Abstract
In inflamed human tissues, we often find intact eosinophilic granules but not eosinophils themselves. Eosinophils, tissue dwelling granulocytes with several homeostatic roles, have a surprising association with fibrinogen and tissue remodeling. Fibrinogen is a complex glycoprotein with regulatory roles in hemostasis, tumor development, wound healing, and atherogenesis. Despite its significance, the functional link between eosinophils and fibrinogen is not understood. We tested IL-5 primed mouse bone marrow-derived and human blood-sorted eosinophil activity against FITC-linked fibrinogen substrates. The interactions between these scaffolds and adhering eosinophils were quantified using 3D laser spectral, confocal, and transmission electron microscopy (TEM). Eosinophils were labeled with major basic protein (MBP) antibody to visualize granules and assessed by flow cytometry. Both mouse and human eosinophils showed firm adhesion and degraded up to 27±3.1% of the substrate area. This co-occurred with active MBP-positive granule release and the expression of integrin CD11b. Mass spectrometry analysis of fibrinogen proteolytic reactions detected the presence of eosinophil peroxidase, MBP, and fibrin alpha, beta, and gamma chains. Eosinophil activity was adhesion-dependent, as a blocking antibody against CD11b significantly reduced adhesion, degranulation and fibrinogenolysis. Though adhered, eosinophils exhibited no proteolytic activity on collagen matrices. Cytolytic degranulation was defined by loss of membrane integrity, cell death and presence of cell-free granules. From TEM images, we observed only fibrinogen-exposed eosinophils undergoing this process. This is the first report to show that fibrinogen is a specific trigger for cytolytic eosinophil degranulation with implications in human disease.
Keywords: Fibrinogen, fibrinogenolysis, fibrin, degranulation, eosinophils, collagen, adhesion, CD11b, metabolism, cytokines
Introduction
Fibrinogen is a complex glycoprotein with multiple regulatory functions in blood clotting, wound healing, cell-matrix interactions, inflammation, tumor development, and atherogenesis (1). Normally, most fibrinogen is produced by the liver and found in plasma. It is also known that in cases of epithelial damage, plasma exudate to the area introduces fibrinogen. It is less appreciated that epithelial cells are also a source of local fibrinogen production and release. Mechanically damaged bronchial epithelial cells have been shown to release fibrinogen independently of plasma proteins (2), and alveolar epithelial cells were found to synthesize and release fibrinogen de novo when stimulated with proinflammatory mediators (3).
Eosinophils are multifunctional granulocytes that are virtually absent in morphogenetically quiescent tissues but infiltrate tissues in significant numbers during morphogenesis (normal development or pathologic remodeling) and Type 2 immune responses (4). Multiple reports have shown that eosinophils interact with fibrinogen in different contexts. Canine bone marrow and tissue-activated eosinophils are specifically attracted to fibrinogen-enriched scaffolds during wound repair (5). Human airway and IL-5 stimulated blood eosinophils in asthma exhibit a hyperadhesive phenotype towards fibrinogen (6). Furthermore, fibrinogen has been suggested as a chemoattractant for eosinophils into areas of inflammation (5). Multiple reports have shown eosinophils to interact with fibrinogen through receptor CD11b (αMβ2, Mac-1), which is known to be a central mediator of leukocyte adhesion, migration, and activation. Numerous studies have highlighted that the CD11b integrin is essential for neutrophil (7), monocyte, and macrophage binding to fibrinogen, and that inhibiting the CD11b receptor will largely prevent adhesion (8). Eosinophils have also been shown to adhere to fibrinogen via receptor CD11b (9).
Eosinophils are thought to contribute to disease pathology by releasing proinflammatory cytokines and cytotoxic granule proteins (10). Human eosinophils contain granules rich in major basic protein 1 (MBP), eosinophil cationic protein (ECP), eosinophil-derived neurotoxin (EDN), and eosinophil peroxidase (EPX). Mouse eosinophils also contain MBP and EPX (11). Though it is well known that human eosinophils degranulate upon stimulation and in inflammatory settings, mouse eosinophil degranulation is hard to achieve and is rarely reported in in vitro and in vivo inflammation models (12). Though studies have shown some amount of mouse eosinophil piecemeal degranulation in response to stimulation by PAF (13) and PMA (14), the evidence of granule protein release is minor (15), and the concentration of stimulus needed to trigger degranulation is much higher than what is needed to degranulate human eosinophils. Cytolytic eosinophil degranulation (a form of cytolytic cell death) occurs when eosinophils lose membrane integrity and cell-free granules are left behind (11). This form of degranulation is often found in human allergic airway and skin disease (16–19).
Thus far, no studies have attempted to demonstrate the direct link between fibrinogen deposition and eosinophil degranulation, in part due to poor understanding of eosinophil-tissue environment interactions. This report: 1) demonstrates the specific cytolytic degranulation and proteolytic activity of mouse and human IL-5-primed eosinophils adhering to fibrinogen (but not collagen) substrates from different species; 2) shows that eosinophils necessitate CD11b upregulation to adhere and degranulate upon exposure to fibrinogen; 3) shows that fibrinogen (and to some extent, Type I collagen) triggers the expression of pro-inflammatory and remodeling (IL-13, IL1a, IL-4, Tnf, Ccl24, and versican) mediators from murine eosinophils; 4) provides evidence that fibrinogen elicits higher metabolic activity (NADH production) in eosinophils compared to collagen substrates; 5) provides a methodological platform for further studies aimed at manipulating, visualizing, and quantifying eosinophil-matrix interactions.
Materials and Methods
Mouse eosinophils.
Murine eosinophils were cultured from bone marrow according to a published protocol (20). In brief, bone marrow was harvested from 10–12 week old wild-type Balb/c mice (Jackson Labs, Bar Harbor, ME, USA) by flushing femurs and tibiae using RPMI-1640 medium with L-glutamine (Corning, 10–040-CV). Cells were cultured in media containing RPMI-1640 with L-glutamine, 1% Penicillin/Streptomycin (HyClone, SV30010), 1% L-glutamine (HyClone, SH30034.01), 1% MEM Non-Essential Amino Acids (Corning, 25–025-CI), 1% Sodium Pyruvate (Corning 25–000-CI), 2.5% HEPES (Corning 25–060-CI), 20% Heat-Inactivated Fetal Bovine Serum (FBS) (Corning, 35–016-CV), and 0.0000035% 2-Mercaptoethanol (Sigma, M6250). Cell media was supplemented with murine 100 ng/mL recombinant SCF (PeproTech, 250–03) and 100 ng/mL recombinant FLT3-L (PeproTech, 250–31L) from days 0 through 4. On day 4, media was replaced with fresh media containing 10 ng/mL murine recombinant IL-5 (PeproTech, 215–15). Media containing IL-5 was changed every other day until day 14, when cells were used for experimentation. Flow cytometry and cytospins were used to confirm maturation and purity of day 14 cells using a Diff-Quick Staining Kit (Electron Microscopy Sciences, 2609650).
Human eosinophils.
Human eosinophils were obtained from laboratory of Dr. Bruce Bochner (Northwestern IRB), which were isolated following a published protocol (21). Briefly, 60 mL of 0.1 M EDTA-anticoagulated peripheral blood was obtained from healthy donors. Blood was diluted with 2 volumes of 1X PBS, and eosinophils were isolated by Percoll density gradient centrifugation using 40 mL of diluted blood and 10 mL of room temperature Percoll Plus (density 1.090) (GE Healthcare, 17544502), centrifuging for 20 min at 300 × g at room temperature, which concentrated and removed mononuclear cells (lymphocytes and monocytes) and basophils at the plasma-Percoll interface. In the remaining pellets, erythrocytes were removed by hypotonic lysis in ice cold water and neutrophils were removed by immunomagnetic selection using a CD16 MicroBead kit (Miltenyi Biotec, 130-045-701). Resulting eosinophil purity was confirmed by Shandon Kwik-Diff staining (ThermoFisher Scientific, 9990700). Once purified, eosinophils were primed with 30 ng/mL of IL-5 (R&D systems) overnight prior to assay.
Fibrinogen interaction assay.
