Abstract
The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), funded by the US National Science Foundation, National Institutes of Health, and Department of Energy, has served structural biologists and Protein Data Bank (PDB) data consumers worldwide since 1999. RCSB PDB, a founding member of the Worldwide Protein Data Bank (wwPDB) partnership, is the US data center for the global PDB archive housing biomolecular structure data. RCSB PDB is also responsible for the security of PDB data, as the wwPDB‐designated Archive Keeper. Annually, RCSB PDB serves tens of thousands of three‐dimensional (3D) macromolecular structure data depositors (using macromolecular crystallography, nuclear magnetic resonance spectroscopy, electron microscopy, and micro‐electron diffraction) from all inhabited continents. RCSB PDB makes PDB data available from its research‐focused RCSB.org web portal at no charge and without usage restrictions to millions of PDB data consumers working in every nation and territory worldwide. In addition, RCSB PDB operates an outreach and education PDB101.RCSB.org web portal that was used by more than 800,000 educators, students, and members of the public during calendar year 2020. This invited Tools Issue contribution describes (i) how the archive is growing and evolving as new experimental methods generate ever larger and more complex biomolecular structures; (ii) the importance of data standards and data remediation in effective management of the archive and facile integration with more than 50 external data resources; and (iii) new tools and features for 3D structure analysis and visualization made available during the past year via the RCSB.org web portal.
Keywords: electron microscopy, macromolecular crystallography, micro‐electron diffraction, Mol*, open access, PDB, Protein Data Bank, RCSB Protein Data Bank, web‐native molecular graphics, Worldwide Protein Data Bank
1. INTRODUCTION
The Protein Data Bank (PDB) has been serving global science for more than 50 years. It was established on October 20th, 1971 as the first open‐access digital data resource in biology with just seven protein structures. 1 Thanks to the generosity of more than 50,000 structural biologists working on every inhabited continent, the archive has grown to more than 180,000 structures of proteins and nucleic acids (DNA and RNA). Today, the PDB archive is jointly managed by the Worldwide Protein Data Bank (wwPDB, wwpdb.org) partnership, 2 , 3 which was founded in 2003 by the US‐funded RCSB Protein Data Bank (RCSB PDB), the Protein Data Bank in Europe (PDBe), and Protein Data Bank Japan (PDBj). Current wwPDB members also include the Electron Microscopy Data Bank (EMDB) and the Biological Magnetic Resonance Bank (BMRB). Millions of PDB data consumers worldwide working in fundamental biology, biomedicine, bioengineering, biotechnology, and energy sciences enjoy no‐cost access to 3D biostructure information with no limitations on data usage.
The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB; RCSB.org) 4 , 5 is jointly funded by the National Science Foundation, the National Institutes of Health, and the US Department of Energy. Safeguarding and nurturing the PDB archive and providing open access to PDB data are the responsibility of four coordinated RCSB PDB “services,” encompassing data deposition; archive management and access; data exploration; and outreach and education. RCSB PDB, like its wwPDB partners, is committed to the FAIR (Findability, Accessibility, Interoperability, and Reusability) 6 and FACT (Fairness, Accuracy, Confidentiality, and Transparency) 7 Principles emblematic of responsible data stewardship in the modern era. It is no exaggeration to state that the PDB was “walking the walk” decades before people began “talking the talk” regarding these important concepts.
Service 1—Data deposition: The global wwPDB OneDep 8 software system manages deposition, validation, expert biocuration, and remediation of macromolecular crystallography (MX), 3D electron microscopy (3DEM), nuclear magnetic resonance (NMR) spectroscopy, and micro‐electron diffraction (microED) structures, experimental data, and related metadata. Within the wwPDB, RCSB PDB supports PDB data depositors working in the Americas and Oceania ensuring completeness and accuracy of the ever‐growing body of 3D structure data.
Service 2—Archive management and access: In its role as the wwPDB‐designated Archive Keeper, RCSB PDB safeguards the PDB archive and maintains the PDBx/mmCIF data dictionary 9 , 10 that enables organization and searching of archived data. Programmatic access to PDB data is available via FTP and application programming interfaces (APIs). Strict adherence to the PDBx/mmCIF data standard enables facile integration of 3D structure information with >50 trusted external data resources.
Service 3—Data Exploration: Tools for data searching, browsing, visualization, custom report generation, and analysis are made freely available on our research‐focused RCSB.org web portal with no limitations on usage.
Service 4—Outreach and Education: RCSB PDB has a long history of delivering outreach and education resources focused on structural biology and its impact across the sciences via its introductory PDB101.RCSB.org web portal (reviewed in this issue 11 ).
Two other important elements of RCSB PDB operations are the Customer Service Help Desk, responsible for supporting 3D structure depositors and PDB data consumers around the world, and the Infrastructure Team, which works to ensure >99% 24 × 7 × 365 service availability uptime. The status of RCSB PDB servers, microservices, and application programming interfaces (APIs) is monitored by NS1 (NS1.com) and publicly available on a real time basis at status.rcsb.org.
Recent redesign of RCSB PDB data architecture and overhaul of our research‐focused RCSB.org web portal with many added new features have been described previously. 5 , 12 This invited Protein Science Tools Issue contribution describes the unprecedented growth of the PDB archive during 2020, continued evolution of the PDBx/mmCIF data standard, archive‐wide remediation of carbohydrate‐containing structures, and deployment of new RCSB.org tools and features completed during 2021.
2. RESULTS
2.1. PDB data metrics and trends
The first year of the COVID‐19 pandemic witnessed unprecedented growth in the PDB archive. This section presents impressive structure deposition and release metrics for 2020 and describes the ongoing impact of the 3DEM resolution revolution. 13
PDB structures are contributed annually by tens of thousands of structural biologist depositors worldwide through the wwPDB OneDep software system (deposit.wwpdb.org) for structure deposition, 8 rigorous validation, 14 , 15 and expert biocuration. 16 OneDep currently supports 3D macromolecular structures determined using MX, 3DEM, NMR, and microED experimental methods. Incoming structures are processed at regional wwPDB data centers allocated on the basis of the depositor's geographic location.
During calendar year 2020, wwPDB partners processed a record 15,436 experimental structure depositions to the PDB archive. During this same period, RCSB PDB processed ~47% of the global depositions (primarily from the Americas and Oceania, and GroupDep 17 users), PDBe processed ~31% of global depositions (from Europe and Africa), and PDBj processed ~22% of global depositions (from Asia and the Middle East) (Figure 1a,b). Consequently, the number of new structures publicly released into the PDB reached another record high of 14,031 during 2020. Among these newly deposited structures, more than are 1,000 SARS‐CoV‐2 (causative agent of the COVID‐19 pandemic) related protein structures, reflecting enormous efforts made by the structural biology community to understand and fight the pandemic. A comprehensive enumeration of freely available SARS‐CoV‐2 related resources provided by RCSB PDB can be found at rcsb.org/covid19.
FIGURE 1.
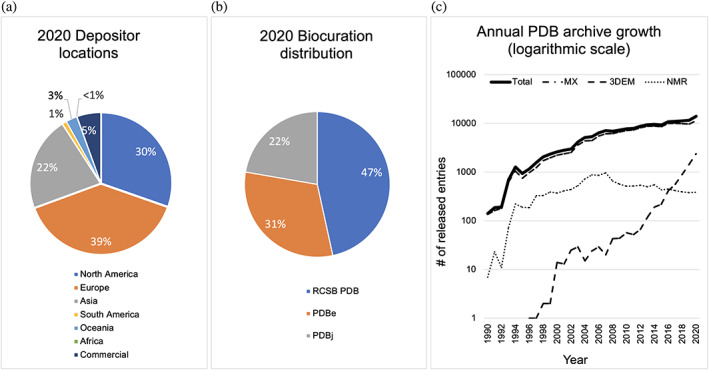
PDB data deposition and release metrics. (a) Depositor geographic locations in 2020. (b) Structure deposition processing by wwPDB regional data centers in 2020. (c) Annual rates of PDB archive growth (logarithmic scale) for 3DEM (dashed line), NMR (dotted line), MX (dashed‐dotted line), and all methods (total, solid line)
Figure 1c illustrates annual growth of the PDB archive broken down by experimental method. 3DEM and related technologies continue to evolve rapidly bringing new capabilities to the structural biology community. 3DEM structures in the archive increased by ~60% between 2019 and 2020 (from 1,452 to 2,390). Equally impressive, 3DEM median structure resolution limits improved from 9 Å in 2010 to 3.5 Å in 2020. microED structure depositions have also increased with median resolution limits improving in recent years (from 8 Å to 1.2 Å). New NMR structures released annually have plateaued at ~400 over the past few years. Notwithstanding rumors of the imminent demise of protein crystallography, the number of newly deposited MX structures continues to grow annually. The much‐heralded success of Google DeepMind AlphaFold 2 18 for protein structure prediction will only increase the efficiency of experimental structural biologists, yielding more PDB depositions of protein–ligand complexes and large multi‐protein assemblies, neither of which can be predicted today at accuracy levels comparable to experiment.
3. PDB ARCHIVE MANAGEMENT
Calendar year 2020 saw considerable improvements in the integrity of PDB archive management and archival data content. This section presents recent advances in data architecture, data remediation, and data integration.
3.1. PDB data hierarchy
Data stored in the PDB archive are categorized according to the following definitions:
Entry: All data pertaining to a particular structure deposited into the PDB constitute an archival Entry, identified with a unique PDB ID (currently four alphanumeric characters, e.g., 1q2w).
-
Entity: Each chemically unique molecule constituting a PDB Entry is defined as an Entity (including Polymer, Branched, or Non‐polymer), and labeled with a numeric Entity ID.
- Polymer Entities are composed of smaller chemical building blocks linked together by covalent bonds (e.g., proteins or polypeptides, DNA or polydeoxyribonucleotides, RNA or polyribonucleotides), which are identified by individually numbered amino acids or nucleotides covalently linked in the order defined by the polymer sequence.
- Branched Entities are either linear or branched carbohydrates and are composed of saccharide units covalently linked via one or more glycosidic bonds.
- Non‐polymer Entities are small chemicals (enzyme cofactors, ligands, water molecules, etc.). Every Non‐polymer Entity has a unique wwPDB Chemical Component Dictionary (CCD) 19 ID (one to three character alphanumeric code). The CCD provides nomenclature standards and chemical descriptions for all small‐chemical ligands and biopolymer components represented in the PDB archive.
