Significance
Myelodysplastic syndromes (MDS) are the most commonly diagnosed malignancy of the elderly in the United States, but what causes severe anemia in MDS has been unknown. Our findings provide a detailed mechanism underlying the origins of severe anemia and other cardinal phenotypes in MDS patients harboring SF3B1 mutations, which are found in about a quarter of all MDS patients. In addition, we define a role of MAP3K7 and a MAP3K7-p38 MAPK axis in human terminal erythroid differentiation and created a SF3B1 cell model that recapitulates many of the terminal erythroid differentiation events of mutant SF3B1 MDS patients, which should prove valuable for developing novel drugs and therapies.
Keywords: spliceosome, p38 MAPK, GATA1, erythropoiesis, cancer
Abstract
SF3B1 is the most frequently mutated RNA splicing factor in cancer, including in ∼25% of myelodysplastic syndromes (MDS) patients. SF3B1-mutated MDS, which is strongly associated with ringed sideroblast morphology, is characterized by ineffective erythropoiesis, leading to severe, often fatal anemia. However, functional evidence linking SF3B1 mutations to the anemia described in MDS patients harboring this genetic aberration is weak, and the underlying mechanism is completely unknown. Using isogenic SF3B1 WT and mutant cell lines, normal human CD34 cells, and MDS patient cells, we define a previously unrecognized role of the kinase MAP3K7, encoded by a known mutant SF3B1-targeted transcript, in controlling proper terminal erythroid differentiation, and show how MAP3K7 missplicing leads to the anemia characteristic of SF3B1-mutated MDS, although not to ringed sideroblast formation. We found that p38 MAPK is deactivated in SF3B1 mutant isogenic and patient cells and that MAP3K7 is an upstream positive effector of p38 MAPK. We demonstrate that disruption of this MAP3K7-p38 MAPK pathway leads to premature down-regulation of GATA1, a master regulator of erythroid differentiation, and that this is sufficient to trigger accelerated differentiation, erythroid hyperplasia, and ultimately apoptosis. Our findings thus define the mechanism leading to the severe anemia found in MDS patients harboring SF3B1 mutations.
Myelodysplastic syndromes (MDS) are a heterogeneous group of blood malignancies that originate from hematopoietic stem cells (HSCs). MDS is characterized by ineffective hematopoiesis, dysplasia, peripheral blood cytopenias, and an increased risk of transformation to acute myeloid leukemia (1). Although MDS is the most commonly diagnosed malignancy of the elderly in the United States (2), few effective therapies are available. In MDS, SF3B1 is the most frequently mutated gene (3), with a frequency of 20 to 29% in all MDS cases and 65 to 83% in the subtype refractory anemia with ring sideroblasts (RARS) (4–6). SF3B1 mutations have also been found in other cancers (5). These mutations cause missplicing of several hundreds of introns due to usage of aberrant 3′ splice sites (3′ss), resulting from abnormal recognition and selection of the branch site (7, 8).
In an effort to elucidate how SF3B1 mutations cause MDS, knockin mice carrying the K700E hotspot mutation were generated (9, 10). These mice displayed anemia but not the anemic features described in mutant SF3B1 MDS (erythroblast hyperplasia/bone marrow [BM] hypercellularity) (5, 11–13) or RARS patients [BM erythroid hypercellularity (14, 15) and apoptotic erythroblasts (14, 16, 17)]. In addition, the mice did not display other hallmarks of mutant SF3B1 MDS, such as ringed sideroblasts (4, 5) and an HSC competitive repopulating advantage, which is indicative of clonal hematopoiesis (18). Additionally, there was minimal overlap of misspliced transcripts between MDS patients and the mutant mice (∼5%) (9, 10), most likely due to the poor conservation of intronic sequences (∼30%) between human and mouse (19) and to significant species-specific differences in alternative splicing (20), suggesting that mice may be less useful in modeling mutant SF3B1 MDS. Among the genes whose expression is affected by mutant SF3B1-induced missplicing, none has been implicated as a cause of severe anemia.
One of the known mutant SF3B1 target transcripts encodes MAP3K7, a serine/threonine kinase and an upstream regulator of the mitogen-activated protein kinase (MAPK) signal transduction pathway (21). MAP3K7-deleted mice displayed massive apoptosis of hepatocytes, HSCs, and committed hematopoietic progenitor cells, and significant reductions in nucleated cells in BM, spleen, and thymus (22). Red blood cells and hemoglobin levels were slightly reduced compared to control mice but were not statistically significant and could simply be secondary effects of massive apoptotic HSCs and committed progenitor cells (22). MAP3K7 genetic aberrations have been found in pediatric T lymphoblastic leukemia (23) and in prostate cancer (24), although their significance is unclear.
In this report, we describe a previously unrecognized role of MAP3K7 in human erythropoiesis, which entails controlling proper terminal erythroid differentiation, and also show that the reduced levels of the kinase that arise from missplicing are at least in part responsible for the severe anemia, but not ringed sideroblast formation, observed in MDS patients harboring SF3B1 mutations. We show that p38 MAPK (p38), a known regulator of various biological processes, such as cell differentiation and apoptosis (25), including erythropoiesis (26), is deactivated in SF3B1 mutant isogenic and patient cells. We demonstrate that MAP3K7 is an upstream positive effector of p38 MAPK and that disruption of this MAP3K7-p38 signal transduction axis leads to premature down-regulation of GATA1, a key regulator of erythroid differentiation, and that this triggers accelerated differentiation, accumulation, and apoptosis of erythroblasts. Thus, with the exception of ring sideroblast formation, our work has provided important insights into SF3B1-mutated MDS, including a detailed mechanism that offers an explanation for the severe anemia that often characterizes the disease.
Results
K562/SF3B1 K700E Cells Exhibit Accelerated Differentiation and Increased Erythroid Cell Death Compared to WT Cells.
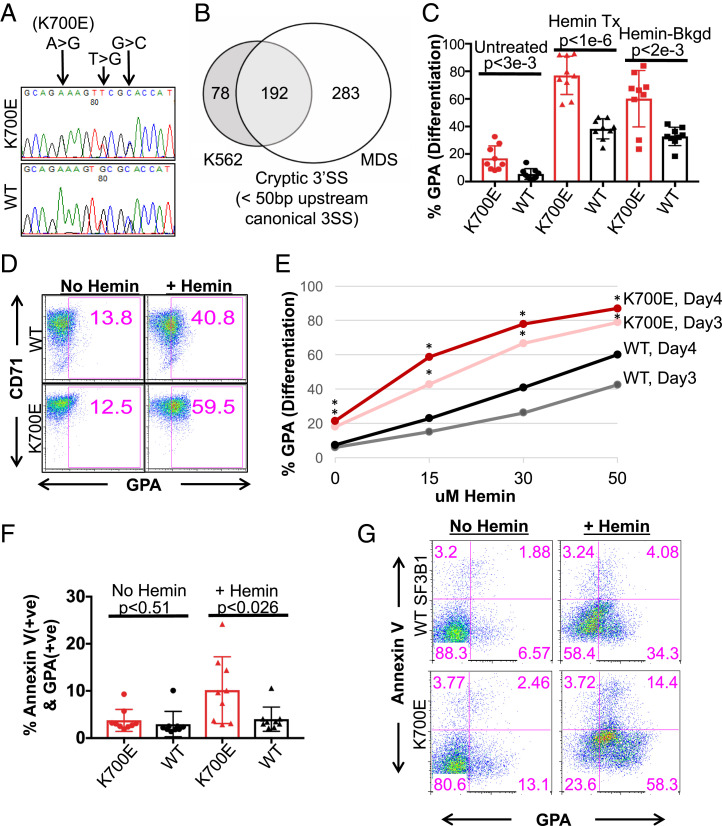
To investigate how SF3B1 mutations might lead to anemia or other features of MDS, we used CRISPR/Cas9 technology to introduce the most common hotspot mutation found in MDS patients, K700E, into human erythroleukemia K562 cells, which were selected for their ability to differentiate along the erythroid lineage upon treatment with various inducers, including hemin (27) (Fig. 1A). To avoid possible clonal or off-target effects, we isolated nine independent K700E mutant and nine independent WT clones for this study. SF3B1 mutant K700E mRNA (SI Appendix, Fig. S1A) and protein (SI Appendix, Fig. S1B) were stably expressed. Total SF3B1 protein levels were equivalent between mutant and WT cells, and all the mutant cells expressed similar levels of SF3B1 K700E, as determined by western blot with a K700E-specific antibody (SI Appendix, Fig. S1B). RNA-sequencing (RNA-seq) analysis of mutant cells revealed usage of cryptic 3′ss that were ∼10 to 25 nt upstream of the canonical 3′ss (SI Appendix, Fig. S1 C and D). This is consistent with our previous analysis of mutant SF3B1 MDS patient cells (7) and with previous observations in other cancers (8). In contrast to the mouse models mentioned in the Introduction, 71% of the unique cryptic 3′ss in the mutant SF3B1 K562 cells overlapped with those in mutant SF3B1 MDS patient cells (Fig. 1B and SI Appendix, Tables S1 and S2). In addition, 9 of the top 10 enriched pathways, derived from the cryptic 3′ss transcript list of the mutant K562 cells, overlapped with the top 10 enriched pathways from a similar list of MDS patient cells (SI Appendix, Fig. S1E). These results suggest that the mutant K562 CRISPR cells will likely model some of the phenotypes of mutant SF3B1 MDS disease.
