Abstract
The Protein Data Bank (PDB) archive is a rich source of information in the form of atomic‐level three‐dimensional (3D) structures of biomolecules experimentally determined using macromolecular crystallography, nuclear magnetic resonance (NMR) spectroscopy, and electron microscopy (3DEM). Originally established in 1971 as a resource for protein crystallographers to freely exchange data, today PDB data drive research and education across scientific disciplines. In 2011, the online portal PDB‐101 was launched to support teachers, students, and the general public in PDB archive exploration (pdb101.rcsb.org). Maintained by the Research Collaboratory for Structural Bioinformatics PDB, PDB‐101 aims to help train the next generation of PDB users and to promote the overall importance of structural biology and protein science to nonexperts. Regularly published features include the highly popular Molecule of the Month series, 3D model activities, molecular animation videos, and educational curricula. Materials are organized into various categories (Health and Disease, Molecules of Life, Biotech and Nanotech, and Structures and Structure Determination) and searchable by keyword. A biennial health focus frames new resource creation and provides topics for annual video challenges for high school students. Web analytics document that PDB‐101 materials relating to fundamental topics (e.g., hemoglobin, catalase) are highly accessed year‐on‐year. In addition, PDB‐101 materials created in response to topical health matters (e.g., Zika, measles, coronavirus) are well received. PDB‐101 shows how learning about the diverse shapes and functions of PDB structures promotes understanding of all aspects of biology, from the central dogma of biology to health and disease to biological energy.
Keywords: education, PDB, Protein Data Bank, protein structure and function, structural biology, training
1. INTRODUCTION
The Protein Data Bank (PDB) is the central archive of experimentally determined three‐dimensional (3D) atomic‐level structures of biological macromolecular machines. The PDB resource is essential to multiple communities: It provides a permanent and expertly curated resource for structural biologists to archive and disseminate their work, provides mechanisms to ensure reproducibility in the structural biology literature, and makes biomolecular structures freely and easily available to a wide community of researchers across scientific disciplines. 1 PDB data are also broadly used in science, technology, engineering, and mathematics (STEM)‐related education, including textbook illustrations, coursework, and learning activities. 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 Images of 3D PDB structures are frequently used to tell scientific stories in mainstream news media, starting with the familiar double helix of DNA to more recent examples of CRISPR and coronavirus. 12
The PDB was established in 1971 at Brookhaven National Laboratory as the first open‐access, digital‐data resource in biology. 13 In 1999, Research Collaboratory for Structural Bioinformatics (RCSB) PDB became responsible for PDB management. 14 Since 2003, the archive has been managed by the Worldwide PDB (wwPDB; wwPDB.org) partnership. 1 , 15 wwPDB members validate, curate, and make data deposited by scientists around the globe publicly which are freely available. 16 wwPDB members also host services providing access to PDB data. 17 , 18 , 19 , 20 , 21
RCSB PDB is responsible for US PDB operations; the research‐focused web portal at RCSB.org supports millions of users worldwide. 14 , 20 This website provides specialized tools and services that support users across scientific disciplines access, analyze, and visualize rich, up‐to‐date structural views of fundamental biology, biomedicine, and energy sciences. 2 , 22
Historically, RCSB PDB has been committed to building educational materials that highlight the power of PDB data and streamline access and understanding of PDB holdings. The inaugural and most enduring activity has been the Molecule of the Month (MOTM) series. Every month since 2000, a new MOTM column has told stories about PDB structures, focusing on their functions and diverse roles within living cells, and highlighting growing connections between biology, medicine, and nanotechnology. 23 This series was the main focus of RCSB PDB online educational development for many years. Over time, materials were added to the “educational resources” section of RCSB.org, such as the first edition of the Molecular Machinery poster (2002) that provides a tour of PDB structures (Figure 1).
FIGURE 1.
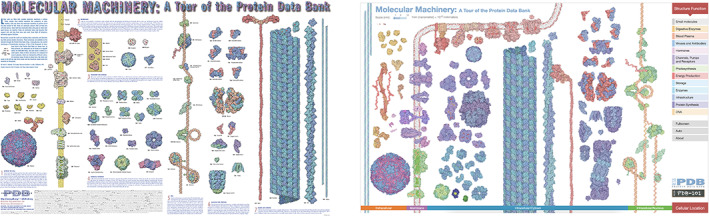
Molecular Machinery posters produced in 2002 (left) and 2014 (right). The enormous range of molecular shapes and sizes in the Protein Data Bank (PDB) is illustrated by these molecular machines. The first poster was one of the earliest outreach projects from the Research Collaboratory for Structural Bioinformatics PDB, in addition to the Molecule of the Month (MOTM) series. This printed poster was distributed to thousands of PDB users, teachers, and students over the years. The graphic was updated in 2014 to celebrate the milestone of 100,000 released PDB structures. The updated version reflects the increase in structural complexity in the archive and demonstrates PDB‐101's emphasis on storytelling. Structures are depicted relative to the cellular membrane and organized in various categories related to function. A scale bar provides a sense of molecular size in nanometers. The 2014 edition is available as a poster, flyer, and interactive animation; most of the structures shown are highlighted in MOTM articles. All related materials are available from https://pdb101.rcsb.org/learn/flyers‐posters‐and‐other‐resources/flyer/molecular‐machinery‐a‐tour‐of‐the‐protein‐data‐bank
With the release of the 100th MOTM feature on Adrenergic Receptors in 2008, RCSB PDB had established a modest collection of educational content. However, features were not easily accessible unless visitors were familiar with PDB structures or knew about specific molecules of interest. RCSB PDB assigned categories to MOTM articles (Protein Synthesis, Enzymes, Health and Disease, Biological Energy, Infrastructure and Communication, and Biotechnology and Nanotechnology) so that users could approach the archive with a question (how are genes replicated?) rather than an answer (DNA polymerase).