N.B.: Every PDB Entry contains at least one Polymer Entity or one Branched Entity (either linear or branched oligosaccharides).
- Instance: There can be multiple Instances of any particular Entity within a PDB structure.
- Each Instance or “copy” of a Polymer Entity is labeled with a unique Chain ID (one or more alphanumeric characters, e.g., A, AA, …).
- Each Instance of a Branched Entity is similarly labeled with a unique Chain ID.
- Each Non‐polymer Entity is identified with the Chain ID of the spatially nearest Polymer Entity. Their Instances are distinguished with unique numbering.
Assembly: Polymer Entity Instances (or Chains) commonly occur in nature as components of larger macromolecular Assemblies, ranging in size and complexity. Each Assembly in a PDB structure is assigned a unique numeric Assembly ID.
3.2. Recent PDBx/mmCIF data standard improvements
The semantic foundation for PDB data architecture is defined in the PDBx/mmCIF dictionary. 10 , 20 PDBx/mmCIF is the macromolecular extension of an earlier community data standard, the Crystallization Information Framework (cif.iucr.org), 21 developed under the auspices of the International Union of Crystallography for description of small molecule X‐ray diffraction studies.
The PDBx/mmCIF data standard is maintained by the wwPDB organization in collaboration with wwPDB PDBx/mmCIF Working Group domain experts recruited from the scientific community (hereafter Working Group, wwpdb.org/task/mmcif). Content dictionaries and Working Group discussions are hosted on the GitHub platform (github.com/pdbxmmcifwg). The PDBx/mmCIF web resource (mmcif.wwpdb.org) supports browse and search access to standard terminology. Because the Working Group includes developers for many of the widely used structure determination software systems, this group plays a vital role in ensuring that data produced by these programs comply with the PDBx/mmCIF data standard, generating complete and correct data files for PDB deposition.
The wwPDB and the Working Group collaborate on developing terminologies for new and rapidly evolving methodologies (e.g., X‐ray Free Electron Laser Serial Crystallography and 3DEM), and improving representations for existing data content (e.g., carbohydrate remediation). Most recently, the Working Group has focused on modernizing content descriptions for processed X‐ray diffraction data, including extensions describing anisotropic diffraction limits, unmerged reflection data, and new quality metrics of anomalous diffraction data (wwpdb.org/news/news?year=2021#60638da1931d5660393084c3). Deposition and delivery of this extended content will significantly enhance our ability to assess experimental data quality, thereby improving every PDB data consumer's ability to Find and Reuse relevant PDB Entries.
3.3. Recent RCSB PDB data architecture improvements
In 2020, RCSB PDB launched a significant upgrade of its data delivery service delivery architecture 12 at RCSB.org. 5 This web resource upgrade transformed a legacy monolithic data delivery application into a distributed deployment of individual microservices, each with a single responsibility. Within the new architecture, data access services (data.rcsb.org) provide both Representational State Transfer (REST) and GraphQL (graphql.org) API access to a data warehouse hosted in a MongoDB document‐oriented database (mongodb.com). In the initial release, Advanced Search Query Builder functionality encompassed text, PDB data attributes, 3D structure, sequence, biopolymer sequence motif, and chemical similarity. Every search function is implemented as an independent service. A separate search aggregation service is responsible for launching each search function, and combining and delivering their integrated search results to front‐end services and public programmatic search APIs (search.rcsb.org). Various operational benefits accrue from reliance on a data architecture in which each service has a single responsibility, notably greater flexibility in scaling the deployment of services in response to changes in user load and significant reductions in the time required to develop, test, and deploy new features.
Since re‐launch of our RCSB.org web portal, we have continued to develop the new data architecture and augment website features. The Sequence Motif search function has been extended with a new 3D Structure Motif search capability. 22 Our Chemical Search function has also been extended with the ability to perform exhaustive substructure searching (eyesopen.com) across the small molecules represented in the PDB archive. Both of these new search services are also managed by the Search Aggregator service and available through our public search APIs. An important feature of the Search Aggregator is the capability to deliver search results at different levels of molecular granularity. For example, in our first deployment, search results could be shown as either deposited structure Entries, Assemblies, or distinct Polymer Entities. These search result types have been augmented to include distinct Non‐polymer molecular constituents, plus the chemical and molecular definitions from the CCD.
To monitor the new service architecture, we recently developed a system for processing service logs and indexing the time course of successful and failed access requests, while respecting PDB data consumer privacy. These statistics are essential for monitoring the health of RCSB PDB production services. They also provide simple analytic information, such as data file downloads or service access. The new metrics provide a rigorous basis for refining and extending our new search and data access services to enable greater FAIRness. This work built atop widely used open‐source telemetry tools (e.g., ElasticSearch, Kibana, Metricbeat, and Filebeat), 23 using the open‐source Elastic Common Schema Standard 24 for encoding log data. N.B.: Historically, when open source tools have become unavailable they were replaced with other open source or low‐cost/no‐cost proprietary tools.
3.4. Ongoing PDB data remediation
As PDB holdings grow in size and related science and technology evolve, 3D structures in the archive require ongoing improvements in representation and remediation to ensure consistency, accuracy, and high overall quality. Routine remediation efforts help ensure that PDB data are FAIR. The wwPDB regularly reviews data processing procedures and coordinates remediation efforts. In 2020, routine remediation efforts focused on the PDBx/mmCIF dictionary, the CCD, and 3D structure files. Various improvements were made to the PDBx/mmCIF dictionary, including introduction of new controlled vocabularies and encoding of hard limits for certain metadata items (particularly 3DEM data). Changes to the PDBx/mmCIF dictionary necessitated remediation of select data files pertaining to individual 3D structures in the archive. Within the CCD, ~5,000 ligand synonyms were re‐organized into a new PDBx/mmCIF category, _pdbx_chem_comp_synonyms, with source provenance provided in machine readable table format. Finally, ~700 SARS‐CoV‐2 protein structures were standardized for macromolecular names, taxonomy identifiers, and Enzyme Classification numbers once relevant UniProt (uniprot.org) reference information became available.
3.5. PDB carbohydrate data remediation
In addition to ongoing remediation efforts, a major remediation project focused on carbohydrates was recently completed by the wwPDB. 25 Understanding carbohydrate structure and organization is critical to comprehending their biological roles in fundamental biology, human health and disease, and bioenergy. Previously, representation of carbohydrates in the PDB was not uniform. The PDBx/mmCIF data standard did not fully reflect the complex nature of carbohydrates, which exhibit stereo‐isomers, anomeric configurations, and branched chains. As a result, glycoscientists and other experts in the carbohydrate community were unable to fully utilize the rich structural information available for glycans and glycoprotein structures archived in the PDB.
Over the past 5 years, wwPDB partners worked with expert members of the glycoscience community, the Working Group, and other key stakeholders to develop a new data representation for carbohydrates. Carbohydrate‐specific annotation and validation tools were developed and implemented within OneDep. 15 These new software tools provide standard nomenclature and consistent oligosaccharide representation that can be easily translated to other representations commonly used by glycobiologists.
wwPDB carbohydrate remediation involved the following technical developments and new features:
Standardized sugar nomenclatures for the CCD, following IUPAC‐IUBMB recommendations (qmul.ac.uk/sbcs/iubmb/);
Uniform Branched Entity representation for oligosaccharides;
Linear descriptors commonly used by the glycoscience‐community;
Annotated glycosylation sites within PDB structures;
Improved carbohydrate structure validation using the Branched Entity representation; and
Two‐dimensional (2D) and 3DSymbol Nomenclature for Glycans (SNFG) representation.
Remediation of carbohydrates in the PDB enabled development of an entirely new search system for glycoproteins and non‐covalently bound carbohydrates, which is described below in the section entitled “Improved Searching for Carbohydrates.”
3.6. Recent advances in RCSB PDB data integration
To make PDB data more Findable and Interoperable, RCSB PDB integrates the content of each expertly biocurated Entry with information from more than 50 external data resources (rcsb.org/docs/general-help/data-from-external-resources-integrated-into-rcsb-pdb). Items of external data content are integrated into a data schema that defines the organization of the RCSB PDB data warehouse. External data integration represents an integral part of the weekly new Entry release workflow, which manages loading of new data into the PDB archive and our data warehouse. From there, it becomes available to RCSB PDB front‐end services, public data access APIs, and our text search indexing service. 26 In response to community input, we have continued to integrate new trusted external data resources (Table 1).
TABLE 1.
Trusted external data resources/data content recently integrated within the RCSB.org web portal
| Data content type | Resource name |
|---|---|
| Protein domain structure annotations | Structural classification of proteins (SCOP2) 27 |
| Evolution‐related annotations | Evolutionary classification of protein domains (ECOD) 28 |
| Antibiotic resistance annotations | Comprehensive antibiotic resistance database (CARD) 29 |
| Immunology‐related annotations | International ImMunoGeneTics information system (IMGT) |
| Structural antibody database (SAbDab) 30 | |
| Small molecule correspondences and annotations | Cambridge structural database (CSD) 31 |
| Crystallographic open database (COD) 32 | |
| Chemical entities of biological interest (ChEBI) 33 | |
| ChEMBL 34 | |
| NCBI PubChem 35 | |
| Glycan correspondences and annotations | GlyGen 36 |
| GlyTouCan 37 | |
| GlyCosmos 38 | |
| Membrane protein annotations | Database of membrane proteins embedded in lipid bilayers (MemProtMD) 39 |
| Protein data bank of transmembrane proteins (PDBTM) 40 | |
| Orientations of proteins in membranes database (OPM) 41 |
4. NEW RCSB PDB WEB PORTAL TOOLS
In April 2020, following a multi‐year effort, the RCSB PDB released an extensively revised version of its research‐focused RCSB.org web portal. 5 This section presents new web portal features and tools introduced in 2021.
