Abstract
Background and Aim:
The periodontal microbiome being complex, this study was aimed to detect and quantify the prevalence of Filifactor alocis in various stages of periodontitis and to evaluate its prospect as a diagnostic marker for periodontal disease.
Settings and Design:
Sixty subjects were selected (20 healthy controls, 20 with chronic periodontitis, and 20 with aggressive periodontitis) for the study.
Materials and Methods:
Clinical parameters probing depth and the level of clinical attachment was recorded, subgingival plaque samples were collected. The F. alocis 16srDNA was cloned, sequenced, and used as the standard for real-time quantification of bacterial load using SYBR green chemistry.
Statistical Analysis:
Clinical, microbiological, and quantitative polymerase chain reaction (PCR) data were analyzed using ANOVA and Pearson's coefficient correlation.
Results:
(a) Real-time PCR analysis showed the highest average F. alocis count in chronic periodontitis subjects (32,409.85), which was followed by count in healthy controls (3046.15) and the least count in aggressive periodontitis subjects (939.84). The bacterial count was statistically significant at P = 0.005. (b) An intra-group comparison reveals that there was a statistically significant increase in the bacterial count with age and mean probing pocket depth at P = 0.0005.
Conclusion:
F. alocis population in aggressive periodontitis was lower compared to chronic periodontitis and healthy controls. The F. alocis population surge in healthy controls may be due to geographical variations and the ethnicity of the subjects. A higher population of F. alocis in chronic periodontitis proves its high pathogenic potential to invade the host tissues to aid in further periodontal destruction.
Keywords: Chronic periodontitis, Filifactor alocis, microbial load, real-time quantitative polymerase chain reaction
Introduction
Periodontitis is an intricate interplay of bacteria-induced infection and host counteraction, modified by systemic factors that prompt the obliteration of the encompassing periodontal structures.[1] The three keystone pathogens Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola initiate the disease.[2,3,4,5] P. gingivalis is the most extensively studied organism, but P. gingivalis alone does not suffice to cause infection.[6] Pathobiont is a critical factor in infection and disease establishment; it exist in the normal oral microbiota and act opportunistically during inflammation.[7] Pathobionts ameliorate the microenvironment for other periodontal pathogens and aid in host immunity manipulation.[7] F. alocis, a periodontal pathogen identified through novel culture-independent techniques such as polymerase chain reaction (PCR), Next-Gen Sequencing, DNA hybridization is emerging as a critical pathobiont due to its increased presence in periodontally diseased patients.[7] F. alocis is obligately anaerobic, fastidious, Gram-positive asaccharolytic bacilli that are slow-growing and difficult to culture.[8]
F. alocis is found along with P. gingivalis qualifying as a microbial diagnostic marker.[8]
Technological advances such as immunoassays, biochemical tests, and nucleic acid hybridization have been used for microbial examination. Since bacterial culture is a prerequisite, the above techniques are mostly laborious and time-consuming. Advanced molecular biology techniques have largely evolved over the past few years to engage in culture-independent methods; PCR is a robust, rapid, and sensitive technique for the identification of periodontal pathogens. The 16S RNA (16SrDNA) gene of the small ribosomal subunit is often used as a target for PCR for its highly conserved regions through evolution and species-specific variable sequences. The conserved sequences can offer PCR primers for amplification of 16SrDNA from all the bacterial species and species-specific regions for the identification of unique bacterial species.[9]
F. alocis is emerging as a pathobiont, gaining more relevance in the world of periodontal microbiology. There is very little done to report F. alocis prevalence in periodontitis in the Indian ethnic population. This study aims to detect the presence and estimate the absolute count of F. alocis in periodontal health and disease in the South Indian ethnic population.
