similar to its counterpart in L. pneumophila, the Dot/Icm system of C. burnetii is essential for its virulence (11). This system functions to transfer virulence factors called effectors from the bacterial cytoplasm into host cells to construct the Coxiella-containing vacuole (CCV) that is permissive for bacterial replication (12). A large number of effectors (>150) have been identified in C. burnetii by using genetics and bioinformatic tools coupled with various reporters designed to determine intercellular protein transfer (12). In many cases, the loss of a single effector gene leads to pronounced defects in bacterial intracellular replication (13–15), which is in sharp contrast to the lack of discernable phenotypes for most L. pneumophila effector genes (16).
Dot/Icm effectors of C. burnetii have been shown to modulate cellular processes such as cell death (17), vesicle trafficking (18), gene expression (19), and autophagy (20). C. burnetii was long considered an obligate bacterial pathogen before the development of a cell-free culture protocol (21). Like other obligate pathogens, infection by phase II C. burnetii strains does not trigger robust immune responses, likely because of immune suppression by its Dot/Icm effectors (12). In support of this notion, the effector IcaA has been shown to inhibit caspase-11–mediated activation of the NLRP3 inflammasome trigged by cytosolic LPS from gram-negative bacteria such as Escherichia coli and L. pneumophila (22). Signaling events regulated by the nuclear transcription factor NF-κB are important in immunity, cell proliferation, and apoptosis and thus, are common targets of pathogens (23). Earlier studies suggest that C. burnetii induces robust NF-κB activation, but such activation appears to be suppressed at later phases of infection, likely by proteins injected by the Dot/Icm transporter (24). In line with this hypothesis, a recent study shows that the Dot/Icm effector NopA (Cbu1217) interferes with the nuclear import of NF-κB components by directly binding to Ran GTPase (25). Yet, a transposon insertion mutant of NopA did not display detectable defects in intracellular bacterial replication (25), suggesting the existence of additional effectors involved in modulating the NF-κB signaling pathway. In this study, we identified the C. burnetii protein Cbu0513 (designated as a Coxiella inhibitor of NF-κB, CinF) as an inhibitor of NF-κB activation triggered by immune stimuli by functioning as a protein phosphatase that dephosphorylates IκBα, thus making it resistant to proteasome-mediated degradation.
Results
Identification of Cbu0513 as an Inhibitor of NF-κB Activation.
To gain insights into the mechanism of how C. burnetii interferes with host immune response, we performed a screen to identify Dot/Icm proteins capable of inhibiting NF-κB activation. Genes coding for established Dot/Icm effectors or for proteins without known roles in virulence were individually inserted into a vector that allows the expression of 4xFlag-tagged proteins in mammalian cells (26). Each construct of this library was cotransfected into HEK293T (human embryonic kidney) cells with a plasmid-based luciferase reporter that responds to NF-κB activity (27). Transfected cells were treated with phorbol 12-myristate 13-acetate (PMA), which specifically activates group A and group B protein kinase Cs (28), leading to the activation of the transcriptional activator NF-κB, the activity of which was evaluated by measuring luciferase activity.
From 115 candidates, we found that Cbu0513 effectively blocked PMA-induced NF-κB activation (Fig. 1). This gene codes for a protein of 382 residues with a predicted molecular weight of ∼42 kDa (Fig. 1). Sequence analysis by the HHpred algorithm (29) revealed that Cbu0513 is similar to ST0318 from S. tokodaii (30) and TnFBPAP from Thermoproteus neutrophilus (31). In both cases, the identity is ∼39%, and the similarity is about 65% (SI Appendix, Fig. S1). Because of its activity in inhibiting NF-κB activation, we designated this protein as CinF (Coxiella inhibitor of NF-κB activation).
Fig. 1.
Identification of cbu0513 (CinF) as an inhibitor of NF-κB activation. (A) Identification of Coxiella proteins capable of inhibiting NF-κB activation. HEK293T cells transfected with plasmids carrying the indicated Coxiella genes and a luciferase-based NF-κB reporter were induced with PMA for 4 h. Luciferase activity was measured, and the fold of induction was calculated using control samples transfected with the empty vector (Upper). A fraction of the cell lysates separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) were probed for the expression of the testing genes by immunoblotting with a Flag-specific antibody (Lower). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a loading control. Results shown were one of two independent experiments done in triplicate. (B) Translocation of CinF into host cells. C. burnetii strains harboring Flag-TEM1-CinF were used to infect Vero cells for 24 h. The delivery of the fusion into host cells was monitored with the fluorescence substrate CCF2-AM. Images were acquired using an IX-83 fluorescence microscope 2 h after adding the substrate. (Scale bar: 100 μm.) The ratio of blue cells was determined by enumerating cells (n = 400) from three randomly taken images. Data shown were from three independent experiments. The expression of the TEM1-CinF fusion in the testing C. burnetii strains was detected by immunoblotting with the Flag-specific antibody. Results shown were representative from two independent experiments with similar results.
An earlier study has shown that CinF is required for optimal intracellular growth by C. burnetii (13), but little is known about its mechanism of action and cellular targets. Furthermore, CinF has been shown to be translocated into host cells by the Dot/Icm system of C. burnetii (14). Because of its high-level similarity to enzymes involved in basic metabolism, we further confirmed its translocation by the Dot/Icm transporter using the CCF2-AM– and β-lactamase–based translocation assay (32). In samples infected with wild-type bacteria expressing the β-lactamase–CinF fusion, ∼62% of cells emitted blue fluorescence, indicating protein transfer. In contrast, no blue cells were detected in samples infected with an icmD mutant (11) similarly expressing the fusion (Fig. 1B). These results indicate that CinF indeed is a substrate of the Dot/Icm system with potential ability to blunt NF-κB activation.
