Abstract
In contrast to artificial intelligence and machine learning approaches, KEGG (https://www.kegg.jp) has relied on human intelligence to develop “models” of biological systems, especially in the form of KEGG pathway maps that are manually created by capturing knowledge from published literature. The KEGG models can then be used in biological big data analysis, for example, for uncovering systemic functions of an organism hidden in its genome sequence through the simple procedure of KEGG mapping. Here we present an updated version of KEGG Mapper, a suite of KEGG mapping tools reported previously (Kanehisa and Sato, Protein Sci 2020; 29:28–35), together with the new versions of the KEGG pathway map viewer and the BRITE hierarchy viewer. Significant enhancements have been made for BRITE mapping, where the mapping result can be examined by manipulation of hierarchical trees, such as pruning and zooming. The tree manipulation feature has also been implemented in the taxonomy mapping tool for linking KO (KEGG Orthology) groups and modules to phenotypes.
Keywords: BRITE hierarchical classification, genome annotation, KEGG, KEGG mapper, KEGG module, KEGG orthology, KEGG pathway map
1. INTRODUCTION
The KEGG database resource has been developed as a reference knowledge base for uncovering cellular and organism‐level functions from genome sequences and other molecular datasets. 1 This is accomplished by the procedure of KEGG mapping, especially with the concept of functional orthologs. When KEGG was first released in 1995, it consisted of just four types of data contents: manually drawn metabolic pathway maps, gene catalogs taken from genome sequences, and enzymes and chemical compounds found in enzymatic reactions. The EC number in Enzyme Nomenclature 2 was the identifier for linking genomes to metabolic pathways, the original concept of KEGG mapping through functional orthologs. Reference (generic) metabolic pathway maps were drawn as networks of EC number nodes and KEGG mapping was enabled by assigning EC numbers to enzyme genes in the genome, thus computationally generating (reconstructing) organism‐specific metabolic pathways with gene product nodes.
By 2000 the EC number was replaced by the ortholog identifier, 3 later called the KO (KEGG Orthology) identifier, for its role of KEGG mapping, in order to include signaling and other non‐metabolic pathways. Now all the KEGG pathway maps, as well as BRITE protein family classifications and KEGG modules, are created as networks of KO identifiers, also called K numbers, and KEGG mapping is enabled by assigning K numbers to genes in the genome. Direct KEGG mapping without conversion through functional orthologs is also available, including metabolites mapped to metabolic pathways, drugs and diseases mapped to BRITE hierarchical classifications, and cellular organisms and viruses mapped to NCBI taxonomy. 4 This article reports KEGG Mapper Version 5, an updated collection of KEGG mapping tools as a sequel to Version 4 reported in the previous article. 5
2. OVERVIEW OF KEGG
KEGG is an integrated database consisting of 16 databases in four categories as shown in Table 1. Fourteen databases excluding GENES and ENZYME are original databases that are all manually created. The sequence data in GENES are taken from RefSeq, GenBank, and other public sequence databases, and given original annotation of gene/protein functions represented by KOs. The EC numbers in ENZYME are taken from ExplorEnz, 2 the official database of Enzyme Nomenclature, and given annotation of enzyme sequence data links.
TABLE 1.
KEGG database contents and identifiers
| Category | Database | Content | KEGG identifier | Expanded prefix |
|---|---|---|---|---|
| Systems information | PATHWAY | KEGG pathway maps | map number | <org>, ko/ec/rn |
| BRITE | BRITE hierarchies and tables | br/ko number | <org> | |
| MODULE |
KEGG modules Reaction modules |
M number RM number |
<org>_M | |
| Genomic information | KO | KO groups for functional orthologs | K number | |
| GENES | Genes and proteins | <org>:<entry> | ||
| GENOME | KEGG organisms and viruses | T number, gn:<org> | ||
| Chemical information | COMPOUND | Small molecules | C number | |
| GLYCAN | Glycans | G number | ||
| REACTION | Biochemical reactions | R number | ||
| RCLASS | Reaction class | RC number | ||
| ENZYME | Enzyme nomenclature | Ec:<entry> | ||
| Health information | NETWORK |
Network variation maps Disease‐related network elements |
nt number N number |
|
| VARIANT | Human gene variants | hsa_var:<entry> | ||
| DISEASE | Human diseases | H number | ||
| DRUG | Drugs | D number | ||
| DGROUP | Drug groups | DG number |
Abbreviations: <org>, KEGG organism code such as hsa for Homo sapiens; <entry>, entry identifier.