FITC-labeled fibrinogen, murine (Oxford Biomedical, FB01) and porcine (Oxford Biomedical, FB04), non-labeled murine fibrinogen (Haematologic Technologies, MCI-5150), FITC-labeled type 1 bovine collagen (Sigma, C4361), and non-labeled type 1 murine collagen (BioRad 2150–1425) were diluted to 100 μg/mL in 1x TBS, pH 8.0 (Corning, 46–012-CM). 100 μL/well of fibrinogen or collagen was plated on a 96-well plate (Greiner, 655090), which was left lightly shaking overnight at 4°C. The following day, solutions were aspirated and 100 μL FBS was added to each well. The plate was incubated for 1.5–2 hours at 37°C. Meanwhile, eosinophils were prepared in media suspensions at 1,000 cells/μL in fresh media with IL-5 (10 ng/mL). Cells were incubated for 30 minutes with either 10 μg/uL of anti-mouse/human CD11b clone M1/70 (Biolegend, 101201) or rat IgG2b κ isotype control (Biolegend, 400602), or were left untreated. After the plate was incubated, FBS was aspirated and 100 μL of eosinophil suspension was added to each well. After 4 hours at 37°C, cell suspensions were removed and wells were washed 2–3 times with 1x TBS to remove non-adherent eosinophils. 100 μL of 2% paraformaldehyde (ThermoFisher, 28908) diluted in PBS was added to each well. rMBP was from LSBio (LS-G14093) and was tested at 1 ug/ml in fibrinogen interaction assays.
Imaging.
Plates were imaged using fluorescent confocal microscopy at both 10x and 20x on Olympus DSU Confocal microscope (phase contrast, FITC channel) or a Keyence BZX-800 microscope, laser spectral microscopy at 20x on Nikon A1R Laser Scanning Confocal microscope (Nikon Imaging Core, Northwestern University), and/or transmission electron microscopy (TEM) on the FEI Tecnai Spirit G2 (Nikon Imaging Core, Northwestern University). At least two to four representative field images were taken per well to assess fibrinogen degradation and eosinophil granule dispersion. For major basic protein (MBP) immunofluorescence staining, plates were fixed with 2% paraformaldehyde for 15 minutes at room temperature. Samples were then blocked with 5% goat serum and 0.3% Triton X-100 in 1x PBS for one hour before incubating with primary antibody diluted in antibody dilution buffer (1% BSA and 0.3% Triton X-100 in 1x PBS). Afterward, samples were incubated with secondary antibody diluted in the same buffer. Primary staining was with rat anti-mouse MBP (Jamie Lee labs, Mayo Clinic Arizona) at a 1:500 dilution, and secondary staining was with goat anti-rat IgG Texas Red (Life Technologies, T6392) at a 1:500 dilution. DRAQ5 (BioLegend) staining was done in at a 1:200 dilution in 1x PBS for 15 minutes prior to assay. DAPI (Cell Signaling Technologies, 4083) staining was performed on fixed cells at 1 μg/mL in 1x PBS for 5 minutes. For TEM, samples were fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer prior to embedding in a resin mixture of Embed 812 kit. Samples were then sectioned on a Leica Ultracut UC6 ultramicrotome and collected on 200 mesh copper grids and stained with 3% uranyl acetate and Reynolds lead citrate.
Quantification of fibrinogen degradation.
ImageJ (22) was used to quantify degraded fibrinogen area on 2D images taken with fluorescent confocal microscopy. Images were converted to greyscale and cropped to the lower right quadrant (250 × 250 pixels). Threshold was adjusted such that only the degraded area pixels were selected, and the particle analysis tool was used to calculate the both the total percentage selected and the diameter of each selected area. Areas touching the field edges were excluded.
Flow Cytometry.
Cell suspensions from culture or cells from fibrinogen interaction assays (cells from supernatants combined with cells detached from fibrinogen surface with Trypsin/EDTA (Lonza, CC-5012)) were stained with a Zombie Aqua Fixable Viability Kit (BioLegend, 423102) for 20 minutes at room temperature, blocked with anti-CD16/CD32 (BD Biosciences, 553142) for 10 minutes in flow staining buffer at 4°C, and incubated with either APC-Cy7 conjugated Siglec-F (BD Biosciences, 561039) or AF647 conjugated CD11b (BD Biosciences, 562680) in flow staining buffer for 30 minutes at 4°C. Cells were washed in 1x PBS and fixed in 2% paraformaldehyde. Samples were run on an LSRII flow cytometer (BD Biosciences) and data were analyzed on FlowJo v10 software (Tree Star, Ashland, OR).
Mass Spectrometry.
Supernatants from fibrinogen interaction assays were removed from plates and cells were spun out at 300 × g for 5 minutes. Proteins were purified by Acetone/TCA precipitation, reduced, and alkylated. Digested peptides were desalted on C18 spin columns prior to analysis on the Mass Spec. MS1 peptide intensity was measured by searching the data against a mouse database using MaxQuant database search engine and Label-free quantification (LFQ) pipeline. Proteomic analysis was performed by the Northwestern University Proteomics Core Facility (Robert H Lurie Comprehensive Cancer Center).
Quantitative PCR.
RNA was isolated from cells using the Qiagen RNeasy mini kit (Qiagen, 74136). cDNA was synthesized using a qScript cDNA synthesis kit (Quanta BioSciences, 95047) and analyzed by real-time PCR on a 7500 real-time PCR system (Applied Biosystems) using primers/probes from Integrated DNA Technologies and PrimeTime Gene Expression Master Mix (IDT, 1055771).
RNA-Seq.
All cells were lysed and RNA was extracted using the RNeasy Plus Mini Extraction Kit (Qiagen, 74136). RNA quality was assessed using an Agilent High Sensitivity RNA ScreenTape System (Agilent Technologies). The NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) was utilized for full-length cDNA synthesis and library preparation. The sequencing of cDNA libraries was done on an Illumina NextSeq 500 instrument (Illumina) at a target read depth of approximately 10 million aligned reads per sample. The pool was denatured and diluted to create a 2.5 pM DNA solution. The PhiX control was spiked at 1%. The pool was then sequenced by 1×5 cycles using the NextSeq 500 High Output Kit (Illumina).
Eosinophil viability testing.
Characterization of eosinophils as live or necrotic was determined by FITC Annexin V Apoptosis Detection Kit with Propidium Iodide (PI) (BioLegend 640914) for the non-adherent fraction or trypan blue dye exclusion test (Invitrogen T10282) for the adherent fraction. After 4 hours of assay, non-adherent eosinophils were removed and stained with Annexin V and PI and analyzed via flow cytometry. Annexin V(–)PI(–) eosinophil populations (live cells) were determined by non-stained controls. Adherent eosinophils were stained in situ with trypan blue dye diluted 1:1 with cell media. Samples were visualized on a Life Technologies EVOS XL Core microscope at 10x. Multiple fields of view within each well were counted to determine the number of cells that stained blue (dead) relative to those with intact membranes that did not take up the dye (live).
Biolog metabolism assay.
To determine metabolic differences, 20,000 eosinophils per well were seeded onto blank 96-well Biolog plates (Biolog, 30311) coated with fibrinogen or collagen in media made with phenol red-free RPMI. Biolog MB (Biolog, 74352) redox dye was added, and cells were then incubated in an OmniLog automated incubator-reader (Biolog, Hayward, CA, 12111) at 37°C. Dye reduction (measuring NAD(P)H-dependent oxidoreductase activity) was measured at 590 nm absorbance. Kinetic background values were manually subtracted and initial rates were measured between the beginning of each run and when the uptake reached its maximum (between 0.25 to 1.25 hours).
Bioinformatical and Statistical Analysis.
RNA-Seq sequenced reads were demultiplexed using bcl2fastq (v 2.17.1.14). Quality control was performed using FastQC. Low quality reads were discarded using trimmomatic (v 0.33). Reads were aligned using STAR aligner. Read counts were generated using htseq. Differential expression analysis was done using the DESeq2 R/Bioconductor package (23). All computational analysis was performed on genomic nodes of Quest, Northwestern’s High Performance Computing Cluster. Gene ontology (GO biological process) analysis was performed in GOrilla (24). Statistical significance of all data was determined by t-tests or ANOVA tests followed by Tukey’s post-hoc pairwise testing, or Mann-Whitney or Kruskal-wallis tests followed by Dunn’s multiple comparison’s test whenever data did not fit parametric test criteria. All data are represented as mean ± S.E.M. Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software, Inc. La Jolla, CA). An alpha level of 0.05 was used as a significance cut-off.