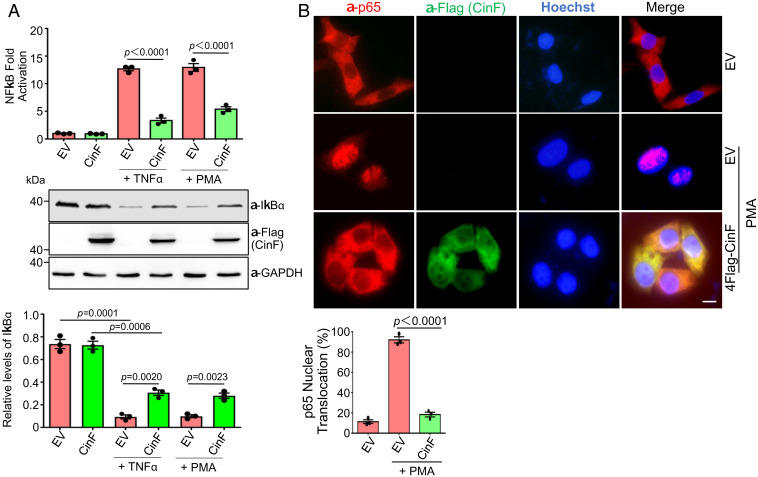
To further analyze the activity of CinF in inhibiting NF-κB activation, we examined its impact on cells stimulated with TNF-α, which activates immune response by engaging a specific receptor (33). Similar to the inhibition of PMA-induced activation, CinF effectively shunted NF-κB activation induced by TNF-α. In each case, the inhibition is accompanied by the accumulation of IκBα in inducer-treated samples that expressed CinF (Fig. 2 A and B). One prominent feature of NF-κB activation is the translocation of components of the transcriptional activator such as p65 (RelA) into the nuclei of induced cells. We thus determined the cellular localization of p65 in cells expressing CinF by immunostaining upon PMA stimulation. As expected, in samples not treated with PMA, p65 resided in the cytoplasm of virtually every cell in the samples, and treatment with this agonist resulted in nuclear localization of p65 in almost all cells (Fig. 2B). Consistent with its ability to inhibit NF-κB activation, in cells expressing CinF, p65 nuclear translocation occurred in less than 20% of the cells upon PMA treatment (Fig. 2B). The better protection by CinF in the p65 nuclear translocation assay than that seen in the IκBα degradation assay likely arose from untransfected cells in the latter.
Fig. 2.
CinF inhibits NF-κB activation induced by multiple stimuli. (A) HEK293T cells were transfected with a mixture of plasmids for CinF, the NF-κB reporter, and the Renila luciferase for 16 h. Samples were treated with TNF-α or PMA for 4 h prior to measuring luciferase activity (Upper). Fractions of cell lysates separated by SDS-PAGE were probed with antibodies specific for IκBα and Flag (to detect CinF). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a loading control (Middle). The levels of IκBα were quantitated by measuring its band intensity in relative to that of GAPDH (Lower). Data shown are one representative done in triplicate, and similar results were obtained in at least three independent experiments. Statistical analysis was performed by the Student’s t test. (B) CinF blocks nuclear translocation of p65 induced by the NF-κB agonist PMA. MLE-12 cells transfected to express Flag-CinF were treated with PMA. Fixed cells were sequentially stained with antibodies specific for Flag (to detect Flag-CinF) and p65 followed by Hoechst to identify the nuclei. Images were acquired using an IX-83 Olympus fluorescence microscope (Upper). (Scale bar: 5 μm.) The rates of nuclear localization were determined by counting at least 300 cells from each sample. Data shown were from three independent experiments (Lower). Statistical analysis was performed by the Student’s t test.
Residues Important for the Putative Activities in Sugar Metabolism in CinF Are Required for Its Inhibition of NF-κB Activation.
Bioinformatic analysis revealed that CinF is highly similar to the fructose-1,6-bisphosphate (FBP) aldolase/phosphatase (FBPAP), a metabolic enzyme found in diverse prokaryotes, particularly certain Achaea species. For example, the E-value is 5.7e−142 when compared to ST0318 (3R1M) from Sulfurisphaera tokodaii strain 7 (30) (SI Appendix, Fig. S1). These unique enzymes use a single active site to catalyze two distinct biochemical reactions: the reversible formation of FBP from dihydroxyacetone phosphate and glyceraldehyde-3-phosphate by aldol condensation and the production of fructose-6-phosphate (F6P) by dephosphorylating FBP (30). Structural modeling revealed that Tyr362 in CinF is corresponding to Tyr347 in ST0318, which is important for both enzymatic activities (30) (SI Appendix, Fig. S1). We thus examined the role of Tyr362 in CinF activity by replacing it with Ala. The CinFY362A mutant has lost its activity in inhibiting NF-κB activation induced by PMA in experiments that measured by luciferase reporter or by IκBα degradation (Fig. 3A), and as expected, ectopic expression of this mutant cannot sequester p65 in the cytoplasm of cells treated with NF-κB agonists such as PMA (Fig. 3 B and C).
Fig. 3.