Each entry of the entire KEGG database is uniquely identified by specifying the KEGG identifier (Table 1), which takes the form of a prefix followed by a five‐digit number, called map number, K number, etc., or the combination of a database name and an entry identifier in the form of “db:entry”. Each entry can be retrieved by entering the KEGG identifier in the search box of the KEGG top page or by specifying a simple URL shown in Table 2. In addition to the general viewer (www_bget), specialized viewers are available for KEGG pathway maps (show_pathway), BRITE hierarchies (show_brite), KEGG modules (show_module), and network variation maps (show_network). The pathway map viewer and the BRITE hierarchy viewer shown in Figure 1 allow KEGG mapping as a client‐side operation, which can be initiated by clicking on the plus sign in the side panel to add a query dataset.
TABLE 2.
KEGG database content viewers
| Viewer | Content | URL | Example of <id> |
|---|---|---|---|
| www_bget | All database contents | https://www.kegg.jp/entry/<id> | K09708, hsa:59272 |
| show_pathway | KEGG pathway maps | https://www.kegg.jp/pathway/<id> | map00140, hsa00140 |
| show_brite | BRITE hierarchies | https://www.kegg.jp/brite/<id> | br08303, ko00199, hsa00199 |
| show_module | KEGG modules | https://www.kegg.jp/module/<id> | M00107, hsa_M00107 |
| show_network | Network variation maps | https://www.kegg.jp/network/<id> | nt06019 |
Note: The URL for the KEGG main site (www.kegg.jp) may be changed to the GenomeNet mirror site (www.genome.jp).
Abbreviation: <id>, KEGG identifier.
FIGURE 1.
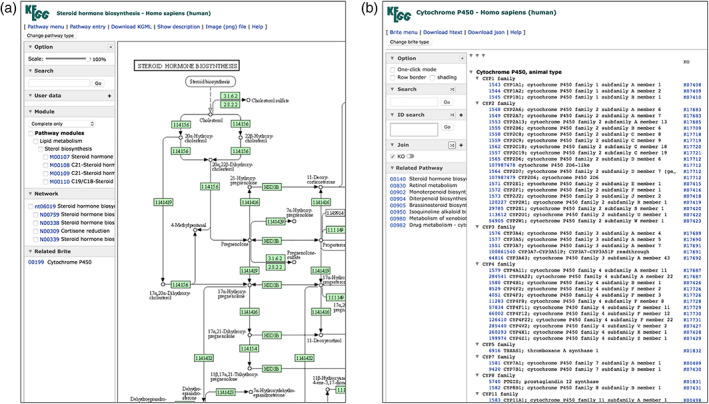
In the new versions of (a) the KEGG pathway map viewer and (b) the BRITE hierarchy viewer, the side panel is available for various client‐side operations, including KEGG mapping operations. The User data section of the pathway map viewer corresponds to the Color tool of KEGG Mapper applied to a single pathway map. The ID search and Join sections of the BRITE hierarchy viewer correspond to the Search and Join tools applied to a single hierarchy file. The plus sign in each section is used to open an window for user data input
There is a convention of expanding the prefix of KEGG identifiers with the organism code <org>, when organism‐specific versions are computationally generated, for the map number of pathway maps, the ko number of BRITE hierarchies, and the M number of KEGG modules (Tables 1 and 2). For example, map00140 is the manually created reference pathway map for steroid hormone biosynthesis and hsa00140 with the organism code “hsa” for Homo sapiens is the corresponding human pathway map with coloring of green for the nodes linked to human genes (Figure 1a). Similarly, ko00199 is the manually created Brite hierarchy for cytochrome P450, and hsa00199 is the corresponding hierarchy for H. sapiens (Figure 1b).