Results
Eosinophils degrade fibrinogen substrate
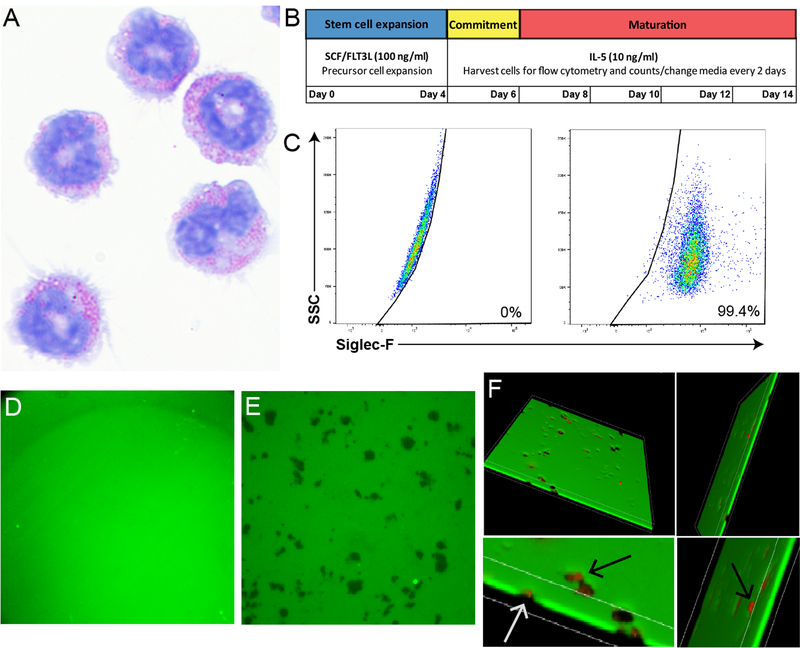
After the 14 day culture protocol (Figure 1A and 1B), eosinophils showed a mature morphology, as identified by red eosin stained granules and segmented nuclear morphology (Figure 1A). These cells were >99% pure at day 14, as determined by Siglec-F expression (Figure 1C). Wells coated with FITC-linked fibrinogen substrate could be visualized using fluorescent confocal microscopy and maintained a homogenous coating in the absence of eosinophils (Figure 1D). The addition of eosinophils resulted in significant fibrinogen degradation (Figure 1E, dark areas). 3D imaging confirmed the complete fibrinogen scaffold loss in these areas (Figure 1F). Furthermore, major basic protein 1 (MBP) staining shows that eosinophils and eosinophil granules are localized to areas of fibrinogen loss (Figure 1F). This fibrinogen degradation was dependent on eosinophil intracellular activity, as cells that had been fixed with 2% paraformaldehyde prior to assay did not degrade the substrate (Figure S1).
Figure 1. Bone marrow-derived murine eosinophils degrade fibrinogen substrates.
A) Day 14 IL-5-cultured eosinophils stained with a Diff-Quick Staining Kit and photographed with a 60x oil objective. Note mature red granular morphology. B) Schematic showing timeline of bone marrow-derived murine eosinophil culture protocol. C) Purity of cultured eosinophils, all cells were >99% Siglec-F positive (right) by non-stained control (left). D) FITC-linked fibrinogen substrate (green) without eosinophil addition visualized using fluorescent confocal microscopy. The substrate maintains a homogenous composition when eosinophils are not added, as is seen in this image. E) Dark spots represent areas of degraded fibrinogen after addition of eosinophils. F) Dark spots depict areas of total fibrinogen degradation, as visualized by using 3D stack images. Anti-MBP stained cells (red) localize to degraded areas and completely penetrate fibrinogen. Bottom two images are top images magnified, arrows point to anti-MBP stained cells penetrating the fibrinogen substrate. Note that cells tend to be smaller than the surrounding empty areas.
Fibrinogen degradation occurs in a species specific manner and increases with cell density
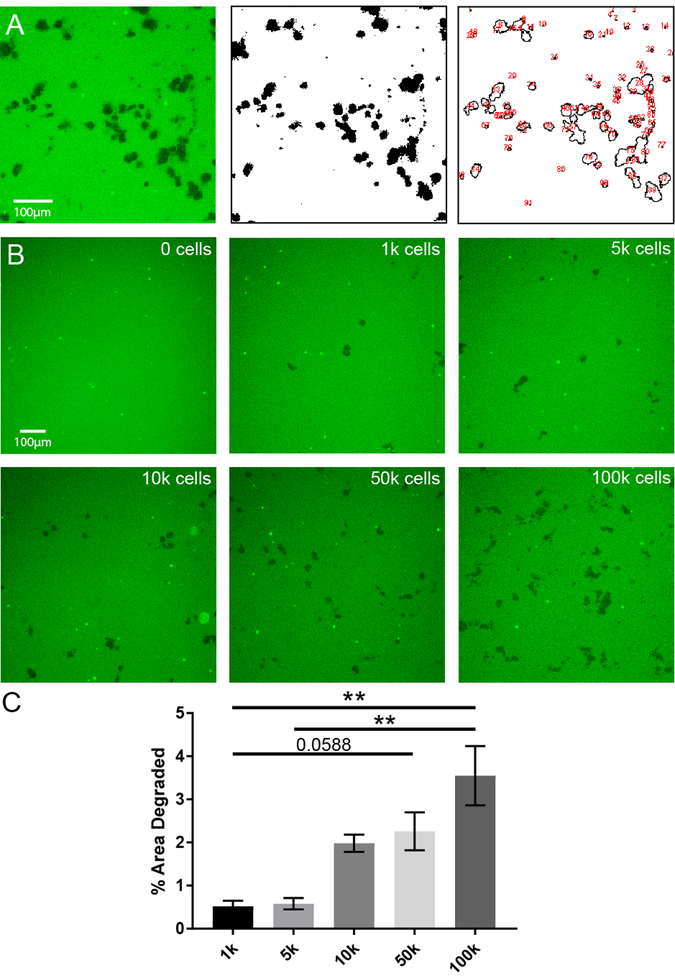
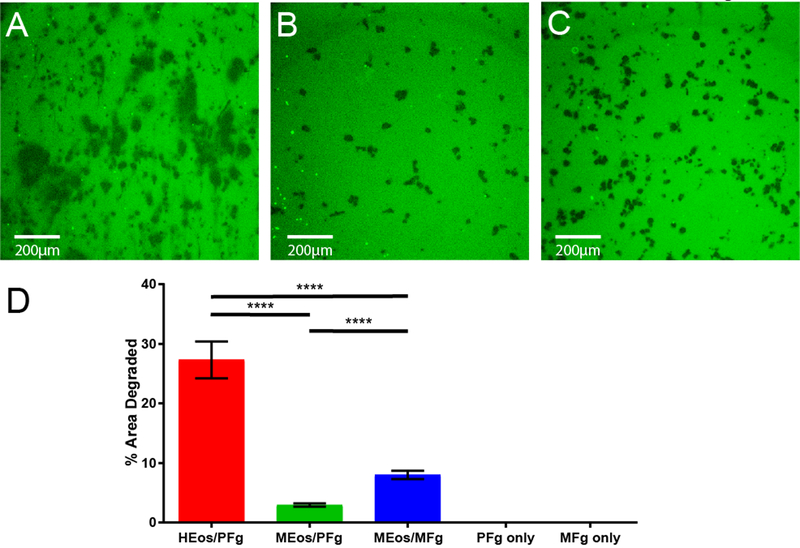
We developed an image analysis protocol to quantify the degree to which eosinophils degrade the fibrinogen substrate under different experimental conditions. By using ImageJ, we first converted the fluorescent image to black and white, and then quantified the total degraded area, represented as the percentage of the image that was degraded (black) relative to that which remained intact (white) (Figure 2A). Using this technique, we were able to quantify eosinophil proteolytic activity as percent area of degraded fibrinogen. The percent area degraded increased significantly with number of cells plated (Figure 2B–C). Fibrinogenolysis occurred in a species-specific manner (Figure 3). Mouse eosinophil fibrinogenolytic activity was the highest on mouse fibrinogen substrate, and human eosinophils had higher activity on porcine fibrinogen than mouse eosinophils. However, the data also showed significant species cross-reactivity, revealing the conserved nature of this eosinophil-fibrinogen interaction (Figure 3).