Inhibition of NF-κB by CinF requires residues important for the putative fructose 1,6-bisphosphate phosphatase activity. (A) Tyr362 is important for the activity of CinF. HEK293T cells were transfected with combinations of plasmids, and the induction of NF-κB was determined after PMA treatment (Upper). The expression of Flag-CinF and its mutant was probed, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control (Middle and Lower). The relative intensity of the bands representing p-IκBα or IκBα was quantitated using the bands of GAPDH as reference. Data shown were from three independent experiments. (B and C) Inhibition of p65 nuclear translocation by CinF requires Tyr362. Samples were prepared and processed as described in Fig. 2B, and the rates of nuclear p65 were scored from samples transfected to express CinF or its mutants. At least 300 cells were counted for each sample. Results shown were from the average of three independent experiments (B). Representative images of samples from one experiment (C). (Scale bar: 5 μm.) Statistical analysis was performed by Student’s t test.
CinF Attacks IκBα by Direct Dephosphorylation.
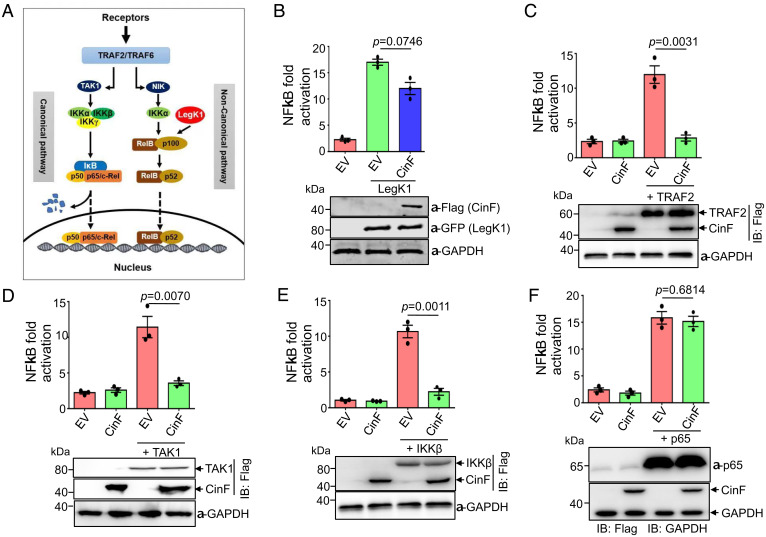
Upon engaging a ligand, NF-κB can be activated via either the canonical or the noncanonical signaling cascade; each requires the participation of multiple protein complexes (23) (Fig. 4A). To address which of these two branches is attacked by CinF, we utilized LegK1, a kinase from L. pneumophila that directly phosphorylates p100 in the noncanonical NF-κB pathway and induces its maturation into p52 (23). As expected, expression of LegK1 robustly induced NF-κB activation. Yet, coexpression of CinF did not lead to significant inhibition of LegK1 activity (Fig. 4B), suggesting that CinF targets the canonical branch of the pathway.
Fig. 4.
Determine the target of CinF in the NF-κB signaling pathway by epistasis analysis. (A) A diagram depicting the canonical and noncanonical branches of the NF-κB signaling pathway. The point of action for LegK1, the L. pneumophila effector that activates NF-κB, was indicated. (B) CinF does not significantly inhibit NF-κB activation induced by LegK1. HEK293T cells transfected with the reporter plasmid together with those that direct the expression of LegK1 and CinF were measured for luciferase activity. The expression of the relevant proteins was detected by immunoblotting with the appropriate antibodies, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was probed as a loading control. Results shown were one representative done in triplicate from three independent experiments with similar results. (C–F) HEK293T cells were transfected with the indicated combinations of plasmids harboring the reporter, CinF, the internal control, and the expression of luciferase driven by NF-κB was measured. Note that CinF effectively inhibits the activation induced by TRAF2 (C), TAK1 (D), and IKKβ (E) but does not affect the activation induced by p65 (F). In each case, results shown were one representative done in triplicate from three independent experiments. Statistical analysis in each panel was performed by Student’s t test. The expression of the relevant proteins was detected by immunoblotting, and GAPDH was probed as a loading control.
One important feature of the canonical branch of the NF-κB pathway is that ectopic expression of some critical components in its signaling knots often leads to its activation in a manner that bypasses the requirement of a ligand (34). We thus attempted to determine CinF’s point of action in this signaling cascade by examining its ability to antagonize NF-κB activity induced by overexpression of several key enzymes in the pathway. Overexpression of the TNF receptor-associated factor 2 (TRAF2), an E3 ubiquitin ligase involved in the formation of K63-type polyubiquitin chains (34), led to NF-κB activation, and such activation can be effectively suppressed by CinF (Fig. 4C). Similarly, activation caused by overexpression of TAK1 and IKKβ, two kinases that are immediately and two steps downstream of TRAF2, respectively (35) (Fig. 4A), can still be inhibited by CinF (Fig. 4 D and E), suggesting that the point of action for CinF is downstream of IKKβ. In contrast, activation of the NF-κB reporter by overexpression of p65, which is one component of the complex directly involved in transcription of target genes (35), cannot be blocked by CinF (Fig. 4F). Together, these results suggest that the target of CinF likely is IκB, the inhibitor of NF-κB that is situated between the IKK kinase and p65.
