Abstract
The antimicrobial peptide database (APD) has served the antimicrobial peptide field for 18 years. Because it is widely used in research and education, this article documents database milestones and key events that have transformed it into the current form. A comparison is made for the APD peptide statistics between 2010 and 2020, validating the major database findings to date. We also describe new additions ranging from peptide entries to search functions. Of note, the APD also contains antimicrobial peptides from host microbiota, which are important in shaping immune systems and could be linked to a variety of human diseases. Finally, the database has been re‐programmed to the web branding and latest security compliance of the University of Nebraska Medical Center. The reprogrammed APD can be accessed at https://aps.unmc.edu.
Keywords: antimicrobial peptides, APD, database, microbiota, peptide design, peptide prediction
1. INTRODUCTION
Decades ago, Hans Boman asked a basic question of why invertebrates such as insects can survive without adaptive immune systems. His study led to the discovery of cecropins, which re‐ignited the interest in search of new antimicrobial peptides (AMPs). 1 , 2 The work of Hoffmann and colleagues went further and elucidated the Toll and Imd pathways that lead to the expression of different AMPs in response to the type of invading pathogens. 3 With the discovery of daptomycin, AS‐48, apidaecins, magainins, α‐defensins, cathelicidins, and histatins from bacteria, insects, amphibians and humans in the 1980s, 4 , 5 , 6 , 7 , 8 , 9 , 10 it is clear that such peptides are widely distributed in nature. Significantly, the Toll receptor is also involved in the expression of human cathelicidin LL‐37 via the vitamin D receptor. 11
Human defensins, cathelicidin LL‐37, and frog magainins show broad activity and inhibit the growth of bacteria, viruses, fungi, and parasites. 12 , 13 , 14 However, some AMPs have narrow‐spectrum activity. For instance, nisin inhibits only certain Gram‐positive bacteria, while proline‐rich AMPs kill primarily Gram‐negative bacteria such as Escherichia coli. 15 , 16 , 17 A classic view is that cationic AMPs kill bacteria by punching a hole in membranes. Barrel‐stave and carpet models have been proposed to explain the mechanisms of action of membrane‐targeting AMPs. 18 , 19 , 20 However, some peptides do not cause membrane leakage and may work via lipid clustering and domain formation. 21 Since all the events occur in the water‐membrane interface, an interfacial model was also proposed. 22 In our opinion, these models reflect different stages of bacterial killing and may be merged into a dynamic phase model that covers all the scenarios on the membranes due to the peptide action. The formation of a particular membrane phase is determined by the types of bacteria, peptides, ratio between them, and environmental conditions. Three basic membrane‐damaging phases are: (a) initial attachment to the membrane surface, which may lead to lipid domain formation; (b) membrane leakage due to the formation of pore or undefined membrane defects; and (c) membrane lysis into smaller pieces, leading to a clear solution. However, the action of AMPs is not limited to the classic membrane target. Some peptides, such as nisin and human α‐defensin HNP1, inhibit bacteria cell wall synthesis, 23 , 24 while proline‐rich AMPs can inhibit protein synthesis via binding to ribosomes. 16 , 17
Structurally, a linear cationic peptide may fold into an α‐amphipathic helix, ideal to associate with anionic bacterial membranes via both hydrophobic and electrostatic interactions. 1 , 13 Defensins are known to form a β‐sheet structure stabilized by three disulfide bonds. In addition, β‐defensins (average length in the APD: 44 amino acids) have an αβ fold since its sequence is slightly longer than that of α‐defensin (average length: 34 amino acids). However, proline‐rich peptides apparently adopt an extended non‐αβ structure in complex with ribosomes. 16 Therefore, AMPs are diverse in biological source, structure, activity, and mechanism of action.
Remarkably, AMPs can kill bacteria rapidly with a low chance of resistance development. In addition, AMPs are capable of eliminating antibiotic‐resistant pathogens, including persisters and biofilms. 25 , 26 , 27 Such features made them attractive for developing a new generation of antimicrobials to combat multidrug resistant pathogens. Indeed, a few peptide antibiotics such as daptomycin and colistin are already used clinically. In addition, nisin A has been utilized as a food preservative by over 50 countries. 28 These successful examples inspire the development of additional peptides into antimicrobials. While most of the developments focus on topical applications, some AMPs may be suitable for systemic treatment. 27 The synergistic effects of AMPs were noticed some time ago. 29 It is proposed that such an effect plays an important role in preventing host infection since AMPs can work below lab determined minimal inhibitory concentrations (MIC). 30 In the presence of an antimicrobial peptide, the efficacy of traditional antibiotics may be enhanced, opening a practical avenue to potential applications of AMPs. 28 , 29 , 30
The benefits of AMPs can extend beyond antimicrobial effects since they may also boost the immune response to further combat pathogen infection. 31 , 32 The immune regulation results from the interaction of AMPs (usually below MIC) with host cell receptors. A combined antimicrobial and host immune regulatory effect makes AMPs outstanding candidates as a new class of antimicrobials. In addition, AMPs may possess other biological functions in determining hair color and egg fertilization. 33 , 34
Our antimicrobial strategies are expanding. Cumulative evidence reveals that commensal bacteria play a critical role in shaping our immune system. In this process, bacteriocins are important components of host microbiota, since they may be able to selectively eliminate a particular type of invading pathogens and leave the community intact. Based on this idea, different methods, such as highly purified bacteriocins, probiotics, and fecal microbiota transplants, are utilized to restore the gut ecosystem. 35 , 36
Databases would be a useful tool to help organize such rich peptide information. In 2002, we started to construct the antimicrobial peptide database (APD) as a tool for designing antimicrobial and anticancer peptides. 37 There was an unanticipated competition as witnessed by the side‐by‐side publication of the APD 37 and ANTIMIC 38 in the 2004 database issue of Nucleic Acids Research. Users preferred the APD built by the antimicrobial peptide lab, establishing its leading position in the field. In the previous Protein Science Tool article, 39 Wang briefly described the three versions of the APD, published in 2004, 2009, and 2016, 37 , 40 , 41 respectively, and summarized the parameter space nature uses to build AMPs with different scaffolds. This invited article describes the major events and milestones of the APD in the past 18 years that evolved to the current status. We then compare the peptide statistics between 2010 and 2020. We also highlight recent database additions. Importantly, the APD has been annotating the AMPs from defined host microbiota (e.g., skin and gut). Our continued successful additions and expansion of new peptide features in the APD reinforce our original idea that this database provides a flexible platform for research and education of antimicrobial peptides, the key components of host innate immunity.
2. MAJOR EVENTS AND MILESTONES OF THE APD
The construction of the original APD during 2002–2003 had a clear goal to facilitate peptide search, prediction, and design (Figure 1a). Starting the second version, 40 the APD stepped on the journey to expand peptide search functions (e.g., antibacterial and hemolytic), leading to a significant increase from five in the first version 37 to 19 in the third version of the APD (APD3). 41 The database is now dedicated to glossary, nomenclature, classification, information search, prediction, design, and statistics of AMPs and beyond (Figure 1b). In 2021, the APD was modernized to the web branding and the latest security standard of the University of Nebraska Medical Center (UNMC; Figure 1c). Table 1 summarizes the major developments and milestones of the APD from 2003 to 2021, which we highlight below.
FIGURE 1.
