Abstract
The new field of synthetic biology aims at the creation of artificially designed organisms. A major breakthrough in the field was the generation of the artificial synthetic organism Mycoplasma mycoides JCVI‐syn3A. This bacterium possesses only 452 protein‐coding genes, the smallest number for any organism that is viable independent of a host cell. However, about one third of the proteins have no known function indicating major gaps in our understanding of simple living cells. To facilitate the investigation of the components of this minimal bacterium, we have generated the database SynWiki (http://synwiki.uni-goettingen.de/). SynWiki is based on a relational database and gives access to published information about the genes and proteins of M. mycoides JCVI‐syn3A. To gain a better understanding of the functions of the genes and proteins of the artificial bacteria, protein–protein interactions that may provide clues for the protein functions are included in an interactive manner. SynWiki is an important tool for the synthetic biology community that will support the comprehensive understanding of a minimal cell as well as the functional annotation of so far uncharacterized proteins.
Keywords: essential genes, genome annotation, synthetic biology, SynWiki
1. INTRODUCTION
The creation of artificial cells that contain only those genes that are essentially required to sustain the major cellular functions is one of the aims of the emerging field of synthetic biology. This aim can be achieved in two complementary approaches: the top‐down approach identifies essential genes and functions, predicts the minimal gene set, and deletes all non‐essential genes in a consecutive manner thus resulting in genome reduced strains. This approach has so far been most successful for the Gram‐positive model bacterium Bacillus subtilis. For this bacterium, the essential gene set has been identified and the genes required for a minimal genome have been predicted 1 , 2 , 3 and the genome has been reduced by about 40%. 4 Alternatively, in the bottom‐up approach genes predicted or identified as components of a minimal genome are synthesized in vitro, assembled and introduced into a living cell. The original DNA will then be lost, and the cellular activities will be driven by proteins encoded by the artificial DNA molecule. This approach has been used to synthesize Mycoplasma mycoides JCVI‐syn3A, an artificial minimal bacterium with as few as 452 protein‐coding genes, less than in any other known natural independently viable bacterium. 5 , 6 Of the proteins encoded by M. mycoides JCVI‐syn3A, about one third has no known function, indicating how far we still are from a comprehensive understanding of even a very simple and minimal living cell. The investigation of M. mycoides JCVI‐syn3.0 by the scientific community will certainly help to get a deeper appreciation for the principal requirements of a minimalistic form of life, and thus touches one of the most basic and most important aspects of biology.
To facilitate the investigation of M. mycoides JCVI‐syn3A, we have developed SynWiki, a database centered around the genes and proteins of this bacterium. This database is based on the platform of the database SubtiWiki which is designed for the functional annotation of B. subtilis. 7 , 8 SynWiki presents the available information on the genes and proteins of M. mycoides JCVI‐syn3A in an easy and highly intuitive manner. One focus of SynWiki is the presentation of links between different genes and proteins that allow the researcher to develop novel hypotheses. The information provided is based on the publications describing the construction of the organism and the reconstruction of the minimal metabolism 5 , 6 and on the published specific information available on the proteins.
2. DESCRIPTION OF THE DATABASE
SynWiki is a relational database for the functional annotation of the synthetic minimal microorganism M. mycoides syn3A and uses the data describing the original JCVI‐syn3.0 bacterium as well as the most recent iteration of this organism, JCVI‐syn3A. 5 , 6 When referring to SynWiki data, we always consider the latter organism.
In SynWiki, the central biological element is the gene with its underlying relationships to proteins and functional RNAs of JCVI‐syn3A. With this in mind, it is clear that most pages focus on a specific gene element and its functional annotation. To access any of these elements, the front page provides a search box and access to several features of different types of data (Figure 1). To view information on any gene, it is only necessary to query for a specific gene's identifier (see more below). Alternatively, it is possible to go directly to a random gene page using a button on the top of the page.
FIGURE 1.
Home page of SynWiki. The main element of this page is the search bar that allows for searching of biological elements. Further down there is a list of SynApps and other helpful links such as a list of labs that work with this organism
In addition to providing scientific information, SynWiki also serves as a hub for the scientific community interested in the minimal organism. On each gene page, there is a banner that gives important information such as upcoming conferences and other events, and a link to labs working on this organism.