4.1. Mol* molecular graphics visualization
The most common use of PDB data is to visualize or “see” the 3D shapes and interactions of biological macromolecules in order to understand biochemical and biological function. In 2020, Mol* 42 was deployed as the default RCSB.org molecular graphics tool for visualizing macromolecules, carbohydrates, and small‐molecule ligands. This open‐source molecular graphics software system was developed as a community project, co‐led by RCSB PDB and the Protein Data Bank in Europe (PDBe; PDBe.org). Mol* is a web‐native graphics tool for interrogating 3D macromolecular structure data from the PDB or computed structure models. It works entirely within the PDB data consumer's internet browser, obviating the need to download, install, or maintain any external software. Importantly, Mol* supports integration of information from other bioinformatics resources to provide new insights (e.g., about active site amino acids, known mutations, locations of post‐translational modifications). In turn, these insights can help develop new hypotheses for research and facilitate analysis and/or interpretation of observations and experimental results.
Mol* is a versatile tool driven by an intuitive graphical user interface (GUI). It enables users to easily visualize entire polypeptide or nucleic acid chains, whole biological Assemblies (some including millions of non‐hydrogen atoms), or specific atoms or groups of atoms in a particular biological macromolecule. With a few mouse clicks, it can present 3D structure data in a variety of commonly used molecular representational styles. It can also display molecular surfaces, and non‐covalent interactions with bound ligands, ions, drugs, and inhibitors. The GUI enables rapid display of specific biomolecular features, comparison of related structures, and launch of archive‐wide queries for Instances containing specified 3D structure motifs of amino acids or nucleotides (see below).
We exemplify some new Mol* features in Figure 2a using a 3DEM structure of SARS‐CoV‐2 spike protein (PDB ID 6vxx). 43 The spike protein interacts with its human cell‐surface receptor, angiotensin converting enzyme 2 or ACE2, to facilitate cellular invasion/infection. It is the target of antibodies and T‐cells produced by the host immune system in response to viral infection or vaccination. Various engineered anti‐spike protein antibodies have received emergency use authorization for treatment of mild to moderate COVID‐19 infections in adults and pediatric patients (e.g., bamlanivimab and etesevimab, Eli Lilly and Co.). Disturbingly, substitutions that change only a few amino acids on the spike protein yielded more transmissible SARS‐CoV‐2 variants that in turn spread rapidly around the world. Such variants have put the enormous number of individuals not yet fully vaccinated and the smaller, but not insignificant, number of individuals who cannot be vaccinated (for medical or religious reasons) at risk of serious illness requiring hospitalization or death.
FIGURE 2.
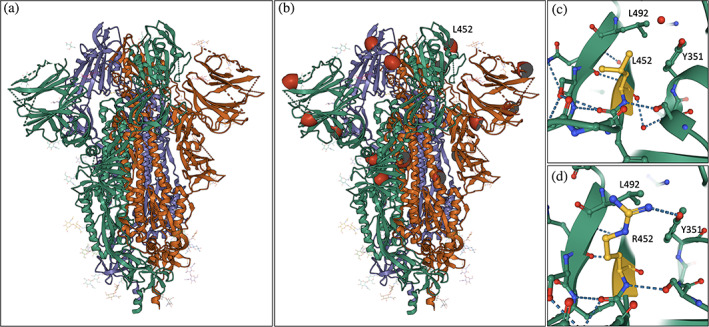
Exploring PDB structures of spike proteins from SARS‐CoV‐2 variants. (a) Colored ribbon drawing representation of the spike protein trimer (individual polypeptide chains are depicted with different colors, and attached carbohydrates are depicted as atomic stick figures; PDB ID 6vxx). 43 (b) Locations of selected substitutions seen in the Delta variant (T95, G142, A222, L417, L452‐labeled, and D950) are indicated with red hemispheres. (c) Close up view of L452 in the structure of the ACE2‐binding domain of the original viral isolate (PDB ID 7ora). 44 (d) Close up view of R452 in a structure of the R452 variant (PDB ID 7orb). 44 (Atom color coding: C‐green or dark yellow; N‐blue; O‐red)
Locations of some of the substitutions seen in the SARS‐CoV‐2 spike protein of the B.1.617.2 or Delta “variant of concern” designated by the US Centers for Disease Control and Prevention (cdc.gov/coronavirus/2019-ncov/variants/variant-info.html) are marked with red hemispheres in Figure 2b. The full tally of Delta variant substitutions includes T19R, V70F, T95I, G142D, E156‐, F157‐, R158G, A222V, W258L, K417N, L452R, T478K, D614G, P681R, and D950 (where “–” indicates an amino acid deletion) (cdc.gov/coronavirus/2019-ncov/variants/variantinfo.html). One particular amino acid substitution at position 452 of the spike protein that changes a leucine to an arginine (L452R) has been detected in more than five SARS‐CoV‐2 variants. Mol* enables quick and easy examination of differences in molecular interactions for native versus variant spike protein structures (Figure 2c,d). Similar analysis of key inter‐ and intra‐molecular interactions of different viral variants can facilitate both basic research and design of new diagnostic tools and therapeutic interventions.
4.2. Improved searching for carbohydrates
New tools available on the RCSB.org web portal enable access, analysis, and visualization of the recently remediated PDB structures containing carbohydrates. Simple text‐based searches for carbohydrates can be performed using the main search bar located at the top of every RCSB.org webpage by entering the name of a component sugar (monosaccharide) or oligosaccharide, such as “sucrose,” “FRU,” “heparin,” “cellulose,” etc. Such text‐based searches return lists of Entries that include the carbohydrate and Entries related to the carbohydrate. PDB data consumers can narrow the search further using the “Unique Ligands” and “Unique branched monosaccharides” fields in the search result list to identify relevant PDB archival information.
The full impact of the carbohydrate remediation is apparent when PDB data consumers take advantage of the “Advanced Search Query Builder” capabilities. Examples of the power of RCSB PDB Advanced Search for carbohydrates are listed below:
Monosaccharides can be searched using the “Chemical Components” category, as for any other ligand;
Oligosaccharides can be searched using the “Oligosaccharide/Branched Molecular Features” category, including options for branch type, sugar components, and common oligosaccharide descriptors; and
Glycosylation annotation can be accessed at the “Polymer Molecular Features” category, enabling search for a specific glycosylation type or for all Entries containing glycosylated proteins.
Ligand Summary Pages from Chemical search results utilize standardized monosaccharide descriptors, IUPAC‐IUBMB nomenclature, and well‐organized synonyms. All PDB structures containing a queried monosaccharide are grouped into one or more of three categories, including non‐covalently bound, covalently linked to proteins (glycosylation), and branched oligosaccharide, with total counts for each category provided. Users can access the sub‐divided list of searched structures by simply clicking on the count for any of the three categories.
Inclusion of glycoscience community‐developed linear descriptors during the carbohydrate remediation enabled use of graphical tools to translate them into 2D and 3D SNFG representation, which is supports efficient visualization of glycan structures.
Tabular Reports for Advanced Search results include an oligosaccharide summary report, which contains PDB ID and Entity identifiers, glycosylation type, 2D SNFG image, linear descriptors, standardized molecular name, and a list of unique monosaccharide components. PDB data consumers can download these reports in CSV or JSON formats for further analysis.
Every PDB structure is described in detail on a corresponding Structure Summary Page (SSP) on RCSB.org. The new “Oligosaccharides” section of the SSP includes a brief summary of information about oligosaccharides occurring in the structure. Use of this feature is exemplified in Figure 3, showcasing a 3DEM structure of the SARS‐CoV‐2 spike protein bearing the common D614G substitution (PDB ID 7krr). 45 Figure 3a shows the Oligosaccharides section of the SSP for PDB ID 7krr, with the full name of the Branched Entity, Chain IDs, Chain Length, 2D diagram of the Branched Entity (in this case a linear oligosaccharide), and the type of Glycosylation (in this case N‐linked). Clicking the Oligosaccharides Interaction button positioned on the far right of the section (under 3D Interactions) automatically generates the Mol* visualization depicted in Figure 3b. This close up view of the N‐linked disaccharide reveals its location within the 3D structure near the interface between two spike protein chains (colored green and orange). The carbohydrate is depicted using 3D SNFG representation. In addition to branched oligosaccharide representation and search capabilities available on RCSB.org, links are provided to external glycosylation resources, including GlyTouCan (glytoucan.org), 46 GlyCosmos (glycosmos.org), 38 and GlyGen (glygen.org) 36 wherever possible. These links provide PDB data consumers with access to external data for each Branched Entity for further exploration of the glycans themselves.
FIGURE 3.
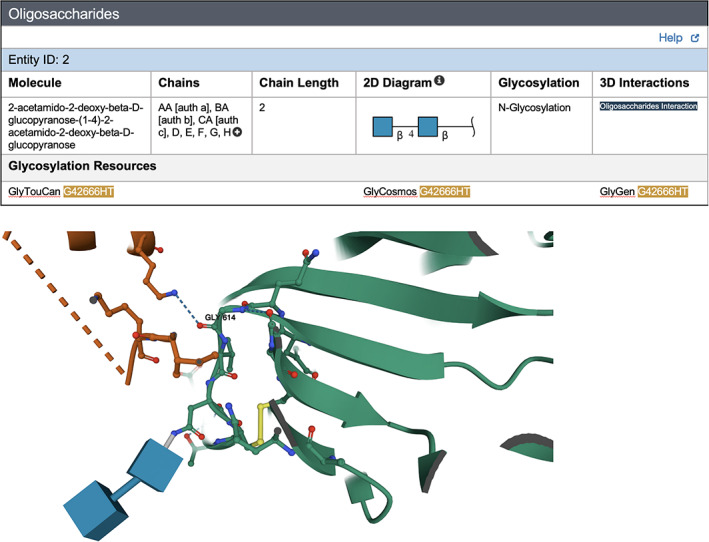
RCSB.org web portal exploration of a glycosylated form of SARS‐CoV‐2 spike protein D614G variant (PDB ID 7krr). 45 (a) Oligosaccharide section of the SSP for PDB ID 7krr. (b) Mol* 3D interaction view of the corresponding oligosaccharide in 3D SNFG representation (two blue cubes) at a glycosylation site in the vicinity of the location of a common amino acid substitution (residue 614). The protein is shown with green ribbon representation. Amino acid G614 and nearby residues are shown in ball‐and‐stick representation. (Atom color coding: C‐green or orange, denoting the neighboring spike protein; N‐blue; O‐red; S‐yellow)
4.3. Structure motif search with the Mol* graphical user Interface
Structural motifs are characteristic three‐dimensional arrangements of amino acid residues that frequently contribute to biochemical or biological function. 47 Perhaps the best characterized structure motif is the Ser‐His‐Asp catalytic triad common to phylogenetically related serine proteases (reviewed in 48 ). Comparable structural motifs also occur in phylogenetically unrelated enzymes that catalyze chemically similar reactions. Such cases are often referred to as products of convergent evolution. Support for Structure Motif searching was recently added to the RCSB.org web portal. 22 PDB data consumers can input a Structure Motif search within the RCSB.org Advanced Search Query Builder. But the easiest way to do so is to use the Mol* GUI to select relevant amino acid residues (or nucleotides) from within the displayed biopolymer sequence or the 3D structure itself.