Members of the IκB family are inhibitors of NF-κB, which function to retain the transcriptional factor in the cytoplasm by forming stable multiprotein complexes (35). The most common mechanism of relieving the inhibitory effects of IκB is by phosphorylation induced by the IKK kinase, which primed it for subsequent ubiquitination and proteasome-mediated degradation (35). Because CinF is similar to sugar phosphatases involved in metabolism (SI Appendix, Fig. S1), we first examined whether it shares similar phosphatase activity toward FBP. To this end, we prepared recombinant proteins of CinF, ST0318, and its catalytically inactive mutant ST0318Y347T (30) and measured their ability to dephosphorylate FBP. As expected, we observed robust FBP phosphatase activity for ST0318 (Km = 0.124 mM) but not for the ST0318Y347T mutant, which is consistent with previous results (30). Intriguingly, CinF did not display detectable phosphatase activity against FBP (Fig. 5A). We also examined the phosphatase activity of these proteins with p-Nitrophenyl phosphate (pNPP), a commonly used nonproteinaceous substrate for alkaline and acid phosphatases. As a control, WipA, an established protein phosphatase from L. pneumophila that perturbs host actin cytoskeleton by dephosphorylating several host proteins (36), exhibited robust activity, and a mutation in its active site abolished such activity (Fig. 5B). Yet, neither ST0318 nor CinF can detectably remove phosphate from pNPP (Fig. 5B). Thus, CinF and ST0318 cannot utilize pNPP as substrate and, despite the high-level similarity to ST0318, CinF does not display phosphatase activity toward FBP. We also examined the ability of ST0318 to inhibit NF-κB activation. Although Flag-ST0318 was expressed at a level similar to that of Flag-CinF, it did not inhibit NF-κB activation induced by PMA (Fig. 5C). Thus, although CinF and ST0318 share high levels of similarity, they are functionally distinctive.
Fig. 5.
CinF is a protein phosphatase that targets IκBα. (A) CinF does not detectably dephosphorylate FBP. FBP dephosphorylation was measured with recombinant CinF, ST0318, or its enzymatically inactive mutant ST0318Y347T. The reaction measures fructose-6-phosphate formation by coupling the reaction with exogenous phosphoglucose isomerase and glucose-6-phosphate dehydrogenase that reduces NADP+ to NADPH. The values of Km and Kcat were obtained by plotting the plot Lineweaver–Burk equation. (B) CinF and ST0318 are unable to hydrolyze the alkaline phosphatase substrate p-nitrophenyl phosphate (pNPP). Testing proteins were individually assayed for their ability to remove phosphate from pNPP, and the amount of free phosphate was determined by the malachite green assay. The established protein phosphatase WipA and its enzymatically inactive mutant were included as controls. (C) ST0318 cannot inhibit NF-κB activation. HEK293T cells transfected with a NF-κB reporter plasmid and constructs that express ST0318 or CinF are shown. Luciferase activity indicative of NF-κB activation was measured after PMA induction. The expression of the relevant proteins was probed by immunoblotting with a Flag-specific antibody (Lower). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a loading control. Data shown were one of three independent experiments done in triplicate with similar results. (D and E) Dephosphorylation of IκBα by CinF. Recombinant CinF was incubated in reactions that contain p-IκBα or p-IKKβ for 60 min. The phosphorylation status of these proteins was detected by immunoblotting with phospho-specific antibodies. The amounts of IκBα, IKKβ, and CinF in the reactions were determined by immunoblotting with the indicated antibodies, respectively (D). Dose-dependent activity of CinF toward p-IκBα. The indicated amounts of CinF or its CinFY362A mutant were incubated with aliquoted p-IκBα and dephosphorylation was evaluated by a phospho-specific antibody are shown. Note that CinFY362A had no impact on p-IκBα (Upper). Reaction time-dependent activity of CinF was measured by incubating 40 ng CinF with equal amounts of p-IκBα, and the dephosphorylation activity was similarly determined. Note that a similar amount of ST0318 did not detectably dephosphorylate p-IκBα even after extended incubation (E, Lower). (F) CinF does not remove phosphate from p-IκBβ. The indicated amounts of CinF were incubated with aliquots of p-IκBβ, and its phosphorylation status was determined by immunoblotting. A fraction of the reactions was probed for IκBβ and CinF, respectively.
We next determined whether CinF can dephosphorylate IκBα. To prepare phosphorylated IκBα, we transfected HEK293T cells to express Flag-IκBα and treated the samples with PMA for 4 h prior to isolating phosphorylated Flag-p-IκBα (p-IκBα) by immunoprecipitation with the Flag antibody. Incubation of recombinant CinF with p-IκBα led to its dephosphorylation in a dose-dependent manner. In contrast, the phosphorylation status of IKKβ did not change upon CinF treatment (Fig. 5D). Further analysis indicated that under our experimental conditions, treatment with 8 ng for 1 h detectably removed phosphate from p-IκBα. Consistent with results from assays in transfected cells, recombinant CinFY362A has lost the protein phosphatase activity toward p-IκBα (Fig. 5 E, Upper). Similarly, recombinant ST0318 did not detectably dephosphorylate p-IκBα, which is consistent with its inability to inhibit NF-κB activation (Fig. 5 C and E, Lower). We further examined the substrate specificity of CinF by testing its activity against phosphorylated IκBβ (p-IκBβ) isolated from PMA-treated cells. Incubation of as much as 200 ng CinF did not detectably remove phosphate from p-IκBβ (Fig. 5F). In our attempts to determine the binding of CinF to IκBα, we did not detect interaction between these two proteins by immunoprecipitation (SI Appendix, Fig. S2). This observation is not surprising because the affinity between virulence factors and their host targets often is low. Nevertheless, these results indicate that CinF is a protein phosphatase that exhibits high-level specificity toward IκBα.