3. KEGG MAPPER VERSION 5
The Version 5 of KEGG Mapper (https://www.kegg.jp/kegg/mapper) released in July 2021 consists of four tools, Reconstruct, Search, Color, and Join, as summarized in Table 3. In comparison to the previous version, 5 the names of the first three tools were changed from Reconstruct Pathway, Search Pathway, and Search&Color Pathway, and the Join tool was improved to be compatible with the other tools. Each tool accepts query data as a set of KEGG identifiers through the web form, searches multiple target databases for entries containing matching KEGG identifiers, and returns the result as the lists of found entries in multiple tabs. When an entry is selected from the list, one of the viewers in Table 2 is used to show the actual mapping result.
TABLE 3.
KEGG Mapper tools
| Tool | Search mode | Target database | Query data (KEGG identifier) |
|---|---|---|---|
| Reconstruct | Reference |
Pathway Brite hierarchy Brite table Module |
K number |
| Search | Reference |
Pathway Brite hierarchy Brite table Module |
K/R/EC number C/G/D/H number KEGG organism code |
| Human‐specific |
Pathway (hsa) Brite hierarchy (hsa) Module (hsa) Network Disease |
Human gene identifier C/G/D number |
|
| Other organism‐specific |
Pathway (org) Brite hierarchy (org) Module (org) |
Gene identifier C/G/D number |
|
| Color | Reference | Pathway |
K/R/EC number C/G/D number |
| Organism‐specific | Pathway (org) |
Gene identifier C/G/D number |
|
| Join | Reference |
Brite hierarchy Brite table |
K number C/G/D/H number KEGG organism code |
3.1. Reconstruct
The Reconstruct tool is the basic tool for KEGG mapping through functional orthologs, which is widely utilized for interpretation of genome and metagenome sequences. As described in the previous article, 5 this tool accepts the output file of the automatic annotation servers 6 , 7 , 8 available at the KEGG and GenomeNet websites. Alternatively, the Assign KO tool in the KEGG Mapper page may be used to quickly assign KOs when closely related genomes are already in KEGG. The annotation output file contains the user's gene identifiers in the first column and the assigned K numbers in the second column. The Reconstruct tool uses only the K numbers to perform mapping against KEGG pathway maps, BRITE hierarchies and tables, and KEGG modules with completeness checks.
One improvement of the new Reconstruct tool is that the KEGG modules representing conserved units of metabolic functions are integrated into the metabolic pathway maps. The list of complete modules, as well as one block missing modules and other incomplete modules, in the Module tab appears in the side panel of the pathway map viewer for individual metabolic pathways selected from the Pathway tab. Furthermore, the global and overview maps (map numbers 01100s and 01200s) may be viewed either in the normal mode with links to KOs or in the module mode with links to modules. The global map of metabolic pathways (map01100), the largest KEGG pathway map, can be treated as consisting of 4400 KOs or as consisting of 370 modules, the latter being more convenient to characterize metabolic capacities in specific organisms or environmental samples. 9
3.2. Search
The Search Pathway tool existed from the beginning of the KEGG database for searching map objects of rectangles (gene products) and circles (chemical compounds) in KEGG pathway map diagrams. As the contents of KEGG expanded, so did the variety of searches. The Search tool is for direct mapping of objects, including genes and proteins, chemical compounds and reactions, and drugs, as they appear in KEGG pathway maps, BRITE hierarchies and tables, and KEGG modules, as well as in network variation maps and disease entries for human datasets. The mapped objects are marked in red. In contrast to the Reconstruct tool, which is limited to genomics data, the Search tool has much wider applications in omics data including transcriptomics, proteomics, metabolomics, and glycomics, and also other data such as drugs and diseases.
The current version is basically the same as the previous version 5 except the treatment of aliases. The use of gene symbols (in the Gene name field of GENES entry) as aliases is no longer supported, because many‐to‐many relationships may result in erroneous links to KEGG identifiers. However, widely used HGNC symbols 10 for hsa (Homo sapiens) are accepted as primary identifiers using the correspondence to KEGG human gene identifiers updated every 3 months of RefSeq releases.
3.3. Color
The Color tool is another traditional tool, used to be called Search&Color Pathway. It works in the same way as the Search tool except that the mapped objects may be colored in any combination of background and foreground colors in order to distinguish, for example, up‐regulated and down‐regulated genes. In the current version the target database is limited to KEGG pathway maps only, and the automatic conversion of outside gene identifiers is no longer supported. The Convert ID tool in the KEGG Mapper page may be used to do the same conversion.