Our new Structure Motif search tool is exemplified in Figure 4, showcasing the first structure of the SARS‐CoV‐2 main protease (PDB ID 6lu7). 49 Figure 4a illustrates the Mol* window ready to launch a three‐residue Structure Motif search based on three residues lining the main protease active site (His:A‐41, Cys:A‐145, and Glu:A‐166). Clicking “Submit Search” in the “Structure Motif Search” section of the control panel triggers the search. Figure 4b shows how to launch the same search from the Advanced Search Query Builder (clicking the “magnifying glass” icon triggers the search). Using the Mol* GUI reduces the number of mouse clicks and input text data required versus the Advanced Search Query Builder. Either means of launching Structure Motif searches invokes the same API call, which can also be accessed programmatically. Because query amino acids may be present in more than one Polymer Entity, search results are presented as Assemblies. On July 7, 2021, search execution identified 1,523 assemblies in the PDB archive containing similar arrangements of the three residues. Approximately one third (577) of the search hits correspond to structures of riboviria proteins, most of which are other SARS‐CoV‐2 main protease structures containing different bound ligands. Approximately 90 of the riboviria search hits correspond to structures of the SARS‐CoV main protease, which was extensively studied following the 2003 epidemic (e.g., PDB ID 1q2w, 50 His:B‐43, Cys:B‐147, Glu:B‐168).
FIGURE 4.
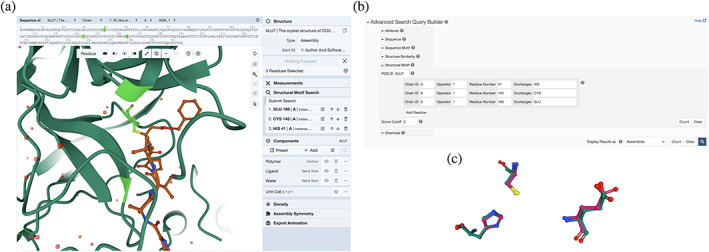
Structure motif search for SARS‐CoV‐2 main protease active site residues (PDB ID 6lu7). 49 (a) Query construction using the Mol* GUI. (b) Query construction using the Advanced Search Query Builder. (c) 3D visualization of a structure motif search hit using Mol*
Structure Motif search hits are initially listed in order of structural similarity versus the query structure (root‐mean‐square deviation or r.m.s.d. for equivalent non‐hydrogen atoms detected in the query). The same list can be reordered according to various criteria, as for any search result. Each search hit can be visualized in 3D by clicking on the “View” button. This functionality delivers a pairwise structure motif superposition of query and search hit residues using a quaternion‐based characteristic polynomial that generates the r.m.s.d. value. 51 The superposition is presented in a Mol* visualization window (Figure 4c), wherein the motifs of interest are shown in ball‐and‐stick representation. Surrounding structural features can be added to the view (using the Mol* controls visible on the right side of Figure 4a). The comparison shown in Figure 4c illustrates the striking similarity of the SARS‐CoV‐2 and SARS‐CoV main protease active site residues (r.m.s.d. ~ 0.4 Å). The phylogenetically‐related enzymes are ~96% identical in amino acid sequence, and extremely similar in overall 3D structure (r.m.s.d. ~ 0.7 Å for 295 equivalent ‐carbon atom pairs). Moreover, the amino acid sidechains lining the two active sites are chemically identical and virtually identical in 3D structure (r.m.s.d. <0.5 Å for non‐hydrogen atoms). 52 At the time of writing, SARS‐CoV‐2 main protease is the target of multiple structure‐guided drug discovery efforts aided by open access to more than 350 publicly disclosed PDB structures. Multiple small‐molecule main protease inhibitors have entered phase 1 clinical trials (clinical trials.gov), including PF‐07304814 (intravenous dosing) 53 and PF‐07321332 (oral dosing). 54
All Structure Motif search hits share geometric similarity but may not be biologically relevant, particularly those with high r.m.s.d. values. To aid identification of true positives, we enabled filtering of Structure Motif search hit r.m.s.d. values <1 Å for hits meriting closer inspection. For the riboviria search hits described above, 472 of the 577 that gave r.m.s.d. values <1 Å corresponded primarily to SARS‐CoV‐2, SARS‐CoV, and other coronavirus main protease structures in the PDB archive.
4.4. Pairwise structure comparison and alignment
Our newly‐developed Pairwise Structure Comparison tool generates alignments of 3D structures. This feature can be accessed from any RCSB.org web page by mousing over the Analyze tab at the top of the page and clicking on “Pairwise Structure Alignment” to reach the rcsb.org/alignment webpage. Once there, the PDB data consumer can provide either a PDB ID or upload a file in PDBx/mmCIF or legacy PDB format and then select Chain IDs (for each of the molecules to be aligned). Alignments are generated in real time using a web service available at alignment.rcsb.org. Currently supported structure comparison methods include rigid body, flexible, and topology‐independent alignments employing Java implementations of well‐established tools (e.g., Combinatorial Extension or CE, 55 CE with Circular Permutations, 56 FATCAT, 57 TM‐align, 58 and superposition guided by Smith‐Waterman sequence alignment 59 ) provided by the BioJava project. 60 The results page displays the superposed structures interactively in 3D using the Mol* viewer, together with a 1D sequence alignment corresponding to the 3D structure alignment. For both sequence and structure alignments, individual macromolecules are highlighted in orange and blue, respectively. Aligned regions appear in a darker hue than non‐aligned regions. Additionally, Mol* provides various presets for visualizing aligned residues, chains, or full content of the PDB Entry including all biopolymer chains, ligands, and solvent molecules.
Use of this new feature is exemplified in Figure 5, illustrating alignment results for spike protein structures from SARS‐CoV (PDB ID 5x5b, 61 Chain A) and SARS‐CoV‐2 (PDB ID 6vsb, 62 Chain A). 3D structure superposition of these two phylogenetically‐related proteins reveals r.m.s.d. ~ 1.5 Å for 917 equivalent ‐carbon atom pairs (~78% amino acid sequence identity). The SARS‐CoV spike protein structure was made public in 2017 and was freely available from the PDB with no restrictions on usage to designers of SARS‐CoV‐2 mRNA vaccines in early 2020 (reviewed in 63 ). Various results can be downloaded, including the aligned sequences in FASTA format and PDBx/mmCIF files for the superposed atomic coordinates.
FIGURE 5.
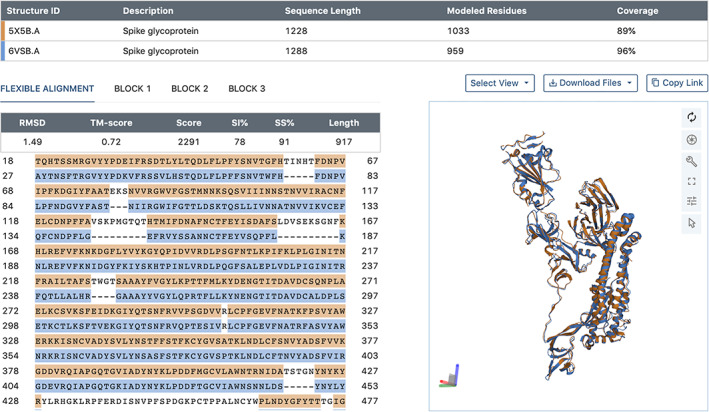
jFATCAT‐flexible 3D comparison and alignment of SARS‐CoV (PDB ID 5x5b, 61 Chain A; orange) with SARS‐CoV‐2 (PDB ID 6vsb, 62 Chain A; blue) spike protein structures
4.5. New tools for understanding and visualizing membrane proteins
Phospholipid bilayer membranes envelope cells and cellular organelles. Membrane proteins of various types can be found embedded in or closely associated with biological membranes. There, they play critical roles in cell survival, cell communication, transport of solutes and proteins across bilayers, and endo‐ and exocytosis. They are also the targets of more than 50% of US Food and Drug Administration (FDA) approved drugs (proteinatlas.org/humanproteome/tissue/druggable). Enveloped viruses, including SARS‐CoV‐2, utilize lipid bilayers to package and protect their nucleic acid genomes. The coronaviridae, of which SARS‐CoV‐2 is a member, typically possess four structural proteins that make the virion together with the RNA genome and the lipid bilayer. With the exception of the nucleocapsid protein, the spike protein (Figures 2, 3, 4), the M protein, and the E protein (Figure 6) are integral membrane proteins.
FIGURE 6.
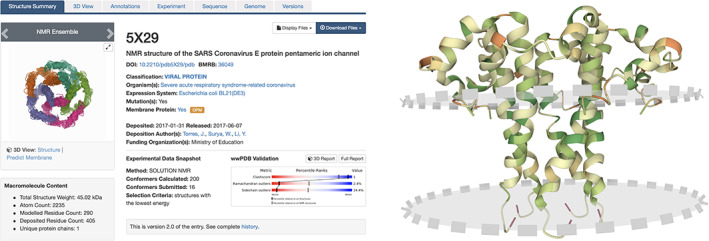
SSP for a SARS‐CoV E protein (PDB ID 5x29) 66 (left) with a view of the homopentamer looking down the molecular pore and Mol* visualization with predicted membrane location depicted using pink circles with dashed grey border (right). Each E protein monomer is shown in ribbon representation (color coded by hydrophobicity: dark green hydrophobic, dark red polar) and viewed nearly parallel to the plane of the membrane bilayer with the extracellular portion of each monomer in the upper portion of the image
Annotations of membrane proteins enhance the value of structural information housed in the PDB archive. Until recently, the RCSB.org web portal included only external reference data for membrane proteins from the mpstruc database. 64 RCSB.org users now have access to membrane protein‐related annotations from the MemProtMD, 39 PDBTM, 65 and OPM 41 resources. Integration of PDB structure data with these additional trusted external resources has nearly doubled the number of PDB Entries annotated as membrane proteins. In parallel, we substantially improved the functionality of the RCSB.org web portal by supporting user access to membrane protein annotations and visualization of related data at the levels of 1D sequence and 3D structure.