Figure 2. Quantification of fibrinogen degradation by eosinophils.
A) Original image used for quantification using ImageJ (left), thresholded version of image within ImageJ (middle), and quantification view of the image within ImageJ (right). Black outlines demarcate areas where degradation occurred, red numbers quantify number of areas of degradation. B) Fibrinogen substrate (green) exposed to increasing numbers of eosinophils. Images are representative of at least two independent experiments. C) Quantification of images from part B using ImageJ strategy from A. **p<0.01 by Kruskal-Wallis. Fields from three replicates were counted per condition from two independent experiments.
Figure 3. Eosinophils process fibrinogen in a species-specific manner.
A) Human IL-5-primed blood eosinophils on porcine fibrinogen substrate. B) Murine bone marrow-derived eosinophils on porcine fibrinogen. C) Murine bone marrow-derived eosinophils on murine fibrinogen. D) Quantification of percent area of fibrinogen degradation across different species. HEos: human eosinophils; MEos, mouse eosinophils; PFg, porcine fibrinogen; MFg, mouse fibrinogen. ****p<0.0001 by ANOVA. Six fields were counted per condition from at least two independent experiments for mouse eosinophils. One experiment was conducted using human eosinophils in triplicate.
Fibrinogen induces cytolytic eosinophil degranulation
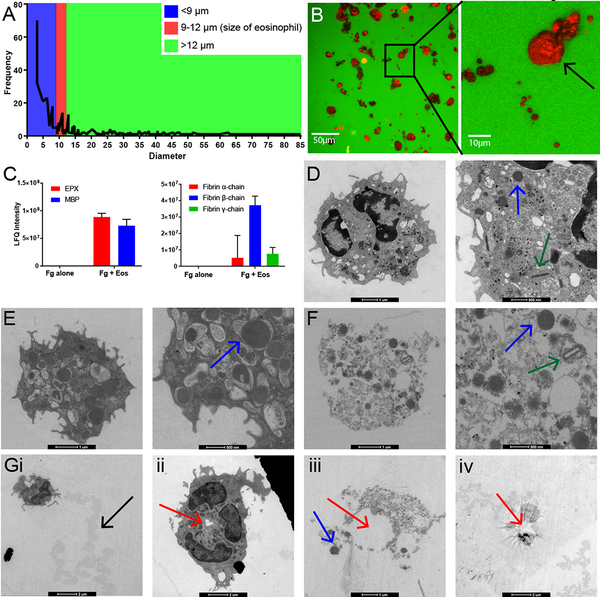
The majority of eosinophil-degraded fibrinogen areas were less than 9 μm in diameter, and only a small percentage corresponded to areas between 9 and 12 μm, or the size of a single intact eosinophil (Figure 4A). This suggested that something other than the cell itself, possibly eosinophil granule proteins, was creating the smaller holes. Further evidence for eosinophil degranulation can be seen in the detection of MBP positive staining (red) in all degraded areas, suggesting that MBP-rich granule release from cells is followed by substrate degradation (Figure 4B). As additional confirmation of degranulation, we performed proteomic analysis of the products released in the fibrinogen assay supernatants (after removing the cellular fraction). Using untargeted mass spectrometry analysis, we detected significant levels of degranulation products MBP and EPX in assay supernatants, confirming that eosinophils are degranulating upon exposure to fibrinogen (Figure 4C left, also Supplementary Table S1). Moreover, proteome analysis also showed the presence of fibrin monomers when eosinophils were allowed to interact with fibrinogen, showing that fibrinogen is being broken down into its fibrin subunits (α-,β- and γ-chains) (Figure 4C right) and supporting eosinophil-mediated fibrinogenolysis.
Figure 4. Cytolytic degranulation of eosinophils adhering to fibrinogen.
A) Diameter of fibrinogen degraded areas versus their frequency. Red interval represents the diameter of a single intact eosinophil (9–12 μm); blue corresponds to smaller areas fitting the diameter of eosinophil granules (< 9 μm). Most degraded areas on images correspond to the size of eosinophil granules. B) MBP-stained murine eosinophils interacting with fibrinogen scaffold. Large red spots are intact eosinophils; smaller red dots are eosinophil granules. The arrow points to a spot the size of an intact eosinophil. Images are representative of at least two independent experiments. C) Eosinophil degranulation products detected in mass spectrometry analysis of assay supernatants (left) and fibrinogen degradation products present in assay supernatants (right). Fg alone: Fibrinogen substrate without eosinophils added to assay; Fg + Eos: Fibrinogen substrate with eosinophils added to assay. Samples were analyzed in duplicate. D-F) Transmission electron microscopy images of eosinophils. Green arrows point out compact rod-like granules with electron lucent crystalline cores. Blue arrows denote representative eosinophil granules lacking a detectible crystalline core. Red arrows denote loss of membrane integrity beginning at the center of the eosinophil. D) Representative image of an activated eosinophil on collagen control substrate with an intact cytoplasmic membrane. E) Activated eosinophil on fibrinogen substrate. Note the increase in number and size of granules. F) Cell-free granules on fibrinogen substrate. This image shows loss of membrane integrity characteristic of cytolytic degranulation. G) Degranulation of eosinophils on fibrinogen substrates via cytolytic degranulation: i) Intact eosinophil with a trail of degraded substrate behind it, noted by the black arrow. ii) Granule-packed eosinophil with a center hole beginning to form in the membrane. iii) Cell-free granules with membrane fragments exhibiting a circular pattern in the center. iv) “Suction cup” imprint left on the fibrinogen substrate with a circular pattern in the center.
We performed transmission electron microscopy on eosinophils exposed to either fibrinogen or a control collagen substrate to ensure that substrate degradation by eosinophils and cytolytic degranulation was specific to fibrinogen and not a general property of adhering eosinophils. When activated eosinophils were exposed to collagen substrates, they retained typical eosinophil morphology (Figure 4D). These cells appeared intact with visible granules inside the cells (Figure 4D arrows). Eosinophils exposed to fibrinogen had a different appearance than the collagen exposed eosinophils. One form we observed was intact cells that were exceptionally granule-packed (Figure 4E, arrows pointing to granules). The other form we observed was eosinophils with fragmented membranes and cell-free granules (Figure 4F, 4Giii). We were able to observe eosinophils in various stages of degranulation on the fibrinogen substrate, as demonstrated in the image sequence in Figure 4G. From left to right, we visualized: (1) intact eosinophils packed with granules leaving behind areas of visibly degraded substrate (Figure 4Gi arrow), (2) followed by initial loss of membrane integrity beginning from the center of the cell (Figure 4Gii arrow). (3) Next, we note the presence of cell-free granules, often with circular patterns in the middle (Figure 4Giii red arrow). (4) Finally, we would observe a “suction cup” imprint of degranulated eosinophils on the fibrinogen substrate with few cell remnants, again with a circular pattern in the middle (Figure 4Giv arrow). By all indications, eosinophils exposed to fibrinogen substrate in our assays undergo cytolytic degranulation.