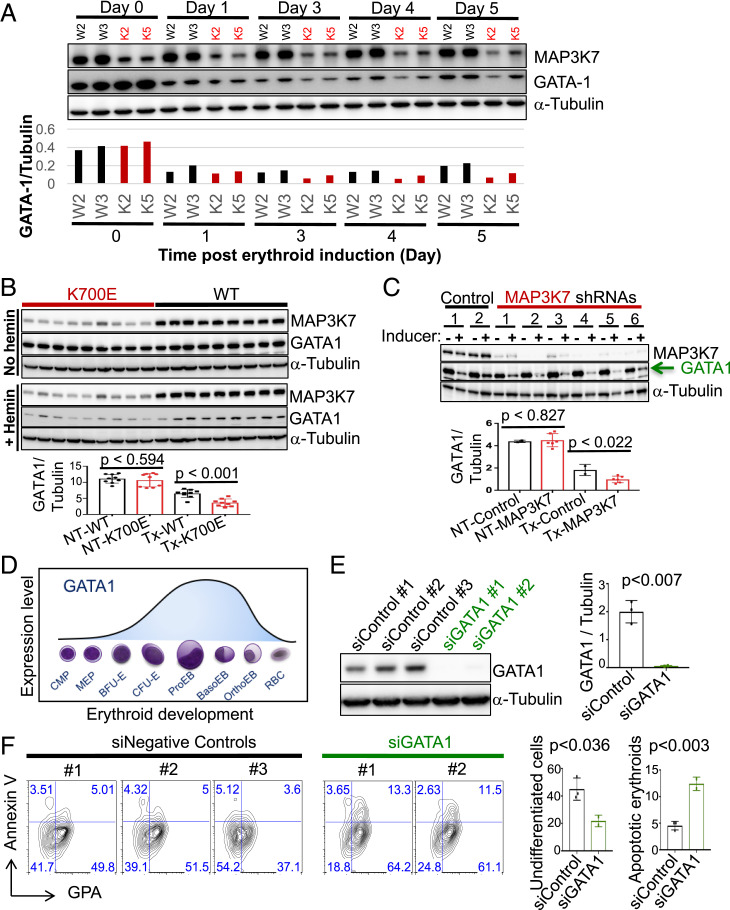
Fig. 1.
K562/SF3B1 K700E cells exhibit accelerated differentiation and increased erythroid cell death as compared to WT cells. (A) DNA chromatograms of representative K562/SF3B1 mutant and WT clones, showing K700E and two silent mutations in the SF3B1 gene. (B) Venn diagram showing the overlap of cryptic 3′ss (cutoff at q-value < 0.05, distance < 50 nt to the associated canonical 3′ss) that were utilized significantly more in SF3B1 mutants than in SF3B1 WT K562 cells and MDS patients. (C) Bar graph quantifying the percentage of surface GPA+ cells as analyzed by flow cytometry (FACS) with cells that were treated or not with the erythroid inducer hemin (50 μM) for 3 d. Data shown represent n = 6 independent experiments. Each dot represents an independently derived cell clone. P values obtained from t tests are shown. (D) FACS plots of representative mutant and WT clones from C with similar background, displaying percent CD71 (transferrin receptor) and GPA+ cells following hemin treatment, as indicated. GPA positivity was gated based on unstained WT and mutant cells. (E) Line graph displaying percent GPA positive cells vs. various concentrations (μM) of hemin for mutant and WT clones as measured by FACS after 3 and 4 d of hemin or no treatment. *P < 0.05. (F) Bar graph specifying the percentages of AnnexinV vs. GPA expressed on the surface of K700E and WT cells as measured by FACS after hemin (50 μM) or no treatment for 4 d. Representative data from n = 4 independent experiments. P values from t tests are shown. (G) FACS plots (lower left quadrants of each plot: GPA-AnnexinV− as undifferentiated cells; Lower Right quadrants: GPA+AnnexinV− as nonapoptotic erythroids; Upper Right quadrants: GPA+AnnexinV+ as apoptotic erythroids; Upper Left quadrants: GPA−AnnexinV+ as apoptotic cells) of representative mutant and WT clones from G showing the percentages of AnnexinV vs. GPA expression under 4 d of hemin or no treatment.
We next investigated whether the K700E mutant isogenic cells display phenotypes characteristic of MDS. We initially observed that while the mutant cells displayed a small growth defect compared to WT cells, there was no statistically significant difference in apoptotic cell death under normal growth conditions between K700E and WT cells (SI Appendix, Fig. S1 F and G). We next examined the ability of the K562 CRISPR cells to differentiate along the erythroid lineage by treating the cells with the erythroid differentiation inducer hemin (27). Unexpectedly, the mutant cells differentiated to erythroid cells to a greater extent than did the WT cells, as quantified by FACS analysis measuring glycophorin A (GPA), a well-characterized surface marker of erythroid differentiation (Fig. 1 C and D). While it has been reported that normal K562 cells undergo low levels of spontaneous erythroid differentiation (25), the mutant clones had on average higher GPA levels (Fig. 1C), indicative of an increased frequency of spontaneous differentiation. The higher percentage of overall hemin-induced erythroid differentiation, however, was not due to the increased spontaneous differentiation, as some mutant clones had similar GPA+ background as WT but had higher differentiation capacity (Fig. 1D). In fact, mutant cells differentiated more rapidly and completely to erythroid cells than did WT clones (Fig. 1E and SI Appendix, Fig. S1G). However, following differentiation, more K700E cells underwent apoptosis as measured by the apoptotic surface marker AnnexinV, as the percentage of apoptotic erythroid GPA+ AnnexinV+ cells was significantly increased compared to the WT cells (10.2 ± 6.7 in K700E vs. 4.0 ± 2.4 in WT) (Fig. 1 F and G). Our results thus suggest plausible explanations for two key features of RARS MDS cells, aberrant erythroid differentiation and increased erythroid cell death.
p38 MAPK Is Specifically Deregulated and Only in Mutant SF3B1 Cells.
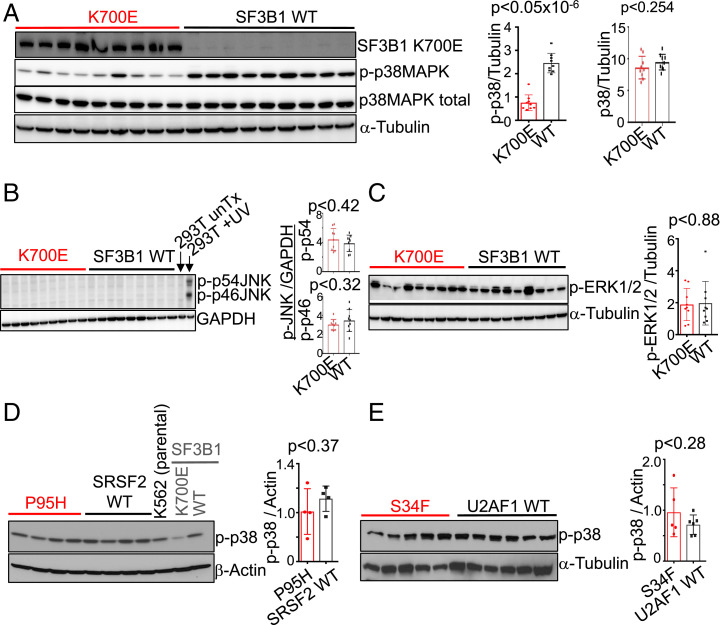
While investigating pathways that may be of pathophysiologic significance to MDS, we observed that the p38 pathway was deactivated in mutant cells. The p38 pathway is known to be involved in governing hematopoietic cell differentiation and apoptosis (25), including a pathway central to erythrocyte differentiation (26). Specifically, western blot analysis revealed that phospho-p38 (p-p38) levels were significantly reduced in K700E cells compared to WT cells, although total p38 protein levels were similar (Fig. 2A). Notably, of the three major MAP kinase pathways, only p38 was deactivated in K700E cells: we observed no differences in levels of activated JNK or ERK between mutant and WT cells (Fig. 2 B and C).