This top‐down approach to packaging educational content led to the first official release of the educational online portal PDB‐101 in 2011 that was accessible by clicking on the new blackboard‐inspired logo on the RCSB PDB website. 24 The name “101” denotes an entry‐level course and is meant to signal the intended audience: STEM undergraduates and high school students. The focus of PDB‐101 was the improved Structural View of Biology browser that supported contextual browsing of MOTM articles, augmented with separate collections of posters and other introductory materials. All resources were integrated within the main RCSB PDB website architecture with the intent of fostering even tighter connections and interactions between educational content and the scientific data principally consumed by researchers.
Creation of a better home for RCSB PDB's educational materials worked, but soon, the design of the initial launch proved insufficiently flexible for ready integration of new content. The website architecture limited content exploration to browsing static HTML pages; any kind of searching was not an option. Adding new content, and particularly nonstandard web page content, required implementation by a software developer. While the main RCSB PDB website focused on providing many tools for interactive query, visualization, and analysis, PDB‐101 offerings were flat, limited, and difficult to find.
The main RCSB PDB website regularly releases new tools and features for use by scientific researchers such as pairwise structure alignment, advanced search on a wide range of structure attributes, and a tool for mapping structure annotations to a sequence. 20 Review of web access to the PDB‐101 content available from the RCSB PDB website at the time made it clear that there was enough of an audience that a new and separate website for PDB beginners could be established. Between MOTM articles and other features, there was enough content to serve users interested in the biological stories contained in the PDB. To better engineer PDB‐101, the concept of a separate education‐focused portal was revisited and released in 2015. New website technology was needed to make the website searchable, easily updated and extensible to new content, mobile‐device friendly, and enable simple access to 3D views of PDB structures. These advances helped create an educational website for PDB data that could run in parallel to the more expert RCSB.org.
An organizational framework for content was developed to support access to different types of features from the top menu and the home page and from the new search box. Assignment of keywords and scientific categories to all features helped to expose access to all PDB‐101 content. A 3D molecular browser was added to the home page that enabled interactive views of the current MOTM. The resulting resource boasted user‐friendly navigation, keyword searching, and a Browse feature providing access to content organized by scientific concept, all still in use today. 25
This evolution has enabled PDB‐101 to provide educational resources centered on 3D biomolecular structure data for learning about biological concepts. The majority of materials are designed to target high school and undergraduate students, while also appealing to a broad range of users across expertise and discipline. PDB‐101 materials demonstrate the value of structural biology in understanding topics across the sciences by providing direct access to primary PDB structure data and providing multimodal resources for access and understanding.
2. PDB‐101 FEATURES
PDB information is regularly streamlined for students and teachers at PDB‐101 by a team of scientists and outreach specialists at the RCSB PDB. The goal is to expand and improve the collection of materials that explore biomolecular structure and function and highlight their application to human health and welfare. New content is developed with multiple audiences in mind and is designed to make PDB shapes and functions familiar to target audiences. Many PDB‐101 pages feature topics commonly used in classrooms such as hemoglobin function and evolution. More topical features help students and the public understand current events through the lens of structural biology such as the use of coronavirus protein structures 26 in the discovery and development of vaccines and therapies to fight the COVID‐19 pandemic (https://pdb101.rcsb.org/browse/coronavirus). PDB‐101 features also serve as an introduction to help nonexpert users understand and navigate the PDB archive, and highlight the new, cutting‐edge technologies (such as X‐ray free‐electron lasers) that are being used to determine biomolecular structures. To make these materials accessible to a wide range of users, PDB‐101 materials are clearly illustrated, written in an accessible, story‐driven language, and provide multiple pathways to introduce biomolecular concepts and then empower users to explore the topics in more detail at the main PDB archive. 3
Visitors to PDB101.rcsb.org are offered multiple modes for finding materials relevant to their interests. Some of these search options are based on biological topics. The main search box at the top of each page queries content by title and keyword (e.g., a search for vaccine will return content related to human papillomavirus, measles, SARS‐CoV‐2, and more). PDB‐101 materials are also accessible by selecting Browse from the top menu or home page widget to explore categories related to biological function, relevance to health, biotechnology, and topics related to the science of structural biology (Figure 2). The menu at the top of every PDB‐101 web page offers quick entry to all content organized by type: MOTM; Learn (instructional materials and articles, guides, videos, activities, 3D printing, and “other” features like structural biology playing cards and a board game); Global Health (disease‐based articles); Teach (curricular modules), SciArt (digital galleries); and Events. A selection of these items will be described here.
FIGURE 2.
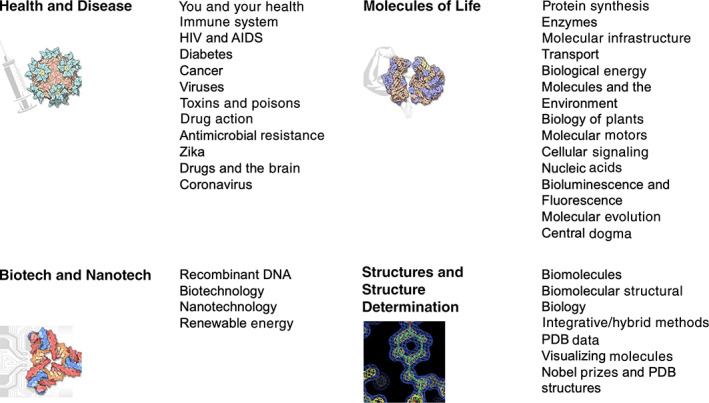
PDB‐101 categories. Users can browse these categories to find corresponding content and activities at https://pdb101.rcsb.org/browse
The PDB‐101 website is built around a modern, extensible database technology. All activities are supported by the robust RCSB PDB infrastructure that ensures 24/7/365 support of millions of PDB data depositors and users worldwide.