Materials and Methods
Study samples
Sixty systemically healthy individuals visiting the Department of Periodontics underwent a comprehensive periodontal examination that included probing depth (PD) and level of clinical attachment (CAL). The subjects were assigned into three groups: periodontally healthy (Group A), aggressive periodontitis (Group B), and chronic periodontitis (Group C) [AAP 1999] based on the following inclusion criteria: group A-Clinically healthy gingiva without bleeding on probing, CAL and PD ≤ 3 mm [Figure 1a], Group B-aggressive periodontitis, at least four sites showing ≥3 mm CAL and PD ≥5 mm including rapid attachment loss and bone destruction, radiographically visible vertical or arc-shaped bone loss, particularly in central incisors and first molars, amount of destruction does not commensurate with local factors [Figure 1c] and Group C-Chronic periodontitis, at least four sites showing ≥3 mm CAL and probing pocket depth (PPD) ≥5 mm, amount of destruction consistent with local factors, radiographically visible bone loss in the horizontal direction, frequent detection of subgingival plaque and calculus [Figure 1b].[10] The patients who had any systemic diseases like diabetes mellitus, HSV or HIV infection, patients with a history of either smoking or chewing tobacco, and patients who underwent periodontal treatment or antimicrobial therapy in the duration of the pst 6 months were excluded from the study.
Figure 1.
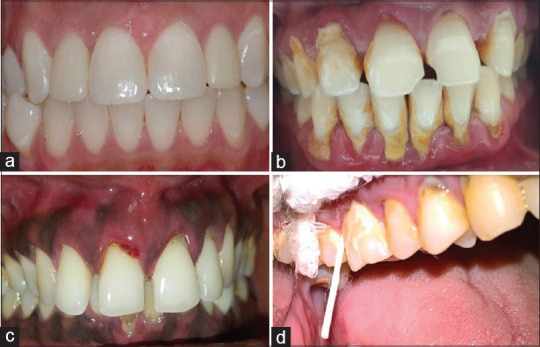
Representation of the various clinical specimens involved in the study. (a) Healthy periodontium, (b) chronic periodontitis, (c) aggressive periodontitis, and (d) sample collection using paper points
Sample collection
The plaque samples were collected the next day following clinical examination to avoid contamination of the samples by bleeding induced by probing. The samples were collected from four deep pockets using sterile paper points of size 35, pooled and transferred immediately to Eppendorf tubes containing 1 mL of Tris-EDTA buffer [Figure 1d].[11] This ensures high sensitivity and accurate representation of the subject. The samples were freeze stored at −80°C until the bacterial count.
16srDNA cloning and sequencing
From the total sample collected, 1 μl of the sample was used to amplify 594 bp of 16srDNA region of F. alocis by PCR using the primer pair FP-CTAATACCGCATACGTCCTAAG/RP-CTACTAAGCAATCAAGTTGCCC. The PCR conditions used were 95°C for 3 min (initial denaturation) followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s and a final extension at 72°C for 3 min. The PCR product was gel purified, cloned into the pGEM-T easy vector, transformed into Top10 cells and confirmed by DNA sequencing. The plasmid was used as a standard for the absolute quantification of F. alocis.
Absolute quantification by quantitative polymerase chain reaction
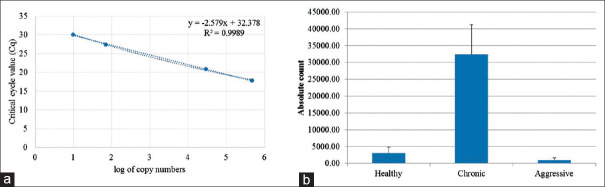
A 20 μl reaction was set using 2.5 pm each of primers, with known concentrations of standard plasmid DNA. Absolute quantification was performed using the following program: 95°C, 7 min, 35 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s, melt analysis consisting of 95°C for 1 min, −65°C for 1 min, −95°C continuous acquisition. Fluorescence data was captured during the extension stage of each cycle. The plasmid containing the 16srDNA fragment was used as a standard [Figure 2a]. A standard graph was computed using log of the copy number of the plasmid against Cq value.
Figure 2.