Inhibition of NF-κB Activation by CinF Is Important for C. burnetii Virulence.
In a study aiming at analyzing phenotypes associated with transposon-induced mutants of C. burnetii, an insertional mutant of cinF was shown to display significant defects in intracellular replication (13). Our attempts to use a recently developed method (37) to create a deletion mutant of cinF was not successful despite extensive efforts by two laboratories over a period of more than 2 y. We thus employed alternative strategies to analyze its role in NF-κB signaling during bacterial infection. We first attempted to knockdown (KD) the expression of cinF in C. burnetii by expressing the antisense sequence of its entire coding region by the promoter of cbu1169 from C. burnetii on a multicopy plasmid (the KD strain) (38). This manipulation reduced the level of CinF protein in C. burnetii by ∼90% but did not affect its growth in the defined acidified citrate cysteine medium (ACCM-D) (39) (SI Appendix, Fig. S3). More importantly, this reduction can be complemented to wild-type levels by expressing a version of cinF whose codon had been optimized to that of E. coli (Fig. 6A and SI Appendix, Fig. S4). We next examined intracellular growth of our KD strain. In a 7-d experimental duration, we observed a significant growth defect for the KD strain. Importantly, the defect can be complemented by the wild-type gene but not the CinFY362A mutant (Fig. 6C). These results are consistent with those from a Tn insertion mutant (13), thus validating the effectiveness of our KD method.
Fig. 6.
The protein phosphatase activity of CinF is important for its role in C. burnetii virulence. (A) KD of cinF in C. burnetii and complementation with a codon-changed allele. The level of CinF was probed in bacteria grown in a bacteriological medium. An ∼80-kDa protein nonspecifically recognized by the CinF antibodies was used as a loading control (Upper). The relative abundance of CinF was determined by the intensity ratio between the target band and the nonspecific band (Lower). Results shown were from three independent experiments. (B) KD of cinF affects intracellular replication of C. burnetii. The indicated bacterial strains were used to infect HeLa cells at a multiplicity of infection (MOI) of 100, and the growth of the bacteria was determined by measuring genome equivalents. Note that KD of cinF led to less replication, a defect that can be restored by expressing a wild-type allele of cinF. (C and D) Kinetics of IκBα, p-IκBα, and CinF translocation at the third day (C) and fourth day (D) after bacterial uptake. THP-1 cells infected with the indicated C. burnetii strains were probed for p-IκBα, IκBα, and translocated CinF. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a loading control. The insoluble fractions were used to probe for CinF associated with the bacteria with the metabolic enzyme isocitrate dehydrogenase (ICDH) as a loading control. (E and F) The translocation of p65 in cells infected with relevant C. burnetii strains. THP-1 cells infected with the indicated strains as described in C were stained for p65, and nuclear localization of the protein was scored under a fluorescence microscope (refer to SI Appendix, Fig. S5A for representative images) (E). Results shown were from three independent experiments done in triplicate. Statistical analysis was performed by Student’s t test. Similarly infected cells were subjected to fractionation to obtain the nuclear fractions, which were probed for C. burnetii and p65, respectively (F). GAPDH and histone H3 were detected as controls. Results shown were one representative from three independent experiments with similar results. (G) Knocking out p65 allows the cinF KD strain to replicate intracellularly at rates similar to the wild-type strain. The wild-type and cinF KD strains were used to infect HeLa cells (solid lines) or the p65−/− line (dashed lines) at an MOI of 100. The growth of the bacteria was monitored by measuring genome equivalents. Note that the KD strain grew at rates similar to those of the wild-type strain in the p65−/− cell line. Data shown were one representative from three independent experiments with similar results. (H) Expression of CinF in L. pneumophila led to inhibition of NF-κB activation induced by bacterial infection. The indicated L. pneumophila strains were used to infected HEK293 cells transfected with the luciferase reporter plasmids. The induction was measured 6 h after infection (Upper). The level of IκBα in infected cells was probed by immunoblotting. CinF or CinFY362A translocated into infected cells was also probed, and GAPDH was detected as a loading control (Lower). When applicable, statistical analysis was performed by Student’s t test.
We then used these strains to probe the role of CinF in NF-κB signaling during C. burnetii infection. In cells infected with the wild-type strain, translocated CinF in samples infected with wild-type bacteria was detectable only at the third and fourth days postinfection, which corresponded well with the higher levels of IκBα. In contrast, in cells infected with the KD strain, IκBα decreased significantly at the third and fourth days (Fig. 6 C and D). Expression of the wild-type but not the enzymatically inactive CinFY362A mutant in the KD strain complemented the accumulation of IκBα (Fig. 6 C and D). We also examined the impact of CinF on NF-κB activation at day 3 by scoring p65 nuclear translocation and by detecting its presence in the nuclear fraction of infected cells. Results from these two experiments are highly consistent with the IκBα levels in these samples (Fig. 6 E and F and SI Appendix, Fig. S5A). In contrast, the differences in IκBα levels were barely detectable in the first 2 d of infection among samples infected with these strains, which is in line with the observation that translocated CinF was undetectable at these time points (SI Appendix, Fig. S5 B and C). At the fifth day postinfection, although translocated CinF was only detectable in samples infected with the KD strain expressing active CinF, the levels of IκBα in samples infected with the KD strain or the strain complemented with the CinFY362A mutant were detectably lower than those infected with the wild-type strain or the KD strain expressing active CinF (SI Appendix, Fig. S5D), suggesting that CinF is active at the time of infection. At day 6 postinfection, the effects of CinF in these strains became undetectable (SI Appendix, Fig. S5E). Thus, although less significant, CinF appears functional after the infection has proceeded for 5 d but became undetectable at the sixth day.