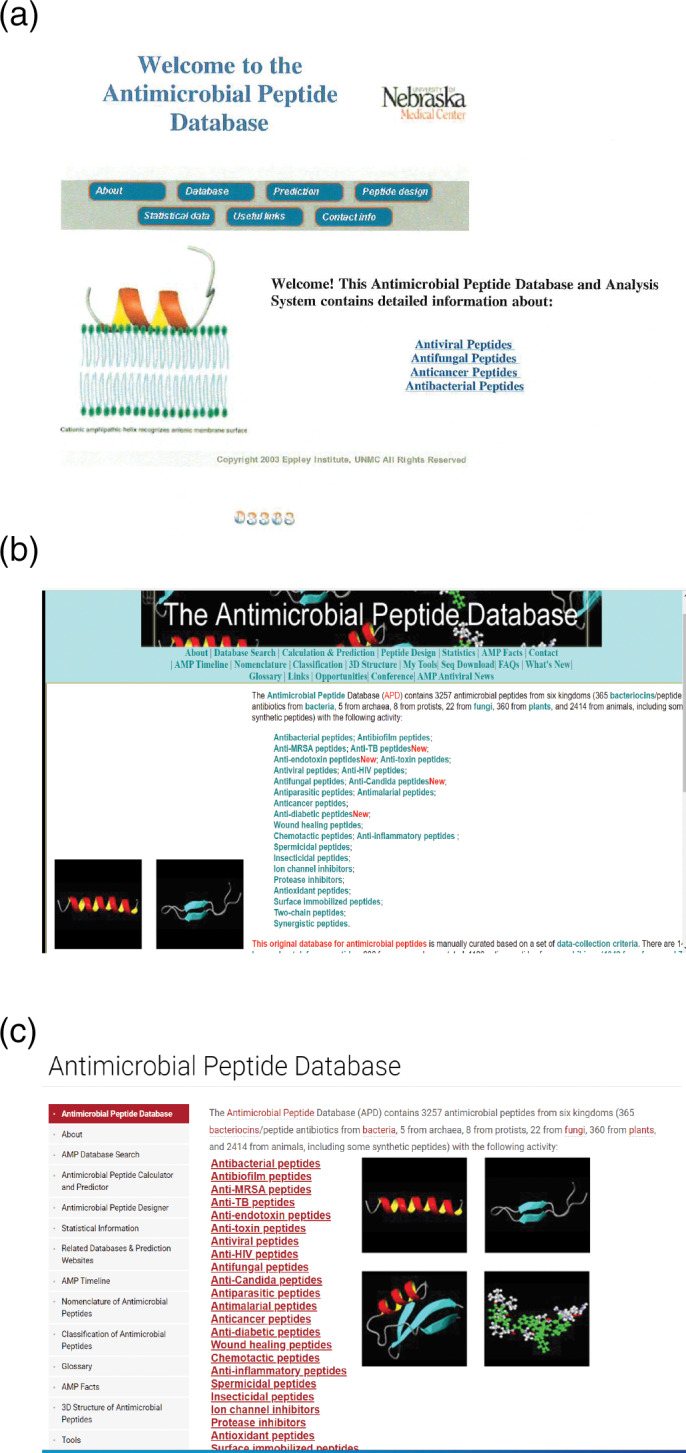
Web main pages for the Antimicrobial Peptide Database (APD) in 2003 (a), between 2008 and 2021 (b), and starting June 2021 (c)
TABLE 1.
Major events/milestones of the antimicrobial peptide database (APD)
| Year | Key additions and milestones | Education and applications |
|---|---|---|
| 2002 | Zhe Wang met Dr. Guangshun Wang and started the database project. | |
| 2003 | The database was online (Figure 1a) | Birth place: URL: http://aps.unmc.edu/AP |
| Activity (bacteria, viruses, fungi, cancer) and hemolysis search | ||
| NMR and crystal structure search | ||
| Peptide calculation and prediction | ||
| Peptide design interface | ||
| Peptide statistics (sequence, structure, and activity) | ||
| 2004 | APD paper was published 37 | Birth certificate: Nucleic Acids Res. 2004 |
| 2005 | First database‐aided peptide design 42 | |
| 2006 | Linguistic model for peptide design 43 | |
| 2007 | First machine learning prediction 44 | |
| 2008 | Searchable AMP activities (ZZ code, outdated), first expansion | Antiparasitic, insecticidal, spermicidal, anti‐HIV |
| Chemical modifications (XX code) | CD, a third peptide structure indicator | |
| Clinical trials indicated | AMP glossary | |
| The AMP count reached 1,000 | Database screening 45 | |
| Database new web design (Figure 1b) | ||
| 2009 | APD2 was published 40 | Definition of frequently occurring amino acids and their use in peptide design design 40 |
| Synergistic effect (JJsn) | ||
| Peptide binding targets (BB code) | ||
| Peptide family and source classification | ||
| Author/publication year search | ||
|
2010 |
APD‐aided book editing 46 | |
| Structural classification into α, β, αβ, and non‐αβ 46 | Non‐αβ includes turns, bends, spiral, extended structures, and so forth. | |
| Defined amino acid‐rich AMPs >25% | New‐rich families: Asp‐rich, Ser‐rich, Ala‐rich, Lys‐rich, and so forth. | |
| 2011 | Searchable AMP functions, second expansion | Antioxidant, protease inhibitor, antimalarial |
| Peptide registration criteria defined (mature peptide, known activity <100 μM, less than 100 amino acids, and natural peptides) | ||
| Animal models | ||
| Amphibian geographic sources | ||
| 2012 | Database filtering technology 47 | |
| The AMP count reached 2000 | AMP discovery timeline | |
| Collected human antimicrobial proteins <200 amino acids | ||
| Programmed additional AMP calculation capabilities | Mol Wt, Mol formula, Ex. Coeff., GRAVY, Wimley‐White hydrophobicity | |
| Updated prediction interface based on the AMP parameter space | Established multiple education web pages | |
| Search AMPs found in multiple species | ||
| 2013 | Species‐specific activity | Search format (e.g., Escherichia coli, Staphylococcus aureus) |
| 2014 | Searchable AMP functions, third expansion | Anti‐inflammatory; anti‐oxidant; protease inhibitory; antimalarial |
| “Tools,” “What's new” established | ||
| 2015 | Universal peptide classification based on peptide bond patterns 48 | |
| Peptide sources: Five life kingdoms or three domains | ||
| Searchable AMP functions, fourth expansion | Antibiofilm, wound healing | |
| 2016 | APD3 was published 41 | APD in education described |
| Six life kingdoms for peptide sources | ||
| 2017 | AMP news (including antiviral news) | |
| Search surface‐immobilized AMPs | ||
| 2018 | The AMP count reached 3,000 | |
| Searchable AMP functions, fifth expansion | Anti‐toxin, anti‐MRSA, channel inhibitor | |
| Search AMPs in use | ||
| 2019 | Low cationicity of database designed peptides for systemic efficacy 49 | |
| APD update described 39 | ||
| 2020 | Searchable AMP molecular forms | Two‐chain; synergistic peptides |
| Searchable AMP functions, sixth expansion | Anti‐diabetes, anti‐TB, anti‐sepsis (LPS), anti‐candida | |
| Salt/pH/serum effects on peptide activity | Pho‐length and net charge‐length correlations in frog AMPs 50 | |
| Microbiota | ||
| Database shutdown in October due to a cyberattack to UNMC | ||
| In December, a temporary APD site established in a commercial cloud | URL: https://wangapd3.com (closed 2021) | |
| 2021 | In June, the reprogrammed APD online | https://aps.unmc.edu (open) |
2.1. Database scope: Natural, synthetic, and predicted peptides
The initial APD covered natural, synthetic, and predicted AMPs with a particular emphasis on the structure–activity relationship (SAR). 37 By the time this manuscript was completed, there were 2,991 natural peptide sequences with experimental activity data, 210 predicted peptides without activity data (128 from animals and 82 from plants), and 72 synthetic peptides. Natural peptides are made either ribosomally or non‐ribosomally. Most of the AMPs in the current APD are gene‐encoded and synthesized in ribosomes. In this database, the term “synthetic peptides” is used to refer to peptide derivatives synthesized in laboratories in order to better understand the SARs of natural peptides. Predicted antimicrobial peptides include all sequences that are identified in the genomes or isolated from organisms but do not have antimicrobial activity data. Such an all‐inclusive data collection tradition was retained in the APD2. 40 It was in the third version that we started to establish a set of criteria for data registration. 41 Due to uncertainty in antimicrobial activity, we postponed the collection of predicted peptides without experimental data. We kept 210 “predicted” peptides isolated from different organisms. They are very likely to be true AMPs as we illustrated recently. 51 To avoid the distortion of the picture for natural AMPs, we also restricted the registration of synthetic peptides at this stage (only 72 entries listed separately and more are listed as derivatives of the parent peptides). Thus, naturally occurring AMPs account for 97.8% of the current APD. Our practice generates a core data set for natural AMPs, which facilitates the development of the database filtering technology where each step points at peptides and parameters derived from natural peptides. 47 Peptides designed based on natural templates are more likely to be patentable than those derived from synthetic peptides designed and characterized by other laboratories.