Absolute quantification of Filifactor alocis in healthy, chronic and aggressive subjects of periodontitis. The 16srDNA cloned plasmid was used as a standard to quantify Filifactor alocis in the subjects (a). The average absolute count of the bacteria in the various subjects were estimated by pooling of paper points of individual subjects and absolute count quantified in triplicates using quantitative polymerase chain reaction (b)
Statistical analysis
The data were analyzed with IBM. SPSS statistics software 23.0 Version (SPSS statistics software produced by SPSS Inc , Chicago, Illinois USA). Descriptive statistics were performed using mean and standard deviation, while multivariate analysis was computed using one-way ANOVA with Tukey's Post-Hoc test at P = 0.05. To assess the relationship between the variables, Pearson's Correlation was used.
Results
PPD and CAL correspond to the severity of periodontitis
The mean age of Group A, Group B, and Group C was 36.85 ± 8.55, 26.20 ± 3.32, and 43.05 ± 9.29, respectively [Supplementary Table 1]; Group B subjects were younger than Group C and Group A subjects. In addition, there was no significant difference in gender-wise distribution among the three groups. Periodontal examination revealed that PPD and CAL were high in chronic subjects, but was maximum in aggressive subjects. The mean PPD for Group A, Group B, and Group C was 2.45 ± 0.65, 6.75 ± 0.75, and 5.71 ± 0.98, respectively [Supplementary Table 1]. The mean CAL for Group A, Group B, and Group C was 0, 7.28 ± 1.47 and 6.23 ± 1.23, respectively [Supplementary Table 1]. The mean PPD and mean CAL were statistically significant at P ≤ 0.05 (0.0005) [Supplementary Table 2].
Supplementary Table 1.
Summarises the mean, standard deviation, maximum and the minimum values of count, age, PPD and clinical attachment level in each group
| Mean | SD | SE | Minimum | Maximum | |
|---|---|---|---|---|---|
| Count | |||||
| Aggressive | 939.84 | 670.03 | 149.82 | 127.37 | 2796.95 |
| Chronic | 32,409.85 | 88,735.34 | 19,841.83 | 96.23 | 379,451.37 |
| Healthy | 3046.15 | 1784.16 | 398.95 | 337.07 | 6473.92 |
| Total | 12,131.95 | 52408.83 | 6765.95 | 96.23 | 379,451.37 |
| Age | |||||
| Aggressive | 26.20 | 3.32 | 0.74 | 21.00 | 33.00 |
| Chronic | 43.05 | 9.29 | 2.08 | 27.00 | 60.00 |
| Healthy | 36.85 | 8.55 | 1.91 | 24.00 | 53.00 |
| Total | 35.37 | 10.20 | 1.32 | 21.00 | 60.00 |
| Mean PPD | |||||
| Aggressive | 6.75 | 1.09 | 0.24 | 5.00 | 9.00 |
| Chronic | 5.71 | 0.98 | 0.22 | 4.25 | 7.50 |
| Healthy | 2.45 | 0.65 | 0.14 | 1.00 | 3.50 |
| Total | 4.97 | 2.06 | 0.27 | 1.00 | 9.00 |
| Mean CAL | |||||
| Aggressive | 7.28 | 1.47 | 0.33 | 5.00 | 10.00 |
| Chronic | 6.23 | 1.23 | 0.28 | 5.00 | 9.00 |
| Healthy | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Total | 4.50 | 3.42 | 0.44 | 0.00 | 10.00 |
n denotes the number of subjects in each group (20). CAL: Clinical attachment level; PPD: Probing pocket depth; SD: Standard deviation; SE: Standard error
Supplementary Table 2.