Fig. 2.
p38 MAPK is specifically deregulated and only in mutant SF3B1 cells. (A) Representative western blot images showing expression of total p38 MAPK, phospho-p38 MAPK (p-p38MAPK; p-p38), SF3B1 K700E and α-Tubulin in the independent mutant and WT K562/SF3B1 clones. n = 5 independent experiments. Representative western blot analysis of (B) phospho-JNK (p-p46 and p-p54) and GAPDH, and (C) phospho-ERK1/2 (p-ERK12) and α-Tubulin in K562/SF3B1 K700E and WT clones. Representative western blot analysis of p-p38 in (D) K562/SRSF2 mutant (P95H) and WT and in (E) K562/U2AF1 mutant (S34F) and WT independent clones. For B and C, n ≥ 3, and for D and E, n = 2 independent experiments. Bar graphs are shown next to all western blot images, displaying results of ImageJ-quantified, loading control-normalized protein band intensities and P values from t tests.
Mutations in genes encoding RNA splicing factors occur in a mutually exclusive manner in MDS (6), suggesting that the mutations could act on the same pathway. We therefore examined p38 activation in cells with the next two most frequently mutated splicing factor genes, SRSF2 and U2AF1. As with the SF3B1 cells, we used CRISPR to introduce appropriate hotspot mutations (28) (SI Appendix, Fig. S2). Interestingly, there were no differences in p-p38 levels between mutant SRSF2 P95H or U2AF1 S34F cells and their WT counterparts (Fig. 2 D and E), suggesting that the p38 pathway defect is specific to SF3B1 mutant cells.
MAP3K7 Transcripts Are Misspliced in Mutant SF3B1 Cells and This Is Responsible for p38 Deactivation.
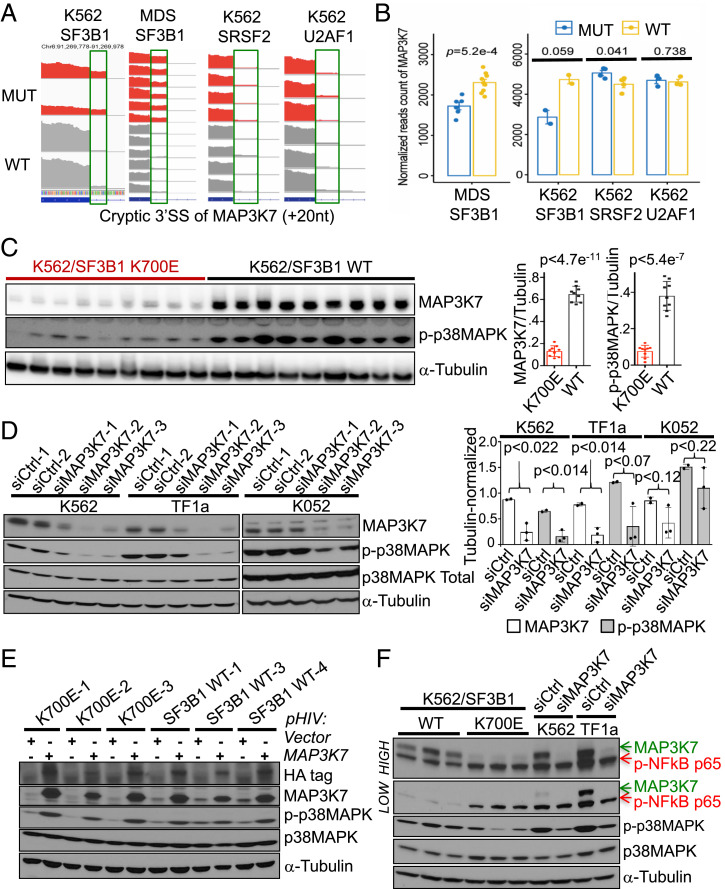
We hypothesized that a mutant SF3B1-induced splicing error was involved in p38 deactivation in the SF3B1 mutant cells. We therefore examined misspliced transcripts encoding kinases or phosphatases that could directly or indirectly regulate p38. MAP3K7 is a known upstream regulator of p38 (21), and its transcript has been shown previously to be misspliced in MDS mutant cells (29). Analysis of RNA-seq data from our SF3B1 mutant K562 cells and from MDS patient cells revealed extensive cryptic 3′ss usage of MAP3K7, which was not observed in the SRSF2 and U2AF1 mutant K562 cells (Fig. 3A and SI Appendix, Fig. S3A). MAP3K7 is also misspliced in other cancers with SF3B1 mutations (8) (SI Appendix, Fig. S3B), indicating that MAP3K7 is a common target of mutant SF3B1 regardless of tissue type. Reported as a target of nonsense-mediated RNA decay (29), MAP3K7 transcript levels were decreased in mutant SF3B1 K562 and in MDS cells (Fig. 3B), but as expected, not in the mutant SRSF2 and U2AF1 K562 cells (Fig. 3B). MAP3K7 protein levels were sharply reduced in all SF3B1 mutant K562 clones (Fig. 3C).
Fig. 3.
MAP3K7 transcripts are misspliced in mutant SF3B1 cells, and this is responsible for p38 deactivation. (A) RNA-seq coverage plots of MAP3K7 cryptic 3′ss transcripts in WT and mutated samples from (left to right) K562/SF3B1, MDS patients, K562/SRSF2, and K562/U2AF1 cells. (B) Comparison of total MAP3K7 mRNA levels (normalized reads counts) in WT and mutated RNA-seq samples from (left to right) MDS patients, K562/SF3B1, K562/SRSF2, and K562/U2AF1 cells. P values from t tests are shown. (C) Representative western blot images showing expression of MAP3K7, p-p38 MAPK, and α-Tubulin proteins in the independent mutant and WT K562/SF3B1 clones. n ≥ 6 independent experiments. (Right) Bar graphs displaying the results of ImageJ-quantified, α-Tubulin-normalized band intensities and P values from t tests. (D) Representative western blot analysis of the effects on p-p38 MAPK and total p38 MAPK expression in MAP3K7 KD K562, TF1a, and K052 cells using two different negative control siRNAs (siCtrl-1/2) and three different siRNAs specific for MAP3K7. n ≥ 3 independent experiments. (Right) Bar graphs displaying the results of ImageJ-quantified, α-Tubulin–normalized band intensities and P values from t tests. (E) Representative western blot analysis of the effects on p-p38 MAPK and total p38 MAPK expression in three WT and three mutant K562 clones expressing HA-tagged MAP3K7. n ≥ 3 independent experiments. (F) Representative western blot analysis of the effects on phospho-NF-κB p65 (p-NF-κB p65), p-p38 MAPK and total p38 MAPK expression in MAP3K7 KD K562 and TF1a cells using negative control siRNA #1 and siMAP3K7 #2 (see D). Expression of p-NF-κB p65 in three mutant and three WT K562/SF3B1 is also shown. “Low” and “High” represent different exposures of the same gel. n = 3 independent experiments.
We next set out to determine whether the reduced MAP3K7 levels were in fact responsible for the lower p38 activation. To this end, we knocked down (KD) MAP3K7 using three different small-interfering RNAs (siRNAs) in parental K562 cells and in two other leukemic cell lines, TF1a and K052, and measured p-p38 levels (Fig. 3D). We observed significant reductions in p-p38, but not p38 levels. Importantly, when we reintroduced MAP3K7 into SF3B1 mutant cells, p-p38 abundance was restored (Fig. 3E). Thus, MAP3K7 is a positive regulator of p38 in these cells.