Eosinophil degranulation and proteolytic activity is specific to fibrinogen and is CD11b-dependent
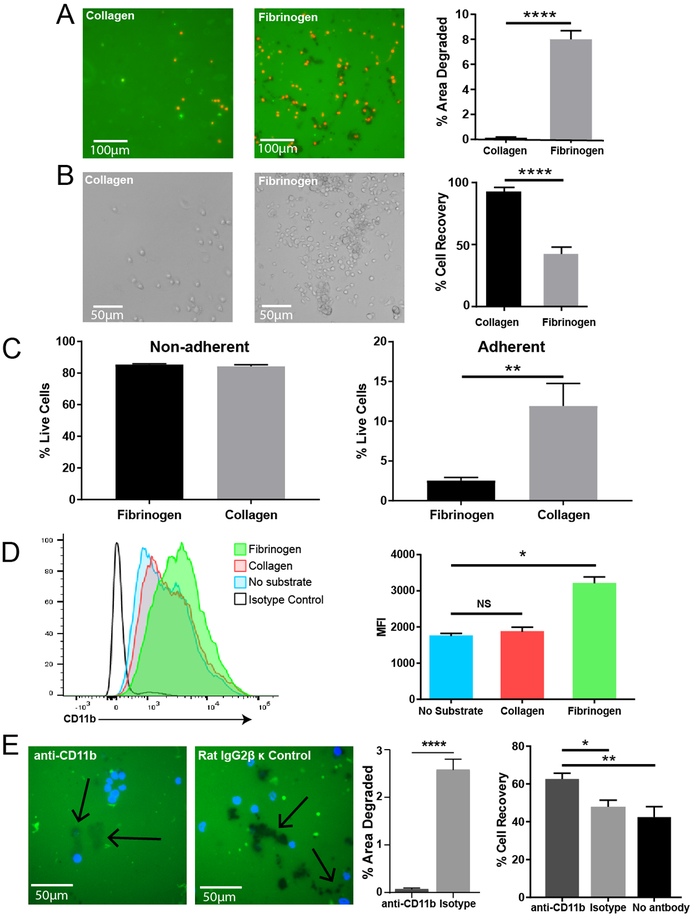
To further understand the interaction between eosinophils and fibrinogen, we compared eosinophil exposure to collagen and fibrinogen substrates. Eosinophils exposed to collagen from two different species, murine and bovine, were similarly inactive relative to fibrinogen exposure (Figure S2). We found that, unlike in the fibrinogen interaction, eosinophils exposed to collagen will adhere to but do not degrade the substrate. This shows that it is a fibrinogen-specific process (Figure 5A). Furthermore, bright field images showed that eosinophils adherent to collagen have a rounded cell-intact phenotype as opposed to those adhered to fibrinogen (Figure 5B). This adds evidence supporting that eosinophils adherent on collagen remain fully intact and are not degranulating. Eosinophils more readily adhere to fibrinogen than collagen, as seen by the larger percentage of cells recovered (the non-adherent cell fraction) after 4 hours of incubation (Figure 5B). In analyzing the two fractions, adherent and non-adherent, we observed no difference in the viability of the non-adherent fraction after 4 hours of substrate exposure, but eosinophils exposed to fibrinogen showed lower viability after 4 hours than eosinophils exposed to collagen (Figure 5C).
Figure 5. Eosinophil proteolytic activity is specific to fibrinogen and is CD11b-dependent.
A) Fluorescent images of FITC-linked Type I collagen substrate remaining intact and fibrinogen substrate being degraded (green = substrate, black = absence of substrate). Eosinophils can be visualized by DRAQ5 staining (red). Images are representative of at least six independent experiments. Quantification of area degraded on collagen and fibrinogen substrates (right). ****p<0.0001 by ANOVA. B) Representative brightfield images of eosinophils adhered to collagen control substrate and fibrinogen substrate after washing. Bar graph on right quantifies percentage of cells recovered after washing on collagen and fibrinogen. **p<0.01 by t-test. Images and graph are representative of three separate experiments. C) Viability of eosinophils that are non-adherent (left) or adherent (right) after 4 hours of incubation on fibrinogen or collagen. **p<0.01 by t-test. Graphs are representative of four separate experiments. D) Histogram quantifying CD11b receptor levels on eosinophils exposed to substrates by flow cytometry (left), and bar graphs showing the Median Florescence Intensity of cells exposed to substrates (right). *p<0.05 by Kruskal-Wallis. Graphs are representative of two independent experiments. E) Images of fibrinogen substrate when eosinophils were blocked with anti-CD11b or isotype control antibody (left). Arrows point to degraded areas. Eosinophils can be visualized by DAPI staining (blue). Images are representative of three independent experiments. First bar graph quantifies degraded area between the two treatments (middle). ****p<0.0001 by Mann-Whitney test. Second bar graph quantifies percentage of cells recovered after washing on fibrinogen with different antibody treatments (right). *p<0.05, **p<0.01 by Kruskal-Wallis.
Though eosinophils constitutively express CD11b, those exposed to fibrinogen display a significant upregulation of this integrin (Figure 5D). Blocking CD11b receptors on eosinophils prior to adding them to fibrinogen resulted in a significantly diminished amount of substrate degradation compared to eosinophils treated with isotype control antibody (Figure 5E). Interestingly, anti-CD11b treated eosinophils appear to be capable of removing fibrinogen, but to a much lesser extent than isotype control eosinophils. This can be seen as the anti-CD11b condition resulted in faint dark spots on the FITC-linked substrate as opposed to the much darker spots seen in the isotype control condition. Faint spots represent the shallow removal rather than complete penetration of fibrinogen in those areas (Figure 5E). Furthermore, eosinophils treated with anti-CD11b do not adhere to fibrinogen as readily as controls. This supports the notion that CD11b is at least in some part responsible for eosinophil adherence to fibrinogen and that activation of the CD11b receptor by fibrinogen is necessary for complete eosinophil degranulation in this system.
Shared and unique eosinophil gene expression profiles elicited by adhesion to fibrinogen vs. Type I collagen
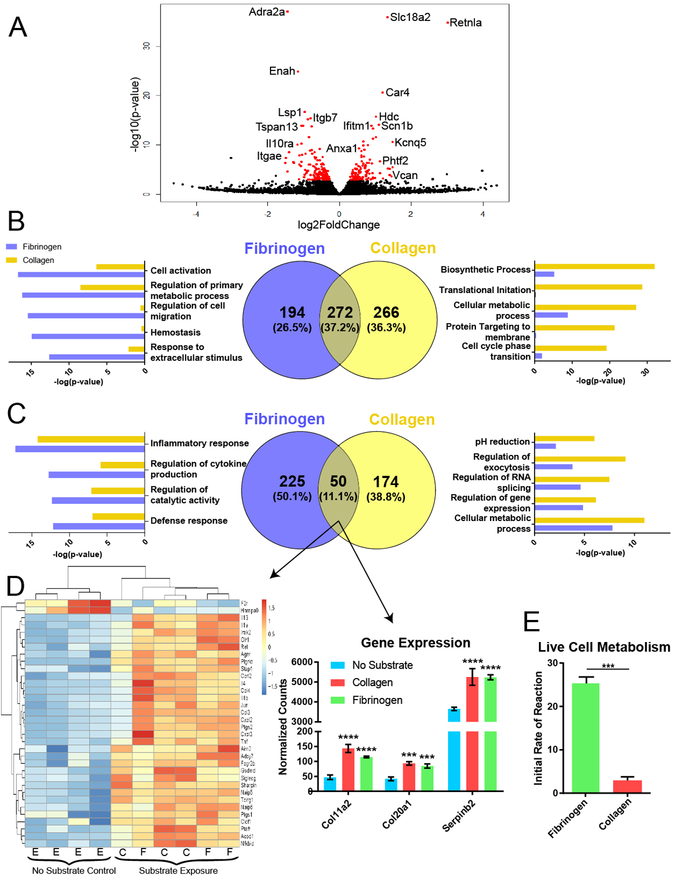
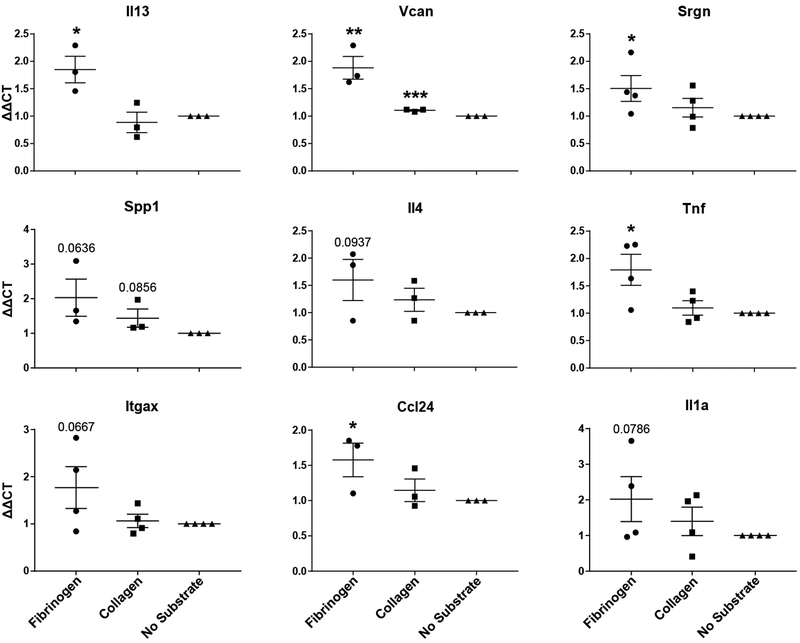
To further interrogate the biological processes taking place in eosinophils exposed to fibrinogen vs. collagen, we performed RNA-seq analysis on eosinophils isolated from our assays. 276 eosinophil genes were differentially regulated between cells exposed to fibrinogen and those exposed to collagen (113 upregulated, 163 downregulated) (Figure 6A). When eosinophils adhered to fibrinogen were compared to eosinophils without substrate exposure, 746 genes were significantly differentially expressed (471 upregulated, 275 downregulated). Interestingly, the same was true for eosinophils interacting with Type I collagen: 770 genes were significantly differently expressed (545 upregulated, 225 downregulated) compared to eosinophil no substrate controls. We further compared these two differential expression lists (no substrate vs. fibrinogen and no substrate vs. collagen) and found that 26.5% downregulated genes were unique to fibrinogen and 36.3% downregulated genes were unique to collagen. The remaining 37.2% were shared by the two (Figure 6B). We examined which GO biological processes were dominated by each substrate and found that fibrinogen exposed cells were downregulating processes related to primary metabolic processes and cell migration, while those exposed to collagen were downregulating processes related to biosynthetic process, cellular metabolic process, translation, and cell cycle transitions. We performed similar analysis for upregulated genes and found that 50.1% genes were exclusive to fibrinogen and 38.8% exclusive to collagen (Figure 6C). However, the consensus upregulated signature, at only 11.1% of the overall gene signature, showed a striking alignment in inflammatory response genes, such as the upregulation of Il-4, Il-13, Cxcl2, and Cxcl3. Furthermore, specific collagens, Col11a2 and Col20a1, and Serpinb2 showed consensus upregulation with fibrinogen and collagen interaction (Figure 6D). To further examine differences in metabolic processes from the GO analysis, we performed a live cell functional Biolog metabolism assay on eosinophils and found that eosinophils interacting with fibrinogen had significantly higher metabolic activity than those interacting with collagen (Figure 6E), confirming that there is a substantial metabolic difference triggered by substrate exposure. To validate genes from the RNA-seq analysis, qPCR analysis was run on four separate experiments. In each experiment, two to four replicates per treatment were averaged to determine ΔΔCT values. This analysis validated the significant expression of inflammatory genes in eosinophils activated by fibrinogen. Expression of these genes triggered by collagen showed a similar trend, although at a lower fold change level and not reaching statistical significance in most cases (Figure 7).