Intergroup and intragroup comparison of count, age, mean probing pocket depth, and mean clinical attachment level using analysis of variance with P value at a significance of 0.05
| Sum of squares | df | Mean square | F | Significant | |
|---|---|---|---|---|---|
| Count | |||||
| Between groups | 12,380,164,074 | 2 | 6,190,082,037 | 2.357 | 0.104 |
| Within groups | 149,674,263,552 | 57 | 2,625,864,273 | ||
| Total | 162,054,427,626 | 59 | |||
| Age | |||||
| Between groups | 2905 | 2 | 1453 | 25.566 | 0.0005 |
| Within groups | 3238.700 | 57 | 56.819 | ||
| Total | 6143.933 | 59 | |||
| Mean PPD | |||||
| Between groups | 201.402 | 2 | 100.701 | 116.881 | 0.0005 |
| Within groups | 49.109 | 57 | 0.862 | ||
| Total | 250.511 | 59 | |||
| Mean CAL | |||||
| Between groups | 618.525 | 2 | 309.263 | 251.918 | 0.0005 |
| Within groups | 69.975 | 57 | 1.228 | ||
| Total | 688.500 | 59 | |||
| P | Highly significant at P≤0.01 | ||||
| P | Not significant at P>0.05 | ||||
Intergroup comparison of the bacterial count shows that it is statistically not significant. A large F value indicates a varied proportion of mean squares among the three groups. ANOVA: Analysis of variance; PPD: Probing pocket depth; CAL: Clinical attachment level
Cloning and absolute quantification of Filifactor alocis
The 16srDNA of F. alocis was PCR amplified, cloned, and sequence confirmed (MT573543) for use as a standard to quantify the pathogen in healthy, aggressive, and chronic patient groups. The standard curve was prepared with varying copy numbers of the 16srDNA containing plasmid (r2 = 0.9989), and the bacteria were quantified in all three groups [Figure 2a].
Filifactor alocis colonization and growth depends on the periodontal microbiome
Absolute count of F. alocis was determined using real-time quantitative PCR (qPCR). The average bacterial count in Group A (healthy) was 3046.15; 40% of the samples showed more than the average bacterial count, whereas the average bacterial count in Group B (aggressive) was only 939.84; 40% of the samples showed more than the average. The mean bacterial count in Group C (chronic) was 32,409.85, with 15% of the samples having an absolute count more than the group's average count [Figure 2b].
Inter- and intra-group comparison for clinical parameters and absolute count
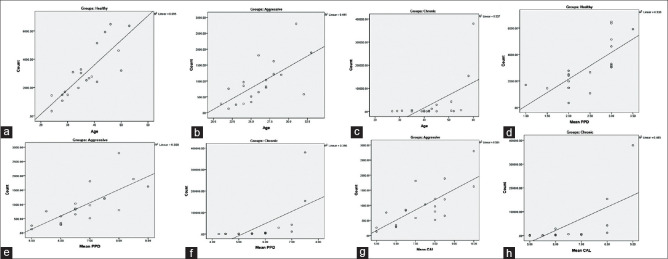
Analysis of variance showed a significant difference between age, PPD, CAL, and the count of F. alocis in healthy, aggressive, and chronic patients [Supplementary Table 1]. The correlation within the groups for the absolute bacterial count with age, mean PPD, and mean CAL was studied using intra-group Pearson's coefficient relation [Supplementary Tables 2–5] and Figure 3]. An intra-group comparison across the three groups for the bacterial count showed a significant increase with age, mean PPD, and mean CAL increased in healthy, aggressive, and chronic periodontitis groups (P ≤ 0.01).
Supplementary Table 5.
Intragroup correlation of count with age, mean probing pocket depth, and mean clinical attachment level in Group C by Pearson coefficient correlation with P value significant at ≤0.01
| CorrelationsC | |||
|---|---|---|---|
| Count | Age | Mean PPD | Mean CAL |
| Pearson correlation | 0.580** | 0.629** | 0.682** |
| Significant (two-tailed) | 0.007 | 0.003 | 0.001 |
| n | 20 | 20 | 20 |
| P | **Highly significant at P≤0.01 | ||
**Correlation is significant at the 0.01 level (two-tailed). Group C: Chronic; PPD: Probing pocket depth; CAL: Clinical attachment level
Figure 3.
The correlation between age and bacterial count in Group A, Group B, and Group C shows a linear increase in the count as age increases with a positive linear association of 0.695 (a), 0.441 (b), and 0.337 (c) respectively. The correlation between mean probing pocket depth and bacterial count in Group A, Group B, and Group C shows a linear increase in the count as mean probing pocket depth increases with a positive linear association of 0.538 (d), 0.568 (e), and 0.396 (f). The correlation between mean clinical attachment and bacterial count in Group B and Group C shows a linear increase in the count as mean clinical attachment increases with a positive linear association of 0.581 (g), and 0.465 (h) respectively
Supplementary Table 3.