In addition to p38, previous studies showed that MAP3K7 can also regulate NF-κB under certain circumstances (21). However, KD of MAP3K7 by siRNA (Fig. 3F) or six different short-hairpin RNAs (shRNAs) (SI Appendix, Fig. S3C) did not alter levels of phospho–NF-κBp65 in parental K562 or TF1a cells, suggesting that under unstimulated, normal growth conditions, MAP3K7 does not regulate NF-κBp65 activation in these cells. Interestingly, under the same normal growth conditions, we observed that SF3B1 mutant K562 cells displayed modestly hyperactivated NF-κB (Fig. 3F), indicating another altered pathway of possible pathological significance to MDS and that reduced MAP3K7 levels were not responsible for the NF-κB activation under these conditions, suggesting that another missplicing event likely underlies this response.
KD of MAP3K7 in Parental K562 and Normal Human CD34+ Cells Causes Increased Erythroid Differentiation and Cell Death.
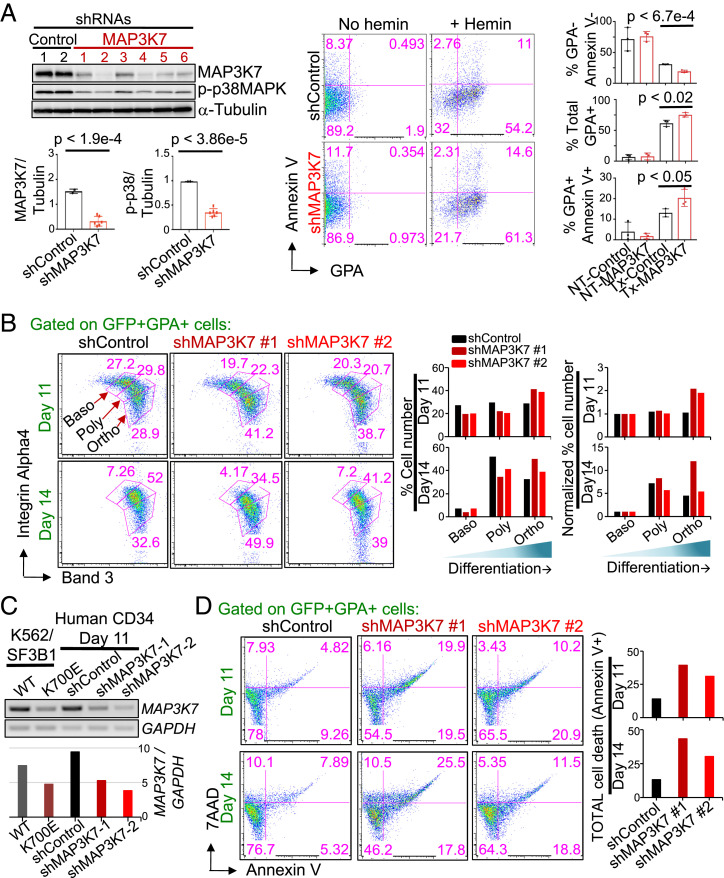
We next determined whether reduced levels of MAP3K7 are sufficient to induce the aberrant erythrocyte differentiation and apoptosis observed in the SF3B1 mutant K562 cells. KD of MAP3K7 in parental K562 cells had no effects on cell proliferation or, similar to the mutant cells, apoptosis under normal growth conditions (SI Appendix, Fig. S4). However, KD of MAP3K7 during hemin-induced erythroid differentiation in these cells led to an increase in differentiation, as shown by an elevated percentage of total erythroid (GPA+) cells and a subsequent reduction in undifferentiated GPA−AnnexinV− double-negative cells (Fig. 4A). In addition, MAP3K7 KD also induced erythroid cell death (Fig. 4A), as similarly observed in the SF3B1 mutant isogenic cells.
Fig. 4.
KD of MAP3K7 in parental K562 and normal human CD34+ cells causes increased erythroid differentiation and cell death. (A) Images (from Left to Right) of representative western blot analysis of GFP-sorted, shRNA-mediated MAP3K7 KD (two different negative control and six different MAP3K7 shRNAs) in parental K562 cells, representative two-color FACS plot of AnnexinV vs. GPA from a control and shMAP3K7, and bar graphs showing the averages and SDs of (from Top to Bottom) % GPA−Annexin− (undifferentiated cells), % total GPA+ (nonapoptotic erythroids), and % GPA+AnnexinV+ cells (apoptotic erythroids) after 3 d of treatment with 50 μM hemin from three independent experiments. Bar graphs displaying the results of ImageJ-quantified, α-Tubulin–normalized MAP3K7 and p-p38 band intensities in Western blot analysis are shown below the Western blot. P values from t tests are labeled on all bar graphs. (B) Representative FACS plots showing surface expression of Integrin α-4 and band 3 from day 11 and day 14 shRNA-mediated MAP3K7 KD (one negative control and two different MAP3K7 shRNAs) in human erythropoietin (EPO)-induced CD34+ cells that were CD45− and double-positive GFP+GPA+ (= erythroblasts cells). Erythroblast stages are depicted and labeled (Baso, basophilic normoblast; Poly, polychromatophilic normoblast; Ortho, othrochromatic normoblast). (Right) Bar graphs, quantifying the percentage of erythroblasts in each stage, are shown, as well as Baso-normalized percent erythroid cells for comparison. n = 2 independent experiments. (C) RT-PCR showing day 11 shRNA-mediated MAP3K7-KD expression in day 11 EPO-induced CD34+ cells. Abundance of GAPDH-normalized MAP3K7 expression is depicted in the bar graph below. (D) Representative FACS analysis of cell death via 7AAD and AnnexinV from (C) day 11 and day 14 MAP3K7 KD, human EPO-induced, CD45−GFP+GPA+ erythroblasts. Bar graphs, specifying the percentage of total erythroblast cell death, are shown. n = 2 independent experiments.
The above experiments utilized primarily our K562 CRISPR cell lines. While these cells are ideal in many ways for evaluating the consequences of SF3B1 mutation on erythroid cell growth and differentiation, they are a transformed leukemia cell line. We therefore next sought to confirm our findings concerning the role of MAP3K7 with normal primary human blood cells, and to this end evaluated the effects of MAP3K7 KD on erythroid differentiation in CD34+ blood stem/progenitor cells. We induced these cells to differentiate along the erythroid lineage with erythropoietin, and then employed a refined, three-erythroid surface marker (GPA, Integrin α-4, band 3) method that allows for determination of the four stages of erythroblast maturation: proerythroblast, basophilic erythroblast, polychromatic erythroblast, and orthochromatic erythroblast (30). Elucidating these four stages was done by FACS analysis by first gating on the GPA marker that is present on cells during all four erythroblast stages and then examining for surface levels of Integrin α-4 and band 3; expression of Integrin α-4 decreases with development while surface expression of band 3 increases with maturation (30). Importantly, MAP3K7 KD again resulted in accelerated differentiation: Fewer KD CD34+ cells were present at early-staged erythroblasts and more at late-stages compared to the negative control shRNA (Fig. 4 B and C). Notably, we also observed more erythroid cell death in the two MAP3K7 KD cells than in the negative control cells (Fig. 4D).
Premature Down-Regulation of GATA1 Underlies the Accelerated Differentiation and Erythroid Cell Death.
We next investigated the mechanism by which reduced levels of MAP3K7 caused accelerated erythroid differentiation followed by apoptosis. We examined the expression levels of major erythroid transcription factors by western blot (Fig. 5A) or qPCR (SI Appendix, Fig. S5) in mutant and WT cells during erythroid differentiation. Based on the expression patterns of these transcription factors and their connection, or lack thereof, to p38, we hypothesized that the transcription factor GATA1—known to be a master regulator of erythroid differentiation and a possible downstream target of p38 (31)—is misregulated in mutant SF3B1 cells. We therefore examined GATA1 expression during a time course of hemin-induced erythroid differentiation by western blot analysis, using two representative K700E and WT K562 clones (Fig. 5A). The two WT clones showed that GATA1 expression decreased during differentiation (Fig. 5A), which is consistent with previous studies showing that during erythroblast differentiation, GATA1 expression is required to be down-regulated for erythroid maturation (32). Importantly, GATA1 expression in the two K700E clones was also down-regulated, but to a greater extent and more rapidly than in the two WT clones (Fig. 5A). We then examined our full panel of nine K700E and nine WT clones after 3 d of differentiation, and observed that GATA1 was indeed more down-regulated in K700E than in WT cells (Fig. 5B). To demonstrate that the reduced MAP3K7 levels in the mutant K562 cells were responsible for the greater down-regulation of GATA1 during differentiation, we examined GATA1 levels in MAP3K7 KD parental K562 cells, and again observed a greater decrease in GATA1 expression than in the negative control cells (Fig. 5C).