2.2. Peptide family classification
In version 2, the APD started to annotate peptide families in the name field directly after the peptide name. This addition enables users to identify a family of AMPs from all sources (below). Some common peptide families are defensins, cathelicidins, histatins, temporins, and aureins. In the current database, there are 349 defensins and 134 cathelicidins. A statistical analysis reveals a higher percentage of arginine than lysine in cathelicidins in both 2008 and 2020. The same trend retains in defensins (R% > K%) as well. Some family names are dedicated to peptides from a defined source (e.g., bacteriocins from bacteria, cyclotides from plants, and temporins from amphibians). A detailed list of different amphibian AMP families can be found elsewhere. 50 Overall, linear amphibian AMPs in the current database differ from defensins and cathelicidins in that lysines are dominant. 40 Some peptides are rich in certain amino acids. 1 The classic families include tryptophan‐rich (12 Trp‐rich, on average 27.2% W), histidine‐rich (29 His‐rich, 23.3% H), arginine‐rich (31 Arg‐rich, 27.7% R), glycine‐rich (48 Gly‐rich, 49.6% G), and proline‐rich (71 Pro‐rich, 29.6% P). The shortened form in parentheses (e.g., Gly‐rich) can be searched in the name field of the APD. It is evident that the content of a “rich” amino acid depends on the type in the range of 23.3% for His‐rich AMPs to 49.6% for Gly‐rich peptides. Based on these analyses, we made the first attempt to define the word “rich” as “greater than 25%” in 2010. This definition led to the recognition of other less‐known rich peptide families, including alanine‐rich (six Ala‐rich, 31.1%), lysine‐rich (nine Lys‐rich, 35.6%), aspartic acid‐rich (three Asp‐rich, 85.7%), serine‐rich (three Ser‐rich, 29.6%), and cysteine‐rich (19 Cys‐rich ɵ‐defensin, 33.3%). However, this definition (>25%) may not apply to multiple domain AMPs although each domain is clearly rich in certain amino acids such as serine, proline, or cysteine. 39 For such AMPs, it will be useful to define also the amino acid‐rich region.
AMPs can also be grouped based on net charge. In the APD, 2887 peptides (88%) have a net charge greater than zero. These are cationic peptides. One hundred and ninety peptides (6%) happen to have a net charge of zero. They are neutral peptides. Another small population even (6%) has net charges less than zero. Such peptides are referred to as anionic peptides.
2.3. Antimicrobial peptides have been discovered in six life kingdoms covering both prokaryotes and eukaryotes
Our classification of peptide sources evolved with time as well. This information is absent in version 1. The annotation started in version 2 directly after peptide name. 40 For instance, nisin A (AP00205) is found from Gram‐positive bacteria (prokaryotes), while LL‐37 (AP00310) is made in human (primates, mammals, and animals). Our annotations enabled users to search AMPs from bacteria, plants, ants, fish, cow, insects, crabs, toads, and frogs. In the APD2, the number of AMPs from frogs was 398, which accounted for 32% of all the peptides. In the current APD, we found 1,049 frog peptides (32%) out of a total of 3,273 AMPs. This indicates that amphibians continue to occupy an important niche in AMP discovery, probably due to their wide distributions on different continents of Earth. Likewise, AMPs from insects increased from 118 (9.6%) in the APD2 to the current 325 (9.9%), while bacterial AMPs increased from 112 (9.1%) in the APD2 to the current 369 (11.3%). It is notable that the numbers of AMPs from frogs, insects, and bacteria increased proportionally from the APD2 to the current version. Their sum accounts for ~50% of the total AMPs registered in the APD in the past decade. Since AMPs from bacteria, plants, insects, and amphibians are abundant in the APD, we compared their amino acid composition profiles (signatures) between 2008 and 2020 (Figure 2). Remarkably, their signatures remain essentially the same despite a substantial increase in peptide count in each group. We initially defined the frequently occurring amino acids in the APD2 as ≥10%. 40 Based on this definition, the frequently occurring residues also remained the same: A and G in bacterial AMPs, C and G in plant AMPs, A, G, K in insect AMPs, and L, A, G, and K in amphibian peptides (Figure 2), further confirming the significance of these abundant amino acids.
FIGURE 2.
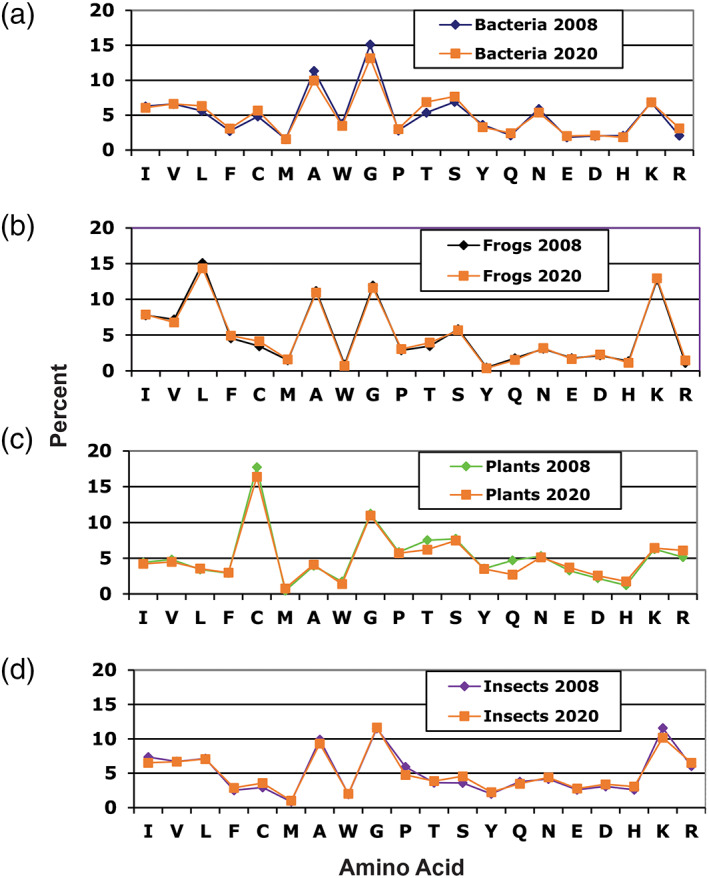
Comparison of amino acid signatures for the major sources of the antimicrobial peptides (AMPs) from (a) bacteria (115 bacteriocins in 2008 and 365 in 2020), (b) frogs (453 frog AMPs in 2008 and 1,042 in 2020), (c) plants (214 AMPs in 2008 and 360 in 2020), and (d) insects (142 AMPs in 2008 and 323 in 2020) collected in the antimicrobial peptide database (http://aps.unmc.edu/AP)
The biological sources of antimicrobial peptides were further systematically classified in the APD in 2010 during the AMP book editing. 46 AMPs have been discovered in both prokaryotic and eukaryotic sources. In 2010, prokaryotes included eubacteria (118 entries) and archaea (two entries). Eukaryotes were further split into protozoa (five entries), fungi (four entries), plants (217 entries), animals (1,089 entries), and humans (54 entries). In 2016, these groups were formally merged into the six life kingdoms in the APD3: bacteria, archaea, protists, fungi, plants, and animals. Table 2 compares the AMPs from the six kingdoms in 2010 and 2020. In 2010, protists were represented by protozoa. Also, human AMPs were separated from animal peptides. Clearly, the dominance of animal sources (74%) persists. Started in the APD2, this database has a detailed classification of AMPs from a variety of animal sources, ranging from invertebrates (e.g., insects, spiders, molluscs, and crustaceans) to vertebrates (e.g., fish, amphibians, reptiles, and birds). The increase in the counts of AMPs from such animals in 2010 and 2020 can be viewed in Table 2.