2.1. MOTM series
For more than 20 years, the MOTM series has inspired and educated readers around the world. It is a regular read for students, educators, and researchers alike. 23 Consistently, more than 50% of all PDB‐101 traffic each year is focused on MOTM content. MOTM content received more than 1 million visits in 2020, with the article on Coronavirus Proteases the most popular article with more than 85,000 views. Each installment includes an introduction to the structure and function of the molecule, a discussion of the relevance of the molecule to human health and welfare, and suggestions for how visitors might continue exploration in the main PDB archive. A consistent style of bright, cartoon‐like illustrations presents molecular “portraits” in a non‐photorealistic rendering that highlights the shape and form of the atomic structures throughout the series. 27 This same style is used in related watercolor paintings that provide a visual context for molecules inside the cell in the Goodsell Molecular Landscapes SciArt gallery at PDB‐101. Additionally, a public web interface and source code for the Illustrate software program used to create MOTM images are available (ccsb.scripps.edu/illustrate) 27 ; users can also use the “Illustrative” representation in the Mol* 3D viewer at RCSB.org. 28
New articles are highlighted on the PDB‐101 and RCSB PDB home pages, announced in a monthly MOTM electronic newsletter, and published on social media.
Given the diversity of MOTM content, visitors are provided with effective methods to search the archive of entries for topics of interest and in turn guide visitors to related materials. Readers do not need to read the column each month in order to find features of interest. The entire MOTM archive can be explored by title, year, or category, which is useful in cases where visitors click through a topic hierarchy. For visitors who have a specific molecule in mind, relevant articles are returned by keyword or title searches. Individual MOTM articles link to related PDB‐101 Browse categories, connect to specific PDB structure entries highlighted in an article, and suggest additional content for further exploration. Integration of MOTM articles with other content aims to provide accessible pathways for novice users to begin exploring PDB data.
MOTM topics are selected with many criteria in mind. Foremost, topics are chosen to promote understanding of the biostructural basis of fundamental biology, biomedicine, biotechnology, and biological energy (Figure 3). Throughout the year, molecules are selected to cover the entire range of these topics, to ensure that the varied interests of our audience are addressed. MOTM topics also support the PDB‐101 biennial health focus, for example, with a recent focus on drugs and the brain, and a planned focus on the molecular bases of cancer. Other articles highlight emerging trends or interesting structures as they are released in the PDB (e.g., 3D structures resulting from the ongoing resolution revolution in cryo‐electron microscopy 29 and topics related to Nobel Prizes), and topics of societal interest, such as the COVID‐19 pandemic and Designed Proteins and Citizen Science. In this way, PDB‐101 grows and matures along with the PDB archive.
FIGURE 3.
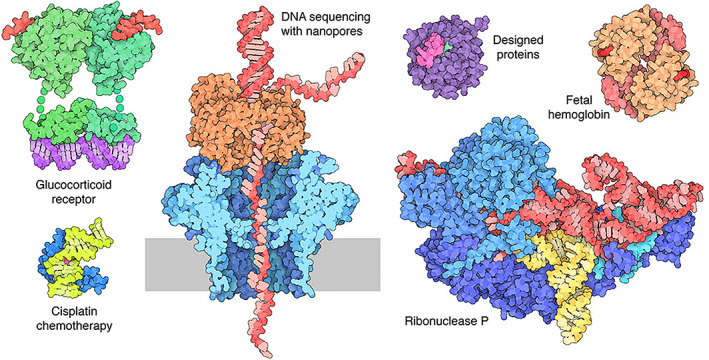
Selected Molecule of the Month (MOTM) topics from 2021. These articles were written and illustrated by students in collaboration with PDB‐101 as part of a week‐long boot camp on “Science Communication in Biology and Medicine” for undergraduate and graduate students hosted by the Rutgers Institute for Quantitative Biomedicine in January 2021. MOTM articles organized by year can be accessed at https://pdb101.rcsb.org/motm/motm‐by‐date
2.2. Materials for training and education
Steady growth of structural biology and the open‐access PDB archive are increasing the impact of 3D biostructure data on basic and applied research across disciplines. This represents an audience of users who need resources to help learn the particulars of PDB data and structural biology. Within PDB‐101, one of our goals is to provide training materials for users who are not versed in the technical aspects of structural biology to enable the use of biostructural PDB data in research and education across scientific disciplines. More formal technical documentation for expert users is available at RCSB.org. As with other PDB‐101 initiatives, multiple approaches aim to support the differing needs of our user communities.
2.2.1. Training the next generation of PDB users
PDB‐101 offers an evolving Guide to Understanding PDB Data that delves into issues specific to the PDB archive surrounding data representation, visualization, and experimental methods. These materials are written at a general level, guiding users first through the basic structural and biological principles that underlie the archive and the structures therein, and then describing tools that users will need to explore the archive themselves. For example, materials pertaining to macromolecular crystallography, nuclear magnetic resonance (NMR), and electron microscopy (3DEM), the major experimental methods used to elucidate PDB structures, are available throughout the Guide and PDB‐101. In 2020, Guide materials were accessed more than 208,000 times, with the most‐accessed entry the Introduction to Biological Assemblies and the PDB Archive. The Guide is also updated as the PDB archive grows and transforms, such as the recent article on carbohydrates that accompanies a remediation project that reviewed and improved the representation of over 14,000 carbohydrate‐containing entries accounting for nearly 10% of the archive. 30
2.2.2. Materials for education and learning
The PDB archive holds structural entries that underpin all of the basic principles of biomolecular structure and function, making it an unparalleled resource for biological education. PDB represents an important example of a body of scientific knowledge from which educators can directly use the primary data from scientists as part of lessons. PDB‐101 includes multiple resources to assist educators and students in using PDB data for learning.