These improvements are exemplified in Figure 6 with the SARS‐CoV E protein (PDB ID 5x29), 66 which is a homopentameric viroporin and part of the virus particle itself. It is essential for viral replication within infected cells. 66 The RCSB.org SSP for PDB ID 5x29 is depicted in Figure 6 (left). The orange OPM button, located immediately to the right of the small graphic of the pentamer, links to the E protein webpage at the OPM database web portal. The “Predict Membrane” link located under the image on the SSP (Figure 6, left) launches a Mol* visualization window showing the homopentameric ion channel formed by the hydrophobic, transmembrane helix of the E protein with two translucent circular planes depicting the predicted positions of the leaflets of the membrane bilayer (Figure 6, right). Prediction of membrane locations utilizes the ANVIL algorithm. 67 The SSP Sequence tab (visible in Figure 6, left) provides information about the boundaries of the transmembrane segment in the Protein Feature View. 68 The OPM database employs a custom hierarchy (accessible from the SSP Annotations tab, Figure 6, left). Therein, SARS‐CoV E Protein superfamily are classified as “Bitopic viroporins” and “Coronavirus E protein,” respectively. RCSB PDB search infrastructure 12 supports searching for Polymer Entities sharing these terms (e.g., by clicking the links provided in the SSP Annotations tab).
4.6. Improved chemical component data searching
The previously introduced RCSB PDB Advanced Search Query Builder provides powerful options for constructing complex searches and managing results. The new Chemical Attribute option illustrated in Figure 7 upper (red box) enables searching of chemical reference data that describe small molecules defined in the CCD and molecules defined the Biologically Interesting molecule Reference Dictionary (BIRD). 69 Approximately one third of the BIRD molecules are also represented in the CCD, and have both CCD IDs and BIRD IDs (format PRD_######). Remaining BIRD molecules are only identified using PRD_######.
FIGURE 7.
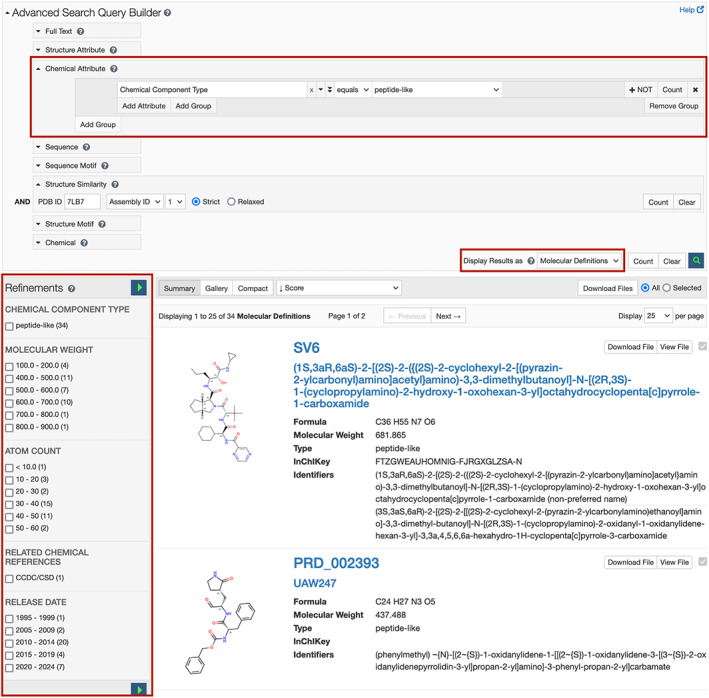
Chemical Attribute searching from the Advanced Search Query Builder. The executed. Search (upper) identified 33 peptide‐like ligands similar to ligand PRD_002214 occurring in PDB ID 7lb7 70 (lower). N.B.: Result count from search includes ligand PRD_002214. Search results can be narrowed by selecting from the “Refinements” menu (lower left, red box)
The “Display Results as” menu in the lower right corner of the Advanced Search Query Builder manages how search results are presented, as shown in Figure 7 middle right (red box). A new option for “Molecular Definitions” returns a unique list of chemical definitions matching the search criteria. The search results area displays name and formula of each small molecule together with a link to the ligand summary page with detailed descriptions of each chemical component. Molecular definition search results (Figure 7, lower) can be further refined using the Refinements panel shown in Figure 7 lower (left, red box), which features categories relevant to chemical reference, data such as molecular weight, atom count, component type, etc. Resulting molecular definitions can be exported in .sdf and .mol2 file formats.
N.B.: The Chemical Attribute search option can be combined readily with macromolecular data and chemical structure searches.
4.7. 3D visualization of bound ligands with Mol*
Once a PDB structure containing a relevant ligand has been identified, Mol* supports visualization of (i) the spatial location and environment of the ligand; (ii) how the ligand interacts with macromolecule(s) and other small molecule ligands in the structure; and (iii) how well the experimental data support the positioning of the ligand. For large PDB structures and those containing multiple ligands, the easiest way to examine the ligand structure and the immediate environment of the ligand is to click the “Ligand Interactions” button within the Small Molecules section of the SSP for each ligand. Doing so automatically opens a Mol* visualization window centered on the selected ligand, which displays any non‐covalent interactions with the ligand (within 5 Å). The ligand is denoted with a halo, while any covalent and/or noncovalent interactions involving the ligand are shown as dashed lines. Various types of non‐covalent interactions can be selectively displayed and/or hidden by clicking on the Options icon (located to right of “+ ADD” in the Control Panel) and expanding the “Non‐covalent Interactions” options. For MX structures, the experimental electron density map can be visualized by clicking the “Density” button in the Control panel and enabling the option. Finally, the location of the ligand in the context of the entire PDB structure can be viewed by clicking on the Reset Camera button in the vertical menu of buttons on the top right corner of the white 3D canvas.
To explore binding of ligand/inhibitor TTT to the active site of SARS‐CoV‐2 papain‐like proteinase (PDB ID 7jir), 71 the “Ligand Interactions” button on the SSP opens a Mol* visualization window (Figure 8, left). The “Density” option in the Mol* Control Panel can be used to display experimental electron density maps and examine how well these data support the positioning of the ligand in the active site. Electron density maps can be displayed as semi‐transparent volumes or meshes by using options made available by clicking on the + icons in the Control Panel, next to the various maps displayed (e.g., 2Fo‐Fc, Fo‐Fc). Finally, selectively changing the coloring of carbon atoms in the ligand can make it easier to view and explore (Figure 8, right). PDB ID 7jir is one of more than 30 publicly‐disclosed structures of the SARS‐CoV‐2 papain‐like proteinase that are contributing to structure‐guided drug discovery efforts against this promising enzyme target.
FIGURE 8.
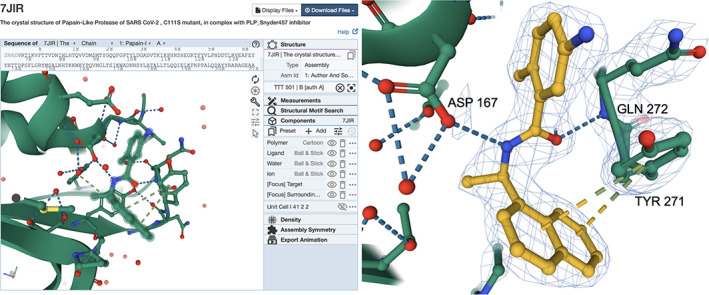
Examining the 3D structure of a SARS‐CoV‐2 papain‐like proteinase enzyme inhibitor (CCD ID TTT) in PDB ID 7jir. 71 (left) Mol* view generated by clicking on the “Ligand Interaction” button (right). Portions of the macromolecule in the neighborhood of the ligand/inhibitor are shown using ribbon representation (green), while residues participating in non‐covalent interactions within 5 Å of the ligand/inhibitor are shown in ball‐and‐stick representation with the ligand denoted by the presence of a light green halo surround. (Atom color coding: C‐green; N‐blue; O‐red; S‐light yellow.) Mol* view of ligand TTT, displaying a 2|Fobserved|‐|Fcalculated| difference electron density map as a mesh, contoured at 1.5σ. Carbon atoms of the ligand are colored dark yellow for ease of visualization. Non‐covalent interactions between the ligand and protein are highlighted with dashed lines. (Interaction color coding: Hydrogen bonds‐blue; π–π Interactions: dark yellow‐light green)
4.8. PDB ligand structure quality assessment
More than 70% of the macromolecular structures archived in the PDB include small‐molecule ligands. PDB structure depositors can designate one or more “Ligands of Interest” (LOI) within the wwPDB OneDep system. Such LOIs are typically the focus of the structural study (e.g., enzyme cofactors, inhibitors, and activators; substrate analogs; reaction intermediates and products; and their analogs). All designated LOIs are highlighted in wwPDB validation reports. A subset of small‐molecule ligands occurring in PDB structures include US FDA‐approved therapeutics and investigational agents. 72 Analyses of US FDA drug approvals between 2010 and 2018 revealed (i) that open access to PDB data facilitated discovery and development of the vast majority of these new drugs, 73 and (ii) that structure‐guided drug discovery has been particularly effective in generating new anti‐cancer agents. 74 , 75
Given the importance of co‐crystal structures of protein–ligand complexes for biological and biomedical research, ligand quality is extensively characterized by the OneDep validation software. 14 Structural biologists (~1% of PDB data consumers) should have little if any difficulty understanding overall structure quality and ligand quality information presented in wwPDB validation reports. Ligand quality metrics for co‐crystal structures include the local electron density goodness‐of‐fit indicators of real space R factor (RSR) 76 and real space correlation coefficient (RSCC) 77 ; chemical structure quality indicators of Root‐Mean‐Square deviation Z‐score for bond lengths (RMSZ‐bond‐length) and bond angles (RMSZ‐bond‐angle) generated by Mogul 78 ; and a measure of interatomic clashes computed using MolProbity. 79 For an LOI, recently incorporated visual displays provide additional at‐a‐glance summary information suitable for experts relating to ligand chemistry and goodness‐of‐fit between the atomic structure and the experimental data. 15
RCSB PDB recently introduced new tools to help all PDB data consumers (including individuals not expert in structural biology) easily understand ligand structure quality. 80 Figure 9 exemplifies the new RCSB.org ligand quality review feature for the co‐crystal structure of an inhibited form of the SARS‐CoV‐2 papain‐like proteinase (PDB ID 7jir) 71 in which an inhibitor (CCD ID TTT) was designated as an LOI during structure deposition. Figure 9 Upper Left schematically presents the quality of ligand TTT in PDB ID 7jir (compared to all ligands in the PDB archive) using a simplified 2D colored‐slider plot (horizontal axis: goodness‐of‐fit of the atomic structure of the ligand versus experimental data; vertical axis: geometry quality of the ligand 3D structure versus chemical reference bond lengths and bond angles). The fit of the inhibitor structure to the experimental data is better than ~86% of all PDB ligands and geometry quality is better than ~26% of all PDB ligands. The remaining 2D plots compare the quality of ligand TTT in PDB ID 7jir to the same ligand occurring in other PDB structures of the SARS‐CoV‐2 papain‐like proteinase (Figure 9, upper middle) and the same ligand in PDB structures of other proteins (Figure 9, upper right). With this information, PDB data consumers can easily discern that the ligand TTT structure in PDB ID 7jir has a superior fit to experimental data versus ~86% of ligands across the entire archive, while appreciating the fact the geometry quality of the ligand TTT structure in PDB ID 7jir is not the best example available in the archive. The 2D slider plots displayed on RCSB.org are interactive. A single mouse click on the ligand Instance symbol in the 2D plot launches a 3D view of the ligand comparable to that shown in Figure 8 (right). An accompanying tabular report presented in Figure 9 (lower) provides detailed statistics extracted from wwPDB validation reports for multiple Instances of ligand TTT.