Intragroup correlation of count with age and mean probing pocket depth in Group A by Pearson coefficient correlation with P value significant at ≤0.01
| CorrelationsA | ||
|---|---|---|
| Count | Age | Mean PPD |
| Pearson correlation | 0.834** | 0.733** |
| Significant (two-tailed) | 0.0005 | 0.0005 |
| n | 20 | 20 |
| P | **Highly significant at P≤0.01 | |
**Correlation is significant at the 0.01 level (two-tailed). Group A: Healthy; PPD: Probing pocket depth
Supplementary Table 4.
Intragroup correlation of count with age, mean probing pocket depth, and mean clinical attachment level in Group B by Pearson coefficient correlation with P value significant at ≤0.01
| CorrelationsB | |||
|---|---|---|---|
| Count | Age | Mean PPD | Mean CAL |
| Pearson correlation | 0.664** | 0.754** | 0.749** |
| Significant (two-tailed) | 0.001 | 0.0005 | 0.0005 |
| n | 20 | 20 | 20 |
| P | **Highly significant at P≤0.01 | ||
**Correlation is significant at the 0.01 level (two-tailed). Group B: Aggressive; PPD: Probing pocket depth; CAL: Clinical attachment level
Discussion
Periodontitis is characterized by chronic inflammation of the surrounding periodontal structures leading to alveolar bone loss and destruction of gingival and periodontal ligament attachments to the teeth.[12,13,14]
Advances in technology have redefined the oral microbiome into a synergistic and dysbiotic microbial community rather than by select “periopathogens,” such as the “redcomplex.”[5] Certain newer organisms, such as F. alocis, TM7, Selenomonas, Desulfobulbus, and Synergistes, have been implicated in periodontal pathogenesis. Culturing F. alocis, the third-most prevalent pathogen in aggressive periodontitis, the second most prevalent in chronic periodontitis is difficult owing to its fastidious and anaerobic nature, prevalent at mild to moderate infections in healthy controls.[15,16,17]
Here, we have used qPCR to quantify F. alocis in clinically categorized healthy, aggressive and chronic periodontal subjects; the pathogen was detected in all the samples analyzed. The prevalence of F. alocis is high in periodontitis compared to gingivitis and healthy; more common in sites with periodontitis than in healthy sites.[18,19] We observed the highest average bacterial count in the chronic subjects. The prevalence of F. alocis vary from14%,[17] 20%,[16] 30%,[20] 44%[21] and up to 60%[15] in chronic periodontitis. In some other parts of the world, it varies from 30% in Sweden,[18] 66.7% in Germany[22] to 83% and 36%, respectively, in deep and shallow sites of periodontal region in Korea.[23]
Periodontal therapy reduced F. alocis occurrence in patients with chronic periodontitis conditions in the Thai population;[24] however, in another study, non-surgical periodontal treatment failed to reduce F. alocis.[25] Therefore, we believe that the microbiome is complex and might be region or population specific with more factors implicated in the constitution of the microbiome. Furthermore, the prevalence and virulence of one bacteria influence other members of the microbiota. F. alocis and P. gingivalis co-exist together and have a symbiotic relationship; P. gingivalis is more dominant in chronic periodontitis, it can enhance the multiplication and virulence potential of F. alocis. Coexistence and resistance to oxidative stress might be the critical cause for the increased incidence of F. alocis in chronic periodontitis.[26]
The F. alocis prevail at low to moderate levels in healthy controls.[16,17,22,23,27,28] In our study, about 40% of the healthy controls showed more than the average bacterial count of 3046. Although the incidence of F. alocis varied among healthy controls, a maximum of 29% has been detected in healthy controls.[15] The variation in the incidence of F. alocis may be due to strain variability and difference in PD; less virulent strain of F. alocis ATCC 35896 was shown to be less invasive than the isolate D-62D.[26] Since only PCR was used to detect F. alocis, the magnitude of prevalence was unknown, our report is the first to report real-time detection and quantification of F. alocis in various subjects.