TABLE 2.
Antimicrobial peptides from different kingdoms, vertebrates, and invertebrates in 2010 and 2020 in the APD
| 2010 | 2020 | |
|---|---|---|
| Total peptide count | 1,528 | 3,257 |
| Kingdom | ||
| Bacteria | 118 | 365 |
| Archaea | 2 | 5 |
| Protists | 5 | 8 |
| Fungi | 4 | 22 |
| Plants | 217 | 360 |
| Animals | 1,143 | 2,414 |
| Vertebrate | ||
| Mammals | 328 | |
| Fish | 50 | 136 |
| Amphibians | 592 | 1,120 |
| Reptiles | 8 | 45 |
| Birds | 27 | 43 |
| Invertebrate | ||
| Insects | 156 | 323 |
| Spiders | 24 | 43 |
| Scorpions | 88 | |
| Mollusca | 6 | 47 |
| Crustaceans | 11 | 71 |
The AMP sources can also be grouped into three life domains (eukarya, archaea, and eubacteria). 52 The eukaryotes will include fungi, protists, plants, and animals. Therefore, the total numbers of AMPs in the three life domains were 365 in eubacteria, five in archaea, and 2,804 in eukarya in 2020.
Since each continent has different life forms, the APD made one more step in annotating geographic source information for each species directly after the scientific name (Source Organism). This enabled the first comparison of the amphibian peptide signatures from different continents in 2020. 39 It appears that frog AMPs from Europe contain a higher content of leucine, while those from South America are higher in alanine.
2.4. The expanding biological functions of antimicrobial peptides
AMP functions expanded with time in the APD (Figure 3). In 2003, only antibacterial, antiviral, antifungal, anticancer, and hemolytic peptides were searchable. In 2008, antibacterial peptides with a narrow activity spectrum were split into anti‐Gram‐positive (G+) and anti‐Gram‐negative (G−) AMPs. Antiviral peptides with anti‐HIV activity were annotated due to the research need of our database‐guided design of such peptides. 53 Also annotated are peptides that neutralize lipopolysaccharides (LPS, or endotoxin), the outer membrane components of Gram‐negative bacteria. Some of the peptide function searches were enabled initially by creating a special string search (starting ZZ). 40 The ZZ system was subsequently replaced by the more user‐friendly icon search. In 2015, nine additional activities/functions were enabled in the APD3: antiparasitic, antimalarial, spermicidal, insecticidal, antibiofilm, antioxidant, protease inhibitory, chemotactic, and wound healing activities. In 2018, AMPs with anti‐toxin, anti‐inflammatory, anti‐MRSA, channel inhibitor activities became searchable. In 2020, the icon search for antiTB, anti‐candida, and anti‐diabetic activities was also enabled. A detailed record of the peptide functions and count changes in the three versions of the APD and current database is provided in Table 3.
FIGURE 3.
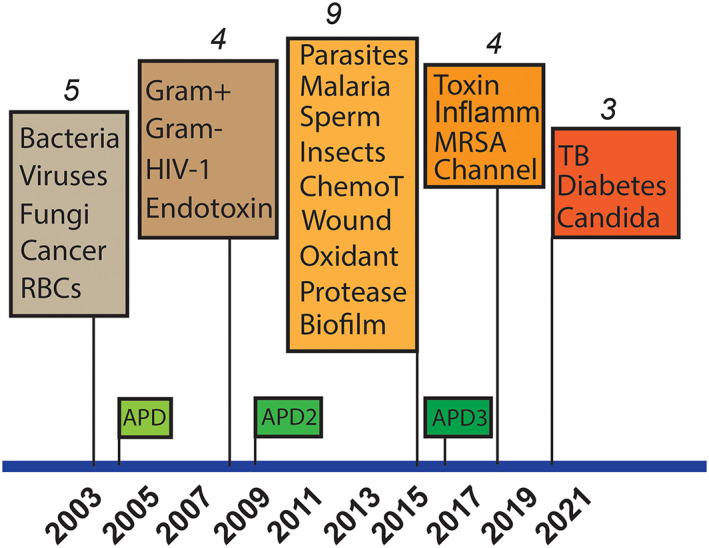
Timeline for the expansion of searchable peptide functions in the antimicrobial peptide database from 2003 to 2020
TABLE 3.
Peptide search function expansion in the APD
| Parameter | APD (2004) | APD2 (2009) | APD3 (2016) | APD current (2021) a |
|---|---|---|---|---|
| Total peptides | 525 | 1,228 | 2,619 | 3,273 |
| Antibacterial | 498 | 944 | 2,169 | 2,743 |
| AntiGram+ | 267 | 426 | 558 | |
| AntiGram− | 202 | 323 | ||
| Anti‐MRSA | 182 | |||
| Anti‐TB | 14 | |||
| Antibiofilm | 16 | 66 | ||
| Anti‐toxin | 15 | |||
| Anti‐endotoxin | 61 | 88 | ||
| Antifungal | 155 | 327 | 961 | 1,212 |
| Anti‐candida | 676 | |||
| Antiviral | 28 | 76 | 172 | 193 |
| Anti‐HIV | 53 | 105 | 109 | |
| Antiparasitic | 80 | 138 | ||
| Antimalarial | 16 | 33 | ||
| Anticancer | 18 | 65 | 185 | 252 |
| Hemolytic | 64 | 108 | 307 | 313 |
| Spermicidal | 11 | 14 | ||
| Insecticidal | 27 | 39 | ||
| Chemotactic | 53 | 62 | ||
| Anti‐inflammatory | 27 | |||
| Wound healing | 10 | 23 | ||
| Ion channel inhibitor | 7 | |||
| Antioxidant | 19 | 28 | ||
| Protease inhibitor | 12 | 31 | ||
| Antidiabetic | 16 |
As of July 2021. Obtained from https://aps.unmc.edu.
Figure 4 compares the amino acid frequency changes for antibacterial, antifungal, antiviral, and anticancer peptides in the APD between 2008 and 2020. While the plots are rather similar for antibacterial, antifungal, and anticancer peptides, there are clear deviations for antiviral peptides. The frequency of cysteine dropped from above 15% to below 10%. Meanwhile, the frequency of arginine decreased from ~10% to ~6%. Such changes may be attributed to the increase in linear antiviral peptides resulting from a collaborative project on database screening and design of anti‐HIV peptides between us and ImQuest Biosciences. 45 , 53
FIGURE 4.