Figure 6. Differential gene expression in eosinophils interacting with fibrinogen vs. collagen.
A) Volcano plot showing differential gene expression analysis of RNA-seq data comparing eosinophils exposed to collagen versus those exposed to fibrinogen. Red represents genes with adjusted p values < 0.05. B) Venn diagram comparing the downregulated gene signatures from eosinophils without substrate exposure to those exposed to either collagen or fibrinogen (center). Bar graphs on the left and right show GO biological processes dominated by fibrinogen (blue) or collagen (yellow). C) Venn diagram and GO biological processes of the upregulated genes from the data shown in B. D) Heat map showing differential gene regulation of the consensus upregulated signature from part C that correspond with genes related to inflammatory response (left) and select remodeling genes identified in the bar graphs on the right. E = eosinophils without substrate, C = collagen substrate, F = fibrinogen substrate. ***p<0.001, ****p<0001. N = 4 replicates for no substrate exposure, N = 3 replicates for collagen and fibrinogen exposure. E) Bar graph showing initial rate of reaction in a Biolog metabolic assay of eosinophils exposed to fibrinogen vs collagen. ***p<0.001 by t-test.
Figure 7. qPCR validation of select RNA-seq genes.
Data presented as ΔΔCT of gene values compared to no substrate controls. TBP was used as a housekeeping gene. Cells were either exposed to fibrinogen, collagen, or no substrate. *p<0.05; **p<0.01, ***p<0.001 by one-tailed T-tests. Values were calculated as fold change relative to “No Substrate” control from three to four independent experiments. Each data point represents an average expression value of 2–4 replicates within a single experiment. 3–4 independent experiments are summarized in each graph.
Discussion
Unlike human eosinophils, murine eosinophils are notoriously difficult to degranulate in vitro and show only piecemeal degranulation in response to inflammatory stimuli (15). Higher concentrations of exogenous stimuli are typically necessary to induce even piecemeal degranulation of mouse eosinophils when compared with studies of human cells. This is the first report to demonstrate both visually and quantitatively that adhesion to fibrinogen is sufficient to trigger potent cytolytic degranulation of murine IL-5-primed eosinophils. This confirms the necessity of a “two-hit” system: the presence of both immune and specific tissue adhesive cues to trigger degranulation.
Eosinophil activity in our assays was entirely adhesion-dependent. This is in line with previous reports showing that human eosinophils exhibit a CD11b-dependent hyperadhesive phenotype towards fibrinogen in asthma (6) and degranulate in a CD11b-adhesion dependent manner (25, 26). In humans, ECP and EDN granule product release is also adhesion-dependent and has been demonstrated to increase upon binding to fibrinogen with or without IgG (27, 28). Likewise, in mice, we detected MBP and EPX release in the supernatants of cultures where eosinophils were allowed to adhere to fibrinogen. Similarly, a previous report showed that eosinophils from mice will not degranulate until they have adhered to the extracellular matrix (14). In keeping with these findings on the necessity of adhesion, our results show that the ability of murine eosinophils to degranulate and degrade fibrinogen also acts through a CD11b-adhesion dependent manner. When eosinophils were treated with anti-CD11b antibody prior to assay, we observed a significant decrease in percentage of adherent cells and fibrinogen degradation relative to non-blocked controls. Interestingly, fibrinogen degradation was not entirely ablated by treatment with anti-CD11b, as faint dark spots were left on fibrinogen and some smaller percentage of eosinophils did still remain adherent to the substrate (Figure 5E). They may have been able to dock via a different integrin, such as CD11c, which has also been shown to adhere eosinophils to fibrinogen (9).
Interestingly, in our RNA-seq analysis we did not find expression of metalloproteinases (MMPs) or other proteases in eosinophils interacting with fibrinogen substrates. MMPs are known to be produced by eosinophils (12) and may promote pathologic changes in asthmatic airways by degrading the extracellular matrix. Specifically, MMP-9 has been shown to be overexpressed by eosinophils from asthmatic patients (29), and MMP-9 levels are significantly increased in individuals with severe asthma relative to those with mild or moderate asthma (30). It is worth noting that this protein, when produced in eosinophils, has been found to localize to perinuclear spaces but not granules (29). Not finding protease expression in our assays gives credence to the fact that the fibrinogen degradation seen in this study is not due to de novo synthesis of MMPs or other ECM modulatory proteins, such as cathepsins or ADAMs. We also conclude that free MBP is not sufficient to trigger the potent fibrinogenolysis observed in our assays (Figure S3). It is possible that another pre-stored proteolytic mediator, likely released in a contact-dependent manner (visualized by TEM) at lower concentrations, may account for substrate removal prior to or during degranulation.
There are three different major modes of release of eosinophil granule content: classic exocytotic degranulation, piecemeal degranulation (previously reported in mice), and cytolysis with the release of cell-free granules (11). In this report, we confirmed the presence of cell-free granules from eosinophils exposed to fibrinogen via electron microscopy. Cell-free granules have been reported to retain an intact outer membrane and can secrete their granular content after activation by other stimuli (17, 31). Remarkably, TEM indicated that several events precede cytolytic degranulation: (1) movement of eosinophils around substrate accompanied with some level of substrate removal; (2) initiation of loss of membrane integrity from the center of the adhered eosinophil; (3) complete loss of cellular membrane integrity and deposition of cell-free granules; (4) typical “suction cup” imprint of adhered eosinophil on substrate supporting necessity for focal adhesion This suggests that the induction of cytolytic degranulation by fibrinogen is a tightly regulated process. Additionally, we detected MBP and EPX granular protein release in assay supernatants. Interestingly, this very closely resembles a novel form of cell death and degranulation termed pyroptosis, which is characterized as caspase-1-mediated cell death with accompanying mediator release. This mode of degranulation was recently reported for mouse eosinophils infiltrating liver tissue in response to necrotic liver injury (32). Although the degranulation trigger was not identified in that study, fibrinogen is a likely candidate given that the liver is a major and constitutive source of this protein (33). This manner of free eosinophil granule deposition and subsequent content release is a prominent feature of human allergic airway and skin disease (16–19). Therefore, research elucidating the triggers of cell-free granule release was identified as a high priority by the NIH Taskforce on the Research of Eosinophil-Associated Diseases (34).