These results imply that CinF promotes intracellular replication of C. burnetii by blocking p65 nuclear import, which suggests that the KD mutant may regain the ability to grow in cells lacking p65. We tested this hypothesis by constructing a p65−/− line from HeLa cells with the CRIPSR/Cas9 technology and tested its ability to support relevant C. burnetii strains (Fig. 6 G, Upper). Indeed, the KD strain grew similarly to the wild-type strain in p65−/− cells but not in its parent cell line (Fig. 6 G, Lower).
Immunity by NF-κB signaling is mediated in part by the induction of a large cohort of cytokines. We thus probed the role of CinF in blocking immune response by measuring the expression of the genes coding for IL-1β, IL-6, IL-12, and TNF-α, four cytokines involved in antibacterial activity (40) by quantitating their messenger RNA and secreted cytokine proteins with enzyme-linked immunosorbent assays. Our results indicate that wild-type C. burnetii effectively suppressed the expression of these cytokines at the third and fourth days after bacterial uptake, and the knocking down of cinF led to robust expression of each of these genes. Expression of CinF but not the CinFY362A mutant from a plasmid allowed the KD strain to suppress the expression of these cytokine genes at these two time points (SI Appendix, Fig. S6). Such suppression became diminished at the fifth day and was barely detectable when the infections were extended to 6 d (SI Appendix, Figs. S7 and S8).
Finally, we used L. pneumophila as a heterologous infection system to further probe the impact of CinF on NF-κB signaling. Similar to earlier studies (41), infection by L. pneumophila robustly activates NF-κB in a Dot/Icm-dependent manner, and such activation was dampened by expressing CinF but not CinFY362A (Fig. 6 H, Upper). Consistently, the level of IκBα decreased in cells infected with the wild-type L. pneumophila strain (41) but became accumulated in cells challenged with a strain that expressed CinF but not the CinFY362A mutant, and such accumulation was comparable to samples infected with the dotA mutant (Fig. 6H). Highly comparable amounts of Flag-CinF and Flag-CinFY362A were translocated into the cytosol of infected cells (Fig. 6H), indicating that the enzymatic activity of CinF attributes to the suppression. Taken together, these results establish that CinF contributes to the suppression of NF-κB activation during C. burnetii infection.
Discussion
C. burnetii has evolved effective strategies to escape innate immune recognition, and effectors of its Dot/Icm system play an important role in such evasion. Among these, the effector IcaA inhibits inflammasome activation by a yet undefined mechanism (22). An earlier study indicates that C. burnetii actively modulates the NF-κB pathway potentially with its Dot/Icm effectors (24). This notion was validated by the recent finding that NopA (Cbu1217) interferes with NF-κB signaling by blocking the nuclear import of its components via direct interaction with the Ran GTPase (25). Yet, a nopA mutant does not display detectable defect in intracellular replication, suggesting the participation of other Dot/Icm effectors in this process. By screening a set of C. burnetii genes, we found that CinF (Cbu0513) is a potent inhibitor of NF-κB activation. The strong growth defects associated with strains lacking cinF (Fig. 6B) (13) indicate that it plays a major role in suppressing immunity controlled by this pathway. This notion was further validated by the fact that host cells lacking p65, the key component of the NF-κB pathway, allows the cinF KD strain to grow at rates comparable to those of the wild-type strain. NopA and CinF may function at different phases during C. burnetii infection. Alternatively, NopA may subtly modulate NF-κB in its activity in regulating other cellular processes important for C. burnetii virulence.
One striking feature associated with CinF is its close similarity with glycolytic enzymes from diverse prokaryotes, particularly some Achaea species (SI Appendix, Fig. S1), which suggests that it is evolved from a metabolic enzyme whose gene was likely acquired from a distantly related microorganism by horizontal gene transfer. Despite the high-level similarity, several lines of evidence suggest that CinF has evolved to specifically target host immunity. First, it is injected into host cells by the Dot/Icm system of C. burnetii (14) (Fig. 1). Second, CinF has lost the FBP phosphatase activity. Reciprocally, ST3018, the closest homolog of CinF from S. tokodaii, did not detectably inhibit NF-κB signaling (Figs. 3A and 5C), suggesting that these proteins have diverged to specifically fulfill their respective physiological roles. Third, CinF expressed in L. pneumophila effectively suppressed NF-κB activation induced by the bacterium (Fig. 6H).
The signaling cascade that leads to NF-κB activation is a multistep process, which ultimately causes phosphorylation of members of the IκB family, the inhibitory proteins that prevent NF-κB from entering the nucleus by direct binding (35). Virulence factors that attack critical components of upstream knots of this signaling cascade have been described. For example, UBE2N, the E2 enzyme critical for TAK1 activation by directing the biosynthesis of K63-type polyubiquitin chains, is targeted by OspI of Shigella flexneri and MavC of L. pneumophila, respectively (27, 42). In this regard, CinF may represent the first pathogenic factor that inhibits host immunity by attacking IκBα.