Curriculum Modules, primarily aimed at high school teachers but extensible for other levels, provide lesson plans based on current scientific standards and facile connections to the PDB entries that are relevant to the topic. Accessible from the Teach pull‐down menu, these modules include authentic data from trusted public resources, hands‐on activities, teaching materials, individual and group activities, and assessment suggestions. They were developed through the collaboration and participation of scientists, curriculum design experts, educators, clinicians, and teachers. Topics include biomolecular structures, diabetes, immunology, COVID‐19, and HIV/AIDS. These resources were accessed more than 48,000 times in 2020.
PDB‐101 also provides many activities and materials, found in the Learn pull‐down menu, designed to augment ongoing education. They are designed to be fun and engaging, while remaining tightly coupled to the underlying biomolecular concepts that are being presented. Paper Models provide an easy‐to‐use and cost‐effective method for exploring the 3D nature of biomolecules. For example, paper models of antibodies, green fluorescent protein, and GPCRs may be downloaded as PDF files and printed. Individual pieces for beta‐sheets, alpha‐helices, and loops are then cut out with scissors, assembled first into a long primary chain structure, and then folded into the globular tertiary structure. Similarly, models of Zika and other viruses allow students to explore the role of icosahedral symmetry in the molecular architecture of viral capsids. The collection of paper models were accessed more than 106,000 times in 2020. Other activities include coloring books for future scientists young and old, and a wide range of posters, flyers, and online interactive presentations are available for popular biomolecular topics.
Students and educators are increasingly turning to digital resources, given the pervasive impact of the internet on all aspects of our lives. In recent years, we have found that short videos of molecular animations represent an incredibly effective way to explain 3D structures (and related functions) found in the PDB and make these concepts accessible to a wider audience. 3D animations of molecular stories can show groups of proteins moving around in their cellular context. Videos also illustrate the action of individual proteins in sequential events. Such presentations highlight interdependencies between individual proteins and promote deeper understanding of life processes carried by proteins.
Creating a PDB‐101 video is a multistep process. A storyboard comes first to map out a cohesive molecular story along with the selection of PDB structures needed to illustrate. Sometimes multiple PDB structures must be combined together in an in silico model for visualizing a functional protein in its entirety. Such structure modeling is usually carried out using UCSF Chimera. 31 The structural models are then imported into molecular Maya (mMaya; https://clarafi.com/tools/), a free plug‐in for the hallmark, commercially available 3D animation program Autodesk Maya (https://www.autodesk.com/products/maya/overview). This software enables the creation of complex animations and cellular scenes. Scenes are rendered and combined in video editing software, the narration is added, and the final product is hosted publicly via YouTube at PDB‐101 (https://pdb101.rcsb.org/learn/videos) and the YouTube channel RCSBProteinDataBank. YouTube greatly extends the reach of these animations beyond the PDB‐101 website. Videos were played more than 1.8 million times on YouTube in 2020; PDB‐101 pages that have these videos embedded were only accessed ~110,000 times during the same period.
The video creation process can be complex and time‐consuming, so the topics featured are carefully considered for maximum impact. Selected subjects must be relevant to high school and undergraduate education and ideally represent areas currently lacking 3D‐structure‐driven learning materials. Molecular stories relating to the PDB‐101 health focus, particularly those not well‐covered elsewhere, are prioritized (e.g., Opioids and Pain Signaling).
Currently available videos are centered on Introductions to Protein Structure, Structures and Disease, and Molecular Animation Guides. Recently produced videos have highlighted the 2020–2021 PDB‐101 Drugs and the Brain health focus: Neuronal Signaling and Sodium‐Potassium Pump and Opioids and Pain Signaling (Figure 4).
FIGURE 4.
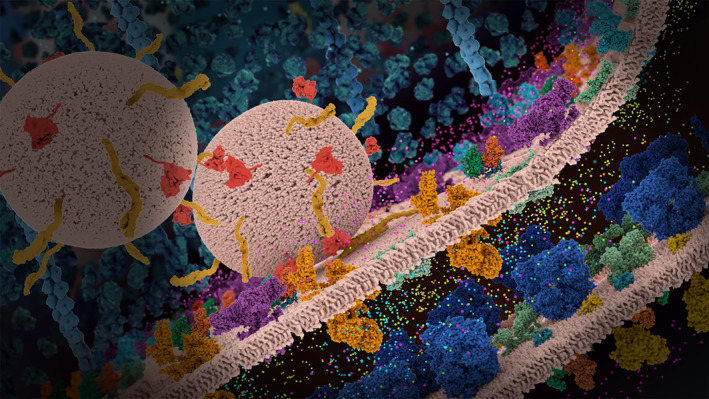
Still image from the video Opioids and Pain Signaling of a neuronal synapse with vesicles and relevant proteins highlighted (https://pdb101.rcsb.org/learn/videos/opioids‐and‐pain‐signaling)
PDB‐101 is also exploring the use of other technologies for education. For example, a 3D Print section offers STL files that have been curated to highlight a molecular story either through the use of color or added materials. A 3D printable model of hemoglobin features pockets for insertable heme groups that can be 3D printed or constructed using other materials, while ferritin is an empty shell that can contain beads to represent iron ions.
2.3. Events
PDB‐101 online materials are the primary vehicle for RCSB PDB outreach and education, enabling broad and freely available usage. On occasion, outreach and education team members work directly with students and educators during programmed events related to PDB‐101 content. These events are useful in many ways; they serve as opportunities to collect direct feedback on PDB‐101 materials and can empower students and educators to create structure‐related educational materials themselves. Two main “events” currently target high school students.