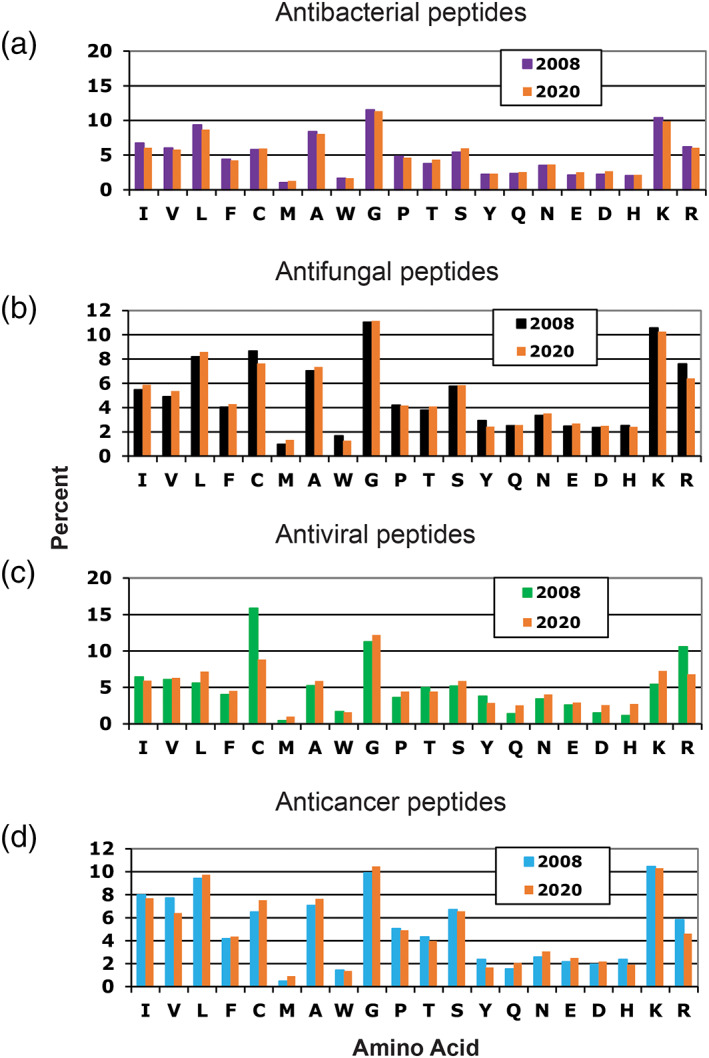
Comparison of amino acid compositions of antimicrobial peptides with activity against bacteria (1,034 AMPs in 2008 and 2,727 in 2020), fungi (388 AMPs in 2008 and 1,210 in 2020), viruses (76 AMPs in 2008 and 190 in 2020), and cancer cells (78 AMPs in 2008 and 251 in 2020). Data obtained from the antimicrobial peptide database (http://aps.unmc.edu/AP)
To facilitate the development of antimicrobials against a specific pathogen, the APD has also enabled the activity search of AMPs against any pathogen. This information can be obtained by entering a shorthand of scientific name (e.g., E. coli, S. aureus, or C. albicans) into the search box for Additional Information followed by search. In the current APD, we found 1,391 peptides with activity against E. coli, 1,324 against S. aureus (Staphylococcus aureus), and 689 against C. albicans (Candida albicans). Hence, the APD is flexible and can search peptide activity for an unlimited number of pathogens as long as they have been tested and entered into the database.
2.5. AMP sequences can be posttranslationally modified in numerous ways
Many AMPs are chemically modified post‐translation. 54 In 2008, we started to annotate post‐translational modifications (PTM) of AMPs in the APD. Each peptide is represented as a string of single‐lettered amino acids. The same set of 20 standard amino acids is also used to represent the amino acid sequences of heavily modified peptides. An annotation system was created in the name field for chemical modifications by commencing with “XX” (like two scissors). For example, XXA implies peptide amidation at the C‐terminus, while XXC indicates peptide cyclization via forming a peptide bond between the N and C‐termini. Seven types of PTMs (C‐end amidation, cyclization, oxidation, phosphorylation, D‐amino acids, glycosylation, and lipidation) were introduced in 2009. The number of PTMs increased to 14 types in 2010. In 2016, additional chemical modification types were included. Table 4 shows the increase of posttranslationally modified AMPs from 2010 to 2020. The current APD contains a total of 26 types of PTMs for AMPs. Note that the APD has taken chemical modification into consideration in calculating the net charge for each peptide. Hence, the net charge of monophosphated and C‐terminally amidated peptides reduce and increase by one, respectively.
TABLE 4.
Post‐translational modifications of antimicrobial peptides in the APD a
| 2010 | 2020 | |
|---|---|---|
| C‐terminal amidation (XXA) | 272 | 622 |
| C‐terminal iron‐binding moieties (XXB) | 4 | 4 |
| Cyclization, N to C peptide bond (XXC) | 139 | 210 |
| D‐amino acids (XXD) | 7 | 37 |
| N‐terminal acetylation (XXE) | 8 | 18 |
| Carboxylic‐acid‐containing group (XXF) | 1 | 3 |
| Glycosylation (XXG) | 7 | 17 |
| Halogenation (XXH) | 1 | 12 |
| N‐formylated met (XXI) | 11 | |
| Sidechain‐backbone bond (XXJ) | 31 | |
| Hydroxylation (XXK) | 1 | 15 |
| Lipidation (XXL) | 41 | |
| Methylation (XXM) | 6 | |
| Nitrosylation (XXN) | 0 | |
| Oxidation (XXO) | 8 | 13 |
| Phosphorylation (XXP) | 2 | 6 |
| N‐terminally cyclic glutamate (XXQ) | 12 | 20 |
| S–S Reduction (XXR) | 2 | |
| Sulfation (XXS) | 1 | 1 |
| Thioether bridge (XXT) | 22 | 61 |
| Rana box S–S (XXU) | 473 | |
| Lantibiotic C–C bridge (XXV) | 2 | |
| Dehydration (XXW) | 36 | |
| ADP‐ribosylation (XXX) | 2 | |
| Citrullination (XXY) | 1 | |
| Carbamylation (XXZ) | 1 |
The chemical modification search system in the APD.
2.6. Binding targets and mechanisms of action of AMPs
This information does not exist in the first version. In the APD2, we created a different information annotation system for us to record peptide binding partners. This system starts with “BB” (Table 5). The following were produced initially: binding to membranes (BBMm), protein/enzyme (BBPP), nucleic acids (BBN), carbohydrates (BBS), lipopolysaccharides (BBL), metal ions (BBII), and peptide aggregation (BBB). Later, peptide aggregation was split into two types: in water (BBBh2o) and in membranes (BBBm). Many AMPs bind to lipid II to inhibit bacterial cell wall synthesis (BBW). More recently, this system has been further refined. Some AMPs, especially proline‐rich peptides that bind to ribosomes can be searched using BBribo in the name field. A more specific protein binding has also been annotated: receptor binding (BBrcp) and bacterial outer membrane proteins (BBomp).
TABLE 5.
Binding partners or targets of antimicrobial peptides in 2010 and 2020 a
| 2010 | 2020 | |
|---|---|---|
| Membranes (BBMm) | 48 | 147 |
| Cell wall (BBW) | 3 | 28 |
| Ribosomes (BBribo) | 9 | |
| Protein receptor (BBrcp) | 10 | |
| Outer membrane protein (BBomp) | 3 | |
| RNA polymerase (BBpol) | 3 | |
| LPS (BBL) | 26 | 92 |
| Nucleic acids (BBN) | 5 | 19 |
| Carbohydrates (BBS) | 21 | 51 |
| Metal (BBII) | 12 | 24 |
| Oligomer in water (BBBh2o) | 6 | 15 |
| Oligomer in membranes (BBBm) | 1 | 5 |
Data obtained from the antimicrobial peptide database (http://aps.unmc.edu/AP).
It is important to note that binding to a molecular partner does not necessarily mean a mechanism of action. Therefore, the APD also annotates the mechanism of action (MOA) of AMPs in the additional information field. At present, 103 AMPs have such information in the APD.
2.7. AMPs can form a variety of three‐dimensional structures
AMPs can form a variety of structural scaffolds. Initially, the APD adopted the Boman classification, which includes α‐helical, disulfide bonded β‐sheet peptides and AMPs rich in proline, histidine, and tryptophan. 1 Because circular dichroism (CD) is an excellent indicator for a helical structure in membrane‐mimicking environments such as trifluoroethanol and micelles, the APD also annotated this information starting 2008. In 2010, we introduced a self‐consistent structural classification (α, β, αβ, and non‐αβ) into the database. 46 These four types of structures have increased in the APD from 2010 to 2020 (Table 6). At present, all the classes of structures have been found in animals and bacteria. Plant AMPs frequently contain disulfide bonds that stabilize β‐sheet structures. 55 There are also exceptions since S–S linked helical structures have been found in plants. 56 However, non‐αβ structures have not been found in plant AMPs.
TABLE 6.
Structural statistics of Antimicrobial Peptides in the APD in 2010 and 2020
| Year | 2010 | 2020 | ||||
|---|---|---|---|---|---|---|
| Kingdom | Bacteria | Plants | Animals | Bacteria | Plants | Animals |
| α‐Helix | 13 | 0 | 73 | 32 | 6 | 152 a |
| β‐Sheet | 4 | 12 | 26 | 15 | 12 | 53 |
| Combined αβ | 2 | 19 | 20 | 5 | 40 | 64 |
| Non‐αβ | 0 | 0 | 3 | 10 | 0 | 7 |
| Total | 19 | 31 | 122 | 62 | 58 | 276 |
It includes 3D structures determined by NMR spectroscopy (142 entries) and X‐ray crystallography (10 entries). After the APD2, secondary structures implicated by circular dichroism (CD) are also annotated in the APD, leading to a total of 382 helical peptides.