Our results reveal that C. burnetii robustly suppresses NF-κB activation in the third and fourth days of infection and became less apparent after the infection was extended beyond 5 d. Intriguingly, translocated CinF became undetectable even in samples infected with C. burnetii strains in which the protein was ectopically expressed from a multicopy plasmid (SI Appendix, Fig. S5 D and E). The decrease in CinF at late phases of infection may be caused by yet unknown regulatory mechanisms of the bacterium or of host cells or a combination of both. Although the antisense method has allowed us to probe the role of CinF in C. burnetii infection, it is not ideal for experiments that require complete absence of a protein. Clearly, a cinF mutant is required to further examine whether it plays a role during the entire infection duration.
It has been long known that the CCV recruits the autophagy marker LC3 in a Dot/Icm-dependent manner (43), and cinF appears to be important for such recruitment (13). It is well established that reciprocal crosstalk between NF-κB and autophagy occurs extensively in development and disease (44). For example, NFκΒ triggers autophagy by inducing the expression of genes or proteins involved in the autophagosome machine (45). Thus, the role of CinF in LC3 recruitment likely is mediated by its impact on NF-κB activation.
Mutational analysis suggests that residues critical for catalysis appear conserved in CinF and ST3018; the divergence in their functions thus likely is due to differences in substrate recognition. Given the vast difference in the size of FBP and p-IκBα, the substrate-binding pocket in CinF evolved to recognize a protein likely cannot effectively engage a small molecule sugar. Future structural studies on CinF and its complex with IκBα clearly will provide the mechanistic explanation for its substrate specificity.
Materials and Methods
Cell Culture, Transfection, NF-κB Luciferase Reporter Assays.
HEK293T, HeLa, MLE-12, and Vero cells purchased from the American Type Culture Collection were grown in Dulbecco’s modified Eagle medium (HyClone) containing 10% fetal bovine serum (FBS) and 2 mM L-glutamine at 37 °C in a 5% CO2 incubator. THP-1 monocytes were cultured in Roswell Park Memorial Institute 1640 containing 10% FBS in 37 °C with 5% CO2. When needed, cells were differentiated into adherent, macrophage-like cells by treating freshly plated cells with 200 nM PMA (Sigma) for 24 h. Lipofectamine 3000 (Thermo Fisher Scientific) was used for plasmid transfection following the manufacturer’s instructions. All cultured cells are regularly examined for potential mycoplasma contamination by PCR-based tests (Sigma, catalogno. MP0025).
To measure NF-κB activation, HEK293T cells seeded (5 × 104 cells/well) in 24-well plates were transfected with 300 ng plasmid DNA derived from pCMV4Flag (26) that harbor the candidate C. burnetii effector genes together with 100 ng NF-κB reporter plasmid pGL4.32 (Promega, E8491) (27, 46). To normalize transfection efficiency, 100 ng pRL-TK Renilla luciferase reporter plasmid was added to each transfection. A total of 16 h after transfection, cells were treated with 20 ng/mL TNF-α or with 50 nM PMA (all final concentrations) for 4 h, and luciferase activities were measured with a dual-specific luciferase assay kit (Promega) according to the manufacturer’s instructions. Firefly luciferase activities were normalized on the basis of Renilla luciferase activities. All reporter assays were performed at least three times. Data shown were average values ± SD from one representative experiment done in triplicate. For NF-κB epistasis analysis, 300 ng DNA of the corresponding plasmids were cotransfected with 300 ng pCMV-Flag-CinF and 100 ng each of the reporter plasmid and pRL-TK, the Renilla luciferase plasmid.
A p65−/− cell line derived from HeLa cells was constructed as follows: a guide RNA that targets the p65 gene was designed using the E-CRISP online service (www.e-crisp.org), and the corresponding DNA fragment was obtained by annealing the two oligos 5′-CACCGG GCGCTTCCGCTACAAGTGCG-3′ and 5′-CACCGGGCTTCCGCTACAAGTGCGAG-3′ and inserted into 2X_pX458_pSpCas9(BB)-2A-GFP (47). The resulting plasmid was transfected into HeLa cells with Lipofectamine 3000 for 24 h. To obtain cells harboring the plasmid, cells that expressed green fluorescence were sorted by fluorescence-activated cell sorting using a BD Influx cell sorter, and single cells were seeded in a 96-well plate. After the clones have expanded to high confluence, cells were sequentially transferred into 24-well and 6-well plates. Cells collected from 6-well plates were used to screen for p65 knockout by immunoblotting using p65-specific antibodies. A total of three independent clones that undetectably express p65 were saved, and one line was used for intracellular replication of C. burnetii strains. More materials and methods are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Robert Heinzen, Stacey Gilk, and Paul Beare for bacterial strains, plasmids, and helpful discussion. This work in part was supported by a research fund from the First Hospital of Jilin University and NIH Grant R01 AI090142-06 (J.E.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110877119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Akira S., Uematsu S., Takeuchi O., Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Brooke R. J., Kretzschmar M. E., Mutters N. T., Teunis P. F., Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect. Dis. 13, 488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madariaga M. G., Rezai K., Trenholme G. M., Weinstein R. A., Q fever: A biological weapon in your backyard. Lancet Infect. Dis. 3, 709–721 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Eldin C., et al. , From Q fever to Coxiella burnetii infection: A paradigm change. Clin. Microbiol. Rev. 30, 115–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vishwanath S., Hackstadt T., Lipopolysaccharide phase variation determines the complement-mediated serum susceptibility of Coxiella burnetii. Infect. Immun. 56, 40–44 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon J. G., Howe D., Heinzen R. A., Virulent Coxiella burnetii does not activate human dendritic cells: Role of lipopolysaccharide as a shielding molecule. Proc. Natl. Acad. Sci. U.S.A. 102, 8722–8727 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moos A., Hackstadt T., Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect. Immun. 55, 1144–1150 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andoh M., et al. , T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect. Immun. 75, 3245–3255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe D., Shannon J. G., Winfree S., Dorward D. W., Heinzen R. A., Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect. Immun. 