Fig. 5.
Premature down-regulation of GATA1 in differentiated K700E or MAP3K7 KD cells underlies the accelerated differentiation and erythroid cell death. (A) Western blot analysis (Upper) and quantifying bar graph (Lower) showing expression of GATA-1 during a 5-d time course of treatment with 50 μM hemin to induce erythroid differentiation in two representative K700E (K2 and K5) and two representative WT (W2 and W3) K562/SF3B1 clones. (B) Representative western blot images showing GATA-1 protein expression in the nine mutant and nine WT K562/SF3B1 clones that were treated or not with 50 μM hemin for 3 d. Bar graph displaying the results of ImageJ-quantified, α-Tubulin–normalized GATA-1 band intensity and P values from t tests. n = 4 independent experiments. (C) Representative western blot image of GATA-1 expression in shRNA-mediated MAP3K7 KD parental K562 cells that were treated or not with 50 μM hemin for 3 d. n = 3 independent experiments. (D) Illustration (adapted from ref. 32) showing GATA1 expression during the course of erythroid development from myeloid progenitor cell (CMP) to mature red blood cell (RBC). BasoEB, basophilic erythroblast/normoblast; BFU-E, Burst-forming unit-erythroid; CFU-(E) colony-forming unit-erythroid; MEP, megakaryocyte-erythroid progenitor; OrthoEB, orthochromatic erythroblast/normoblast; ProEB, Proerythroblast. (E) Western blot image showing GATA-1 KD (two different GATA-1 and three different negative control siRNAs) expression in parental K562 cells and (F) its effects on erythroid differentiation and erythroid apoptosis via FACS analysis of AnnexinV vs. GPA after 3 d of 50 μM hemin treatment. Bar graphs (Right) indicate percent undifferentiated cells (double-negative GPA−AnnexinV−) and percent apoptotic erythroid cells (double-positive GPA+AnnexinV+). P values from t tests are shown. n = 3 independent experiments. GPA positivity was gated based on unstained parental K562 cells.
The requirement for GATA1 expression during erythroid differentiation is biphasic. There is a steadily increasing demand for GATA1 by erythroid-committed progenitor cells that peaks prior to the onset of the erythroblast stages, then during the erythroblast stages, GATA1 expression steadily decreases to enable erythroblasts to properly mature (Fig. 5D, adapted from ref. 32). We hypothesized that GATA1 expression in mutant cells decreases faster and more extensively than in WT cells during the erythroblast stages, thus causing the accelerated differentiation and subsequent apoptosis. To test this hypothesis, we pretreated parental K562 cells with hemin to initiate early erythroid differentiation, and then knocked down GATA1 in the hemin pretreated cells. Strikingly, GATA1 KD after hemin pretreatment resulted in increased erythroid differentiation and subsequent apoptotic cell death compared to the negative control cells (Fig. 5 E and F), analogous to what we observed in SF3B1 mutant cells or following MAP3K7 KD.
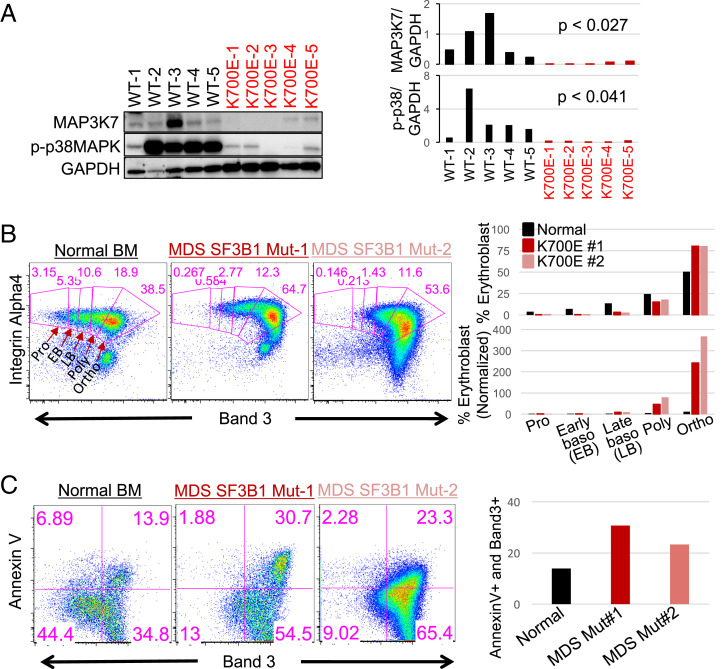
Mutant SF3B1 MDS Patient Cells Display Phenotypes Observed in K562/SF3B1 Mutant Cells and MAP3K7 KD Cells.
An important question is whether the results we have obtained so far are indeed relevant to SF3B1 MDS. To address this important issue, we next investigated whether the phenotypes observed in mutant SF3B1 K562 cells occur in mutant SF3B1 MDS patient cells. We first examined expression of p-p38 and MAP3K7 in SF3B1 MDS BM mononuclear cells (BMMNCs) by western blot analysis (Fig. 6A). Consistent with the SF3B1 mutant K562 cells, p-p38 and MAP3K7 levels were significantly reduced in mutant SF3B1 MDS BMMNCs compared to WT SF3B1 MDS BMMNCs (Fig. 6A). To ascertain the erythroid differentiation and cell death properties of the mutant SF3B1 MDS cells, we examined the erythroblast stages of mutant SF3B1 MDS and normal BM cells using the three-erythroid surface-marker method described above (30). Similar to what we observed with MAP3K7 KD CD34+ cells, mutant SF3B1 MDS patient BM had less early-staged erythroblasts and more late-staged erythroid cells compared to the normal healthy BM control cells (Fig. 6B), indicative of erythroblast expansion and enhanced erythropoiesis. This is consistent with data shown in SI Appendix, Fig. S6A, which was generated by replotting the normal controls and MDS SF3B1 K700E patient data reported previously (33). SI Appendix, Fig. S6A also revealed fewer early-staged erythroblasts and more late-staged erythroid cells in the SF3B1 mutant samples. Additionally, when we measured erythroid cell death by AnnexinV staining, the mutant SF3B1 MDS GPA+ cells showed more total cell death than did those from normal BM (Fig. 6C). Together, our data provide an explanation for the anemic phenotype of mutant SF3B1 MDS patients, involving aberrantly enhanced erythroid differentiation followed by apoptotic cell death, and demonstrate a critical role for the MAP3K7-p38-GATA1 pathway in this process.
Fig. 6.
Mutant SF3B1 MDS patient cells display phenotypes observed in K562/SF3B1 mutant cells and MAP3K7 KD cells. (A) Western blot analysis showing MAP3K7 and p-p38 MAPK expression in MDS BM mononuclear cells from five patients with WT and five patients with K700E SF3B1 mutation. (Right) Bar graphs quantifying the results of western blots and P values from t tests are shown. (B) Representative FACS plots showing the erythroblast profiles of primary BM cells from SF3B1 K700E MDS patients and normal healthy individual. The isolated CD45− BM cells were stained with three erythroid markers: GPA, Integrin α-4, and band 3. FACS plot of Integrin α-4 vs. band 3 on GPA+ BM cells. Erythroblast stages are depicted and labeled. Bar graphs, quantifying the percentage of nucleated erythroblasts in each stage as a total of 100%, are shown as well as proerythroblast (Pro)-normalized percent erythroid cells for comparison. Three SF3B1 K700E MDS patients and three normal healthy individuals were profiled. EB, early basophilic erythroblasts; LB, late basophilic erythroblasts; Ortho, orthochromatic erythroblasts; Poly, polychromatic erythroblasts; Pro, proethroblasts. (C) Representative FACS analysis of erythroblast cell death via AnnexinV vs. band 3 from (B) Bar graph quantifies the percentage of late-stage erythroblast cell death (AnnexinV+Band3+).
Other SF3B1 Mutations Lead to Extensive MAP3K7 Missplicing and Enhanced Erythroid Development.