The coloring of a pathway map is performed on the server side in the Color tool, but the new pathway map viewer (Figure 1a) has the capability to do coloring on the client side. For a selected pathway map, click on the plus sign in the User data section of the side panel to open a widow for query data input. The query data are entered in the same way as the Color tool, KEGG identifiers followed by specification of background and foreground colors. By default the dataset is stored in the local storage of the web browser and there is an option to use the dataset in all pathway maps.
3.4. Join
The Join operation is to combine a BRITE hierarchy or table file with a binary relation file by matching KEGG identifiers, effectively adding a new column to the BRITE file. The Join tool, which was not described in the previous article, 5 has been significantly improved and is now part of KEGG Mapper. The Join tool can be applied to ko‐prefixed BRITE hierarchies for genes and proteins, br‐prefixed BRITE hierarchies for chemical compounds (br numbers 08000s), drugs (08300s), diseases (08400s), cellular organisms and viruses (08600s), and other objects. The Join tool can also be applied to BRITE table files for drugs, which are represented as html table files.
4. BRITE MAPPING
The KEGG mapping against BRITE hierarchies can now be performed in two ways. One is the search operation used in the Reconstruct and Search tools, and the other is the join operation used in the Join tool. The former displays the result by marking (coloring) of nodes in a similar way as the other target databases, and the latter displays the result by adding a new column. These two operations may be compared with the search and color operations against KEGG pathway maps. The search pathway operation accepts a set of KEGG identifiers, while the color pathway operation accepts a set of binary relations between KEGG identifiers and color specification. In fact, coloring of BRITE hierarchies is possible with the Join tool by using html tags for color specification in the additional column.
The newly released BRITE hierarchy viewer allows both types of operations to be performed on the client side using the ID search section and the Join section of the side panel (Figure 1b). In addition, the viewer allows manipulation of hierarchical trees with pruning and zooming functions. The default pruning is to display only the matching nodes and the branches leading to them, which can be applied with a scissor button separately for the keyword Search, ID search, and Join. In the current KEGG Mapper implementation, the pathway map viewer utilizes processed pathway maps sent from the server, while the BRITE hierarchy viewer receives only the query data and performs mapping on the client side. This is a great advantage, for the user data may be included or excluded in tree manipulations especially when combined with the predefined join lists as shown in Figure 2.
FIGURE 2.
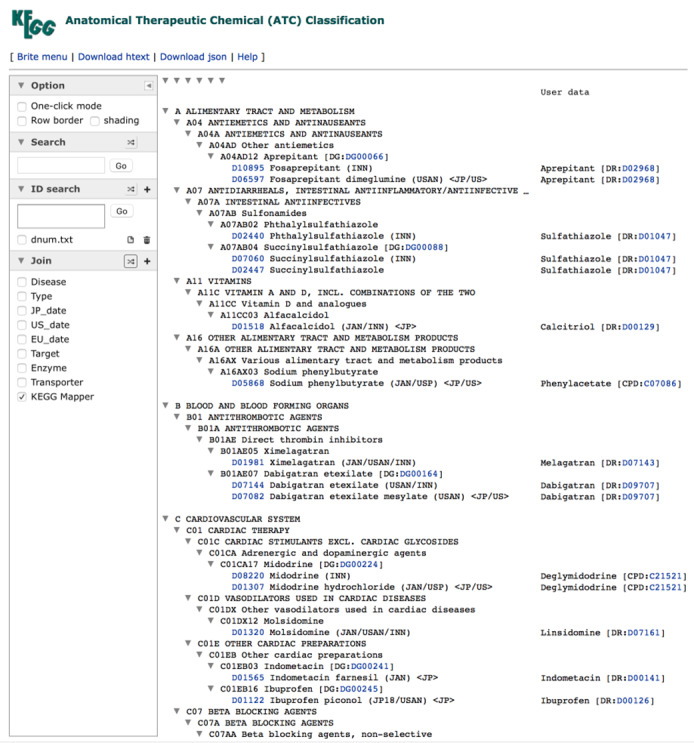
An example of using the Join tool of KEGG Mapper, where the dataset of prodrug to active substance relations is joined with br‐prefixed BRITE hierarchy files. One of the matching BRITE files, br08303 for the ATC drug classification, is shown here. Since the KEGG Mapper result appears in the Join list, it may be examined by combining with other predefined datasets
The predefined list of binary relations for the join operation appears in selected BRITE hierarchy files. Binary relation files are created mostly by extracting specific fields of database entries, such as Target, Metabolism, Disease, and other fields from DRUG entries. With this reorganization, any BRITE hierarchy file no longer contains tab‐separated columns, which now appear in the join list of binary relations.