In collaboration with the Milwaukee School of Engineering, RCSB PDB develops a “Protein Modeling” event held as part of high school Science Olympiads conducted nationally. During this event, student teams demonstrate their understanding of protein structure through 3D visualization and question/answer. 32 , 33 Event materials are based upon a particular MOTM article and focus on protein structures drawn from structures highlighted in the article. Many RCSB PDB staff members have served as Olympiad judges and supervisors over the years.
Annual Video Challenges build upon our broad experience with molecular animation. Participants create short videos to tell molecular stories that connect structural biology and medicine. Previous challenges have focused on topics that could be used to illustrate concepts taught in high school classrooms and relate to the biannual health focus (e.g., HIV/AIDS, diabetes, antimicrobial resistance, and drugs and the brain). Teams are supported with training materials and other PDB‐101 content. The award‐winning videos as judged by an expert panel become part of the PDB‐101 video collection. The health focus for calendar years 2022 and 2023 will be cancer. Since 2014, more than 400 videos have been submitted, 91 of which were entered in 2021. Videos employ a variety of techniques, including team members acting out a premise, stop‐motion animation using 2D images, and cartoon‐like animations. Videos are judged using a rubric that awards points for storytelling 20%, quality of science communication 30%, quality of public health message 10%, originality and creativity 20%, quality of production 10%, and proper accreditation 10% (citation of PDB structures shown, resources cited, and royalty‐free images and sound). Since the launch of the first challenge, technical barriers to creating videos have lowered through demonstrated use of tools such as Animaker (www.animaker.com) and Zoom (zoom.us). As a result, PDB‐101 receives more videos of higher quality.
2.4. SciArt galleries
Structural biology is a visual science. Even before PDB was launched, the combination of symmetrical structures with life‐changing functions had captured the imagination of artists working in many different mediums. The combination of science and art, also known as SciArt, is also a powerful tool for communicating and promoting protein structure. 34 PDB‐101 hosts two digital SciArt galleries that also serve as distribution centers for high‐resolution images (Figure 5).
FIGURE 5.
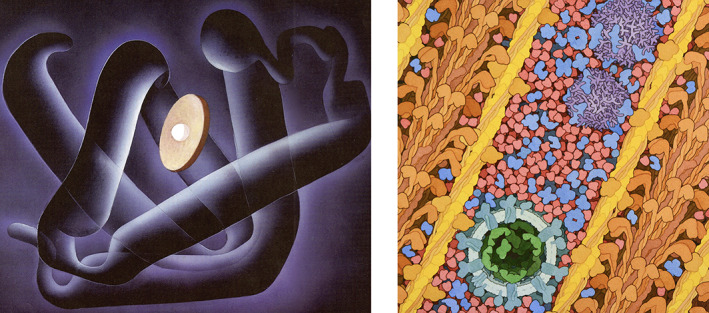
SciArt Gallery Examples. Left: Myoglobin Fold, 1987, by Irving Geis. Available from https://pdb101.rcsb.org/sci‐art/geis‐archive/gallery/rcsb‐0001‐myoglobin‐fold; used with permission from the Howard Hughes Medical Institute (www.hhmi.org). All rights reserved. Right: Myoglobin in a Whale Muscle Cell, 2021, illustration by David S. Goodsell, Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB). doi: 10.2210/rcsb_pdb/goodsell‐gallery‐032. SciArt pages at PDB‐101 were accessed more than 196,000 times in 2020
In the first gallery, PDB‐101 is honored to display many of the seminal works of Irving Geis (1908–1997), a gifted artist who helped announce the field of structural biology with his iconic images of myoglobin 35 and later lysozyme 36 in Scientific American. His textbook illustrations illuminated 3D biostructure for generations of researchers and students. Through a collaboration with the Howard Hughes Medical Institute, owner of the Geis Archives, PDB‐101 hosts ~30 Geis molecular images along with descriptions of the science behind each image, links to corresponding structures in the PDB archive, and interactive displays of these structures rendered in a similar style as the painting.
The second gallery hosts Molecular Landscapes from MOTM articles, plus other work from David S. Goodsell. 37 These images integrate information from structural biology, light microscopy, and molecular biophysics to simulate detailed views of biomolecules at work inside living cells. More than 30 Goodsell paintings reflect work done since 1999, with several landscapes added at the height of the COVID‐19 pandemic. Individual pages in PDB‐101 provide keys and descriptions of many of these paintings and links to PDB structures that are depicted.
Images from both SciArt galleries have been widely used in professional venues such as textbooks 38 and scientific journal covers 39 , 40 and, as announced by users on Twitter, in creative personal applications such as laboratory and classroom decorations, videoconference backgrounds, protective face masks, printed clothing, and jigsaw puzzles.
3. PDB‐101 AND COVID‐19
Given the widespread uncertainty about COVID‐19 at the beginning of the pandemic, and the pressing need for science‐driven information, PDB‐101 development shifted focus to SARS‐CoV‐2 as soon as the first COVID‐19‐related structure was deposited to the PDB archive. When that SARS‐CoV‐2 main protease entry was released (PDB ID 6lu7), 41 it was accompanied by an MOTM article describing coronavirus main proteases. Access to this article elicited an immediate surge in PDB‐101 website traffic. We then presented an integrative painting of a coronavirus virion, which was widely used in mainstream press and garnered yet another surge in PDB‐101 traffic. 40 As new PDB structures were added weekly to a COVID‐19 resources page at RCSB.org/covid19, new PDB‐101 content was developed. 40 Overall, COVID‐related content accounted for 11% of PDB‐101 traffic in 2020 (including >83,000 views of the main protease MOTM). Releasing content early in the pandemic, and in particular, visualizations of coronavirus, at a time when many students and educators were working from home on computers almost certainly contributed to these metrics.