Our study focused on the most common SF3B1 hotspot mutation, K700E. To determine whether other MDS SF3B1 mutants behave similarly to K700E, we analyzed our own and other RNA-seq datasets. We found that other MDS SF3B1 hotspot (E622, R625, H662, and K666) and nonhotspot (D781G) mutations induced extensive MAP3K7 missplicing, ranging from 41 to 90% (10, 34) (SI Appendix, Fig. S6B). Since we demonstrated that reduced levels of MAP3K7 alone, in both K562 and normal CD34+ cells, was sufficient to cause enhanced erythroid development, expansion, and cell death, any SF3B1 mutation that induces extensive MAP3K7 missplicing will almost certainly behave identically to K700E. In support of our hypothesis, we again reanalyzed previously published data (33) and replotted normal controls and MDS samples with non-K700E SF3B1 mutations (three E622, four R625, four H662, two K666, and one D781G) (SI Appendix, Fig. S6C). The resultant plot showed fewer early-staged but more late-staged erythroblasts in the non-K700E SF3B1 mutant cells, consistent with our data (Fig. 6B and SI Appendix, Fig. S6A).
The totality of our data has shown that there was more erythroid development in mutant than in WT SF3B1 cells. To address the basis for this, we examined whether there was a difference in the cell proliferation program between K700E and WT SF3B1 K562 erythorid cells using the proliferation marker Ki67. SI Appendix, Fig. S6D shows that there were indeed more proliferating erythroid cells (GPA+Ki67+) in mutant than in WT cells. These data provide an explanation for the increased number of erythroblasts we detected in mutant SF3B1 MDS patients.
Discussion
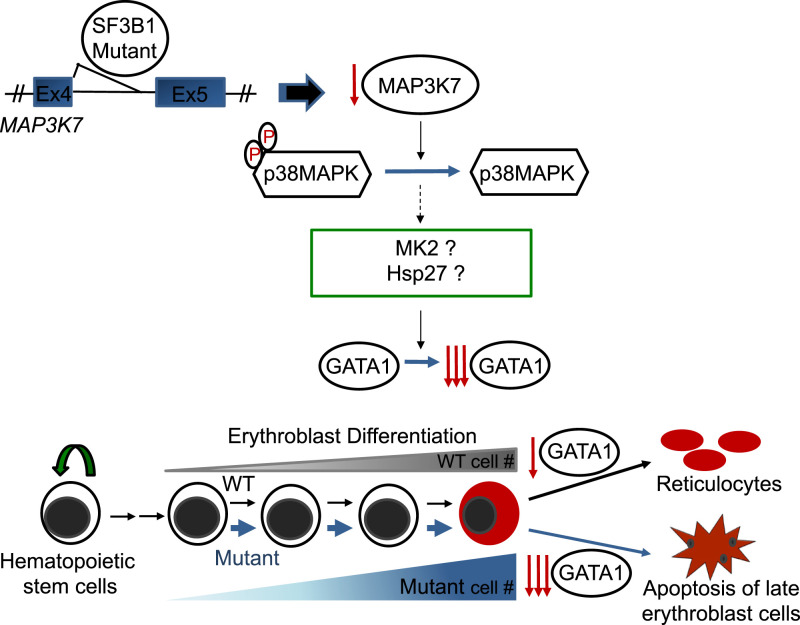
Our findings lead to the following model detailing a mechanism for the origins of severe anemia in SF3B1 mutant MDS patients (Fig. 7): SF3B1 mutant-induced missplicing of MAP3K7 transcripts, and the resultant reduced levels of MAP3K7, is responsible for p38 MAPK deactivation, which may in turn lead to deactivation of p38 downstream targets MAPKAPK2 and HSP27 (SI Appendix, Fig. S7), and ultimately to faster and greater GATA1 down-regulation. This causes accelerated differentiation, expansion, and subsequently apoptotic erythroid cell death, as seen in mutant SF3B1 MDS patient cells. While our findings have not addressed the mechanism of ring sideroblast formation, a key feature of SF3B1 MDS, we have established a direct role for MAP3K7 in regulating proper terminal erythroid differentiation and thereby preventing anemia.
Fig. 7.
A model for how SF3B1 mutations cause anemia. SF3B1 K700E, and other hotspot mutations induce an error in splicing of MAP3K7 transcripts, resulting in reduced levels of MAP3K7. This leads to a reduction of inactivated, phosphorylated p38MAPK (p-p38), which in turn affects downstream potential targets, such as MAPKAPK2 (MK2), HSP27, and HSP70. Ultimately, this causes faster and greater down-regulation of GATA1, resulting in accelerated differentiation and erythroid cell death, thereby explaining the anemia that characterizes MDS patients harboring SF3B1 mutations.
Our collective data provide strong support for the hypothesis that decreased MAP3K7 abundance in SF3B1 mutant cells results in accelerated erythroid development, expansion, and subsequent cell death. This is consistent with previous reports documenting erythroid expansion in RARS patient BMs (14, 15), in which, as mentioned earlier, SF3B1 is mutated in 65 to 83% of this MDS subtype. In fact, it was reported that MDS patients harboring SF3B1 mutations have increased erythroid activity and accumulation of erythroblasts in their BMs (5, 11–13), and that the SF3B1 mutant allele burden positively correlates with the percentage of BM erythroblasts (12). Our findings explain how this hypercellular phenotype arises in the BM of mutant SF3B1 MDS patients through expansion of erythroblasts. During erythroid development from progenitor cells to late-staged erythroblasts, a successive expansion of these nucleated cells occurs as they mature (35). Thus, late-staged erythroblasts are the most numerous nucleated erythroid cells in normal BM, as also shown in our differentiated CD34+ cells and normal BM. However, MAP3K7-KD CD34+ cells and mutant SF3B1 MDS cells display an even higher abundance of late-staged erythroblasts, indicating greater erythroid development and activity. Overall, our findings are in agreement with and provide a mechanistic explanation for reports showing increased erythroid activity (13) and increased erythroblasts in the BM of mutant SF3B1 MDS (5, 11–13), as well as BM erythroid hypercellularity (14, 15) and apoptotic erythroblasts (14, 17) in RARS MDS patients.
In this study, we demonstrated several features of SF3B1-mutated MDS and RARS MDS. We did not, however, detect ringed sideroblasts, one of the hallmarks of SF3B1-mutated MDS (4, 5), in our mutant K562 cells in the presence or absence of hemin-induced differentiation. We note that there is no evidence to support a role for MAP3K7 in ringed sideroblast formation, and we do not believe that MAP3K7 functions in the iron deposition process. In contrast, there is considerable evidence to support a role for ABCB7, a protein involved in the transport of heme from the mitochondria to the cytosol, in ringed sideroblast formation in MDS (34, 36, 37). Supporting this, patients with X-linked sideroblastic anemia and ataxia (XLSA-A) develop ringed sideroblasts, due to germline ABCB7 mutations (38, 39). Furthermore, missplicing of ABCB7 and down-regulation of its RNA levels have been reported in SF3B1-mutated MDS cells at the CD34+ HSC/progenitor cell stage (34, 36). Interestingly, it has been reported that aberrant mitochondrial ferritin was observed in RARS MDS cells that were still CD34+ (37, 40), showing that the events that cause iron accumulation occur early in development, at the CD34+ HSC/progenitor cell stage. Consistent with the absence of ringed sideroblasts in our CD34− progenitor (41) K562 SF3B1 mutant cells, we did not observe significant missplicing of ABCB7 in these cells.
Besides the mechanism for anemia that we have elucidated, iron deposition as seen in XLSA-A patients could also cause anemia in SF3B1-mutated MDS. Although XLSA-A patients have ringed sideroblasts and anemia very early in life, the anemia is typically mild and usually asymptomatic (38, 39, 42), suggesting that iron deposition contributes to the anemic phenotype but is unlikely to cause severe anemia, unless elderly, late-staged erythroblasts in SF3B1-mutated MDS are more sensitive to iron deposition. Thus, while SF3B1 mutations cause iron deposition and ringed sideroblasts in erythroblasts, likely due to ABCB7 missplicing as described above, we believe that this contributes to but is not the main cause of severe anemia in SF3B1-mutated MDS. In support of this hypothesis, the BM of ABCB7-deficient mice is actually hypo-, not hyper-, cellular (43). In addition, ABCB7 KD in normal human CD34+ cells results in a decrease in GPA+ cells (erythroblasts) at days 11 and 14 of erythroid differentiation (37). In contrast, when we knocked down MAP3K7 in normal human CD34+ cells, we observed several phenotypes reported in SF3B1-mutated MDS and RARS MDS: that is, enhanced erythropoiesis, more erythroblast at later stages, and erythroblast apoptosis, that we believe lead to severe anemia. Apoptotic erythroblasts (14, 16, 17), as well as ringed sideroblasts, are features of RARS and believed to cause anemia, but our data support the former as the main cause of the severe anemia that characterizes MDS with SF3B1 mutations.