5. TAXONOMY MAPPING
The KEGG database uses the NCBI taxonomy 4 for classification of cellular organisms and viruses, which are implemented as several Brite hierarchy files. The br08601 file for KEGG organisms is manually created to define the order of organism codes with hsa (Homo sapiens) at the top. The br08610 file is computationally generated using the abbreviated lineage of the NCBI taxonomy for cellular organisms keeping the order of organism codes defined in br08601. The br08611 file is also computationally generated with the fixed number of hierarchy levels for the taxonomic ranks of species, genus and other organism groups. For viruses, the br08620 file is computationally generated from the NCBI taxonomy for viruses, which is based on the ICTV taxonomy, 11 with the traditional Baltimore classification at the top level added by KEGG. 9 The br08610 and br08620 files are used with the Taxonomy and Virus taxonomy buttons, respectively, in KO and module entry pages.
The taxonomy mapping tool linked from the KEGG Mapper page is a special purpose Join tool, designed to integrate taxonomic distributions of KOs and modules with phenotypic features. The taxonomy file used here is br08611 with the fixed number of hierarchy levels, and the tree manipulations are somewhat different from the standard BRITE mapping. First, the pruning involves the display of not only the matching nodes, but also non‐matching sibling nodes under the same parent node. Second, the number of hierarchy levels can be changed, which is called zooming. Figure 3 is an example of viewing the matched organisms with zooming in and out by the greater‐than and less‐than signs, revealing what fraction of organisms are matched (colored in red) under the changing resolution of organism groups.
FIGURE 3.

The taxonomy mapping tool shows the distribution of KEGG organisms for a given set of KOs (K numbers) and modules (M numbers) as well as for user‐defined data. The tool works with a specially organized BRITE hierarchy file, br08611 for KEGG organisms in taxonomic groups. Here the mapping result is shown with (a) zooming in to the species level or (b) zooming out to the genus level, revealing what fraction of organisms are matched (colored in red) under the changing resolution of organism groups
6. CONCLUSION
KEGG pathway maps and BRITE hierarchies have been developed as a computer representation of biological information systems in the cell and the organism, capturing knowledge from literature and manually creating molecular wiring‐diagrams and hierarchies among biological objects. In addition to more basic aspects of KEGG, 1 it has practical values of enabling integration and interpretation of diverse biological datasets. The continuous development of the KEGG Mapper suite is an attempt to meet such practical needs. The new release reported here presents a new type of BRITE mapping with tree manipulation features.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Minoru Kanehisa: Conceptualization (lead); project administration (lead); resources (lead); writing—original draft (lead). Yoko Sato: Resources (supporting); software (equal); validation (equal); visualization (equal). Masayuki Kawashima: Resources (supporting); software (equal); validation (equal); visualization (equal).
ACKNOWLEDGMENTS
The KEGG project is partially supported by the National Bioscience Database Center of the Japan Science and Technology Agency. Computational resources were provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University.
Kanehisa M, Sato Y, Kawashima M. KEGG mapping tools for uncovering hidden features in biological data. Protein Science. 2022;31:47–53. 10.1002/pro.4172
Funding information Institute for Chemical Research, Kyoto University; National Bioscience Database Center, Japan Science and Technology Agency
REFERENCES
- 1. Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McDonald AG, Tipton KF. Fifty‐five years of enzyme classification: advances and difficulties. FEBS J. 2014;281:583–592. [DOI] [PubMed] [Google Scholar]
- 3. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kanehisa M, Sato Y. KEGG mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. [DOI] [PubMed] [Google Scholar]
- 7. Aramaki T, Blanc‐Mathieu R, Endo H, et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. KAAS: An automatic genome annotation and pathway recction server. Nucleic Acids Res. 2007;35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanehisa M, Furumichi M, Sato Y, Ishiguro‐Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tweedie S, Braschi B, Gray K, et al. Genenames.org: the HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021;49:D939–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 2018;46:D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]


