Abstract
Severe acute respiratory syndrome (SARS) is an acute infectious disease with significant mortality. A typical clinical feature associated with SARS is pulmonary fibrosis and the associated lung failure. However, the underlying mechanism remains elusive. In this study, we demonstrate that SARS-associated coronavirus (SARS-CoV) nucleocapsid (N) protein potentiates transforming growth factor-β (TGF-β)-induced expression of plasminogen activator inhibitor-1 but attenuates Smad3/Smad4-mediated apoptosis of human peripheral lung epithelial HPL1 cells. The promoting effect of N protein on the transcriptional responses of TGF-β is Smad3-specific. N protein associates with Smad3 and promotes Smad3-p300 complex formation while it interferes with the complex formation between Smad3 and Smad4. These findings provide evidence of a novel mechanism whereby N protein modulates TGF-β signaling to block apoptosis of SARS-CoV-infected host cells and meanwhile promote tissue fibrosis. Our results reveal a novel mode of Smad3 action in a Smad4-independent manner and may lead to successful strategies for SARS treatment by targeting the TGF-β signaling molecules.
Severe acute respiratory syndrome (SARS)2 is an acute infectious disease with significant morbidity and mortality. SARS-CoV, which has been identified as the etiological agent of this disease, is an enveloped, positive-sense RNA virus with a genome of 29.7 kb in length. A sequence comparison with other known coronaviruses revealed a similar organization of SARS-CoV genes to typical coronaviruses (1, 2). The SARS-CoV nucleocapsid (N) protein is a 46-kDa viral RNA-binding protein and shares little homology with the N proteins of other known coronaviruses (1, 2). Multiple functions have been postulated for N protein throughout the viral life cycle and with pathological changes of SARS patients: its involvement of viral replication and regulation of cellular processes such as gene transcription, actin reorganization, host cell cycle progression, and apoptosis (3, 4, 5, 6).
Transforming growth factor-β (TGF-β) is a well characterized cytokine and controls a variety of biological processes, including cell growth, differentiation, and apoptosis; development; immune homeostasis; and tissue remodeling and repairing (7). Dysregulation of its signaling has been implicated in different kinds of human disorders such as tissue fibrosis, cancer development, and others (7, 8). TGF-β plays a pivotal role in pulmonary fibrosis (8, 9). It increases the production of extracellular matrix proteins, enhances the secretion of protease inhibitors, and reduces secretion of proteases, thus leading to deposition of extracellular matrix proteins. TGF-β can also induce pulmonary fibrosis directly through stimulation of fibroblast chemotactic migration and proliferation as well as fibroblast-myofibroblast transition.
The canonical TGF-β signal transduction is initiated with the ligand binding to its serine/threonine kinase receptors on the cell surface, which leads to the activation of downstream cytoplasmic effectors, the Smad proteins. The receptor-activated Smad2 and Smad3 are involved in TGF-β signaling. Once phosphorylated by the type I receptor, they form heteromeric complexes with Smad4, and the Smad heterocomplexes are accumulated in the nucleus, where they regulate target gene transcription in association with DNA binding partners (10, 11, 12).
A considerable proportion of SARS patients developed severe inflammation of lung, and many of them deteriorated into acute respiratory distress syndrome (13, 14). A typical clinical character of acute respiratory distress syndrome is pulmonary fibrosis and the associated lung failure, which result in a high mortality (14, 15). Diffuse alveolar damage is also common in the lungs of SARS patients. Macrophages and lymphocytes infiltrate into alveolar cavities and the interstitium of lung. Increased apoptotic cells are also detected in SARS patient lungs (13, 14, 16). The roles of the SARS-CoV-encoded proteins in SARS infection and pathology remain obscure. In this study, we report that SARS-CoV N protein can specifically potentiate the Smad3-mediated transcriptional responses of TGF-β such as the expression of plasminogen activator inhibitor-1 (PAI-1), which plays a critical role in fibrosis. Interestingly, N protein interferes with TGF-β-induced and Smad4-mediated pro-apoptotic genes expression and cell apoptosis. Mechanistically, N protein interacts with Smad3 and impairs Smad3-Smad4 heterocomplex formation. Our findings provide evidence of a novel mechanism whereby N protein modulates TGF-β signaling to block apoptosis of SARS-CoV-infected host cells and meanwhile promote tissue fibrosis. These results may also have implication in understanding of other virus-induced tissue fibrosis.
EXPERIMENTAL PROCEDURES
Materials and PlasmidsRecombinant human TGF-β1 was from R&D Systems; mouse anti-HA (F-7), mouse anti-Myc (9E10), mouse anti-GST, goat anti-Smad2/3, rabbit anti-Smad3, rabbit anti-Smad4, rabbit anti-p300 antibodies from Santa Cruz and mouse anti-FLAG antibody (M2) from Sigma, anti-human PAI-1 antibody from American Diagnostica, Inc., and the fluorescein isothiocyanate-labeled antibody against SARS-CoV nucleocapsid protein were described previously (15). The luciferase assay system was from Promega, and the ECL reagent was from Amersham Biosciences. The SARS-CoV N cDNA was cloned into pcDNA3.1(+) at EcoRI and KpnI sites and into pEBG1 at KpnI and ClaI sites. Smad3 and N deletion mutants were generated by PCR and cloned into pCMV5 at KpnI and ClaI sites. We made following short hairpin RNA against green fluorescence protein (GFP) and human Dapper I as nonspecific RNA interference: pSR-shGFP (target sequence AGCGGACTAAGTCCATTGC) and pSR-shhDpr1 (target sequence ATCTGCAGATCTCATAGGATT) (17, 18). pSRG-shSmad3 (target sequence GGATTGAGCTGCACCTGAATG) and pSRG-shSmad4 (target sequence GGATTTCCTCATGTGATCT) were kindly provided by Dr. Xin-Hua Feng (19). All the constructs were confirmed by DNA sequencing.
Cell Culture and Establishment of Stable Cell LinesHEK293T, mouse embryo fibroblasts (MEFs) were maintained in Dulbecco's Modified Eagle's Medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone). HPL1 cells were maintained as described (20). To generate stable cells expressing N protein, HPL1 cells were transfected with pcDNA3.1-N or empty vector using Lipofectamine (Invitrogen), and stable transfectants were selected with 0.6 μg/ml G418 (Invitrogen) for 14 days. Individual clones were then obtained after confirmation of N protein expression by immunoblotting.
Chromatin Immunoprecipitation AssayChIP assay was performed as previously described (21). The primers used in ChIP assay to amplify the human PAI-1 promoter (nucleotide -733 to -484) are 5′-AGCCAGACAAGGTTGTTG-3′ and 5′-GACCACCTCCAGGAAAG-3′.
Luciferase Reporter AssayLuciferase reporter assay was performed as previously described (21). The transfected cells were treated with 50 pm TGF-β1 for 20 h before harvested for reporter assay. Each experiment was performed in triplicate, and the data represent the mean ± S.D. of three independent experiments after normalized to Renilla activity.
Immunoprecipitation and GST Pulldown AssayHEK293T cells were transfected with the indicted plasmids using the phosphate calcium method. At 48 h post-transfection, cells were harvested with lysis buffer (20 mm Tris-HCl at pH 7.4, 2 mm EDTA at pH 8.0, 25 mm NaF, 1% Triton X-100) plus protease inhibitors (Sigma) for 15 min at 4 °C. Total cell lysates were prepared by centrifugation at 12000 × g for 10 min. For co-immunoprecipitation, specific antibody and protein A-Sepharose beads (Zymed Laboratories Inc.) were added into cell lysates. For GST pulldown, Sepharose 4B-glutathione (Amersham Biosciences) beads were added. After incubation for 3 h at 4 °C, beads were washed four times with washing buffer (50 mm Tris-HCl at pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). The bound proteins were then examined by immunoblotting.
RNA Preparation, Reverse Transcription-PCR, and Quantitative Real-time PCRRNA preparation and reverse transcription-PCR have been performed as previously described (21). The cells were treated with 100 pm TGF-β1. PCR and real-time PCR were performed with the following primer sets: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5′-CATCACTGCCACCCAGAAGA-3′ and 5′-GCTGTAGCCAAATTCGTTGT-3′), β-actin (5′-CGAGGACTTTGATTGCAC-3′ and 5′-TATCACCTCCCCTGTGTG-3′), PAI-1 (5′-GAGACAGGCAGCTCGGATTC-3′ and 5′-GGCCTCCCAAAGTGCATTAC-3′), α2 chain of type I collagen (COL1A2) (5′-GTGTAAGCGGTGGTGGGT-3′ and 5′-GCCCGGATACAGGTTT-3′), Bim (5′-GCCTTCAACCACTATCTCA-3′ and 5′-ATCCAGCTCGGTGTCTTCT-3′), and Bax (5′-ATGGACGGGTCCGGGGAGCAG-3′ and 5′-CATGATGGTTCTGATCAGTT-3′). Real-time PCR was performed using Mx3000P™ (Stratagene). The amplified DNAs were quantitated by the comparative cycle threshold (Ct) method for relative quantitation of gene expression, normalized to GAPDH. Post-PCR melting curves confirmed the specificity of single specific target amplification.
Fluorescence-activated Cell SortingHPL1 cells were co-transfected with GFP and other constructs as indicated. One day after transfection, cells were treated with 200 pm TGF-β1 for 48 h. Then cells were harvested and fixed with 75% alcohol at -20 °C overnight. Before loaded to flow cytometry analysis, cells were treated with 100 μg/ml RNase and 5 μg/ml propidium iodide at 4 °C for 30 min. The GFP-positive cells were selected by FACScan flow cytometer (BD Sciences) for cell cycle analysis. Experiments were performed in triplicate.
ImmunohistochemistryThe experiment was performed as previously described (15). Color was developed by using the 3-amino-9-ethylcarbazole or diaminobenzidine tetrahydrochloride substrate kit (Sigma) as indicated in the figures. Collagen was stained using the Masson trichrome method, and slide examinations were performed with a Nikon Eclipse E-800 fluorescence microscope.
Statistic AnalysisStudent's t test was performed to assess the significance of treatments versus controls. Asterisks in the figures represent p values of <0.05 to indicate statistical significance.
RESULTS
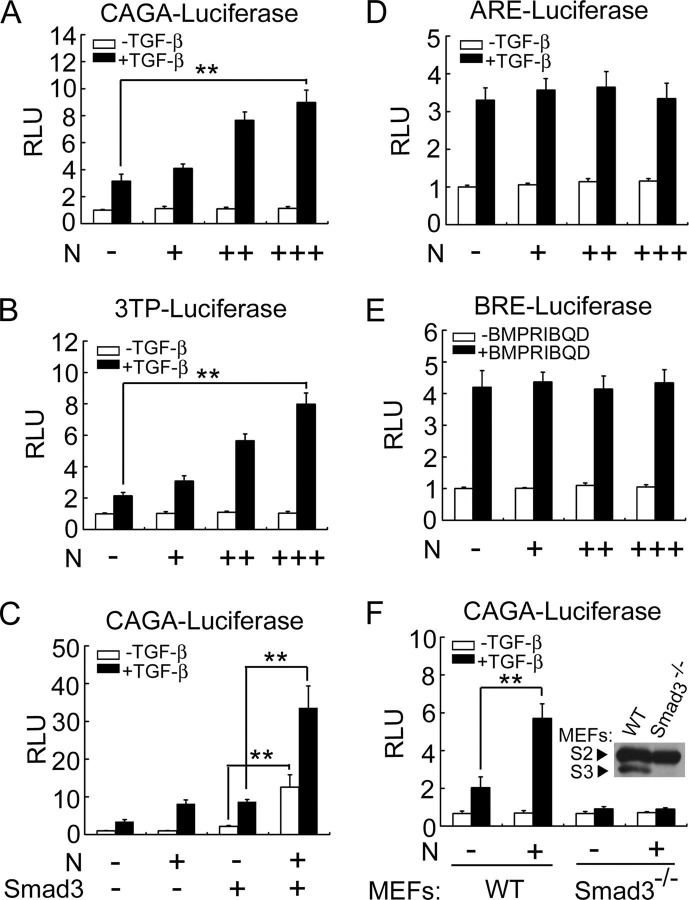
SARS-CoV N Protein Specifically Enhances TGF-β/Smad3-induced Transcriptional ActivationAs one of major clinical characteristics that resulted from SARS-CoV infection is lung fibrosis and TGF-β has been well documented to play a critical role in tissue fibrosis, we attempted to explore whether TGF-β signaling is involved in SARS-CoV-induced lung fibrosis. Among the confirmed SARS-CoV-encoded proteins, N protein is a structural cytosolic protein involved in viral nucleocapsid assembly and regulation of host cell processes. To explore if N protein affects TGF-β signaling, transcriptional responses of TGF-β in the presence of N protein were examined with TGF-β-responsive luciferase reporters in human peripheral lung epithelial HPL1 cells, which retain characteristics of type II pneumocytes and respond to TGF-β1 (20). HPL1 cells were co-transfected with N protein and CAGA-luciferase, which contains the Smad-binding element tetranucleotide (CAGA) sequence and can be specifically activated by Smad3 and Smad4 (22). As shown in Fig. 1A , N protein enhanced the TGF-β-induced reporter expression, and this enhancement was in a dose-dependent manner. Similar results were also obtained in human hepatoma HepG2 cells (data not shown). N protein also enhanced the expression of 3TP-luciferase, a TGF-β-responsive reporter (23), in a dose-dependent manner (Fig. 1B). Furthermore, N protein further enhanced Smad3-promoted expression of CAGA-luciferase (Fig. 1C). ARE-luciferase, which contains activin-response element from Xenopus Mix.2 promoter, is known to be activated by both TGF-β and activin via Smad2, Smad4, and forkhead DNA-binding protein FoxH (24). Interestingly, N protein had no effect on the expression of ARE-luciferase induced by TGF-β (Fig. 1D). BRE-luciferase-containing BMP response element, which was derived from the Xenopus Vent2 promoter, can be specifically activated by Smad1 (25). Again, N protein had no effect on the expression of BRE-luciferase induced by constitutively active form type I receptor of BMP, BMPRIB(QD) (Fig. 1E). These data implied that N protein specifically promotes Smad3-mediated TGF-β signaling.
FIGURE 1.
SARS-CoV N protein specifically up-regulates Smad3-mediated transcriptional response of TGF-β.A, N protein enhances TGF-β-induced CAGA-luciferase expression in a dose-dependent manner. HPL1 cells were co-transfected with CAGA-luciferase reporter (0.5 μg) and pcDNA3.1-N (0.1, 0.3, or 0.5 μg). At 24 h post-transfection, the cells were treated with 50 pm TGF-β. After 20 h, the cells were harvested for determination of luciferase activity. B, N protein enhances TGF-β-induced 3TP-luciferase expression in a dose-dependent manner. HPL1 cells were co-transfected with 3TP-luciferase reporter (0.5 μg) and pcDNA3.1-N (0.1, 0.3, or 0.5 μg). C, N protein synergizes with Smad3 to induce CAGA-luciferase expression. HPL1 cells were co-transfected with CAGA-luciferase (0.5 μg), pCS2-Myc-Smad3 (20 ng), and pcDNA3.1-N (0.5 μg). D, N protein has no effect on the ARE-luciferase expression. HPL1 cells were co-transfected with ARE-luciferase reporter (0.5 μg) plus FoxH1 (0.25 μg) and pcDNA3.1-N (0.1, 0.3, or 0.5 μg). E, N protein has no effect on the BRE-luciferase expression. HPL1 cells were co-transfected with BRE-luciferase reporter (0.5 μg), pCMV5-FLAG-OAZ (0.25 μg), pCMV5-BMPRIB(QD)-HA (0.1 μg), and pcDNA3.1-N (0.1, 0.3, or 0.5 μg). F, N protein enhances the TGF-β-induced expression of CAGA-luciferase in wild-type (WT) MEFs, but has no effect on CAGA-luciferase expression in Smad3-/- MEFs. The endogenous expression of Smad2 (S2) and Smad3 (S3) in WT MEFs and Smad3-/- MEFs were detected by anti-Smad2/3 immunoblotting. The asterisks indicate a statistically significant difference (**, p < 0.01). RLU: relative luciferase units.
To further confirm that N protein has specific effect on the TGF-β/Smad3 pathway, we examined the effect of N protein on CAGA-luciferase expression in Smad3 -/- MEFs. As shown in Fig. 1F, N protein up-regulated the TGF-β-induced expression of CAGA-luciferase in normal MEFs in a dose-dependent manner, whereas neither TGF-β nor N protein activated CAGA-luciferase expression in Smad3 -/- MEFs. Taken together, these results suggested that N protein elevates TGF-β signaling via Smad3 but not Smad2.
N Protein Enhances TGF-β-induced Expression of PAI-1Histopathological studies have demonstrated that SARS-CoV infection causes lung pathological changes in SARS patients such as infiltration of macrophages in the alveolar spaces (13, 14) and elevated levels of pro-inflammatory cytokines, including TGF-β1 in pneumocytes (16). Therefore, it is reasonable to speculate that TGF-β might have an important function in SARS-associated lung pathological changes. The TGF-β-regulated expression of extracellular matrix molecules such as PAI-1 and type I collagen play important roles in lung fibrosis (8, 26). Immunohistochemical analysis showed that SARS-infected lungs expressed a high level of collagen (Fig. 2A , top right panel). Accordingly, PAI-1 expression in the vicinity of pneumocytes was apparently higher in the lung tissue of SARS patients than in the normal lung (Fig. 2A , bottom panels).
FIGURE 2.
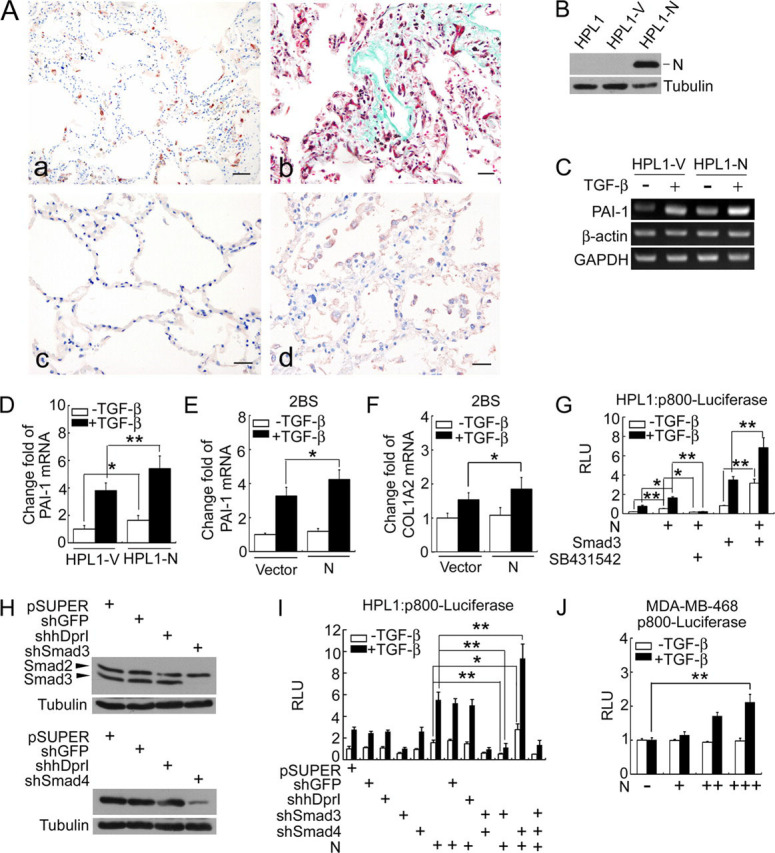
N protein enhances TGF-β-induced PAI-1 expression in a Smad3-dependent manner.A, immunohistochemistry staining shows the expression of N protein in SARS patient's lung (a), PAI-1 in normal lung (c), SARS patient's lung (d), and color developed by the 3-amino-9-ethylcarbazole and Masson's trichrome staining of collagen in SARS patient's lung (b); scale bar:50 μm. B–F, N protein up-regulates TGF-β target gene expression. The expression of N protein in stable HPL1 cells expressing N protein (HPL1-N) cells was determined by anti-N immunoblotting (B). The PAI-1 mRNA levels in HPL1-V or HPL1-N cells were analyzed by reverse transcription-PCR (C), or by real-time PCR (D). The PAI-1 mRNA levels (E) or COL1A2 (F) in 2BS cells transiently transfected with empty vector or N protein were analyzed by real-time PCR. β-Actin and GAPDH served as loading controls. G, N protein cooperates with Smad3 in enhancing the expression of luciferase driven by the PAI-1 promoter. HPL1 cells were co-transfected with p800-luciferase reporter (0.5 μg), pCS2-Myc-Smad3 (20 ng), and pcDNA3.1-N (0.5 μg) as indicated. At 24 h post-transfection, the cells were treated with 50 pm TGF-β with or without 10 μm SB431542. After 20 h, the cells were harvested for determination of luciferase activity. H, siRNAs efficiently knock down the expression of endogenous Smad3 and Smad4 in HPL1 cells. HPL1 cells were transfected with various siRNA constructs as indicated. After 0.2 mg/ml puromycin selection for 4 days, the cell lysates were collected and protein expression was determined by immunoblotting. pSR-shGFP and pSR-human Dapper I served as off-target siRNAs, and tubulin as the loading control. I, knockdown of Smad3 but not Smad4 in HPL1 cells represses the expression of TGF-β-induced p800-luciferase. HPL1 cells were co-transfected with p800-luciferase reporter, pSR-shGFP, pSR-human Dapper I, pSRG-shSmad3, or -shSmad4 and pcDNA3.1-N (0.5 μg for each construct) as indicated. Reporter assay was performed as in G. J, N protein enhances TGF-β-induced p800-luciferase expression in a dose-dependent manner in MDA-MB-468 cells. Cells were co-transfected with p800-luciferase reporter (0.5 μg) and pcDNA3.1-N (0.1, 0.3, or 0.5 μg). The asterisks indicate a statistically significant difference (*, p < 0.05; **, p < 0.01). RLU: relative luciferase units.
To directly test whether N protein has any effect on the TGF-β-induced expression of PAI-1, we established a stable HPL1 cell line expressing N protein (HPL1-N) (Fig. 2B). Reverse transcription-PCR showed that TGF-β stimulated PAI-1 expression in control cells HPL1-V and N protein enhanced the basal and TGF-β-stimulated expression of PAI-1 (Fig. 2C). This result was confirmed by quantitative real-time PCR (Fig. 2D). Transient expression of N protein in human normal lung fibroblast 2BS cells also enhanced the TGF-β-induced expression of PAI-1 and α2 chain of type I collagen (COL1A2) (Fig. 2, E and F). To further study the effect of N protein on TGF-β-induced PAI-1 expression, we utilized the reporter p800-luciferase that contains a fragment of the PAI-1 promoter harboring Smad-binding element and responds to TGF-β (27). Both N protein and Smad3 enhanced the basal and TGF-β-stimulated expression of this reporter (Fig. 2G), and the small molecule SB431542, a specific inhibitor of TGF-β type I receptor, greatly attenuated the effect of N protein on the p800-luciferase expression. Furthermore, there was a synergistic effect between N protein and Smad3. We also investigated whether knockdown of endogenous Smad3 or Smad4 has any effect on the N protein-enhanced reporter expression. As shown in Fig. 2H, both anti-Smad3 and anti-Smad4 siRNA worked efficiently. Knockdown of endogenous Smad3 expression by siRNA blocked N protein-enhanced reporter expression (Fig. 2I). However, knockdown of endogenous Smad4 expression had no effect on TGF-β-induced p800-luciferase expression, but rather promoted N protein enhancement of TGF-β activity. These results together indicated that N protein enhances TGF-β-induced expression of PAI-1 and type I collagen, and the enhancement of N protein is Smad3-dependent but Smad4-independent. To consolidate this, we carried out a p800-luciferase reporter assay in Smad4-/- human breast cancer cell line, MDA-MB-468. As shown in Fig. 2J, N protein also enhanced the TGF-β-induced p800-luciferase expression in a dose-dependent manner.
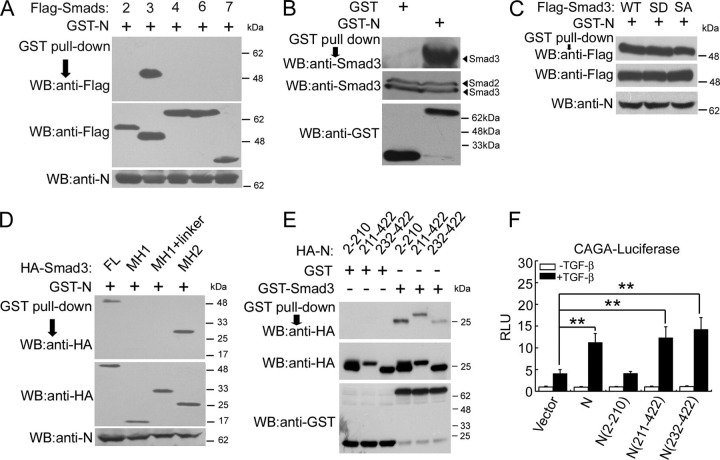
N Protein Interacts with Smad3The above results demonstrated that N protein enhances the TGF-β transcriptional response and collaborates with Smad3 to stimulate PAI-1 expression. To understand the underlying molecular mechanism, we then tested whether N protein interacts with the components of the TGF-β signaling pathway. Glutathione S-transferase (GST)-tagged N protein was co-transfected into HEK293T cells with various FLAG-Smad constructs. GST pulldown and anti-FLAG immunoblotting showed that N protein only interacted with Smad3 but not other Smad proteins tested (Fig. 3A ). N protein also interacted with endogenous Smad3 (Fig. 3B). To investigate whether Smad3 phosphorylation has any effect on Smad3-N protein interaction, we utilized two Smad3 mutants, Smad3(SD), a constitutively active form with replacement of Asp at Ser-422 and Ser-424, and Smad3(SA), phosphorylation-defective form with replacement of Ala at Ser-421, Ser-422, and Ser-424. GST pulldown and anti-FLAG immunoblotting showed that N protein exhibited a similar binding affinity to wild-type Smad3 and Smad3(SD) mutant but had a lower binding affinity to Smad3(SA) (Fig. 3C). To define the interaction domains of Smad3 with N protein, we performed co-immunoprecipitation assay with Smad3 truncation mutants in HEK293T cells and found that N protein interacted with the MH2 domain of Smad3 (Fig. 3D).
FIGURE 3.
N protein interacts with Smad3.A, N protein interacts with Smad3. HEK293T cells were co-transfected with pEBG1-GST-N (2 μg) and FLAG-tagged Smad plasmids (5 μg each) as indicated. Cell lysates were incubated with Sepharose 4B-glutathione beads, and GST-N protein associated Smads were revealed by anti-FLAG immunoblotting (upper panel). The protein expression was confirmed with immunoblotting of total cell lysates (middle and lower panels). B, N protein interacts with endogenous Smad3 in HPL1 cells. After HPL1 cells were transfected with pEBG1 or pEBG1-GST-N for 40 h, the cells were harvested for GST pulldown. GST-N-associated Smad3 proteins (upper panel) and total protein expression (middle and lower panels) were revealed by immunoblotting. C, N protein interacts with Smad3 mutants. HEK293T cells were co-transfected with pEBG1-GST-N (2 μg) and FLAG-tagged Smad3 wild-type (WT) or mutant plasmids (5 μg each) as indicated. Cell lysates were incubated with Sepharose 4B-glutathione beads and GST-N protein-associated Smad3 were revealed by anti-FLAG immunoblotting (upper panel). The protein expression was confirmed with immunoblotting of total cell lysates (middle and lower panels). D, N protein interacts with the Smad3 MH2 domain. HEK293T cells were co-transfected with pEBG1-GST-N (2 μg) and pCMV5-HA-Smad3, MH1 (aa 2–132), MH1 plus linker (aa 2–225), and MH2 (aa 226–425) (5 μg each) as indicted. GST pulldown assay was performed similarly as in A. GST-N protein associated Smad3 was revealed by anti-HA immunoblotting (upper panel). Protein expression was confirmed immunoblotting (middle and lower panels). E, Smad3 interacts with both of the N-terminal and C-terminal domains of N protein. HEK293T cells were co-transfected with N protein and its deletion mutant constructs (5 μg each) as indicted. Cell lysates were incubated with Sepharose 4B-glutathione beads and purified bacteria-expressed GST or GST-Smad3 protein. GST-Smad3-associated N proteins (upper panel) and protein expression (middle and lower panels) were revealed by immunoblotting. F, the C-terminal domain of N protein is important to enhance TGF-β-induced expression of CAGA-luciferase. HPL1 cells were co-transfected with CAGA-luciferase reporter (0.5 μg) and pCMV5 empty vector or pCMV5-HA-N, -N (2–210), -N (211–422), -N (232–422) (0.5 μg each) as indicated. Luciferase activity was determined as in Fig. 1A. The asterisks indicate a statistically significant difference (**, p < 0.01). RLU: relative luciferase units.
The N-terminal domain of N protein binds viral genomic RNA to form a ribonucleoprotein complex, whereas the C-terminal domain is responsible for self-assembly to form a homodimer (28, 29). Several nuclear localization signals have been identified in both the N-terminal and C-terminal domains of N protein, whereas only one putative nuclear export signal is located in the middle (amino acids 220–231) (30, 31). We further mapped the region of N protein interacting with Smad3. GST pulldown assay showed that GST-Smad3 but not GST associated with both of the N- and C-terminal domains of N protein (Fig. 3E). Interestingly, only the C-terminal regions (both N (aa 211–422) and N (aa 232–422)) could up-regulate TGF-β-induced CAGA-luciferase expression while N (aa 2–210) had no effect (Fig. 3F), indicating that the promoting effect of N protein on TGF-β signaling is mediated by its C-terminal domain.
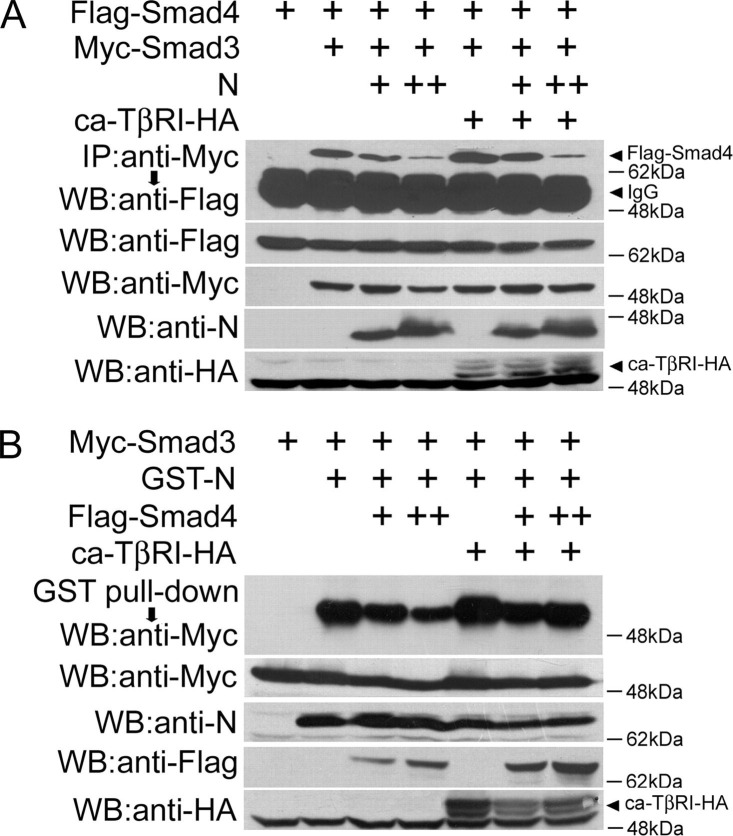
N Protein Interferes with Smad3-Smad4 Complex FormationNext, we examined whether N protein influences Smad3-Smad4 complex formation. HEK293T cells were transfected with Myc-Smad3 and FLAG-Smad4 with or without N protein. The constitutively active TβRI (ca-TβRI) was co-transfected to mimic TGF-β stimulation. When overexpressed, Smad3 interacted with Smad4 independent of ca-TβRI stimulation, but ca-TβRI further enhanced the Smad3-Smad4 interaction (Fig. 4A ). Interestingly, N protein interfered with the interaction between Smad3 and Smad4 in a dose-dependent manner regardless of the presence or the absence of ca-TβRI.
FIGURE 4.
N protein competes with Smad4 to bind Smad3.A, N protein attenuates the Smad3-Smad4 interaction. HEK293T cells were co-transfected with pCS2-Myc-Smad3 (4 μg), pCS2-FLAG-Smad4 (4 μg), pCMV5-ca-TβRI-HA (2 μg), and pcDNA3.1-N (2 μg or 4 μg). The Smad3-associated Smad4 was revealed by anti-Myc immunoprecipitation and anti-FLAG immunoblotting (upper panel). The protein expression was confirmed by immunoblotting of total cell lysates. B, Smad4 competes with N protein to bind Smad3. HEK293T cells were co-transfected with pCS2-Myc-Smad3 (4 μg), pEBG1-GST-N (2 μg), pCMV5-ca-TβRI-HA (2 μg), and pCS2-FLAG-Smad4 (2 μg or 4 μg). Cells lysates were incubated with Sepharose 4B-glutathione beads. The GST-N-associated Smad3 was revealed by anti-Myc immunoblotting (upper panel). The protein expression was confirmed by immunoblotting of total cell lysates.
The above result implied that N protein might compete with Smad4 to bind Smad3. To confirm this, we further examined whether Smad4 could influence Smad3 and N protein complex formation. HEK293T cells were transfected with GST-N and Myc-Smad3 with or without Smad4 and ca-TβRI. GST pulldown and anti-Myc immunoblotting showed that Smad4 interfered with the interaction between Smad3 and N protein in a dose-dependent manner in the absence of ca-TβRI, but this effect was less obvious in the presence of ca-TβRI (Fig. 4B). Ca-TβRI also enhanced the Smad3-N protein interaction, consistent with the above data that N protein exhibited a high binding affinity with phosphorylation-mimicking mutant Smad3(SD). Collectively, these data suggested that N protein competes with Smad4 to interact with Smad3.
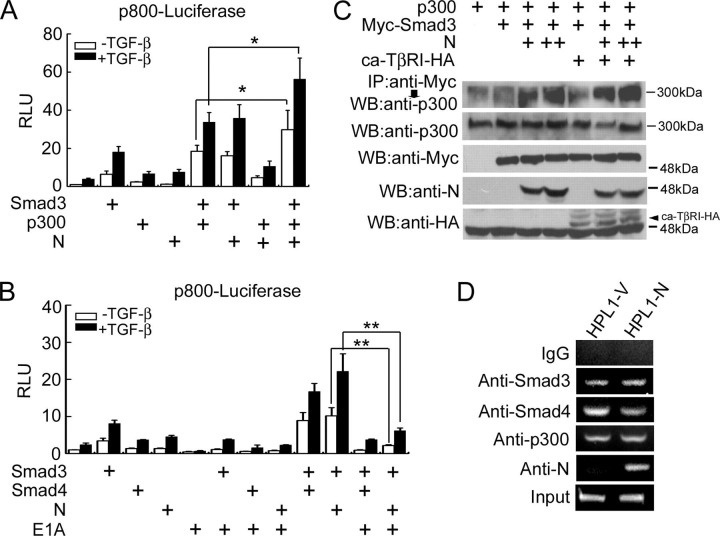
N Protein Enhances the Interaction between Smad3 and p300Transcriptional coactivators p300 and its related protein CBP have been shown to mediate Smad3 transactivation in TGF-β-induced transcriptional activation (32, 33, 34, 35). To test whether p300 mediates the function of N protein in promoting TGF-β-induced expression of PAI-1, we investigated the effect of p300 on p800-luciferase expression. As shown in Fig. 5A , p300 collaborated with N protein and Smad3 to remarkably promote p800-luciferase expression in the presence or the absence of TGF-β.
FIGURE 5.
N protein enhances Smad3-p300 interaction.A, N protein synergizes with Smad3 and p300 to induce p800-luciferase expression. HPL1 cells were co-transfected with p800-luciferase reporter (0.5 μg), pCS2-Myc-Smad3 (20 ng), pCMV-p300 (0.5 μg), and pcDNA3.1-N (0.5 μg). B, E1A inhibits p800-luciferase expression induced by TGF-β and N protein. HPL1 cells were co-transfected with p800-luciferase reporter (0.5 μg), pCS2-Myc-Smad3 (20 ng), pCS2-FLAG-Smad4 (0.5 μg), pXF2F-FLAG-E1A (0.5 μg), and pcDNA3.1-N (0.5 μg). C, N protein promotes Smad3-p300 complex formation. HEK293T cells were co-transfected with pCS2-Myc-Smad3 (4 μg), pCMV-p300 (4 μg), pCMV5-ca-TβRI-HA (2 μg), and pcDNA3.1-N (2 μg or 4 μg). The cells were harvested for anti-Myc immunoprecipitation and the Smad3-associated p300 was revealed by anti-p300 immunoblotting (upper panel). The protein expression was confirmed by immunoblotting of total cell lysates. D, N protein associates with the PAI-1 promoter. HPL1-V and HPL1-N cells were treated with 200 pm TGF-β1 for 2 h. ChIP assay was then performed with anti-Smad3, anti-Smad4, anti-p300, or anti-N antibodies. PCR amplification of the PAI-1 promoter (-733/-484) was performed to detect proteins-bound DNA. Rabbit pre-immune serum served as the negative control. The asterisks indicate a statistically significant difference (*, p < 0.05; **, p < 0.01). RLU: relative luciferase units.
Adenoviral E1A protein, an inhibitor of p300/CBP, can interfere with Smad3 function in TGF-β-induced transcriptional activation (34, 35, 36). To further confirm the role of p300 in mediating the effect of N protein in TGF-β-induced transcriptional activation, we tested whether E1A would interfere with the promoting effect of N protein on the transcriptional responses of TGF-β. As shown in Fig. 5B, E1A alone inhibited TGF-β-induced expression of p800-luciferase. Furthermore, E1A impaired the expression of p800-luciferase enhanced by Smad3 and N protein. These data strongly support the notion that p300 is involved in the promoting effect of N protein on the TGF-β-induced expression of p800-luciferase.
It has been demonstrated that Smad3 can associate with p300, and this association is potentiated by TGF-β-mediated activation of Smad3 (35). As p300 collaborated with N protein and Smad3 to enhance p800-luciferase expression, we then explored whether N protein could influence Smad3 and p300 complex formation. In accordance with a previous report (35), Smad3 had weak interaction with p300 when overexpressed, and this interaction was increased by ca-TβRI (Fig. 5C). Moreover, N protein greatly enhanced the interaction between Smad3 and p300 in a dose-dependent manner even in the absence of ca-TβRI. These results together suggested that N protein potentiates the transcriptional activity of Smad3 by increasing its interaction with p300.
Because early studies reported that N protein can directly bind to DNA to regulate the expression of NF-κB and cyclooxygenase-2 (4, 37), we attempted to investigate whether N protein can bind to the promoter of PAI-1 in vivo. Chromatin immunoprecipitation (ChIP) assay was performed with HPL1 cells stably expressing N protein, and the results showed that the PAI-1 promoter DNA was pulled down by the anti-N protein antibody from HPL1-N cells but not from the control HPL1-V cells (Fig. 5D), indicating that N protein was able to bind the PAI-1 promoter. In agreement with early reports (34), Smad3, Smad4, and p300 bound to the PAI-1 promoter in the presence of TGF-β. Consistent with the above result that N protein competed with Smad4 to bind Smad3, the expression of N protein decreased the Smad4-DNA association in HPL1-N cells (Fig. 5D). Together, these data strongly support the notion that N protein competes with Smad4 to form the transcriptional complex with Smad3 and p300 on the PAI-1 promoter under the physiological condition.
N Protein Inhibits TGF-β-induced Apoptosis of HPL1 CellTGF-β promotes apoptosis of several types of cells such as lymphocytes, pneumocytes, and hepatocytes (7), and the important role of Smad3 has been well established (8). We then investigated whether N protein has any influence on TGF-β-induced apoptosis of HPL1 cells. Transient transfection of Smad3 increased remarkably the number of apoptotic sub-G1 cells, and this effect was further enhanced by TGF-β treatment (Fig. 6A ), in agreement with the previous report that constitutively expression of Smad3 in HPL1 made cells sensitive to TGF-β-induced apoptosis (38). Overexpression of Smad3 and Smad4 further enhanced the sub-G1 cell number. In line with the importance of Smad4 in TGF-β-induced apoptosis, knockdown of endogenous Smad4 expression by siRNA attenuated TGF-β/Smad3-induced apoptosis of HPL1 cells. Interestingly, N protein impaired the pro-apoptotic activity of Smad3 and Smad4. Consistent with that, N protein down-regulated the expression of pro-apoptotic genes Bax and Bim in HPL1-N cells as shown by quantitative real-time PCR (Fig. 6, B and C). These data implicated that N protein can interfere with the proapoptotic activity of Smad3 and Smad4.
FIGURE 6.
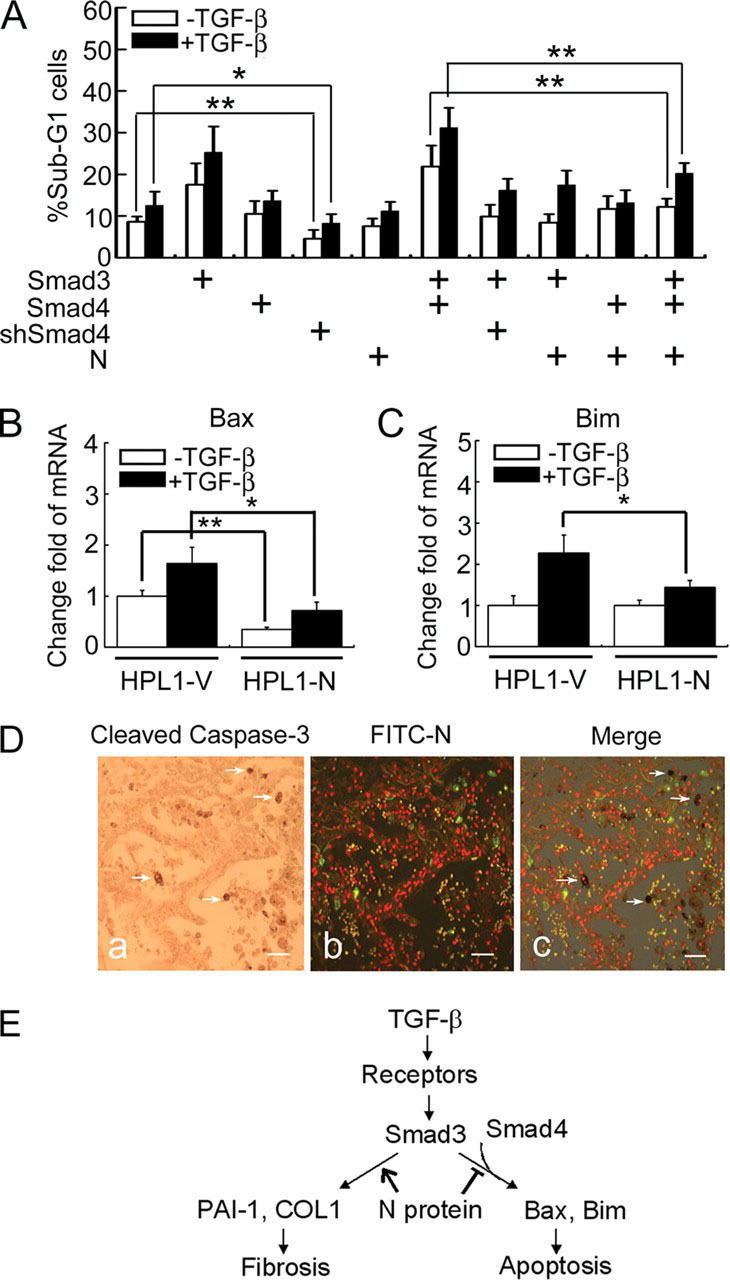
N protein impairs TGF-β-induced HPL1 cell apoptosis.A, N protein inhibits TGF-β-induced HPL1 cell apoptosis. HPL1 cells were co-transfected with GFP plasmid (0.1 μg), pCS2-Myc-Smad3 (1 μg), pCS2-FLAG-Smad4 (1 μg), pSUPER-siRNA against Smad4 (0.5 μg), and pcDNA3.1-N (1 μg) as indicated. Those GFP-positive cells were selected by FACS for DNA contents analysis. The percentage of sub-G1 cells in total GFP-positive cells was accounted for. Each experiment was repeated in triplicate, and the data represent the mean ± S.D. of three independent experiments. B and C, the mRNA levels of endogenous Bim and Bax in HPL1-V and HPL1-N were analyzed by real-time PCR. GAPDH served as a loading control. The asterisks indicate a statistically significant difference between HPL1-V and HPL1-N cells (*, p < 0.05; **, p < 0.01). D, N protein-positive cells are not co-localized with active caspase-3-positive apoptotic cells in SASR patient's lung. Apoptotic cells were detected by anti-cleaved caspase-3 antibody and developed by diaminobenzidine tetrahydrochloride (a). SARS-CoV-infected cells were detected by fluorescein isothiocyanate-labeled anti-N protein antibody and the image was taken with a fluorescence microscopy. The virus-infected cells were green (b). White arrows indicate the apoptotic cells in c. Scale bar:50 μm. E, a working model depicts the role of SARS-CoV N protein in modulating TGF-β/Smad3-induced fibrosis and apoptosis. The asterisks indicate a statistically significant difference (*, p < 0.05; **, p < 0.01).
Apoptotic cells have been detected in different tissues and organs of SARS patients (13, 16), however, N protein was only detected in the early stage of SARS (15). When apoptotic cells were stained with cleaved capase-3 antibody by immunohistochemistry, no co-localization of active capase-3-positive cells with N protein-positive cells was observed (Fig. 6D). This suggested that infection of SARS-CoV might not induce apoptosis in the early stage of SARS due to the high level of N protein.
DISCUSSION
Multiple functions have been postulated for SARS-CoV N protein throughout the viral life cycle and in the progression of the clinical symptom of SARS. N protein binds to and stabilizes viral genomic RNA. It appears to be the major immunogenic antigen, and the immune response to N protein can serve as an early diagnostic marker for SARS infection (39). In our study, we found that N protein specifically interacted with Smad3 via the MH2 domain and competed with Smad4 to bind Smad3. We further showed that N protein enhanced the transcriptional responses of TGF-β by promoting Smad3-p300 complex formation. Finally, our results demonstrated that N protein enhanced TGF-β/Smad3-induced expression of PAI-1, but attenuated Smad3/Smad4-meidated apoptosis.
The multifunctional feature of TGF-β suggests that it may be an important target of viruses to influence host cell fate in favor of virus replication and proliferation. Several viral proteins, including hepatitis B virus pX, hepatitis C virus core protein, NS3 and NS5, adenovirus E1A, human papillomavirus E7, human T-lymphotropic virus Tax, and Epstein-Barr virus LMP1 have been reported to modulate TGF-β signaling (36, 40, 41, 42, 43, 44). The common strategy utilized by viruses to modulate TGF-β signaling is through the direct binding of viral proteins to Smad proteins. Except for pX protein, which has been shown to enhance the transcriptional responses of TGF-β, the other viral proteins are reported to negatively regulate TGF-β signaling by interfering with the Smad transcriptional complex formation. Here, we report that SARS-CoV N protein promotes TGF-β/Smad3-mediated expression of PAI-1 but inhibits Smad3/Smad4-mediated apoptosis.
A typical clinical characteristic of SARS-associated acute respiratory distress syndrome is pulmonary fibrosis and the associated lung failure (13, 14, 16). Pulmonary fibrosis is the final result of many severe lung injuries, and it is characterized by an initial diffuse inflammatory reaction or epithelial injury followed by fibroblast proliferation and extracellular matrix accumulation (45). High levels of pro-inflammatory cytokines, including TGF-β1, are expressed in the SARS-CoV-infected cells (16). TGF-β stabilizes the extracellular matrix by down-regulating the expression of extracellular matrix proteases and stimulating the expression of some extracellular matrix protease inhibitors, including PAI-1, which is the primary inhibitor of both tissue-type and urokinase-type plasminogen activator. PAI-1 is a well established target of TGF-β via the Smad pathway and plays a pivotal role in TGF-β-promoted tissue fibrosis (26, 46). We show here that N protein potentiates TGF-β-induced PAI-1 expression by quantitative PCR and by measuring the expression of the PAI-1 promoter-derived p800-luciferase.
Smad4, the co-Smad, is generally regarded to be essential for the transcriptional responses elicited by the TGF-β family members (11). The expression of endogenous PAI-1 is regulated by Smads, CBP/p300, TFE3, and Sp1 (22, 27, 34, 47, 48). Although the role of Smad3 in regulating PAI-1 expression has been established, the importance of Smad4 has been controversial. Smad4 has been suggested to act as a key coactivator that enhances ligand-induced transcription by stabilizing the association of Smad3 with CBP/p300 in the PAI-1 promoter (34). However, TGF-β might induce PAI-1 expression in Smad4-independent ways. For instance, TGF-β stimulates PAI-1 expression in a dose-dependent manner in both wild-type and Smad4-deficient mouse fibroblasts (49). In Smad4-defcient colon carcinoma SW480 cells, re-introduction of Smad4 reduced PAI-1 expression (50). Our results showed that Smad4 is important for TGF-β-induced apoptosis of HPL1 cells, but not required to mediate TGF-β effect on PAI-1 synthesis. This is in agreement with a recent report that Smad4 is required for the anti-proliferative response but not for the differentiation response of human hematopoietic stem/progenitor cells, although Smad2/3 participate in both responsiveness to TGF-β (51). We further found that N protein potentiates TGF-β/Smad3-induced expression of PAI-1 but attenuates Smad3/Smad4-mediated apoptosis. This observation is consistent with the finding that N protein inhibits the expression of pro-apoptotic genes Bax and Bim. We did not observe that N protein had any obvious influence on the anti-proliferative effect of TGF-β on HPL1 cells, although it down-regulated TGF-β-induced p15 expression (data not shown). Therefore, N protein can modulate TGF-β signaling by selectively activating a subset of target genes and inhibiting the others.
SARS infection results in increased cell apoptosis or necrosis in the lung, liver, and lymphatic tissue (13, 16). SARS-CoV also can induce apoptosis of cultured cells, and N protein, also some non-structural protein like 3a, 3b, and 7a proteins of SARS-CoV have been suggested to be pro-apoptotic (5, 52, 53, 54). Because the information about the early pathological changes of SARS patients is very limited, the contribution of apoptosis to the SARS-associated pathology is unclear. By examining the SARS-CoV-infected lungs at the early stage of infection, we found no obvious co-localization of N-protein-positive cells and active caspase 3-positive cells, indicating that SARS-CoV infection does not cause apoptosis of host cells at the early stage. Interestingly, N protein is detected at the early stages of SARS and diminishes during the progress of the disease development (15). Based on our findings, we postulate that N protein inhibits apoptosis in favor of virus packaging and replication at the early stage of SARS development. Meanwhile, N protein potentiates TGF-β-induced PAI-1 expression leading to development of lung fibrosis at the late stage. This hypothesis is consistent with the notion that virus can promote or inhibit the apoptosis of host cells in favor for its replication and proliferation (55).
In summary, our results demonstrated that SARS-CoV N protein interacts with Smad3 and up-regulates the TGF-β-induced synthesis of the fibrotic promoter PAI-1, leading to tissue fibrosis (Fig. 6E). On the other hand, N protein competes with Smad4 to bind Smad3 and attenuates TGF-β-induced apoptosis. These results provide new insights into our understanding of the molecular mechanism underlying the pathogenesis of SARS-CoV. In addition, our findings also suggest novel therapeutic strategies for SARS treatment, i.e. the molecules involved in TGF-β signaling could be therapeutic targets.
Acknowledgments
We thank Dr. Xiao Yang for Smad3-/- MEF cells, Dr. Xin-Hua Feng for plasmids, Dr. Jianwei Wang for mouse anti-N protein antibody, Dr. Edward B. Leof for rabbit anti-p-Smad3 antibody, and Teng Fei for assistance with the ChIP assay.
Footnotes
- SARS
- severe acute respiratory syndrome
- SARS-CoV
- SARS-associated coronavirus
- ca
- constitutively active
- ChIP
- chromatin immunoprecipitation
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GST
- glutathione S-transferase
- HPL1
- human peripheral lung epithelial cells
- MEF
- mouse embryo fibroblast
- N protein
- nucleocapsid protein
- PAI-1
- plasminogen activator inhibitor-1
- TGF-β
- transforming growth factor-β
- siRNA
- small interference RNA
- aa
- amino acid(s)
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein.
References
- 1.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 2.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. Biochem. J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surjit M., Liu B., Chow V.T., Lal S.K. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel P.M., Massague J. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 8.Roberts A.B., Tian F., Byfield S.D., Stuelten C., Ooshima A., Saika S., Flanders K.C. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Border W.A., Noble N.A. N. Engl. J. Med. 1994;331:1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y., Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 11.Massague J., Seoane J., Wotton D. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 12.Feng X.H., Derynck R. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 13.Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D., Lu Y., Wu D., He L., Yao K. J. Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H., Tsang N.C., Guan Y., Yuen K.Y., Peiris J.S. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls J.M., Butany J., Poon L.L., Chan K.H., Beh S.L., Poutanen S., Peiris J.S., Wong M. PLoS Med. 2006;3:e27. doi: 10.1371/journal.pmed.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L., Ding Y., Zhang Q., Che X., He Y., Shen H., Wang H., Li Z., Zhao L., Geng J., Deng Y., Yang L., Li J., Cai J., Qiu L., Wen K., Xu X., Jiang S. J. Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Ren L., Zhao X., Hung T., Meng A., Wang J., Chen Y.G. J. Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Gao X., Wen J., Ning Y., Chen Y.G. J. Biol. Chem. 2006;281:8607–8612. doi: 10.1074/jbc.M600274200. [DOI] [PubMed] [Google Scholar]
- 19.Lin X., Duan X., Liang Y.Y., Su Y., Wrighton K.H., Long J., Hu M., Davis C.M., Wang J., Brunicardi F.C., Shi Y., Chen Y.G., Meng A., Feng X.H. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda A., Kondo M., Saito T., Yatabe Y., Kobayashi T., Okamoto M., Suyama M., Takahashi T. Cancer Res. 1997;57:4898–4904. [PubMed] [Google Scholar]
- 21.Zhang S., Fei T., Zhang L., Zhang R., Chen F., Ning Y., Han Y., Feng X.H., Meng A., Chen Y.G. Mol. Cell. Biol. 2007;27:4488–4499. doi: 10.1128/MCB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J.M. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carcamo J., Weis F.M., Ventura F., Wieser R., Wrana J.L., Attisano L., Massague J. Mol. Cell. Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Weisberg E., Fridmacher V., Watanabe M., Naco G., Whitman M. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 25.Hata A., Seoane J., Lagna G., Montalvo E., Hemmati-Brivanlou A., Massague J. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 26.Kaminski N., Allard J.D., Pittet J.F., Zuo F., Griffiths M.J., Morris D., Huang X., Sheppard D., Heller R.A. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1778–1783. doi: 10.1073/pnas.97.4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeton M.R., Curriden S.A., van Zonneveld A.J., Loskutoff D.J. J. Biol. Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 28.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surjit M., Liu B., Kumar P., Chow V.T., Lal S.K. Biochem. Biophys. Res. Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowland R.R., Chauhan V., Fang Y., Pekosz A., Kerrigan M., Burton M.D. J. Virol. 2005;79:11507–11512. doi: 10.1128/JVI.79.17.11507-11512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timani K.A., Liao Q., Ye L., Zeng Y., Liu J., Zheng Y., Yang X., Lingbao K., Gao J., Zhu Y. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janknecht R., Wells N.J., Hunter T. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pouponnot C., Jayaraman L., Massague J. J. Biol. Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 34.Feng X.H., Zhang Y., Wu R.Y., Derynck R. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen X., Hu P.P., Liberati N.T., Datto M.B., Frederick J.P., Wang X.F. Mol. Biol. Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishihara A., Hanai J., Imamura T., Miyazono K., Kawabata M. J. Biol. Chem. 1999;274:28716–28723. doi: 10.1074/jbc.274.40.28716. [DOI] [PubMed] [Google Scholar]
- 37.Yan X., Hao Q., Mu Y., Timani K.A., Ye L., Zhu Y., Wu J. Int. J. Biochem. Cell Biol. 2006;38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Yanagisawa K., Osada H., Masuda A., Kondo M., Saito T., Yatabe Y., Takagi K., Takahashi T. Oncogene. 1998;17:1743–1747. doi: 10.1038/sj.onc.1202052. [DOI] [PubMed] [Google Scholar]
- 39.Che X.Y., Hao W., Wang Y., Di B., Yin K., Xu Y.C., Feng C.S., Wan Z.Y., Cheng V.C., Yuen K.Y. Emerg. Infect Dis. 2004;10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng P.L., Chang M.H., Chao C.H., Lee Y.H. Oncogene. 2004;23:7821–7838. doi: 10.1038/sj.onc.1208066. [DOI] [PubMed] [Google Scholar]
- 41.Choi S.H., Hwang S.B. J. Biol. Chem. 2006;281:7468–7478. doi: 10.1074/jbc.M512438200. [DOI] [PubMed] [Google Scholar]
- 42.Lee D.K., Park S.H., Yi Y., Choi S.G., Lee C., Parks W.T., Cho H., de Caestecker M.P., Shaul Y., Roberts A.B., Kim S.J. Genes Dev. 2001;15:455–466. doi: 10.1101/gad.856201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee D.K., Kim B.C., Brady J.N., Jeang K.T., Kim S.J. J. Biol. Chem. 2002;277:33766–33775. doi: 10.1074/jbc.M200150200. [DOI] [PubMed] [Google Scholar]
- 44.Prokova V., Mosialos G., Kardassis D. J. Biol. Chem. 2002;277:9342–9350. doi: 10.1074/jbc.M109099200. [DOI] [PubMed] [Google Scholar]
- 45.Chapman H.A. J. Clin. Invest. 2004;113:148–157. doi: 10.1172/JCI20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eitzman D.T., McCoy R.D., Zheng X., Fay W.P., Shen T., Ginsburg D., Simon R.H. J. Clin. Invest. 1996;97:232–237. doi: 10.1172/JCI118396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta P.K., Blake M.C., Moses H.L. J. Biol. Chem. 2000;275:40014–40019. doi: 10.1074/jbc.C000508200. [DOI] [PubMed] [Google Scholar]
- 48.Hua X., Liu X., Ansari D.O., Lodish H.F. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sirard C., Kim S., Mirtsos C., Tadich P., Hoodless P.A., Itie A., Maxson R., Wrana J.L., Mak T.W. J. Biol. Chem. 2000;275:2063–2070. doi: 10.1074/jbc.275.3.2063. [DOI] [PubMed] [Google Scholar]
- 50.Schwarte-Waldhoff I., Klein S., Blass-Kampmann S., Hintelmann A., Eilert C., Dreschers S., Kalthoff H., Hahn S.A., Schmiegel W. Oncogene. 1999;18:3152–3158. doi: 10.1038/sj.onc.1202641. [DOI] [PubMed] [Google Scholar]
- 51.He W., Dorn D.C., Erdjument-Bromage H., Tempst P., Moore M.A., Massague J. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 52.Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. Virol. J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W., Chan J.Y., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M., Sung J.J., Lo Y.M., Tsui S.K. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 55.Roulston A., Marcellus R.C., Branton P.E. Annu. Rev. Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]