Today, the PDB‐101 Coronavirus category contains MOTM features on the main protease, RNA‐dependent polymerase, and the spike surface glycoprotein; the video Fighting Coronavirus with Soap (accessed >467,000 times and designated as an official selection at the 2020 American Public Health Association Film Festival); seven paintings related to coronavirus biology in the David S. Goodsell SciArt Gallery; a new series on Resources to Fight the COVID‐19 Pandemic; and curricular materials.
Similarly, time‐sensitive materials have been created in response to past global developments, notably with viral outbreaks such as Zika, Ebola, and measles. 42 , 43 Rapid response was enabled by a team that regularly reviews the news and the scientific literature and technically by an internal Content Management System that simplifies the process of publishing content online. We aim to provide scientifically valid information as quickly as possible and thereby expose the PDB‐101 community to the biostructural underpinnings of the fight against important pathogens.
4. CELEBRATING 50 YEARS OF PDB STRUCTURES
Throughout 2021, wwPDB and RCSB PDB commemorated the 50th anniversary of the PDB with special symposia and events. It has been a time for celebration and for introspection. To continue the celebration, unique content has been added to the PDB‐101 collection. Downloadable “Structural Biology Playing Cards” have enhanced the traditional suits with structural themes: infrastructure for clubs, information for hearts, catalysis for diamonds, and energy for spades; face cards highlight careers in science. A historical video animates milestones in PDB's history and highlights PDB‐related experimental techniques. A PDB50 board game can be downloaded or played online, giving participants the opportunity to explore the process of structure discovery. Other PDB‐101 features are planned, including an MOTM feature that celebrates landmark structures from 5 decades of the archive and a 3DEM‐themed calendar for 2022. The PDB50 milestone has also provided an opportunity to evaluate methods that have been effective in RCSB PDB outreach efforts 3 , 44 and document the impact the PDB archive has had on the research community and beyond. 45 , 46 , 47 , 48 , 49 , 50
5. COMMUNITY INTERACTIONS AND IMPACT
PDB‐101 content is primarily developed by RCSB PDB team members. A quarterly Education Corner publishes descriptions of how other researchers and educators (e.g., 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 ) are using PDB data in classrooms, art, and science outreach.
In keeping with our overarching mission of supporting the PDB user community and honoring the spirit of open access, all PDB‐101 content is available under a CC‐BY‐4.0 license for use and reuse with attribution to PDB‐101 and RCSB PDB.
In addition to frequent re‐use, PDB‐101 images have been recognized with awards from the Federation of American Societies for Experimental Biology BioArt Awards, Wellcome Trust, and others. New content is published monthly, with announcements of interest to the PDB‐101 community published regularly on the home page.
Feedback from the community is solicited to ensure that we are covering the most topical developments in structural biology and providing materials that support topics currently being taught in classrooms.
While rigorous analysis of impact remains a challenge, feedback from in‐person activities, reprint requests, and website analytics document that PDB‐101 is well received and widely used. Traffic peaks in the fall and spring months and declines during July–August and the end of December, consistent with substantial use by educators and their students.
PDB‐101 has proven useful in STEM classrooms throughout the world by providing materials that promote understanding of fundamental biology through the lens of 3D structures in the PDB archive. Rapidly providing educational materials that help explain the structural biology behind public health crises has also helped build our global audience. PDB‐101 traffic surged when we provided materials in a timely response to Ebola in 2014 and again for measles in 2019. In 2020, PDB‐101 hosted >850,000 users and >2.6 million page views. Approximately 11% of web page views were directed toward coronavirus‐related content, mostly during the first half of the year when new knowledge concerning the pandemic was at a very high premium. We believe that this level of traffic was largely a consequence of the combination of a pervasive need for information about COVID‐19 and the fact that many were sheltering in place at home. We are striving to retain these new visitors by continuing an active program of new material focused on basic science and current developments. Molecular animations and videos represent a growing area of active views and represent an opportunity to reach a large community of new users. Rather than visits to PDB‐101, most of the video views are served and hosted by third‐party resources. The What is a Protein? video, for example, introduces primary, tertiary, and quaternary structures and has been viewed >1.8 million times on YouTube since its release in 2017.
6. CONCLUSION/PERSPECTIVE
PDB‐101 today is an evolving tool for telling stories about structural biology and the PDB archive. The availability of a treasure trove of MOTM content drove the initial release of PDB‐101 in 2011, but improving the underlying technology and website usability helped establish the resource as the going concern we know today.
Growth and expansion of the PDB archive present a host of new opportunities, and some challenges, for the future of PDB‐101. As PDB data continue to impact research and development across sciences, 46 , 47 , 48 , 49 the need for PDB‐101 content will only grow. Recent advances in 3DEM are providing views of biomolecular assemblies of unprecedented size and complexity, which will require new methods and careful thought to describe and explain to our users. New crystallographic techniques are providing dynamic views of biomolecular processes, and solid‐state NMR methods are revealing the structural basis of certain neurological diseases related to protein aggregation. PDB‐101 will continue to ensure that the PDB structures that capture the imagination of researchers equally inspire and motivate the next generation of scientists.
AUTHOR CONTRIBUTIONS
Christine Zardecki: Project administration (lead); supervision (supporting); visualization (supporting); writing – original draft (lead). Shuchismita Dutta: Writing – review and editing (equal). David Goodsell: Visualization (equal); writing – review and editing (equal). Robert Lowe: Software (lead). Maria Voigt: Visualization (equal); writing – review and editing (equal). Stephen Burley: Funding acquisition (lead); supervision (lead); writing – review and editing (equal).
CONFLICT OF INTEREST
The authors declare no competing financial interests.