78, 3465–3474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel J. P., Andrews H. L., Wong S. K., Isberg R. R., Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Beare P. A., et al. , Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio 2, e00175-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Schaik E. J., Chen C., Mertens K., Weber M. M., Samuel J. E., Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 11, 561–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabill E., Schofield W. B., Newton H. J., Goodman A. L., Roy C. R., Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect. Immun. 86, e00758-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber M. M., et al. , Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J. Bacteriol. 195, 3914–3924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pechstein J., et al. , The Coxiella burnetii T4SS effector AnkF is important for intracellular replication. Front. Cell. Infect. Microbiol. 10, 559915 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu J., Luo Z. Q., Legionella and Coxiella effectors: Strength in diversity and activity. Nat. Rev. Microbiol. 15, 591–605 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Lührmann A., Nogueira C. V., Carey K. L., Roy C. R., Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc. Natl. Acad. Sci. U.S.A. 107, 18997–19001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larson C. L., Beare P. A., Howe D., Heinzen R. A., Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E4770–E4779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber M. M., et al. , Modulation of the host transcriptome by Coxiella burnetii nuclear effector Cbu1314. Microbes Infect. 18, 336–345 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Thomas D. R., Newton P., Lau N., Newton H. J., Interfering with autophagy: The opposing strategies deployed by Legionella pneumophila and Coxiella burnetii effector proteins. Front. Cell. Infect. Microbiol. 10, 599762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omsland A., et al. , Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 106, 4430–4434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunha L. D., et al. , Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat. Commun. 6, 10205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman M. M., McFadden G., Modulation of NF-κB signalling by microbial pathogens. Nat. Rev. Microbiol. 9, 291–306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahapatra S., et al. , Coxiella burnetii employs the Dot/Icm type IV secretion system to modulate host NF-κB/RelA activation. Front. Cell. Infect. Microbiol. 6, 188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burette M., et al. , Modulation of innate immune signaling by a Coxiella burnetii eukaryotic-like effector protein. Proc. Natl. Acad. Sci. U.S.A. 117, 13708–13718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G., Liu H., Luo Z. Q., Qiu J., Modulation of phagosome phosphoinositide dynamics by a Legionella phosphoinositide 3-kinase. EMBO Rep. 22, e51163 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan N., Nakayasu E. S., Hollenbeck P. J., Luo Z. Q., Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N. Nat. Microbiol. 4, 134–143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castagna M., et al. , Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J. Biol. Chem. 257, 7847–7851 (1982). [PubMed] [Google Scholar]
- 29.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fushinobu S., Nishimasu H., Hattori D., Song H. J., Wakagi T., Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase. Nature 478, 538–541 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Du J., Say R. F., Lü W., Fuchs G., Einsle O., Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase. Nature 478, 534–537 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Chen C., et al. , Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 107, 21755–21760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiDonato J. A., Hayakawa M., Rothwarf D. M., Zandi E., Karin M., A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature 388, 548–554 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Rothe M., Sarma V., Dixit V. M., Goeddel D. V., TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 269, 1424–1427 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Li Q., Verma I. M., NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 (2002). [DOI] [PubMed] [Google Scholar]
- 36.He L., et al. , The Legionella pneumophila effector WipA disrupts host F-actin polymerisation by hijacking phosphotyrosine signalling. Cell. Microbiol. 21, e13014 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Beare P. A., Jeffrey B. M., Long C. M., Martens C. M., Heinzen R. A., Genetic mechanisms of Coxiella burnetii lipopolysaccharide phase variation. PLoS Pathog. 14, e1006922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beare P. A., Genetic manipulation of Coxiella burnetii. Adv. Exp. Med. Biol. 984, 249–271 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Sandoz K. M., Beare P. A., Cockrell D. C., Heinzen R. A., Complementation of arginine auxotrophy for genetic transformation of Coxiella burnetii by use of a defined axenic medium. Appl. Environ. Microbiol. 82, 3042–3051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolls J. K., McCray P. B. Jr., Chan Y. R., Cytokine-mediated regulation of antimicrobial proteins. Nat. Rev. Immunol. 8, 829–835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Losick V. P., Isberg R. R., NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 203, 2177–2189 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanada T., et al. , The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Gutierrez M. G., et al. , Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell. Microbiol. 7, 981–993 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Verzella D., et al. , Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 11, 210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nivon M., Richet E., Codogno P., Arrigo A. P., Kretz-Remy C., Autophagy activation by NFkappaB is essential for cell survival after heat shock. Autophagy 5, 766–783 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Li X., et al. , Negative regulation of hepatic inflammation by the soluble resistance-related calcium-binding protein via signal transducer and activator of Transcription 3. Front. Immunol. 8, 709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H. J., et al. , Topological isolation of developmental regulators in mammalian genomes. Nat. Commun. 12, 4897 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.