2.1. SynWiki identifiers
Currently, two unique identifiers can be used for each gene. The first one is a specific gene/ protein name that serves as a mnemonic for its function (such as eno for enolase) or its synonym (such as ktrC with its synonym trkA). The second identifier is an “eternal” locus tag, a gene‐specific identifier that will remain unchanged even though mnemonic gene designation may alter. 6 For this, we have maintained the original locus tags created for JCVI‐syn3A, which abide by the following rules: a “JCVISYN3A_” prefix with a numerical suffix “XXXX” that derives from the initial data. 5 As an example, to retrieve information on the JCVISYN3A_0001 gene, or dnaA, it is possible to search for either the locus tag or gene name. For uncharacterized genes, that yet lack a mnemonic designation, such as JCVISYN3A_0399, only this locus identifier can be used to access the page.
2.2. The gene pages
As mentioned before, SynWiki revolves around the gene entity, meaning that each gene has a fully dedicated page where all documented information can be found. Independent of the data available for each gene, all gene pages share the same structure (Figure 2). At the top, there is a banner highlighting community events, just followed by the gene name accompanied and a short description of its function, if available (Figure 2a). Followed by this, there is a table containing specific information of the searched gene and its product (Figure 2b). It contains information regarding locus tag, protein specific information (molecular weight, isoelectric point, product and function), gene and protein lengths with direct links to their corresponding sequences, a BLAST search, and information on essentiality. Moreover, SynWiki provides links to the respective entries in UniProt, KEGG, KEGG Pathway, and STRING databases (Figure 2b). Below that, an interactive genomic context browser is shown, which is part of the corresponding Genome Browser SynApp (see more below) (Figure 2c). This feature allows to see the gene's position in the genome and to scroll through the genome. Right under this section, information on the annotated functional categories (see below), as well as a link to the protein homology analysis developed in our lab (see below) are provided (Figure 2d). The remaining part of the page presents additional information on the gene/ protein. The gene section lists information regarding the gene element, such as genomic coordinates, phenotypes of mutants if available and other information centered on the gene (Figure 2e). The protein section lists specific information regarding this element, such as protein family and biochemical details (Figure 2f). At the bottom, there is a list of publications annotated for the current biological element (Figure 2g). Although not currently represented in SynWiki due to lack of data, information on operons and regulations can also be displayed in this page. Finally, the right panel of the page provides links to other important pages such as a list of labs working on JCVI‐syn3A, search boxes for SynWiki and PubMed entries, and displays protein structures as well as protein–protein interactions (if any) (Figure 2h).
FIGURE 2.
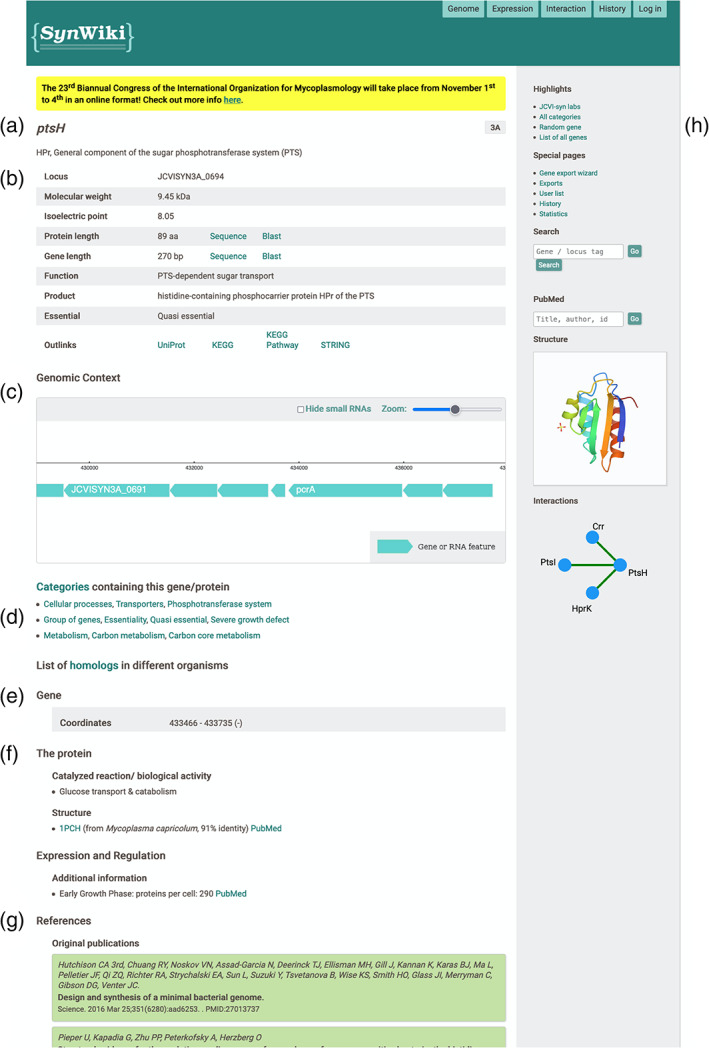
Gene page overview for ptsH. Gene page structure is shared among all genes with differences in displayed data, as it is dependent on its availability. (a) Gene title and description; (b) overview table focusing on different gene/protein details; (c) genomic context of genes with an interactive genome browser; (d) list of functional categories the gene is annotated with and list of homologs in other organisms; (e) section focused on gene centered information; (f) section focused on protein centered information; (g) list of references annotated to the gene; (h) Overview of protein structure and protein–protein interactions
2.3. SynApps
One of the strong points of our family of databases is the integration of different levels of data using multiple browsers under “SynApps” (Figure 1). Among these, we highlight the integration of genomic context (Genome Browser), expression data (Expression Browser), and protein–protein interactions (Interaction Browser).
In the Genome Browser, the user gets access to a scrollable genome to check for gene context and orientation, as well as DNA and protein sequences. As mentioned before, this browser is also partially included in every gene page for easier overview of each gene's genomic context. Importantly, due to evolutionary pressure towards an AT rich genome, Mycoplasmas use the TGA codon for tryptophan, in contrast to other organisms that use it as a stop codon instead. 9 , 10 Therefore, the internal codon table was adjusted accordingly and all protein sequences displayed in Genome Browser are correctly translated.
The Expression Browser provides data on a proteomics analysis during early growth for 428 proteins. 6 These data are also presented on the gene pages (in the paragraph “Expression and Regulation”). However, the Expression Browser allows a direct comparison of the protein amounts for different proteins of choice (see Figure 3a).
FIGURE 3.
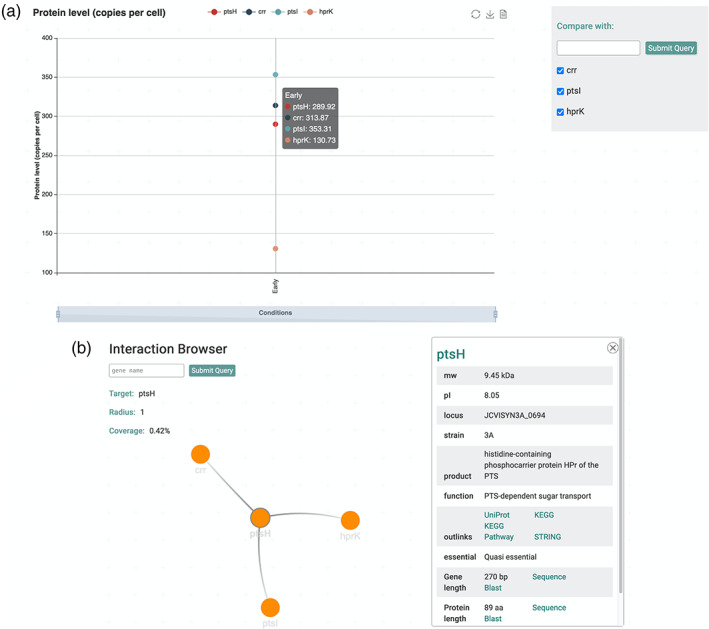
Overview of implemented SynApps. (a) Expression Browser on protein level for PtsH. Shows the number of protein molecules per cell under each experimental condition. Only “Early growth phase” data is available, thus it is the only one being represented. It is possible to add other proteins for comparison on the same view (all interacting proteins shown, Crr, PtsI, and HprK) and it is also possible to download data directly. (b) Interaction browser for PtsH, highlighting direct interaction partners and displaying an overview from its gene page
To identify the functions of so far unknown or poorly studied proteins, data on physical protein–protein interactions are of key importance. We have recently used a combination of cross‐linking, mass spectrometry and cryo‐electron tomography to unravel the in vivo interactome of Mycoplasma pneumoniae. 11 While global interaction data are not yet available, a recent study has shed light on the prediction of complex interactions in M. mycoides JCVI‐syn3A. 12 Protein–protein interactions are displayed on the gene pages (Figure 2h) and in the Interaction browser (Figure 3b). Similar to the Expression Browser, the Interaction Browser provides opportunities of interrogation that could not be reached on the gene pages. As an example, the user can visualize complex interaction patterns and selection one protein highlights direct neighbors and displays an overview from the gene page (Figure 3b).