In our study, the average estimate of F. alocis in the aggressive subjects was low (939.84), the tissue invasiveness and lack of subgingival plaque might be a compelling factor for the lower incidence. Furthermore, F. alocis prevail in consortium with Aggregatibacter actinomycetemcomitans and Streptococcus parasanguinis before the bone loss in localized aggressive periodontitis patients.[29] The incidence and prevalence of F. alocis are complicated by the age of the patient, personal habits, population type, and country of origin.[30,31] Biologically, the population of F. alocis is dependent on the periodontal microbiome; the microbiota varies significantly between chronic and aggressive patients. In an in vitro community model study, Streptococcus gordonii, Gram-negative bacteria of aggressive periodontitis microbiota was antagonistic to F. alocis, and A. actinomycetemcomitans interacted with F. alocis in a strain-specific manner.[32]
The present study observed a statistically significant association between age, clinical periodontal parameters such as PPD and CAL, and the presence of the organism. Intra-group correlation of the age with bacterial count was substantial in all the three groups (P ≤ 0.01). Similarly, intra-group correlation reveals that there was a positive correlation between PPD and CAL with the bacterial population in all three groups (P ≤ 0.01). Geographical locations, ethnicity, food habits, smoking habits influence the distribution, prevalence, and virulence of the oral microbiome; variations prevail in the subgingival microbiota worldwide.[33]
Conclusion
F. alocis is a novel microorganism gaining more relevance in periodontal pathogenesis. It has been implicated to be the second- and third-most prevalent organism in chronic and aggressive periodontitis, respectively. The highest bacterial count was observed in chronic periodontitis patients (32,409.85), followed by healthy controls (3046.15) and least in aggressive periodontitis patients (939.84). There was a statistically significant difference between the bacterial count of three groups with P value at 0.005 (P ≤ 0.05). On intra-group comparison, there was a statistically significant increase in the bacterial count as the age and mean PPD increases with P value at 0.0005 (P ≤ 0.05). The higher prevalence of F. alocis in chronic periodontitis patients in the Indian population indicates that it can be a candidate marker for chronic periodontitis; however, to qualify it as a marker, validation with more longitudinal studies in a larger study group is warranted.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We would like to acknowledge the facilities provided by SRM IST to carry out the work.
References
- 1.Saini R, Marawar PP, Shete S, Saini S. Periodontitis, a true infection. J Glob Infect Dis. 2009;1:149–50. doi: 10.4103/0974-777X.56251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller WD. The Micro-Organisms of the Human Mouth. Philadelphia, PA: The S.S. White Dental MFG. CO; 1890. [PMC free article] [PubMed] [Google Scholar]
- 3.Loesche WJ. Chemotherapy of dental plaque infections. Oral Sci Rev. 1976;9:65–107. [PubMed] [Google Scholar]
- 4.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–71. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 5.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–25. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–19. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014 Jan;35(1):3–11. doi: 10.1016/j.it.2013.09.001. doi: 10.1016/j.it.2013.09.001.Epub 2013 Oct 23. PMID: 24269668; PMCID: PMC3947349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wade WG. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol. 2011;38(Suppl 11):7–16. doi: 10.1111/j.1600-051X.2010.01679.x. [DOI] [PubMed] [Google Scholar]
- 10.Armitage GC. Development of classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Hartroth B, Seyfahrt I, Conrads G. Sampling of periodontal pathogens by paper points: Evaluation of basic parameters. Oral Microbiol Immunol. 1999;14:326–30. doi: 10.1034/j.1399-302x.1999.140510.x. [DOI] [PubMed] [Google Scholar]
- 12.Oliver RC, Brown LJ, Loe H. Variations in the prevalence and extent of periodontitis. J Am Dent Assoc. 1991;122:43–8. doi: 10.1016/s0002-8177(91)26016-x. [DOI] [PubMed] [Google Scholar]