2.8. Animal models for peptide efficacy in vivo
As an important step toward novel antibiotic development, animal models play an important role. Both invertebrate and vertebrate models have been utilized to test the efficacy of AMPs. This information was not annotated in the first two versions of the APD. In the APD3, this can be searched in the “Additional Information” field by entering “animal model.” The animal type is followed after a colon in a format “animal model:type.” A search of animal model: mouse returned 52 entries in the current database. Other testing animals include cattle, rabbit, rat, chicken, zebrafish, fish, and wax moth.
2.9. Potential leads, clinical trials, and peptide antibiotics in use
Some recently identified peptides appeared to be highly promising for antimicrobial development. We decided to label them as “potential leads” (searchable in Additional Information). These include plectasin, 57 teixobactin, 26 taromycin A, 58 horine, verine, 27 lysocin E, 59 darobactin, 60 and lugdunin. 61 In 2008, a few AMPs were subjected to “clinical trials” and the quoted phrase can be searched in the Name field. Some well‐known candidates include magainin 2 (MSI‐78 or pexiganan), 62 indolicidin (MBI226 or Omiganan), 63 PG1 (Iseganan or protegrin IB367), 64 LL‐37 (OP‐145), 65 and human histatin 5 (P113). 66 In 2018, we also annotated “AMPs in use” (search the quoted phrase in Add Info). While gramicidin A, gramicidin S, colistin A, polymyxin B, and cubicin (daptomycin) are in clinical use, lysozyme, nisin A, and pediocin PA‐1 (AcH) are used as food preservatives. 28
3. THE APD FOR EDUCATION
Another important goal of the APD3 is to promote student education. 41 This ranges from glossary, timeline, and nomenclature to classification. The glossary provides frequently used abbreviations and defines some terms. Timeline provides a chronical list of select AMPs discovered annually. Nomenclature summarizes the common methods used to name AMPs. These include biological sources, properties, generation methods, and a combination of the above. To facilitate users' search, the APD includes both old and new, abbreviated and full names for AMPs. Classification documents the major methods utilized to classify AMPs into families. These include peptide synthesis machinery, source, property, chain bonding pattern, 3D structure, activity, and mechanism of action. In addition, FAQ provides a list of frequently asked questions and answers. For example, how many AMPs have we discovered in nature? The current APD estimates over 3,000. My Tools provides the links to numerous web tools useful for AMPs. What's New informs users what has been newly added to the APD. As a consequence of an increased interest in discovering new therapeutics to combat COVID‐19, the APD also enabled “AMP Antiviral News” in 2020. Such additions make the APD unique and more useful.
4. DATABASE REPROGRAMMING AND CURRENT STATUS
4.1. Database reprogramming into a new look
The APD codes were originally written during 2002–2003 (Figure 1a). The APD system used the LAMP software bundle, deployed in the Red Hat Linux operating system using the freeware Apache web server, MySQL relational database management system, and PHP script language. Each peptide was assigned a unique five‐digit ID starting with AP (e.g., AP00301). In 2021, Michael Zietz and Ashok Mudgapalli at UNMC modernized the computer codes of the APD. PHP5 has been upgraded to PHP7 and MySQL has been replaced with MariaDB 10.5. In addition, the entire web site, including user interfaces, backend PHP code, and database has gone through series of security Penetration Test Scanning processes using latest UNMC security tools for vulnerabilities and got approved to operationalize web site in production. All the web pages have been unified in the website style of the UNMC branding. Therefore, we referred to this UNMC‐branded APD as APD2021. This newly programed database maintained the overall design and tradition of the original APD programmed by Zhe Wang and expanded by Guangshun Wang. It replaced the web designed by Shuona Wang in 2008. The original horizontal database interfaces for research and education (e.g., About, Database Search, Calculation & Prediction, Peptide Design, and Statistics) at the top of the main page (Figure 1a,b) have been converted to a vertical format on the left (Figure 1c). This column of database interfaces serves as the left navigation bar as they are always present during information search, thereby facilitating jumping between web interfaces. Another change was also made. The original APD provides five AMPs most similar to users' query. This re‐programmed database has removed this restriction by providing users with many more similar sequences. The additional sequence aligning may be helpful for peptide design.
4.2. New additions to the APD
4.2.1. Peptide entries continued to increase
We reported 3,081 peptides in the previous paper for the Protein Science Tool. 39 By the end of 2020, there were 3,257 AMPs. Based on this set of AMPs, the averaged peptide length was 33.2. The averaged net charge and hydrophobic content were +3.3 and 41.4%, respectively. Of these peptides, over 90% were small peptides with less than 60 amino acids (87% are less than 50 aa). The majority (94%) of peptides had a net charge of zero or greater. This data set was utilized for analysis throughout the text. With the newly programmed database online in June 2021, some new AMPs from 2021 have been added, leading to a total of 3,273 entries (Table 3 updated).
4.2.2. New database search functions for peptide forms and functions
Since the last description of the APD in 2019, 39 several new peptide search functions have been enabled. These include anti‐TB, anti‐endotoxin, anti‐candida, and anti‐diabetic peptides. In the current database, we found 14 AMPs with anti‐TB activity, 85 anti‐endotoxin peptides, 676 anti‐candida peptides, and 16 anti‐diabetic peptides.
Peptide molecular forms also play an important role in combating pathogens. The majority of peptides in the APD contain a single chain. However, some AMPs possess two polypeptide chains and do not show antimicrobial activity when a single chain is tested alone. The first two‐chain peptide was discovered in bacteria in 1992 (AP01151). 67 Moreover, single‐chain AMPs may work synergistically for efficacy. Such molecular form information was annotated in the database some time ago (Table 1). In 2017, we annotated surface‐immobilized peptides. For convenience, we also enabled a single‐click icon search for two‐chained and synergistic peptides in 2020. There are currently 35 peptides with two chains and 47 AMPs with synergistic antimicrobial effects with other peptides.
4.2.3. Antimicrobial robustness
Some AMPs are known to lose activity under certain conditions (e.g., salts, serum, and pH). Despite a challenging task, we found it possible to identify more robust AMPs by conducting in vitro filtering. Interestingly, peptides that retain antimicrobial activity under the above conditions show systemic efficacy in mice. 49 , 68 To facilitate the development of more robust AMPs, we started to include such properties in the Additional Information field. At present, we found 24 salt‐sensitive AMPs and 20 salt‐insensitive peptides in the current APD.
4.2.4. AMP atlas
A single species can express different AMPs under different conditions. A known set of AMPs from a particular species can be searched in the APD by entering its scientific name into the search box for Source Organism. The collections of all the AMPs from a variety of organisms constitute AMP atlas. For instance, we found 10 honeybee AMPs using “Apis mellifera,” 14 fruitfly AMPs using “Drosophila melanogaster,” 69 and 20 wax moth peptides from Galleria mellonella. Based on genomic and proteomic approaches, Zhang and colleagues identified 36 AMPs from the frog “Odorrana andersonii” with demonstrated antibacterial activity. 70 A search of “Homo sapiens” returned 141 human AMPs discovered from a variety of organs or tissues (reviewed in Reference 71). In the future, AMP atlas also includes AMP distribution and kinetics in various tissues of a single species at different seasons and geographic regions. The tissue information for AMPs (e.g., seeds, leaves, stems in the case of plants and skin, lung, and brain for humans and other animals) can be searched in the same Source field. For example, the APD has annotated 324 AMPs from skin and 24 AMPs from the brain of frogs and humans.
4.2.5. Microbiota
Microbiota include a set of microbes in a defined location. Bacteria, archaea, protists, and fungi in microbiota can all produce antimicrobial peptides. Thus, the search of novel AMPs has been extended to host microbiota, including both plants and animals. It is proposed that AMPs in microbiota play an essential role in guarding the exposed surfaces susceptible to pathogen invasion. This observation laid the foundation for restoring normal microbiota by using probiotics. 72 , 73 Recently, the APD has started to annotate such information. A search of microbiota in “Source Organism” produced 24 bacteriocins. A particular organ such as skin and gut can also be searched by entering “:skin or: gut” after “microbiota”. We found 19 bacteriocins from “human microbiota: gut”. Considering the growing interest in microbiota, we anticipate a significant increase in new AMPs from commensal bacteria in the future.