Such dramatic engagement of eosinophils by fibrinogen raises several important questions. Where do such interactions take place in an organism? Which biological processes necessitate fibrinogen processing by eosinophils in health and disease? There is evidence that eosinophils have the ability to produce various molecules associated with coagulation. In the coagulation cascade, a series of factors are activated leading to the conversion of fibrinogen to fibrin by activated thrombin. Tissue factor (TF), a molecule released in response to tissue damage, is released upstream of thrombin activation. Work by Moosbauer et al. (35) showed that blood eosinophils produce and can release TF, which suggests that eosinophils play an upstream role in thrombin activation. Furthermore, eosinophils can generate thrombin in a TF dependent manner when exposed to a procoagulant phospholipid surface enriched in 12/15 lipoxygenase-derived oxidized phospholipids (36).
There could be other modalities of eosinophil-fibrinogen interaction, as studies have shown that fibrinogen can be deposited in the ECM without proteolytic processing in non-vascular injury contexts (37, 38). In particular, extrahepatic fibrinogen produced by the lung and intestinal epithelium during inflammation is thought to play a critical role during localized repair and remodeling to restore tissue homeostasis (39–41). Disordered coagulation and fibrinogen and thrombin accumulation in the airway is a feature of human disease and can contribute to airway hyperresponsiveness, remodeling, and decline in lung function (42–44). Compartmentalization of eosinophil recruitment and degranulation in human and mouse models of airway disease may provide additional clues to the existence and significance of this tissue-driven interaction. After segmental airway challenge in human asthma, only airway (but not blood) eosinophils show the hyperadhesive phenotype for fibrinogen and other ECM proteins (6). The same is true for mouse models of asthma (14). In support, we previously found that the integrin profile of eosinophils in a mouse model of asthma changes upon trans-epithelial transition to the airway, including upregulating CD11c and CD11b – receptors for fibrinogen (9). Moreover, airway eosinophils in asthma patients with increased hyperresponsiveness express urokinase-type plasminogen activator receptor (uPAR or CD87) (45), which may also support a role for eosinophils in tissue fibrinogen/fibrin processing in disease. Interestingly, plasminogen-deficient mice show a dramatic reduction in the number of airway eosinophils in a murine model of asthma (46). Although fibrinogen and Type I collagen deposition both take place in allergic asthma (47, 48), we found that the responses to two different substrates can be different. Eosinophils degranulate only when adhering to fibrinogen, although both substrates may induce eosinophil inflammatory responses to various degrees. This could also be associated with the spatiotemporal compartmentalization of these interactions in tissues. Specific substrates could be deposited in different tissue compartments (fibrinogen in the airway and collagen in the basement membrane) or represent different stages of remodeling.
In summary, our data show that both mouse and human eosinophils promote fibrinogenolysis via CD11b-dependent adhesion to fibrinogen. Moreover, we demonstrated that fibrinogen is, so far, the only known trigger of cytolytic degranulation of murine eosinophils, which features the release of free eosinophil granules and eosinophil granular proteins. Eosinophil activity toward fibrinogen in our assays is consistent with eosinophil biology in wound healing and the tissue remodeling environment of a chronic inflammatory disease. The participation of eosinophils in fibrinogen processing and tissue remodeling warrants further investigation, as this has the significant potential to advance our understanding of tissue eosinophil function and heterogeneity in health and disease.
Supplementary Material
Key Points.
Fibrinogen is a specific trigger for cytolytic eosinophil degranulation
Eosinophils adhere to and degrade fibrinogen substrates in a CD11b-dependent manner
Fibrinogen-interacting eosinophils are more metabolically active than collagen
Acknowledgements
We would like to thank Yun Cao (Northwestern University) for his help in preparing human eosinophils. We also thank Dr. Dina Arvanitis, manager of the Nikon Imaging Center (Northwestern University) and Dr. Farida Korobova, microscopy specialist in the Nikon Imaging Center (Northwestern University) for their personal assistance in visualizing fibrinogen-eosinophil interactions.
Financial Support
This work was supported by the National Institutes of Health (NIH/NIAID) grants R21AI115055 and R01AI127783 to Dr. Sergejs Berdnikovs. Additionally, this study was supported by the Ernest S. Bazley Foundation. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center.
References
- 1.Pulanic D, and Rudan I. 2005. The past decade: fibrinogen. Coll Antropol 29: 341–349. [PubMed] [Google Scholar]
- 2.Perrio MJ, Ewen D, Trevethick MA, Salmon GP, and Shute JK. 2007. Fibrin formation by wounded bronchial epithelial cell layers in vitro is essential for normal epithelial repair and independent of plasma proteins. Clin Exp Allergy 37: 1688–1700. [DOI] [PubMed] [Google Scholar]
- 3.Haidaris PJ 1997. Induction of fibrinogen biosynthesis and secretion from cultured pulmonary epithelial cells. Blood 89: 873–882. [PubMed] [Google Scholar]
- 4.Abdala-Valencia H, Coden ME, Chiarella SE, Jacobsen EA, Bochner BS, Lee JJ, and Berdnikovs S. 2018. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J Leukoc Biol 104: 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riddle JM, and Barnhart MI. 1965. The Eosinophil as a Source for Profibrinolysin in Acute Inflammation. Blood 25: 776–794. [PubMed] [Google Scholar]
- 6.Barthel SR, Jarjour NN, Mosher DF, and Johansson MW. 2006. Dissection of the hyperadhesive phenotype of airway eosinophils in asthma. Am J Respir Cell Mol Biol 35: 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yakubenko VP, Lishko VK, Lam SC, and Ugarova TP. 2002. A molecular basis for integrin alphaMbeta 2 ligand binding promiscuity. J Biol Chem 277: 48635–48642. [DOI] [PubMed] [Google Scholar]
- 8.Flick MJ, Du X, Witte DP, Jirouskova M, Soloviev DA, Busuttil SJ, Plow EF, and Degen JL. 2004. Leukocyte engagement of fibrin(ogen) via the integrin receptor alphaMbeta2/Mac-1 is critical for host inflammatory response in vivo. J Clin Invest 113: 1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdala Valencia H, Loffredo LF, Misharin AV, and Berdnikovs S. 2016. Phenotypic plasticity and targeting of Siglec-F(high) CD11c(low) eosinophils to the airway in a murine model of asthma. Allergy 71: 267–271. [DOI] [PubMed] [Google Scholar]
- 10.Hogan SP, Rosenberg HF, Moqbel R, Phipps S, Foster PS, Lacy P, Kay AB, and Rothenberg ME. 2008. Eosinophils: biological properties and role in health and disease. Clin Exp Allergy 38: 709–750. [DOI] [PubMed] [Google Scholar]
- 11.Weller PF, and Spencer LA. 2017. Functions of tissue-resident eosinophils. Nat Rev Immunol 17: 746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JJ, Jacobsen EA, Ochkur SI, McGarry MP, Condjella RM, Doyle AD, Luo H, Zellner KR, Protheroe CA, Willetts L, Lesuer WE, Colbert DC, Helmers RA, Lacy P, Moqbel R, and Lee NA. 2012. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J Allergy Clin Immunol 130: 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyer KD, Percopo CM, Xie Z, Yang Z, Kim JD, Davoine F, Lacy P, Druey KM, Moqbel R, and Rosenberg HF. 2010. Mouse and human eosinophils degranulate in response to platelet-activating factor (PAF) and lysoPAF via a PAF-receptor-independent mechanism: evidence for a novel receptor. J Immunol 184: 6327–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark K, Simson L, Newcombe N, Koskinen AM, Mattes J, Lee NA, Lee JJ, Dent LA, Matthaei KI, and Foster PS. 2004. Eosinophil degranulation in the allergic lung of mice primarily occurs in the airway lumen. J Leukoc Biol 75: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 15.Lee JJ, and Lee NA. 2005. Eosinophil degranulation: an evolutionary vestige or a universally destructive effector function? Clin Exp Allergy 35: 986–994. [DOI] [PubMed] [Google Scholar]
- 16.Erjefalt JS, Greiff L, Andersson M, Matsson E, Petersen H, Linden M, Ansari T, Jeffery PK, and Persson CG. 1999. Allergen-induced eosinophil cytolysis is a primary mechanism for granule protein release in human upper airways. Am J Respir Crit Care Med 160: 304–312. [DOI] [PubMed] [Google Scholar]
- 17.Neves JS, Perez SA, Spencer LA, Melo RC, Reynolds L, Ghiran I, Mahmudi-Azer S, Odemuyiwa SO, Dvorak AM, Moqbel R, and Weller PF. 2008. Eosinophil granules function extracellularly as receptor-mediated secretory organelles. Proc Natl Acad Sci U S A 105: 18478–18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erjefalt JS, and Persson CG. 2000. New aspects of degranulation and fates of airway mucosal eosinophils. Am J Respir Crit Care Med 161: 2074–2085. [DOI] [PubMed] [Google Scholar]
- 19.Cheng JF, Ott NL, Peterson EA, George TJ, Hukee MJ, Gleich GJ, and Leiferman KM. 1997. Dermal eosinophils in atopic dermatitis undergo cytolytic degeneration. J Allergy Clin Immunol 99: 683–692. [DOI] [PubMed] [Google Scholar]
- 20.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, and Rosenberg HF. 2008. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol 181: 4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim KG, and Weller PF. 2001. Isolation of human eosinophils. Curr Protoc Immunol Chapter 7: Unit 7 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider CA, Rasband WS, and Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie S, and Kita H. 1994. CD11b/CD18 (Mac-1) is required for degranulation of human eosinophils induced by human recombinant granulocyte-macrophage colony-stimulating factor and platelet-activating factor. J Immunol 152: 5457–5467. [PubMed] [Google Scholar]
- 26.Egesten A, Gullberg U, Olsson I, and Richter J. 1993. Phorbol ester-induced degranulation in adherent human eosinophil granulocytes is dependent on CD11/CD18 leukocyte integrins. J Leukoc Biol 53: 287–293. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, and Hakansson L. 2000. Regulation of the release of eosinophil cationic protein by eosinophil adhesion. Clin Exp Allergy 30: 794–806. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko M, Horie S, Kato M, Gleich GJ, and Kita H. 1995. A crucial role for beta 2 integrin in the activation of eosinophils stimulated by IgG. J Immunol 155: 2631–2641. [PubMed] [Google Scholar]
- 29.Ohno I, Ohtani H, Nitta Y, Suzuki J, Hoshi H, Honma M, Isoyama S, Tanno Y, Tamura G, Yamauchi K, Nagura H, and Shirato K. 1997. Eosinophils as a source of matrix metalloproteinase-9 in asthmatic airway inflammation. Am J Respir Cell Mol Biol 16: 212–219. [DOI] [PubMed] [Google Scholar]
- 30.Barbaro MP, Spanevello A, Palladino GP, Salerno FG, Lacedonia D, and Carpagnano GE. 2014. Exhaled matrix metalloproteinase-9 (MMP-9) in different biological phenotypes of asthma. Eur J Intern Med 25: 92–96. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg HF, and Foster PS. 2013. Reply to eosinophil cytolysis and release of cell-free granules. Nat Rev Immunol 13: 902. [DOI] [PubMed] [Google Scholar]
- 32.Palacios-Macapagal D, Connor J, Mustelin T, Ramalingam TR, Wynn TA, and Davidson TS. 2017. Cutting Edge: Eosinophils Undergo Caspase-1-Mediated Pyroptosis in Response to Necrotic Liver Cells. J Immunol 199: 847–853. [DOI] [PubMed] [Google Scholar]
- 33.Pieters M, and Wolberg AS. 2019. Fibrinogen and fibrin: An illustrated review. Res Pract Thromb Haemost 3: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoury P, Akuthota P, Ackerman SJ, Arron JR, Bochner BS, Collins MH, Kahn JE, Fulkerson PC, Gleich GJ, Gopal-Srivastava R, Jacobsen EA, Leiferman KM, Francesca LS, Mathur SK, Minnicozzi M, Prussin C, Rothenberg ME, Roufosse F, Sable K, Simon D, Simon HU, Spencer LA, Steinfeld J, Wardlaw AJ, Wechsler ME, Weller PF, and Klion AD. 2018. Revisiting the NIH Taskforce on the Research needs of Eosinophil-Associated Diseases (RE-TREAD). J Leukoc Biol 104: 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moosbauer C, Morgenstern E, Cuvelier SL, Manukyan D, Bidzhekov K, Albrecht S, Lohse P, Patel KD, and Engelmann B. 2007. Eosinophils are a major intravascular location for tissue factor storage and exposure. Blood 109: 995–1002. [DOI] [PubMed] [Google Scholar]
- 36.Uderhardt S, Ackermann JA, Fillep T, Hammond VJ, Willeit J, Santer P, Mayr M, Biburger M, Miller M, Zellner KR, Stark K, Zarbock A, Rossaint J, Schubert I, Mielenz D, Dietel B, Raaz-Schrauder D, Ay C, Gremmel T, Thaler J, Heim C, Herrmann M, Collins PW, Schabbauer G, Mackman N, Voehringer D, Nadler JL, Lee JJ, Massberg S, Rauh M, Kiechl S, Schett G, O’Donnell VB, and Kronke G. 2018. Correction: Enzymatic lipid oxidation by eosinophils propagates coagulation, hemostasis, and thrombotic disease. J Exp Med 215: 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guadiz G, Sporn LA, and Simpson-Haidaris PJ. 1997. Thrombin cleavage-independent deposition of fibrinogen in extracellular matrices. Blood 90: 2644–2653. [PubMed] [Google Scholar]
- 38.Rybarczyk BJ, Lawrence SO, and Simpson-Haidaris PJ. 2003. Matrix-fibrinogen enhances wound closure by increasing both cell proliferation and migration. Blood 102: 4035–4043. [DOI] [PubMed] [Google Scholar]
- 39.Lawrence SO, and Simpson-Haidaris PJ. 2004. Regulated de novo biosynthesis of fibrinogen in extrahepatic epithelial cells in response to inflammation. Thromb Haemost 92: 234–243. [DOI] [PubMed] [Google Scholar]
- 40.Laurens N, Koolwijk P, and de Maat MP. 2006. Fibrin structure and wound healing. J Thromb Haemost 4: 932–939. [DOI] [PubMed] [Google Scholar]
- 41.Barker TH, and Engler AJ. 2017. The provisional matrix: setting the stage for tissue repair outcomes. Matrix Biol 60–61: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthay MA, and Clements JA. 2004. Coagulation-dependent mechanisms and asthma. J Clin Invest 114: 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagers SS, Norton RJ, Rinaldi LM, Bates JH, Sobel BE, and Irvin CG. 2004. Extravascular fibrin, plasminogen activator, plasminogen activator inhibitors, and airway hyperresponsiveness. J Clin Invest 114: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ierodiakonou D, Portelli MA, Postma DS, Koppelman GH, Gerritsen J, Ten Hacken NH, Timens W, Boezen HM, Vonk JM, and Sayers I. 2016. Urokinase plasminogen activator receptor polymorphisms and airway remodelling in asthma. Eur Respir J 47: 1568–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks AM, Bates ME, Vrtis RF, Jarjour NN, Bertics PJ, and Sedgwick JB. 2006. Urokinase-type plasminogen activator modulates airway eosinophil adhesion in asthma. Am J Respir Cell Mol Biol 35: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swaisgood CM, Aronica MA, Swaidani S, and Plow EF. 2007. Plasminogen is an important regulator in the pathogenesis of a murine model of asthma. Am J Respir Crit Care Med 176: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Millien VO, Lu W, Mak G, Yuan X, Knight JM, Porter P, Kheradmand F, and Corry DB. 2014. Airway fibrinogenolysis and the initiation of allergic inflammation. Ann Am Thorac Soc 11 Suppl 5: S277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura A, Uchida Y, Sakamoto T, Ishii Y, Masuyama K, Morishima Y, Hirano K, and Sekizawa K. 2002. Increases in collagen type I synthesis in asthma: the role of eosinophils and transforming growth factor-beta. Clin Exp Allergy 32: 860–865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.