3. IMPLEMENTATION OF THE DATABASE
3.1. SynWiki architecture
SynWiki's architecture relies on the framework previously developed for the well‐known database SubtiWiki. 8 The framework uses a relational database management system to establish relationships between entities, based on the relation theory, 13 thus allowing the organization and storage of data in tables. This allows for the creation of complex and rich relationships between genes/proteins and their annotation. Another advantage of using SubtiWiki's framework is that it provides the full experience one can find in this database, such as Browser features and the fully‐fledged user system to add and edit data.
3.2. SynWiki data
As mentioned before, SynWiki uses the data from the original JCVI‐syn3.0 and JCVI‐syn3A publications. 5 , 6 According to the authors, the latter iteration of this organism features the addition of 17 genes (14 M. mycoides genes and 3 pseudogenes), making a total of 492 genes, resulting in better growth and improved stability.
In addition to the annotation in the original publications we also went through the literature, checked known functions of homologs, and searched for the availability of protein structures of the JCVI‐syn3A proteins or their homologs from other organisms. All these results from manual data curation were added to the pages. Moreover, the curation of the pages is an ongoing process that will lead to ever‐improved annotation and data presentation.
The success of a biological database strongly depends on the quality of annotation, and for those cases where there is poor or no annotation, we wanted to take a step forward and give our own contribution. With this in mind, and starting from the original annotation, we have also assigned each gene to one or more functional category. We took inspiration from GO term tree 14 and created a tree‐like structure containing all functional categories. Currently, there are four major functional branches, “Cellular processes,” “Group of genes,” “Information processing” and “Metabolism,” which then branch out into more specific categories (see here for an overview: http://synwiki.uni-goettingen.de/v1/category, and Table 1). For example, in “Cellular processes” it first branches out into “Cellular envelope and cell division,” “Homeostasis,” and “Transporters.” Then, “Homeostasis” branches out into “Metal ion homeostasis” and “Coping with stress”; “Transporters” branch out into “ABC transporters,” “ECF transporters,” “Phosphotransferase system” and “Symporter.” The “Group of genes” parent category branches out into “Foreign genes,” “Membrane proteins,” “Poorly characterized enzymes,” “Proteins of unknown function,” “Pseudogenes,” “ncRNAs” and “Essentiality.” As a result of this annotation, we can now dig deeper into the data and assess it according to specific categories. As an example, looking for “Essentiality” allows to identify 46% of genes to be labeled as “Essential,” while only 13.6% are classified as “Non‐essential” (Figure 4). Moreover, we have annotated most genes with over 500 publications to help better understand the potential underlying roles of JCVI‐syn3A genes.
TABLE 1.
Overview of the number of genes in higher‐level functional categories
| Functional category | Number of genes | |
|---|---|---|
| Cellular processes | Cell envelope and cell division | 4 |
| Homeostasis | 13 | |
| Transporters | 39 | |
| Groups of genes | Foreign gene | 2 |
| Membrane proteins | 113 | |
| Poorly characterized enzymes | 14 | |
| Proteins of unknown function | 87 | |
| Pseudogenes | 1 | |
| ncRNA | 3 | |
| Essentiality | 485 | |
| Information processing | Genetics | 42 |
| Protein synthesis, modification and degradation | 183 | |
| Regulation of gene expression | 6 | |
| RNA synthesis and degradation | 22 | |
| Metabolism | Amino acid acquisition | 8 |
| ATP synthesis | 8 | |
| Carbon metabolism | 31 | |
| Cofactor acquisition | 10 | |
| Iron metabolism | 2 | |
| Lipid metabolism | 14 | |
| Nucleotide metabolism | 25 | |
| Phosphate metabolism | 5 |
FIGURE 4.