5. CONCLUDING REMARKS
The construction of a useful database helps to put relevant information in order. The APD is an original database in terms of both platform design and peptide entries. It has defined and unified numerous aspects of AMPs to reach the current status (Table 1). Since its birth in 2003, this database has been updated and expanded for over a decade. As milestones, the peptide entries in the APD reached 1,000 in 2008, 2000 in 2012, and 3,000 natural AMPs in 2018, respectively. Post‐translational modifications annotated in the APD (e.g., N‐formylated methionine, halogenation, C‐terminal amidation, and phosphorylation) increased from seven in the APD2 and 24 in the APD3 to 26 in 2021. The peptide search functions (e.g., anti‐TB, antiviral, anticancer, and anti‐diabetic) increased from the original five in 2003, nine in 2009 to 25 in 2021 (Figure 3). These expansions over a decade greatly enriched the searchable information of antimicrobial peptides in this database. For scientific rigor, the APD3 defined a set of criteria for data registration (natural peptides, a known amino acid sequence, a size less than 100 amino acids, and demonstrated antimicrobial activity). Our definition of data registration criteria produced a core data set for natural antimicrobial peptides. This core data set in the APD laid the foundation for understanding the design principles of natural AMPs. 39 It also allowed us to propose a universal peptide classification method that does not depend on peptide source or activity. 48 Although empirical, our database programmed the first peptide prediction program. 37 The APD also provided one of the first useful data sets for researchers to develop machine‐learning predictions. 44 Moreover, we demonstrated the first database‐aided peptide design.42 In addition, our database is an important platform for developing peptide design methods such as the Grammar approach by others, 43 template‐based design, 42 , 74 database screening, 45 , 75 and database filtering technology 47 by us to identify novel antibacterial and antiviral peptides with topical and systemic efficacies in mice. 27 , 49 , 68 , 76 Our discovery of the relationships between peptide net charge and length as well as hydrophobic content and peptide length based on over 1,000 amphibian AMPs may be useful for designing linear AMPs at different lengths. 50 Our continued expansion of the APD functions for over a decade confirmed the flexibility of this database platform. Consequently, we anticipate that this modernized database will continue to play a role in research and education of antimicrobial peptides. Future research will elucidate the unique role each innate immune peptide plays in its biological niche. Such knowledge is important for developing these peptides into useful molecules for different applications. We also anticipate that future studies will continue to search for novel antimicrobial peptides, which may enrich our antimicrobial arsenal to combat drug‐resistant pathogens.
AUTHOR CONTRIBUTIONS
Guangshun Wang: Conceptualization (equal); data curation (equal); formal analysis (equal); funding acquisition (equal); project administration (equal); supervision (equal); validation (equal); writing – original draft (equal). C. Michael Zietz: Software (equal); writing – review and editing (equal). Ashok Mudgapalli: Project administration (equal); software (equal); writing – review and editing (equal). Shuona Wang: Software (equal); writing – review and editing (equal). Zhe Wang: Data curation (equal); methodology (equal); software (equal); writing – review and editing (equal).
ACKNOWLEDGMENTS
The continued update and development of this database resource would be impossible without the consistent support of Dr. Steve Hinrichs, Professor and Chair, the Department of Pathology and Microbiology, UNMC. This study is supported by grants from the National Institutes of Health in the past and current GM138552, AI137161, as well as the University of Nebraska to GW. We thank Sriram Srinivasan, Deepak Khazanchi (UNO) and Joe Ziskovsky (UNMC) for establishing and maintaining a temporary APD3 website in the Amazon cloud during the reprograming of the APD. The support of the UNMC IT for over a decade is greatly appreciated. We thank Annie Wang for editing the final manuscript.
Wang G, Zietz CM, Mudgapalli A, Wang S, Wang Z. The evolution of the antimicrobial peptide database over 18 years: Milestones and new features. Protein Science. 2022;31:92–106. 10.1002/pro.4185
Funding information National Institute of General Medical Sciences, Grant/Award Number: GM138552
REFERENCES
- 1. Boman HG. Antibacterial peptides: basic facts and emerging concepts. J Intern Med. 2003;254:197–215. [DOI] [PubMed] [Google Scholar]
- 2. Steiner H, Hultmark D, Engström A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann JA, Hetru C. Insect defensins: Inducible antibacterial peptides. Immunol Today. 1992;13:411–415. [DOI] [PubMed] [Google Scholar]
- 4. Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. Primary structures of three human neutrophil defensins. J Clin Invest. 1985;76:1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gálvez A, Maqueda M, Valdivia E, Quesada A, Montoya E. Characterization and partial purification of a broad spectrum antibiotic AS‐48 produced by Streptococcus faecalis . Can J Microbiol. 1986;32:765–771. [DOI] [PubMed] [Google Scholar]
- 6. Eliopoulos GM, Willey S, Reiszner E, Spitzer PG, Caputo G, Moellering RC Jr. In vitro and in vivo activity of LY 146032, a new cyclic lipopeptide antibiotic. Antimicrob Agents Chemother. 1986;30:532–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oppenheim FG, Xu T, McMillian FM, et al. Histatins, a novel family of histidine‐rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans . J Biol Chem. 1988;263:7472–7477. [PubMed] [Google Scholar]
- 9. Romeo D, Skerlavaj B, Bolognesi M, Gennaro R. Structure and bactericidal activity of an antibiotic dodecapeptide purified from bovine neutrophils. J Biol Chem. 1988;263:9573–9575. [PubMed] [Google Scholar]
- 10. Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. Apidaecins: Antibacterial peptides from honeybees. EMBO J. 1989;8:2387–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu PT, Stenger S, Li H, et al. Toll‐like receptor triggering of a vitamin D‐mediated human antimicrobial response. Science. 2006;311:1770–1773. [DOI] [PubMed] [Google Scholar]
- 12. Lehrer RI, Lu W. α‐Defensins in human innate immunity. Immunol Rev. 2012;245:84–112. [DOI] [PubMed] [Google Scholar]
- 13. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. [DOI] [PubMed] [Google Scholar]
- 14. Wang G, Narayana JL, Mishra B, et al. Design of antimicrobial peptides: Progress made with human cathelicidin LL‐37. Adv Exp Med Biol. 2019;1117:215–240. [DOI] [PubMed] [Google Scholar]
- 15. Gharsallaoui A, Oulahal N, Joly C, Degraeve P. Nisin as a food preservative: Part 1: Physicochemical properties, antimicrobial activity, and main uses. Crit Rev Food Sci Nutr. 2016;56:1262–1274. [DOI] [PubMed] [Google Scholar]
- 16. Gagnon MG, Roy RN, Lomakin IB, Florin T, Mankin AS, Steitz TA. Structures of proline‐rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016;44:2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mardirossian M, Grzela R, Giglione C, et al. The host antimicrobial peptide Bac71‐35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem Biol. 2014;21:1639–1647. [DOI] [PubMed] [Google Scholar]
- 18. Matsuzaki K. Membrane Permeabilization mechanisms. Adv Exp Med Biol. 2019;1117:9–16. [DOI] [PubMed] [Google Scholar]
- 19. Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. [DOI] [PubMed] [Google Scholar]
- 20. Huang HW, Charron NE. Understanding membrane‐active antimicrobial peptides. Q Rev Biophys. 2017;50:e10. [DOI] [PubMed] [Google Scholar]
- 21. Epand RM. Anionic lipid clustering model. Adv Exp Med Biol. 2019;1117:65–71. [DOI] [PubMed] [Google Scholar]
- 22. Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiedemann I, Benz R, Sahl HG. Lipid II‐mediated pore formation by the peptide antibiotic nisin: A black lipid membrane study. J Bacteriol. 2004;186:3259–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Leeuw E, Li C, Zeng P, et al. Functional interaction of human neutrophil peptide‐1 with the cell wall precursor lipid II. FEBS Lett. 2010;584:1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gan BH, Gaynord J, Rowe SM, Deingruber T, Spring DR. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem Soc Rev. 2021;50:7820–7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ling LL, Schneider T, Peoples AJ, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lakshmaiah Narayana J, Mishra B, Lushnikova T, et al. Two distinct amphipathic peptide antibiotics with systemic efficacy. Proc Natl Acad Sci U S A. 2020;117:19446–19454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptides as antibiotics: Design and application strategies. Curr Opin Chem Biol. 2017;38:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neter E, Kunz EH, Morgan ED. Synergistic effects of polymyxin B and terramycin on bacteria encountered in urinary tract infections. J Urol. 1952;67:773–775. [DOI] [PubMed] [Google Scholar]