- 13.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. CDC Periodontal Disease Surveillance workgroup: James Beck (University of North Carolina, Chapel Hill, USA), Gordon Douglass (Past President, American Academy of Periodontology), Roy Page (University of Washin). Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–20. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 14.Thornton-Evans G, Eke P, Wei L, Palmer A, Moeti R, Hutchins S, et al. Periodontitis among adults aged >/=30 years - United States, 2009-2010. MMWR Surveill Summ. 2013;62(Suppl 3):129–35. [PubMed] [Google Scholar]
- 15.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 16.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–55. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–73. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cato EP, Moore LV, Moore WE. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int J Syst Bacteriol. 1985;35:475–7. [Google Scholar]
- 19.Maiden MF, Tanner A, Macuch PJ. Rapid characteri-zation of periodontal bacterial isolates by using flurogenic substrate tests. J Clin Microbiol. 1996;34:376–84. doi: 10.1128/jcm.34.2.376-384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlén G, Leonhardt A. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol Immunol. 2006;21:6–11. doi: 10.1111/j.1399-302X.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- 21.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, Berning M, et al. Filifactor alocis Involvement in periodontal biofilms. BMC Microbiol. 2010;10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun JH, Park JE, Kim DI, Lee SI, Choi SH, Cho KS, et al. Identification of putative periodontal pathogens in Korean chronic periodontitis patients. J Korean Acad Periodontol. 2008;38:143–52. [Google Scholar]
- 24.Payungporn S, Arirachakaran P, Poomipak W, Praianantathavorn K, Charalampakis G, Poworawan Y. Identification of bacteria associated with a periodontal disease in Thai patients based on next-generation sequencing. Jundishapur J Microbiol. 2017;10:E13646. [Google Scholar]
- 25.Spooner R, Weigel KM, Harrison PL, Lee K, Cangelosi GA, Yilmaz Ö. In situ anabolic activity of periodontal pathogens Porphyromonas gingivalis and Filifactor alocis in chronic periodontitis. Sci Rep. 2016;6:33638. doi: 10.1038/srep33638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aruni AW, Roy F, Fletcher HM. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun. 2011;79:3872–86. doi: 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-hebshi NN, Al-Alimi A, Taiyeb-Ali T, Jaafar N. Quantitative analysis of classical and new putative periodontal pathogens in subgingival biofilm: A case-control study. J Periodontal Res. 2015;50:320–9. doi: 10.1111/jre.12210. [DOI] [PubMed] [Google Scholar]
- 28.Galimanas V, Hall MW, Singh N, Lynch MD, Goldberg M, Tenenbaum H, et al. Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome. 2014;2:32. doi: 10.1186/2049-2618-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, et al. A consortium of Aggregatibacter actinomycetemcomitans (Aa), Streptococcus parasanguinis and Filifactor alocis are present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. 2013;51:2850–61. doi: 10.1128/JCM.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albandar JM, Thomas ER. Global epidemiology of periodontal diseases: An overview. Periodontology 2000. 2002;29:7–10. doi: 10.1034/j.1600-0757.2002.290101.x. [DOI] [PubMed] [Google Scholar]
- 31.Susin C, Valle P, Oppermann RV, Haugejorden O, Albandar JM. Occurrence and risk indicators of increased probing depth in an adult Brazilian population. J Clin Periodontol. 2005;32:123–9. doi: 10.1111/j.1600-051X.2005.00637.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. Oral community interactions of Filifactor alocis in vitro. PLoS One. 2013;8:e76271. doi: 10.1371/journal.pone.0076271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lafaurie GI, Contreras A, Baron A, Botero J, Mayorga-Fayad I, Jaramillo A, et al. Demographic, clinical, and microbial aspects of chronic and aggressive periodontitis in Colombia: A multicenter study. J Periodontol. 2007;78:629–39. doi: 10.1902/jop.2007.060187. [DOI] [PubMed] [Google Scholar]