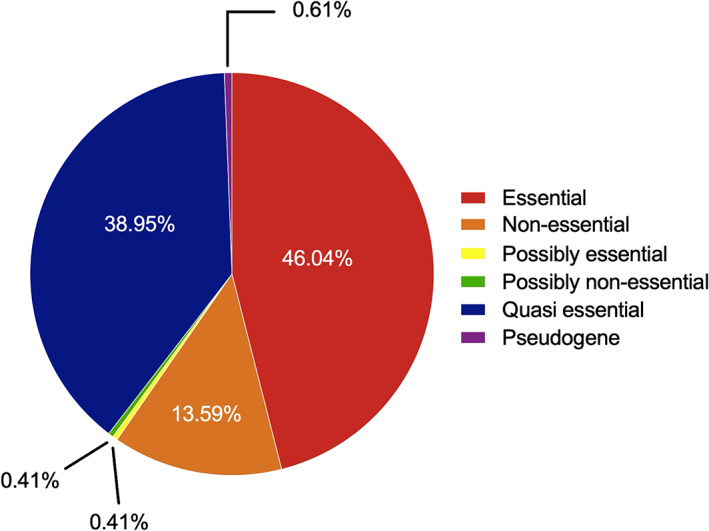
Distribution of essentiality among all genes in SynWiki. While essential genes cannot be removed from the genome without loss of viability, the removal of quasi‐essential genes results in an observable growth disadvantage 6
Additionally, we added an extensive list of potential protein homologs for 16 relevant bacterial species, among them M. pneumoniae and the model organisms Escherichia coli and B. subtilis. The proteomes of these organisms were extracted from UniProt 15 and a set of a bidirectional alignments was performed using the similarity search tool FASTA 16 between the library of proteomes and the proteome of JCVI‐syn3A. The resulting aligned protein pairs in both directions showing an E‐value ≤0.01 and high similarity were considered to be potential bidirectional homologs. The results can be accessed on the gene page (Figure 2d) and they are presented in detail in a table format, showing the respective scores (Figure 5). To complement this, and based on protein structure homology, we have included a protein structure prediction for 327 proteins (Figure 2h).
FIGURE 5.
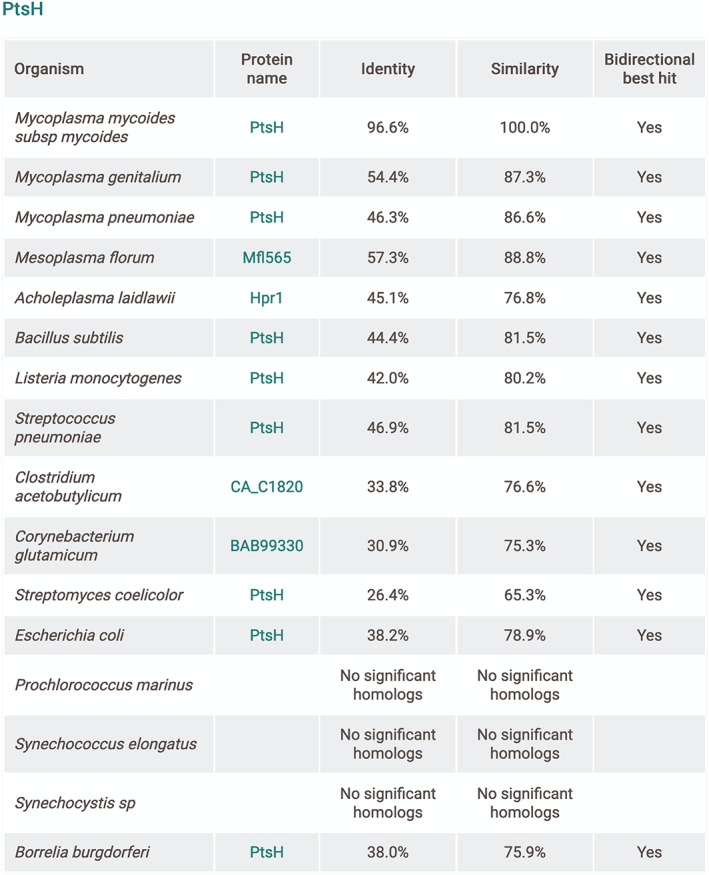
Representation of protein homology analysis for PtsH. Results displayed in a table format for each organism used in this analysis with respective best potential homolog with UniProt link. Identity, similarity and bidirectionality scores also displayed
4. FUTURE PERSPECTIVES
SynWiki is a new database for the recently created minimal genome microorganism M. mycoides JCVI‐syn3A. SynWiki uses the framework of SubtiWiki, which gives it a robust and consistent way of searching and updating data. Although there is scarce information for this organism, we have created a powerful structure to store and display the current data. However, SynWiki is also prepared to include new findings that might arise from emerging studies. Importantly, recent attempts to model the complete metabolism of JCVI‐syn3A 6 and related natural minimal organisms Mesoplasma florum 17 will certainly benefit from the collection of information provided in SynWiki. On the other hand, the information derived from the metabolic models and similar global approaches is important to update SynWiki. As more research groups delve into the unknown and try to unveil more of JCVI‐syn3A, we want to provide the scientific community with a platform where all biological elements of this organism can be updated on a daily basis with curated data. We aim not only to put data together, but also to organize it and give scientists the confidence and the necessary tools to create new approaches for their research.
AUTHOR CONTRIBUTIONS
Tiago Pedreira: Conceptualization (lead); investigation (lead); methodology (lead); project administration (supporting); resources (lead); supervision (lead); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Christoph Elfmann: Methodology (supporting); resources (supporting); visualization (supporting). Neil Singh: Conceptualization (supporting); data curation (supporting); resources (supporting). Jörg Stülke: Conceptualization (equal); data curation (equal); funding acquisition (lead); project administration (lead); supervision (lead); writing – original draft (equal); writing – review and editing (equal).
ACKNOWLEDGMENTS
We are grateful to Hannes Gebauer for the initial work on SynWiki.
Pedreira T, Elfmann C, Singh N, Stülke J. Syn Wiki: Functional annotation of the first artificial organism Mycoplasma mycoides JCVI‐syn3A . Protein Science. 2022;31:54–62. 10.1002/pro.4179
REFERENCES
- 1. Commichau FM, Pietack N, Stülke J. Essential genes in Bacillus subtilis: A re‐evaluation after ten years. Mol Biosyst. 2013;9:1068–1075. [DOI] [PubMed] [Google Scholar]
- 2. Koo BM, Kritikos G, Farelli JD, et al. Construction and analysis of two genome‐scale deletion libraries for Bacillus subtilis . Cell Syst. 2017;4:291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reuß DR, Commichau FM, Gundlach J, Zhu B, Stülke J. The blueprint of a minimal cell: MiniBacillus . Microbiol Mol Biol Rev. 2016;80:955–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reuß DR, Altenbuchner J, Mäder U, et al. Large‐scale reduction of the Bacillus subtilis genome: Consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 2017;27:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hutchison CA 3rd, Chuang RY, Noskov VN, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. [DOI] [PubMed] [Google Scholar]
- 6. Breuer M, Earnest TM, Merryman C, et al. Essential metabolism for a minimal cell. Elife. 2019;8:e36842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michna RH, Commichau FM, Tödter D, Zschiedrich CP, Stülke J. SubtiWiki – A database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 2014;42:D692–D698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu B, Stülke J. SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis . Nucleic Acids Res. 2018;46:D743–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamao F, Muto A, Kawauchi Y, et al. UGA is read as tryptophan in Mycoplasma capricolum . Proc Natl Acad Sci U S A. 1985;82:2306–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inamine JM, Ho KC, Loechel S, Hu PC. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum . J Bacteriol. 1990;172:504–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Reilly FJ, Xue L, Graziadei A, et al. In‐cell architecture of an actively transcribing‐translating expressome. Science. 2020;369:554–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang C, Zheng W, Cheng M, Omenn GS, Freddolino PL, Zhang Y. Functions of essential genes and a scale‐free protein interaction network revealed by structure‐based function and interaction prediction for a minimal genome. J Proteome Res. 2021;20:1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Codd EF. A relational model of data for large shared data banks. Commun ACM. 1983;26:64–69. [PubMed] [Google Scholar]
- 14. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. The UniProt Consortium . UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madeira F, Park YM, Lee J, et al. The EMBL‐EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lachance JC, Matteau D, Brodeur J, et al. Genome‐scale metabolic modeling reveals key features of a minimal gene set. Mol Syst Biol. 2021;17:e10099. [DOI] [PMC free article] [PubMed] [Google Scholar]