- 30. Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: Application informed by evolution. Science. 2020;368:eaau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84:435–458. [DOI] [PubMed] [Google Scholar]
- 32. Hancock RE, Haney EF, Gill EE. The immunology of host defence peptides: Beyond antimicrobial activity. Nat Rev Immunol. 2016;16:321–334. [DOI] [PubMed] [Google Scholar]
- 33. Yu H, Dong J, Gu Y, et al. The novel human beta‐defensin 114 regulates lipopolysaccharide(LPS)‐mediated inflammation and protects sperm from motility loss. J Biol Chem. 2013;288:12270–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Candille SI, Kaelin CB, Cattanach BM, et al. A β‐defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bonar E, Bukowski M, Chlebicka K, et al. Human skin microbiota‐friendly lysostaphin. Int J Biol Macromol. 2021;183:852–860. [DOI] [PubMed] [Google Scholar]
- 36. Yang M, Yang Y, He Q, et al. Intestinal microbiota ‐ a promising target for antiviral therapy? Front Immunol. 2021;12:676232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Z, Wang G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004;32:D590–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brahmachary M, Krishnan SP, Koh JL, et al. ANTIMIC: A database of antimicrobial sequences. Nucleic Acids Res. 2004;32:D586–D589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci. 2020;29:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang G, Li X, Wang Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang G, Li X, Wang Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang G, Li Y, Li X. Correlation of three‐dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J Biol Chem. 2005;280:5803–5811. [DOI] [PubMed] [Google Scholar]
- 43. Loose C, Jensen K, Rigoutsos I, Stephanopoulos G. A linguistic model for the rational design of antimicrobial peptides. Nature. 2006;443:867–869. [DOI] [PubMed] [Google Scholar]
- 44. Lata S, Sharma BK, Raghava GP. Analysis and prediction of antibacterial peptides. BMC Bioinformatics. 2007;8:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang G, Watson KM, Buckheit RW Jr. Anti‐human immunodeficiency virus type 1 activities of antimicrobial peptides derived from human and bovine cathelicidins. Antimicrob Agents Chemother. 2008;52:3438–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang G, editor. Antimicrobial peptides: Discovery, Design and Novel Therapeutic Strategies. Wallingford, England: CABI, 2010. [Google Scholar]
- 47. Mishra B, Wang G. Ab initio design of potent anti‐MRSA peptides based on database filtering technology. J Am Chem Soc. 2012;134:12426–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol. 2015;1268:43–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mishra B, Narayana JL, Lushinikova T, Wang X, Wang G. Low cationicity is important for systemic in vivo efficacy of database‐derived peptides against drug‐resistant gram‐positive pathogens. Proc Natl Acad Sci U S A. 2019;116:13517–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang G. Bioinformatic analysis of 1000 amphibian antimicrobial peptides uncovers multiple length‐dependent correlations for peptide design and prediction. Antibiotics (Basel). 2020;9:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mishra B, Wang X, Lushnikova T, et al. Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides. 2018;106:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains archaea, bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang G, Watson KM, Peterkofsky A, Buckheit RW Jr. Identification of novel human immunodeficiency virus type 1‐inhibitory peptides based on the antimicrobial peptide database. Antimicrob Agents Chemother. 2010;54:1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang G. Post‐translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr Biotechnol. 2012;1:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tam JP, Wang S, Wong KH, Tan WL. Antimicrobial peptides from plants. Pharmaceuticals (Basel). 2015;8:711–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nolde SB, Vassilevski AA, Rogozhin EA, et al. Disulfide‐stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus‐galli). J Biol Chem. 2011;286:25145–25153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mygind PH, Fischer RL, Schnorr KM, et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. [DOI] [PubMed] [Google Scholar]
- 58. Yamanaka K, Reynolds KA, Kersten RD, et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A. 2014;111:1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hamamoto H, Urai M, Ishii K, et al. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat Chem Biol. 2015;11:127–133. [DOI] [PubMed] [Google Scholar]
- 60. Imai Y, Meyer KJ, Iinishi A, et al. A new antibiotic selectively kills gram‐negative pathogens. Nature. 2019;576:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zipperer A, Konnerth MC, Laux C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–516. [DOI] [PubMed] [Google Scholar]
- 62. Jacob L, Zasloff M. Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. Ciba Found Symp. 1994;186:197–216. [DOI] [PubMed] [Google Scholar]
- 63. Sader HS, Fedler KA, Rennie RP, Stevens S, Jones RN. Omiganan pentahydrochloride (MBI 226), a topical 12‐amino‐acid cationic peptide: Spectrum of antimicrobial activity and measurements of bactericidal activity. Antimicrob Agents Chemother. 2004;48:3112–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mosca DA, Hurst MA, So W, Viajar BS, Fujii CA, Falla TJ. IB‐367, a protegrin peptide with in vitro and in vivo activities against the microflora associated with oral mucositis. Antimicrob Agents Chemother. 2000;44:1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. de Breij A, Riool M, Kwakman PH, et al. Prevention of Staphylococcus aureus biomaterial‐associated infections using a polymer‐lipid coating containing the antimicrobial peptide OP‐145. J Control Release. 2016;222:1–8. [DOI] [PubMed] [Google Scholar]
- 66. Mickels N, McManus C, Massaro J, et al. Clinical and microbial evaluation of a histatin‐containing mouthrinse in humans with experimental gingivitis. J Clin Periodontol. 2001;28:404–410. [DOI] [PubMed] [Google Scholar]
- 67. Nissen‐Meyer J, Holo H, Håvarstein LS, Sletten K, Nes IF. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lakshmaiah Narayana J, Golla R, Mishra B, et al. Short and robust anti‐infective lipopeptides engineered based on the minimal antimicrobial peptide KR12 of human LL‐37. ACS Infect Dis. 2021;7:1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Levashina EA, Ohresser S, Bulet P, Reichhart JM, Hetru C, Hoffmann JA. Metchnikowin, a novel immune‐inducible proline‐rich peptide from drosophila with antibacterial and antifungal properties. Eur J Biochem. 1995;233:694–700. [DOI] [PubMed] [Google Scholar]
- 70. Li J, Xu X, Xu C, et al. Anti‐infection peptidomics of amphibian skin. Mol Cell Proteomics. 2007;6:882–894. [DOI] [PubMed] [Google Scholar]
- 71. Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7:545–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Donia MS, Cimermancic P, Schulze CJ, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'Sullivan JN, O'Connor PM, Rea MC, et al. Nisin J, a novel natural Nisin variant, is produced by Staphylococcus capitis sourced from the human skin microbiota. J Bacteriol. 2020;202:e00639–e00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mishra B, Lushnikova T, Golla RM, Wang X, Wang G. Design and surface immobilization of short anti‐biofilm peptides. Acta Biomater. 2017;49:316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Menousek J, Mishra B, Hanke ML, Heim CE, Kielian T, Wang G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin‐resistant Staphylococcus aureus USA300. Int J Antimicrob Agents. 2012;39:402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Su Y, Mainardi VL, Wang H, et al. Dissolvable microneedles coupled with nanofiber dressings eradicate biofilms via effectively delivering a database‐designed antimicrobial peptide. ACS Nano. 2020;14:11775–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]