The expression profile of GATA1 during the development of progenitor cells to reticulocytes indicates that GATA1 expression must decrease for erythroblasts to mature. Thus, our observation that aberrantly faster and greater down-regulation of GATA1 occurs in mutant SF3B1 cells provides an explanation for the accelerated differentiation phenotype in the mutant SF3B1 K562 cells and, more importantly, MDS patient cells. Consistent with this notion, it had been demonstrated that both GATA1 mRNA and protein levels were reduced in purified day 7- and day 14-cultured GPA+ erythroid RARS cells as compared to those in normal BM cells (40). Furthermore, decreased GATA1 expression in erythroblasts has been linked to ineffective hematopoiesis (44).
In normal erythroid development, terminal differentiation is accompanied by cessation of cell proliferation (45). However, we found that the cell proliferation program was more active in mutant SF3B1 K562 cells than in WT cells, explaining the erythroblast expansion. Consistent with this, levels of soluble transferrin receptor, which is related to cellular iron uptake and erythroid proliferation rate (46), have been reported to be elevated in SF3B1 mutant MDS patients (12, 13). Thus, the conflicting signals of proliferation and terminal differentiation, where cell-cycle arrest must occur, could trigger apoptosis (47). Alternatively, the basis for the uncoupling of terminal differentiation from cell-cycle arrest could be a result of the faster and greater down-regulation of GATA1. During erythroid differentiation, GATA-1 has been shown to directly activate expression of prosurvival Bcl-xL (48) and p21 (49), a negative regulator of the cell cycle. The faster and greater down-regulation of GATA-1 thus might not activate p21 and Bcl-xL sufficiently to turn off cell proliferation and confer erythroblast survival, respectively, providing other explanations for the apoptotic erythroid phenotypes we observed in mutant SF3B1 MDS cells.
As mentioned in the Introduction, the knockin K700E mouse models did not recapitulate many phenotypes seen in mutant SF3B1 MDS or RARS MDS patients. This could be attributed to the minimal overlap between aberrantly spliced transcripts in MDS patients and mice (∼5%) (9, 10), suggesting the principal features of mutant SF3B1 MDS/RARS MDS are caused by mutant SF3B1-induced missplicing. Interestingly, one study (29) found more overlap (21%) in missplicing between SF3B1 mutant MDS patients and K700E knockin mice. Notably, one of the overlapped transcripts was MAP3K7 that was not found to be misspliced in the previous reports (9, 10). However, it was only misspliced in the mutant mice by ∼5 to 10% (29), while we observed 51 to 72% missplicing in both our SF3B1 K700E K562 cells and in MDS patient cells. While protein levels were not measured in that study (29), the low abundance of misspliced MAP3K7 mRNA in the mutant mice was most likely insufficient to affect protein levels significantly enough to account for the phenotypes arising from MAP3K7 missplicing that we have characterized here.
As mentioned before, deletion of MAP3K7 in mice led to massive apoptosis of HSCs and committed hematopoietic progenitor cells (22). However, KD of MAP3K7 in transformed K562 cells and primary human CD34 cells did not trigger immediate apoptosis but instead led to accelerated erythroid differentiation and expansion of erythroblasts followed by apoptosis. The discrepancy could reflect differences in hematopoietic cell regulation between mouse and human, since MAP3K7 is 99% homologous between the two species. Alternatively, the disparity could reflect the remaining expression levels of MAP3K7; in the mouse study, MAP3K7 was deleted, but in our systems and in SF3B1 mutant MDS cells, MAP3K7 is down-regulated, leaving some remaining MAP3K7 expression in these cells. In any case, we have found a role of MAP3K7 in human erythropoiesis, specifically in the proper regulation of terminal erythroid differentiation, thereby preventing severe anemia in humans.
In conclusion, our findings have provided a plausible mechanistic explanation for the severe anemia that characterizes mutant SF3B1 MDS, which affects approximately a quarter of the MDS patient population. With few exceptions, such as ring sideroblast formation, our results have also provided insight into several other key features of mutant SF3B1 MDS, and led to a model of terminal erythroid differentiation. Our findings provide new avenues for drug screening as well as targets for developing novel therapeutic approaches to treat MDS.
Materials and Methods
Experimental Model and Subject Details.
K562, TF1a (ATCC), and K052 (DSMZ) cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco, cat # 12440-053) supplemented with 10% fetal bovine serum (FBS; Seradigm, cat # 89510-186) in a 37 °C, 5% CO2 incubator. Adult human peripheral CD34+ cells were purified from donated whole blood (New York Blood Center Blood Bank). MDS BM aspirates were obtained from patients, who were seen at New York Presbyterian Hospital/Columbia University Irving Medical Center and provided informed consent for the study that was approved by the Institutional Review Board of Columbia University and was in accordance with the Declaration of Helsinki. Fresh, normal, healthy human BM mononuclear cells were purchased from PPA Research Group (Cat # 15-00073). Fresh, healthy human BM aspirates were purchased from Lonza (Cat # 1M-105).
Generation of Knockin Cell Lines Using CRISPR/Cas9 Genome Editing.
For generation of knock-in cell lines using CRISPR/Cas9 genome editing, see detailed methods in SI Appendix.
Erythroid Differentiation of K562 Cells.
Logarithmically growing K562 (30,000-50,000/mL) cells were treated with a final concentration of 30, 40, or 50 μM hemin (Sigma, cat # 51280-1G) in IMDM media supplemented with 10% FBS for 3 to 4 d, as previously described (27). The cells were cultured in a 37 °C, 5% CO2 incubator.
siRNA Transfection.
For small-interfering RNA transfection, see detailed methods in SI Appendix.
MAP3K7 Expression Constructs.
For MAP3K7 expression constructs, see detailed methods in SI Appendix.
Viral Production.
For viral production, see detailed methods in SI Appendix.
Purification, Erythroid Induction Culture, and Transduction of Human CD34+ Cells.
For purification, erythroid induction culture, and transduction of human CD34+ cells, see detailed methods in SI Appendix.
Erythroblast Staging Analysis.
For erythroblast staging analysis, see detailed methods in SI Appendix.
FACS of cultured and CD34+ cells.
For FACS of cultured and CD34+ cells, see detailed methods in SI Appendix.
Flow Cytometric Analysis.
For flow cytometric analysis, see detailed methods in SI Appendix.
Western Blot Analysis.
For Western blot analysis, see detailed methods in SI Appendix.
RT-PCR.
For RT-PCR, see detailed methods in SI Appendix.
Identification of Novel Cryptic 3′SS Usage.
In order to annotate novel 3′ss that are not present in current datasets, we adopted the splice junction read output by STAR alignment (50). We adopted the pipeline from DeBoever (8) for novel splice junction identification and usage, as well as identification of associated canonical 3′ss for cryptic 3′ss. To this end, RNASeq.fastq files were aligned using a splice junction database from DeBoever (8). Counts of junction reads from SJ.out.tab file (STAR output) were merged into a unique matrix, with each row indicates one splice junction and column for each sample. We further filtered out junctions with a read coverage of <20 summed across all samples. DEXeq was then applied to detect significantly differential misspliced 3′ss using the code from DeBoever et al. (8) (https://github.com/cdeboever3/deboever-sf3b1-2015). A bar plot of log2 distance in base pair was used to demonstrate cryptic 3′ splicing pattern on significantly differentially used novel 3′ss.
RNA-Seq Coverage Plots of Cryptic 3′ss Visualization.
We used Integrative Genomics Viewer (IGV) (51) to visualize usage of cryptic 3′ss in SF3B1 mutant and WT cells. We downloaded from Gene Expression Omnibus database the MDS RNA-seq (accession no.: GSE128805) (7) data. We also accessed RNA-seq data of K562 CRISPR cells with SRSF2 mutation (GSE128805) (28).
RNA-Seq Expression Analysis.
To generate mRNA expression matrix for transcriptome analysis, we used featureCounts (52) from package ‘Subread’ to call read counts from STAR realigned bam files. Genes with low read depths across the cohort are removed. Then, read counts were transformed into RPKM values, followed by log2 transformation, and quantile normalized on the sample level.
Statistical Analysis.
Data are expressed as mean ± SD. Comparisons were analyzed by using Student two-tailed paired with equal variance t tests. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank Matthew B. Thomsen and Dr. Govind Bhagat for histological advice/analysis; Dr. Jiquang Wang for his advice on computational analysis; Cao Chen, Muhammad Mumtaz, Jacob Hess, and Lena Huang for technical assistance and figure preparation help; Dr. Donald Rio of University of California, Berkeley for critical reading of the manuscript; and Bethyl Laboratories for preparing and providing the anti-K700E SF3B1 specific antibodies. Most of the cell cytometric sorting and analysis were done at the HICCC and C2B2 flow cytometry cores (NIH Grants P30CA013696 and S10OD020056). This work was supported by NIH Grant R35 GM118136 (to J.L.M.) and an Edward P. Evans Foundation grant (to J.L.M. and S.M.). R.R. is supported by NIH Grants R01 CA185486, R01 CA179044, U54 CA193313, and U54 209997 and NSF/SU2C/V-Foundation Ideas Lab Multidisciplinary Team (PHY-1545805). Z.L. is supported by NIH Grant P01CA087497; X.A. is supported by Grant HL140625; and A.R. is supported by a Cures Within Reach grant. Y.K.L., J.Z., J.L.M., and S.M. are supported in part by a grant from Celgene Pharmaceutical Company (currently Bristol Myers Squibb).
Footnotes
Reviewers: S.O., Kyoto Daigaku; and A.V., Albert Einstein College of Medicine.
Competing interest statement: Y.K.L., J.Z., J.L.M., and S.M. are supported in part by a grant from Celgene Pharmaceutical Company (currently Bristol Myers Squibb); R.R. is a member of the AimedBio SAB and a founder of Genotwin. None of the work is directly related to the current manuscript.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111703119/-/DCSupplemental.
Data Availability and Coding Availability.
The K562/SF3B1 mutant and WT RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE187356). All other study data are included in the main text and SI Appendix.
Change History
December 24, 2021: The byline has been updated.
References
- 1.Sperling A. S., Gibson C. J., Ebert B. L., The genetics of myelodysplastic syndrome: From clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer 17, 5–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cogle C. R., Craig B. M., Rollison D. E., List A. F., Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: High number of uncaptured cases by cancer registries. Blood 117, 7121–7125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa S., Genetics of MDS. Blood 133, 1049–1059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malcovati L., et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium and of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative, Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 118, 6239–6246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E., et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium, Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida K., et al. , Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., et al. , Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol. Cell 76, 82–95.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBoever C., et al. , Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLOS Comput. Biol. 11, e1004105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mupo A., et al. , Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia 31, 720–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obeng E. A., et al. , Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patnaik M. M., et al. , Prognostic irrelevance of ring sideroblast percentage in World Health Organization-defined myelodysplastic syndromes without excess blasts. Blood 119, 5674–5677 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ambaglio I., et al. , Inappropriately low hepcidin levels in patients with myelodysplastic syndrome carrying a somatic mutation of SF3B1. Haematologica 98, 420–423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y., et al. , SF3B1-mutated myelodysplastic syndrome with ring sideroblasts harbors more severe iron overload and corresponding over-erythropoiesis. Leuk. Res. 44, 8–16 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Hellström-Lindberg E., et al. , Apoptosis in refractory anaemia with ringed sideroblasts is initiated at the stem cell level and associated with increased activation of caspases. Br. J. Haematol. 112, 714–726 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Malcovati L., Cazzola M., Refractory anemia with ring sideroblasts. Best Pract. Res. Clin. Haematol. 26, 377–385 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Tehranchi R., et al. , Antiapoptotic role of growth factors in the myelodysplastic syndromes: Concordance between in vitro and in vivo observations. Clin. Cancer Res. 11, 6291–6299 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Fontenay-Roupie M., et al. , Ineffective erythropoiesis in myelodysplastic syndromes: Correlation with Fas expression but not with lack of erythropoietin receptor signal transduction. Br. J. Haematol. 106, 464–473 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Jaiswal S., et al. , Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy S. W., Gilbert W., The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Barbosa-Morais N. L., et al. , The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Aashaq S., Batool A., Andrabi K. I., TAK1 mediates convergence of cellular signals for death and survival. Apoptosis 24, 3–20 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Tang M., et al. , TAK1 is required for the survival of hematopoietic cells and hepatocytes in mice. J. Exp. Med. 205, 1611–1619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordas Dos Santos D. M., et al. , MAP3K7 is recurrently deleted in pediatric T-lymphoblastic leukemia and affects cell proliferation independently of NF-κB. BMC Cancer 18, 663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., et al. , Deletion of a small consensus region at 6q15, including the MAP3K7 gene, is significantly associated with high-grade prostate cancers. Clin. Cancer Res. 13, 5028–5033 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Oeztuerk-Winder F., Ventura J. J., The many faces of p38 mitogen-activated protein kinase in progenitor/stem cell differentiation. Biochem. J. 445, 1–10 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Uddin S., Ah-Kang J., Ulaszek J., Mahmud D., Wickrema A., Differentiation stage-specific activation of p38 mitogen-activated protein kinase isoforms in primary human erythroid cells. Proc. Natl. Acad. Sci. U.S.A. 101, 147–152 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutherford T. R., Clegg J. B., Weatherall D. J., K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature 280, 164–165 (1979). [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., et al. , Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. U.S.A. 112, E4726–E4734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S. C., et al. , Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34, 225–241.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J., et al. , Isolation and functional characterization of human erythroblasts at distinct stages: Implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Thonel A., et al. , HSP27 controls GATA-1 protein level during erythroid cell differentiation. Blood 116, 85–96 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Moriguchi T., Yamamoto M., A regulatory network governing Gata1 and Gata2 gene transcription orchestrates erythroid lineage differentiation. Int. J. Hematol. 100, 417–424 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Ali A. M., et al. , Severely impaired terminal erythroid differentiation as an independent prognostic marker in myelodysplastic syndromes. Blood Adv. 2, 1393–1402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolatshad H., et al. , Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 29, 1092–1103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chasis J. A., Mohandas N., Erythroblastic islands: Niches for erythropoiesis. Blood 112, 470–478 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolatshad H., et al. , Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 30, 2322–2331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikpour M., et al. , The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 27, 889–896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allikmets R., et al. , Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8, 743–749 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Bekri S., et al. , Human ABC7 transporter: Gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood 96, 3256–3264 (2000). [PubMed] [Google Scholar]
- 40.Tehranchi R., et al. , Aberrant mitochondrial iron distribution and maturation arrest characterize early erythroid precursors in low-risk myelodysplastic syndromes. Blood 106, 247–253 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Inoue T., Swain A., Nakanishi Y., Sugiyama D., Multicolor analysis of cell surface marker of human leukemia cell lines using flow cytometry. Anticancer Res. 34, 4539–4550 (2014). [PubMed] [Google Scholar]
- 42.D’Hooghe M., et al. , X-linked sideroblastic anemia and ataxia: A new family with identification of a fourth ABCB7 gene mutation. Eur. J. Paediatr. Neurol. 16, 730–735 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Pondarre C., et al. , Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood 109, 3567–3569 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling T., Crispino J. D., Zingariello M., Martelli F., Migliaccio A. R., GATA1 insufficiencies in primary myelofibrosis and other hematopoietic disorders: Consequences for therapy. Expert Rev. Hematol. 11, 169–184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruijtenberg S., van den Heuvel S., Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mast A. E., Blinder M. A., Gronowski A. M., Chumley C., Scott M. G., Clinical utility of the soluble transferrin receptor and comparison with serum ferritin in several populations. Clin. Chem. 44, 45–51 (1998). [PubMed] [Google Scholar]
- 47.Pucci B., Kasten M., Giordano A., Cell cycle and apoptosis. Neoplasia 2, 291–299 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Testa U., Apoptotic mechanisms in the control of erythropoiesis. Leukemia 18, 1176–1199 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Papetti M., Wontakal S. N., Stopka T., Skoultchi A. I., GATA-1 directly regulates p21 gene expression during erythroid differentiation. Cell Cycle 9, 1972–1980 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson J. T., et al. , Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The K562/SF3B1 mutant and WT RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE187356). All other study data are included in the main text and SI Appendix.