FIGURE 9.
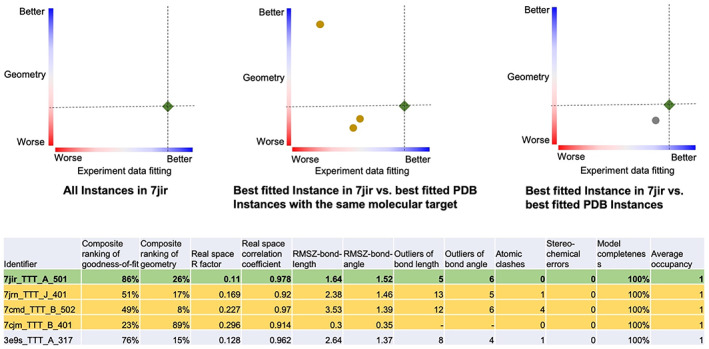
Understanding ligand TTT quality in five coronavirus papain‐like proteinase structures. (upper) Each 2D graph has color coded ranking scales from worst (0%, red) to best (100%, blue) for ligand experimental data fitting quality (horizontal axis) and ligand geometry quality (vertical axis). Each symbol represents an Instance of ligand TTT, showing experimental data fitting quality (horizontal) and denoting geometry quality (vertical). The green diamond symbol in each plot indicates the best‐fitted Instance of ligand TTT in PDB ID 7jir, 71 corresponding to the green‐highlighted row of the tabular report (lower), detailing ligand quality metrics and related information for each instance of ligand TTT. Other rows of the tabular report highlighted in yellow and gray correspond to the same‐color circle symbols in the Upper Middle and Upper Right 2D graphs
4.9. Chemical sketch tool
RCSB.org data consumers wishing to search the PDB archive for a particular small‐molecule ligand, or similar ligands, based on the 2D chemical drawing can now do so. No prior knowledge of chemical descriptors (e.g., SMILES, InChI) is required. The newly introduced Chemical Sketch Tool depicted in Figure 10 uses the ChemAxon Marvin JS web‐based chemical sketcher (generously provided at no charge to the RCSB PDB). RCSB.org users can quickly and conveniently draw 2D chemical structures from scratch, or load structures into the sketch tool using their chemical descriptors or CCD ligand codes. The Chemical Sketch Tool also supports editing the 2D chemical drawing as needed to automatically generate new chemical descriptors. Descriptors generated by this new tool can then be used to query the PDB archive for the specific ligand molecule, or similar compounds using various matching criteria. The descriptors can also be used with the RCSB.org Advanced Search Query Builder. SMILES or InChI descriptors are passed directly as input parameters, and may be combined with other attributes to perform complex queries across the PDB archive.
FIGURE 10.
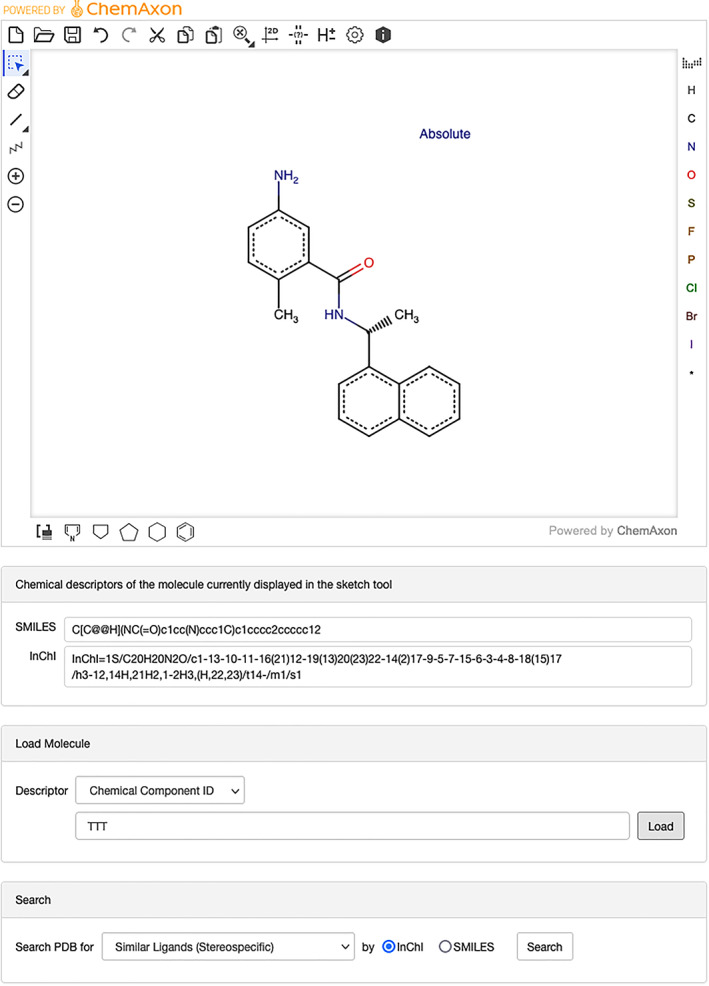
Use of the Chemical Sketch Tool exemplified with CCD ID TTT [5‐amino‐2‐methyl‐N‐[(1R)‐1‐naphthalen‐1‐ylethyl]benzamide]. The Search box at the bottom of the Chemical Sketch Tool page enables single click searching of the PDB archive using various search criteria with either InChI or SMILES chemical descriptors
4.10. Newly integrated annotations
On a weekly basis, annotations from more than 50 external data resources are integrated with every PDB structure. This process ensures that related information delivered to RCSB.org users together with each PDB structure is current. Unlike original publications describing PDB structures, RCSB.org serves as a living data resource that accommodates and integrates new research findings pertaining to each of the more than 180,000 structures archived in PDB. Five external data resources enumerated below were recently incorporated into the weekly update process.
Structural Classification of Proteins (SCOP2; scop.mrc‐lmb.cam.ac.uk): The SCOP2 database 27 aims to provide a detailed and comprehensive description of the structural and evolutionary relationships between proteins whose three‐dimensional structure is known and deposited in the PDB (Figure 11).
Evolutionary Classification of Protein Domains (ECOD; prodata.swmed.edu/ecod): ECOD 28 is a hierarchical classification of protein domains according to their evolutionary relationships. Only experimentally determined protein structures are currently classified in ECOD. Compared with other classification databases, such as SCOP and CATH, ECOD emphasizes on distant evolutionary relationships and updates every week to include new structures released in PDB.
International ImMunoGeneTics Information System (IMGT; imgt.org): The global reference in immune‐genetics and immune‐informatics IMGT is a high‐quality integrated knowledge resource specialized in the immunoglobulins or antibodies, T cell receptors, major histocompatibility proteins of human and other vertebrate species, and related immune system proteins of vertebrates and invertebrates (Figure 12).
Structural Antibody Database (SAbDab; opig.stats.ox.ac.uk/webapps/newsabdab/sabdab): SAbDab 30 contains all of the antibody structures represented in the PDB, biocurated and presented in a consistent fashion. Each structure is annotated with a number of properties including experimental details, antibody nomenclature (e.g., heavy‐light pairings), curated affinity data, and sequence annotations (Figure 12).
Comprehensive Antibiotic Resistance Database (CARD; card.mcmaster.ca/home): CARD 29 is a bioinformatic database of antibiotic resistance genes, their encoded protein sequences and associated phenotypes.
FIGURE 11.
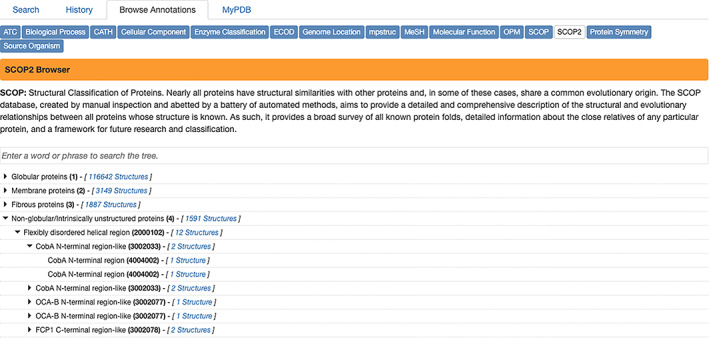
RCSB.org Browse Annotations page showing SCOP2 annotations integrated with large numbers of PDB structures
FIGURE 12.