Supporting information
Figure S1:
Figure S2:
ACKNOWLEDGEMENTS
PDB‐101 would not exist without RCSB PDB members past and present, particularly Cole Christie, Sutapa Ghosh, Rachel Kramer Green, Brian Hudson, Cathy Lawson, Yuhe Liang, Ezra Peisach, Irina Persikova, Chris Randle, Monica Sekharan, Chenghua Shao, Yi‐Ping Tao, Jessie Woo, Jasmine Young, and the guidance and vision of RCSB PDB Director Emerita Helen M. Berman. Special thanks to Gail Bamber, the initial Molecular Machinery graphic designer.
PDB‐101 acknowledges participants of the January 2021 Science Communication in Biology and Medicine boot camp for undergraduate and graduate students hosted by the Rutgers Institute for Quantitative Biomedicine for contributions to the Molecule of the Month series: Steven Arnold, Jenna Abyad, Tanvi Banota, Candice Craig, Megan Diiorio, Samantha Eng, Zachary Fritz, Helen Gao, Jennifer Jiang, Alexandria Lo, Natalie Losada, Changpeng Lu, Jenna Manzo, Katherine Park, Dhyan Ray, Samuel Shrem (Rutgers University), Saai Suryanarayanan, Nithish Selvaraj, Andrew Tkacenko, Kiranmayi Vemuri, and Momodou Camara (University of Nebraska‐Lincoln); and NJ teachers who participated in curricular material development: Anne Sanelli, Arvinder Bhatia, Blair Buck, Henna Sharif, Karen Lucci, Meenakshi Bhattacharya, Michele Witkowski, Richard Tempsick, Subha Eswaran, and William Mott.
Zardecki C, Dutta S, Goodsell DS, Lowe R, Voigt M, Burley SK. PDB‐101: Educational resources supporting molecular explorations through biology and medicine. Protein Science. 2022;31:129–140. 10.1002/pro.4200
Funding information Biological and Environmental Research, Grant/Award Number: DE‐SC0019749; Division of Biological Infrastructure, Grant/Award Number: DBI‐1832184; National Cancer Institute, Grant/Award Number: R01GM133198; National Institute of Allergy and Infectious Diseases; National Institute of General Medical Sciences
REFERENCES
- 1. wwPDB consortium . Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goodsell DS, Zardecki C, Di Costanzo L, et al. RCSB Protein Data Bank: Enabling biomedical research and drug discovery. Protein Sci. 2020;29:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodsell DS, Dutta S, Voigt M, Zardecki C. Molecular storytelling for online structural biology outreach and education. Struct Dyn. 2021;8:020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McLaughlin KJ. Developing a macromolecular crystallography driven cure. Struct Dyn. 2021;8:020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ayella A, Beck MR. A course‐based undergraduate research experience investigating the consequences of nonconserved mutations in lactate dehydrogenase. Biochem Mol Biol Educ. 2018;46:285–296. [DOI] [PubMed] [Google Scholar]
- 6. Bateman RC Jr, Craig P. A proficiency rubric for biomacromolecular 3D literacy. RCSB PDB Newsletter, Spring; 2010;5–7. [Google Scholar]
- 7. Dutta S, Eswaran S, Sanelli A, Bhattacharya M, Tempsick R. Learning biology through molecular storytelling. Sci Teacher. 2018;86:28–33. [Google Scholar]
- 8. Craig PA, Mills JL, Roberts R, et al. Transition to a course‐based undergraduate research experience (cure). FASEB J. 2017;31:588–584. [Google Scholar]
- 9. Brandt GS, Novak WRP. SARS‐CoV‐2 virtual biochemistry labs on bioinformatics and drug design. Biochem Mol Biol Educ. 2021;49:26–28. [DOI] [PubMed] [Google Scholar]
- 10. Roberts R, Hall B, Daubner C, et al. Flexible implementation of the basil cure. Biochem Mol Biol Educ. 2019;47:498–505. [DOI] [PubMed] [Google Scholar]
- 11. Craig PA. Developing and applying computational resources for biochemistry education. Biochem Mol Biol Educ. 2020;48:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ornes S. Science and culture: The evolving portrait of a virus. Proc Natl Acad Sci USA. 2021;118:e2111544118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Protein Data Bank . Crystallography: Protein Data Bank. Nat New Biol. 1971;233:223.20480989 [Google Scholar]
- 14. Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berman HM, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Biol. 2003;10:980. [DOI] [PubMed] [Google Scholar]
- 16. Young JY, Westbrook JD, Feng Z, et al. (2018). Worldwide Protein Data Bank biocuration supporting open access to high‐quality 3D structural biology data. Database. 2018. 10.1093/database/bay002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Armstrong DR, Berrisford JM, Conroy MJ, et al. Pdbe: Improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020;48:D335–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kinjo AR, Bekker GJ, Wako H, et al. New tools and functions in data‐out activities at protein data bank Japan (PDBJ). Protein Sci. 2018;27:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romero PR, Kobayashi N, Wedell JR, et al. Biomagresbank (bmrb) as a resource for structural biology. Methods Mol Biol. 2020;2112:187–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. Nucleic Acid Res. 2021;49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lawson CL, Patwardhan A, Baker ML, et al. Emdatabank unified data resource for 3dem. Nucleic Acids Res. 2016;44:D396–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3d. Protein Sci. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodsell DS, Zardecki C, Berman HM, Burley SK. Insights from 20 years of the molecule of the month. Biochem Mol Biol Educ. 2020;48:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rose PW, Bi C, Bluhm WF, et al. The RCSB Protein Data Bank: New resources for research and education. Nucleic Acids Res. 2013;41:D475–D482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rose PW, Prlic A, Altunkaya A, et al. The RCSB Protein Data Bank: Integrative view of protein, gene and 3d structural information. Nucleic Acids Res. 2017;45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barcena M, Barnes CO, Beck M, et al. Structural biology in the fight against covid‐19. Nat Struct Mol Biol. 2021;28:2–7. [DOI] [PubMed] [Google Scholar]