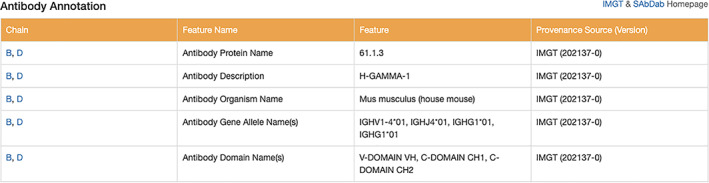
RCSB.org Structure Summary Annotations page showing immunology‐related annotations [IMGT and SAbDab] for PDB ID 1igy 81
These five external resources have been integrated into several existing RCSB.org web pages, including:
Structure Summary Annotations page (SCOP2, ECOD, IMGT and SAbDab, CARD);
Protein Feature View (ECOD); and
Browse Annotations page (SCOP2, ECOD).
5. DISCUSSION
The PDB archive recently celebrated its 50th anniversary, 1 and the RCSB PDB is currently in its 23rd year of continuous operations. Much has changed for the better since the inaugural RCSB PDB publication in 2000. 4 The PDB has grown enormously in size. Structures archived therein a larger and more complex. Many millions of PDB data consumers come to the RCSB.org web portal annually. Subsequent RCSB PDB and wwPDB publications have described our journey as an organization and as a resource. 5 , 8 , 9 , 10 , 12 , 14 , 15 , 16 , 17 , 19 , 20 , 22 , 25 , 42 , 68 , 69 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 89 , 90 , 91 , 92 Additional publications have summarized the impact of the RCSB PDB and the PDB archive on research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. 11 , 63 , 73 , 74 , 75 , 90 , 91 , 92 , 93 , 94 , 95 , 96 , 97 , 98
As the PDB archive enters its 51st year, it is widely appreciated that advances in basic and applied research depend critically on open access to the research findings of the scientific community. It should, and indeed must, be this way because the vast majority of 3D structure data were generated with public or private philanthropic support. Promulgation of the FAIR and FACT principles and the work of non‐governmental organizations such as the CoreTrustSeal (coretrustseal.org) are playing critical roles in raising awareness of the value of open sharing of data. 99 Equally important going forward will be sustainable funding for heavily used open‐access data resources, such as the PDB archive, at levels commensurate with the central roles they play in the global biological and biomedical research and education ecosystems. 100 , 101
AUTHOR CONTRIBUTIONS
Stephen Burley: Conceptualization (lead); funding acquisition (lead); project administration (lead); supervision (lead); writing – original draft (lead). Charmi Bhikadiya: Software (supporting). Chunxiao Bi: Software (supporting). Sebastian Bittrich: Software (supporting); visualization (equal). Li Chen: Software (supporting). Gregg V. Crichlow: Data curation (supporting); validation (supporting). Jose Manuel Duarte: Project administration (supporting); software (lead); supervision (supporting). Shuchismita Dutta: Visualization (equal); writing – review and editing (supporting). Maryam Fayazi: Software (supporting). Zukang Feng: Software (lead); supervision (supporting); validation (supporting). Justin W Flatt: Data curation (supporting); validation (supporting). Sai J Ganesan: Software (supporting). David S. Goodsell: Writing – review and editing (supporting). Sutapa Ghosh: Data curation (supporting); validation (supporting). Rachel Kramer Green: Project administration (supporting). Vladimir Guranovic: Software (lead). Jeremy Henry: Software (supporting). Brian P Hudson: Data curation (supporting); validation (supporting); writing – review and editing (supporting). Catherine L Lawson: Data curation (supporting); validation (supporting). YuHe Liang: Data curation (lead); validation (supporting). Robert Lowe: Software (lead); supervision (supporting). Ezra Peisach: Data curation (supporting); software (supporting); validation (supporting); writing – review and editing (supporting). Irina Persikova: Data curation (lead); validation (lead). Dennis W Piehl: Software (supporting). Yana Rose: Software (lead); visualization (equal); writing – review and editing (supporting). Andrej Sali: Project administration (lead). Joan Segura: Software (supporting). Monica sekharan: Data curation (supporting); validation (supporting). Chenghua Shao: Data curation (supporting); software (supporting); validation (supporting); visualization (equal). Brinda Vallat: Data curation (supporting); software (supporting); validation (supporting). Maria Voigt: Software (supporting). John D Westbrook: Software (lead); supervision (supporting); writing – review and editing (supporting). Shamara Whetstone: Project administration (supporting). Jasmine Young: Data curation (lead); supervision (supporting); validation (lead); visualization (equal); writing – review and editing (supporting). christine zardecki: Project administration (lead); supervision (supporting); writing – review and editing (supporting).
ACKNOWLEDGMENTS
The authors thank the tens of thousands of structural biologists who deposited structures to the PDB since 1971 and the many millions of researchers, educators, and students around the world who consume PDB data. We thank ChemAxon for making Marvin JS available at no cost for use by RCSB PDB. We also gratefully acknowledge contributions to the success of the PDB archive made by past members of RCSB PDB and our Worldwide Protein Data Bank partners (PDBe, PDBj, EMDB, and BMRB). RCSB PDB is jointly funded by the National Science Foundation (DBI‐1832184, PI: S.K. Burley), the US Department of Energy (DE‐SC0019749, PI: S.K. Burley), and the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National Institute of General Medical Sciences of the National Institutes of Health (R01GM133198, PI: S.K. Burley). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, et al. RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D . Protein Science. 2022;31:187–208. 10.1002/pro.4213
Dedicated to the memory of John D. Westbrook.
Funding information Division of Biological Infrastructure, Grant/Award Number: DBI‐1832184; National Cancer Institute, Grant/Award Number: R01GM133198; National Institute of Allergy and Infectious Diseases; National Institute of General Medical Sciences; U.S. Department of Energy, Grant/Award Number: DE‐SC0019749; National Science Foundation
REFERENCES
- 1. Protein Data Bank . Crystallography: Protein data bank. Nat New Biol. 1971;233:223–223.20480989 [Google Scholar]
- 2. Berman HM, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Biol. 2003;10:980. [DOI] [PubMed] [Google Scholar]
- 3. wwPDB consortium . Protein data bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The fair guiding principles for scientific data management and stewardship. Sci Data. 2016;3018:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Aalst WMP, Bichler M, Heinzl A. Responsible data science. Bus Inform Syst Eng. 2017;59:311–313. [Google Scholar]
- 8. Young JY, Westbrook JD, Feng Z, et al. Onedep: Unified wwPDB system for deposition, biocuration, and validation of macromolecular structures in the PDB archive. Structure. 2017;25:536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westbrook J, Henrick K, Ulrich EL, Berman HM. 3.6.2 the protein data bank exchange data dictionary. In: Hall SR, McMahon B, editors. International tables for crystallography. Dordrecht, The Netherlands: Springer, 2005; p. 195–198. [Google Scholar]
- 10. Fitzgerald PMD, Westbrook JD, Bourne PE, McMahon B, Watenpaugh KD, Berman HM. 4.5 macromolecular dictionary (mmCIF). In: Hall SR, McMahon B, editors. International tables for crystallography g definition and exchange of crystallographic data. Dordrecht, The Netherlands: Springer, 2005; p. 295–443. [Google Scholar]
- 11. Zardecki C, Dutta S, Goodsell DS, Voigt M, Burley SK. PDB‐101: Educational resources supporting molecular explorations through biology and medicine. Protein Sci. 31: in press. 10.1002/pro.4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rose Y, Duarte JM, Lowe R, et al. RCSB protein data bank: Architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. J Mol Biol. 2021;443:166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;43:1443–1444. [DOI] [PubMed] [Google Scholar]
- 14. Gore S, Sanz Garcia E, Hendrickx PMS, et al. Validation of structures in the protein data bank. Structure. 2017;25:1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng Z, Westbrook JD, Sala R, et al. Enhanced validation of small‐molecule ligands and carbohydrates in the protein databank. Structure. 2021;29:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young JY, Westbrook JD, Feng Z, et al. Worldwide protein data bank biocuration supporting open access to high‐quality 3D structural biology data. Database. 2018;2018:bay002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burley SK, Berman HM, Bhikadiya C, et al. RCSB protein data bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tunyasuvunakool K, Adler J, Wu Z, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westbrook JD, Shao C, Feng Z, Zhuravleva M, Velankar S, Young J. The chemical component dictionary: Complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the protein data bank. Bioinformatics. 2015;31:1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Westbrook J, Bourne PE. Star/mmCIF: An extensive ontology for macromolecular structure and beyond. Bioinformatics. 2000;16:159–168. [DOI] [PubMed] [Google Scholar]
- 21. Iucr cif specification v1.1. 2005. http://www.iucr.org/resources/cif/spec.
- 22. Bittrich S, Burley SK, Rose AS. Real‐time structural motif searching in proteins using an inverted index strategy. PLoS Comput Biol. 2020;16:e1008502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The ELK stack open source projects: Elasticsearch, Logstash, and Kibana. 2021. https://www.elastic.co/what-is/elk-stack.
- 24. Elastic common schema standard. 2021. https://www.elastic.co/guide/en/ecs/current/index.html.
- 25. Shao C, Feng Z, Westbrook JD, et al. Modernized uniform representation of carbohydrate molecules in the protein data bank. Glycobiology. 2021;31:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elasticsearch – Distributed restful search engine. 2019. https://github.com/elastic/elasticsearch.
- 27. Andreeva A, Kulesha E, Gough J, Murzin AG. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020;48:D376–D382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng H, Liao Y, Schaeffer RD, Grishin NV. Manual classification strategies in the ecod database. Proteins. 2015;83:1238–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dunbar J, Krawczyk K, Leem J, et al. SABDAB: The structural antibody database. Nucleic Acids Res. 2014;42:D1140–D1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Groom CR, Bruno IJ, Lightfoot MP, Ward SC. The Cambridge structural database. Acta Cryst B. 2016;72:171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grazulis S, Daskevic A, Merkys A, et al. Crystallography open database (COD): An open‐access collection of crystal structures and platform for world‐wide collaboration. Nucleic Acids Res. 2012;40:D420–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hastings J, Owen G, Dekker A, et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaulton A, Hersey A, Nowotka M, et al. The ChEMBl database in 2017. Nucleic Acids Res. 2017;45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim S, Chen J, Cheng T, et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. York WS, Mazumder R, Ranzinger R, et al. GlyGen: Computational and informatics resources for glycoscience. Glycobiology. 2020;30:72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tiemeyer M, Aoki K, Paulson J, et al. GlyTouCan: An accessible glycan structure repository. Glycobiology. 2017;27:915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamada I, Shiota M, Shinmachi D, et al. The glycosmos portal: A unified and comprehensive web resource for the glycosciences. Nat Methods. 2020;17:649–650. [DOI] [PubMed] [Google Scholar]
- 39. Newport TD, Sansom MSP, Stansfeld PJ. The MemProtMD database: A resource for membrane‐embedded protein structures and their lipid interactions. Nucleic Acids Res. 2019;47:D390–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tusnady GE, Dosztanyi Z, Simon I. Transmembrane proteins in the protein data bank: Identification and classification. Bioinformatics. 2004;20:2964–2972. [DOI] [PubMed] [Google Scholar]
- 41. Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. [DOI] [PubMed] [Google Scholar]
- 42. Sehnal D, Bittrich S, Deshpande M, et al. Mol* viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021;49:W431–W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181:281–292. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS‐CoV‐2 b.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J, Cai Y, Xiao T, et al. Structural impact on SARS‐CoV‐2 spike protein by d614g substitution. Science. 2021;372:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujita A, Aoki NP, Shinmachi D, et al. The international glycan repository glytoucan version 3.0. Nucleic Acids Res. 2021;49:D1529–D1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nilmeier JP, Meng EC, Polacco BJ, Babbitt PC. 3D motifs. Protein structure to function with bioinformatics. Dordrecht: Springer, 2017;p. 361–392. [Google Scholar]
- 48. Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. [DOI] [PubMed] [Google Scholar]
- 49. Jin Z, Du X, Xu Y, et al. Structure of m(pro) from SARS‐CoV‐2 and discovery of its inhibitors. Nature. 2020;582:289–293. [DOI] [PubMed] [Google Scholar]
- 50. Pollack A. Company says it mapped part of SARS virus. New York Times. 2003. July 30, 2003:C2. (July 30, 2003), C2–C2.14515843 [Google Scholar]
- 51. Liu P, Agrafiotis DK, Theobald DL. Fast determination of the optimal rotational matrix for macromolecular superpositions. J Comput Chem. 2010;31:1561–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burley SK. How to help the free market fight coronavirus. Nature. 2020;580:167. [DOI] [PubMed] [Google Scholar]
- 53. Boras B, Jones RM, Anson BJ, et al. Discovery of a novel inhibitor of coronavirus 3cl protease as a clinical candidate for the potential treatment of COVID‐19. bioRxiv. 2020. 10.1101/2020.09.12.293498 [DOI] [Google Scholar]
- 54. Halford B. Pfizer unveils its oral SARS‐CoV‐2 inhibitor. Chem Eng News. 2021;99:7. [Google Scholar]
- 55. Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatory extension of the optimum path. Protein Eng. 1998;11:739–747. [DOI] [PubMed] [Google Scholar]
- 56. Bliven SE, Bourne PE, Prlic A. Detection of circular permutations within protein structures using CE‐CP. Bioinformatics. 2015;31:1316–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ye Y, Godzik A. FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–W585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Y, Skolnick J. TM‐align: A protein structure alignment algorithm based on the tm‐score. Nucleic Acids Res. 2005;33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. [DOI] [PubMed] [Google Scholar]
- 60. Lafita A, Bliven S, Prlic A, et al. BioJava 5: A community driven open‐source bioinformatics library. PLoS Comput Biol. 2019;15:e1006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yuan Y, Cao D, Zhang Y, et al. Cryo‐EM structures of MERS‐CoV and SARS‐CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goodsell DS, Burley SK. RCSB protein data bank resources for structure‐facilitated design of MRNA vaccines for existing and emerging viral pathogens. Structure. 2021;29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Membrane proteins of known 3d structure (mpstruc). 2014. http://blanco.biomol.uci.edu/mpstruc/.
- 65. Kozma D, Simon I, Tusnady GE. PDBTM: Protein data bank of transmembrane proteins after 8 years. Nucleic Acids Res. 2013;41:D524–D529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Surya W, Li Y, Torres J. Structural model of the SARS coronavirus e channel in lmpg micelles. Biochim Biophys Acta Biomembr. 2018;1860:1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Postic G, Ghouzam Y, Guiraud V, Gelly JC. Membrane positioning for high‐ and low‐resolution protein structures through a binary classification approach. Protein Eng Des Sel. 2016;29:87–91. [DOI] [PubMed] [Google Scholar]
- 68. Bittrich S, Rose Y, Segura J, et al. RCSB protein data bank: Improved annotation, search, and visualization of membrane protein structures archived in the PDB. Bioinformatics. 2021;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dutta S, Dimitropoulos D, Feng Z, et al. Improving the representation of peptide‐like inhibitor and antibiotic molecules in the protein data bank. Biopolymers. 2014;101:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kneller DW, Phillips G, Weiss KL, Zhang Q, Coates L, Kovalevsky A. Direct observation of protonation state modulation in SARS‐CoV‐2 main protease upon inhibitor binding with neutron crystallography. J Med Chem. 2021;64:4991–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Osipiuk J, Azizi SA, Dvorkin S, et al. Structure of papain‐like protease from SARS‐CoV‐2 and its complexes with non‐covalent inhibitors. Nat Commun. 2021;12:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wishart DS, Feunang YD, Guo AC, et al. Drugbank 5.0: A major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Westbrook JD, Burley SK. How structural biologists and the protein data bank contributed to recent FDA new drug approvals. Structure. 2019;27:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Westbrook JD, Soskind R, Hudson BP, Burley SK. Impact of protein data bank on anti‐neoplastic approvals. Drug Discov Today. 2020;25:837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Burley SK. Impact of structural biologists and the protein data bank on small‐molecule drug discovery and development. J Biol Chem. 2021;296:100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jones TA, Zou J‐Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst A. 1991;47:110–119. [DOI] [PubMed] [Google Scholar]
- 77. Brändén C, Jones T. Between objectivity and subjectivity. Nature. 1990;343:687–689. [Google Scholar]
- 78. Bruno IJ, Cole JC, Kessler M, et al. Retrieval of crystallographically‐derived molecular geometry information. J Chem Inf Comput Sci. 2004;44:2133–2144. [DOI] [PubMed] [Google Scholar]
- 79. Chen VB, Arendall WB 3rd, Headd JJ, et al. Molprobity: All‐atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shao C, Westbrook JD, Lu C, et al. Simplified quality assessment for small‐molecule ligands in the PDB archive. Structure. 2021;29. 10.1016/j.str.2021.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Harris LJ, Skaletsky E, McPherson A. Crystallographic structure of an intact igg1 monoclonal antibody. J Mol Biol. 1998;275:861–872. [DOI] [PubMed] [Google Scholar]
- 82. Rose PW, Beran B, Bi C, et al. The RCSB protein data bank: Redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Henrick K, Feng Z, Bluhm WF, et al. Remediation of the protein data bank archive. Nucleic Acids Res. 2008;36:D426–D433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bourne PE, Addess KJ, Bluhm WF, et al. The distribution and query systems of the RCSB protein data bank. Nucleic Acids Res. 2004;32:D223–D225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Westbrook J, Feng Z, Chen L, Yang H, Berman HM. The protein data bank and structural genomics. Nucleic Acids Res. 2003;31:489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Westbrook J, Feng Z, Jain S, et al. The protein data bank: Unifying the archive. Nucleic Acids Res. 2002;30:245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bhat TN, Bourne P, Feng Z, et al. The PDB data uniformity project. Nucleic Acids Res. 2001;29:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Segura J, Rose Y, Westbrook J, Burley SK, Duarte JM. RCSB protein data bank 1D tools and services. Bioinformatics. 2020;36:5526–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shao C, Liu Z, Yang H, Wang S, Burley SK. Outlier analyses of the protein data bank archive using a probability‐density‐ranking approach. Sci Data. 2018;5:180293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shao C, Yang H, Westbrook JD, Young JY, Zardecki C, Burley SK. Multivariate analyses of quality metrics for crystal structures in the PDB archive. Structure, 2017;25(3):458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Korkmaz S, Duarte JM, Prlic A, et al. Investigation of protein quaternary structure via stoichiometry and symmetry information. PLoS One. 2018:13(6);e0197176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Guzenko D, Burley SK, Duarte JM. Real time structural search of the Protein Data Bank. PLoS Comput. 2020;16(7):e1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Goodsell DS, Zardecki C, Di Costanzo L, et al. RCSB protein data bank: Enabling biomedical research and drug discovery. Protein Sci. 2020;29:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Feng Z, Verdiguel N, Di Costanzo L, et al. Impact of the protein data bank across scientific disciplines. Data Sci J. 2020;19:1–14. [Google Scholar]
- 95. Markosian C, Di Costanzo L, Sekharan M, Shao C, Burley SK, Zardecki C. Analysis of impact metrics for the protein data bank. Sci Data. 2018;5:180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Burley SK, Berman HM. Open‐access data: A cornerstone for artificial intelligence approaches to protein structure prediction. Structure. 2021;29:515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sali A. From integrative structural biology to cell biology. J Biol Chem. 2021;296:100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zardecki C, Dutta S, Goodsell DS, Voigt M, Burley SK. RCSB Protein Data Bank: A resource for chemical, biochemical, and structural explorations of large and small biomolecules. J Chem Educ. 2016;93(3):569–575. [Google Scholar]
- 99. Beierlein JM, McNamee LM, Walsh MJ, Kaitin KI, DiMasi JA, Ledley FD. Landscape of innovation for cardiovascular pharmaceuticals: From basic science to new molecular entities. Clin Ther. 2017;39:1409–1425. [DOI] [PubMed] [Google Scholar]
- 100. Anderson WP. Data management: A global coalition to sustain core data. Nature. 2017;543:179. [DOI] [PubMed] [Google Scholar]
- 101. Anderson W, Apweiler R, Bateman A, et al. Towards coordinated international support of core data resources for the life sciences. bioRxiv. 2017. 10.1101/110825. [DOI] [Google Scholar]