- 27. Goodsell DS, Autin L, Olson AJ. Illustrate: Software for biomolecular illustration. Structure. 2019;27:1716–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sehnal D, Bittrich S, Deshpande M, et al. Mol* viewer: Modern web app for 3d visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021;49:W431–W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. [DOI] [PubMed] [Google Scholar]
- 30. Shao C, Feng Z, Westbrook JD, et al. Modernized uniform representation of carbohydrate molecules in the Protein Data Bank. Glycobiology. 2021;31:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goddard TD, Huang CC, Meng EC, et al. UCSF Chimera: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herman T, Morris J, Colton S, et al. Tactile teaching: Exploring protein structure/function using physical models. Biochem Mol Biol Educ. 2006;34:247–254. [DOI] [PubMed] [Google Scholar]
- 33. Goodsell DS, Dutta S, Zardecki C, Voigt M, Berman HM, Burley SK. The RCSB PDB molecule of the month: Inspiring a molecular view of biology. PLoS Biol. 2105;13:e1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goodsell DS. Art as a tool for science. Nat Struct Mol Biol. 2021;28:402–403. [DOI] [PubMed] [Google Scholar]
- 35. Kendrew JC. The three‐dimensional structure of a protein molecule. Sci Am. 1961;205:96–110. [DOI] [PubMed] [Google Scholar]
- 36. Phillips DC. The three‐dimensional structure of an enzyme molecule. Sci Am. 1966;215:78–90. [DOI] [PubMed] [Google Scholar]
- 37. Goodsell DS, Franzen MA, Herman T. From atoms to cells: Using mesoscale landscapes to construct visual narratives. J Mol Biol. 2018;430:3954–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Voet D, Voet JG, Pratt CW. Fundamentals of biochemistry. New York, NY: John Wiley & Sons, Inc., 2002. [Google Scholar]
- 39. Goodsell DS. Painting a portrait of SARS‐CoV‐2. Am Sci. 2021;109:88–93. [Google Scholar]
- 40. Goodsell DS, Voigt M, Zardecki C, Burley SK. Integrative illustration for coronavirus outreach. PLoS Biol. 2020;18:e3000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin Z, Du X, Xu Y, et al. Structure of m(pro) from SARS‐CoV‐2 and discovery of its inhibitors. Nature. 2020;582:289–293. [DOI] [PubMed] [Google Scholar]
- 42. Cohen J. Meet the scientist painter who turns deadly viruses into beautiful works of art. Science. 2019. 10.1126/science.aax6641. [DOI] [Google Scholar]
- 43. Fessenden M. 2016. This painting shows what it might look like when Zika infects a cell. Smithsonianmag.Com. https://www.smithsonianmag.com/science-nature/this-painting-shows-what-might-look-like-when-zika-infects-cell-180959507/. [Google Scholar]
- 44. Richardson JS, Richardson DC, Goodsell DS. Seeing the PDB. J Biol Chem. 2021;296:100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burley SK, Berman HM. Open‐access data: A cornerstone for artificial intelligence approaches to protein structure prediction. Structure. 2021;29:515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burley SK. Impact of structural biologists and the protein data bank on small‐molecule drug discovery and development. J Biol Chem. 2021;296:100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Westbrook JD, Soskind R, Hudson BP, Burley SK. Impact of protein data bank on anti‐neoplastic approvals. Drug Discov Today. 2020;25:837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feng Z, Verdiguel N, Di Costanzo L, et al. Impact of the protein data bank across scientific disciplines. Data Sci J. 2020;9:1–14. [Google Scholar]
- 49. Markosian C, Di Costanzo L, Sekharan M, Shao C, Burley SK, Zardecki C. Analysis of impact metrics for the protein data bank. Sci Data. 2018;5:180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Westbrook JD, Burley SK. How structural biologists and the protein data bank contributed to recent FDA new drug approvals. Structure. 2019;27:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanski J, Zardecki C, Yakovenko A, Eagle C. Transactions from the 70th annual meeting of the American crystallographic association: Structural science—new ways to teach the next generation. Struct Dyn. 2021;8:040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hall BL, Watson KD, Covey T. 2019. Resources for teaching project‐based undergraduate medicinal chemistry courses. Tech Integrat Chem Educ Res. 2019;9:131–142. [Google Scholar]
- 53. Fried DB, Tinio P, Paneque D, Hughes A. Elementary student achievement and teacher perception of advanced chemistry curriculum. Eur J Sci Math Educ. 2019;7:37–148. [Google Scholar]
- 54. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fischetti M, Hays VF, Glaunsinger B, Christiansen J. Inside the coronavirus. Sci Am. 2020;323:32–37. [DOI] [PubMed] [Google Scholar]
- 56. Procko K, Beckham J, Dean D, et al. Building a community of practice for the assessment of biomolecular visual literacy. FASEB J. 2020;34:1–1. [Google Scholar]
- 57. Silverstein TP, Kirk SR, Meyer SC, Holman KL. Myoglobin structure and function: A multiweek biochemistry laboratory project. Biochem Mol Biol Educ. 2015;43:181–188. [DOI] [PubMed] [Google Scholar]
- 58. Iwasa JH. Bringing macromolecular machinery to life using 3d animation. Curr Opin Struct Biol. 2015;31:84–88. [DOI] [PubMed] [Google Scholar]
- 59. McLaughlin KJ. Understanding structure: A computer‐based macromolecular biochemistry lab activity. J Chem Educ. 2017;94:903–906. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1:
Figure S2:


