Abstract
Using the immunomagnetic separation procedure, we isolated sorbitol-fermenting (SF) Shiga toxin-producing Escherichia coli (STEC) O157:H− strains from two patients, one with hemolytic-uremic syndrome and the other with diarrhea, and from a dairy cow epidemiologically associated with the patients. The phenotypic and genotypic characteristics of all isolates were identical or closely related. Moreover, the bovine isolate showed a clonal relatedness to SF STEC O157:H− strains isolated from patients in Germany and the Czech Republic from 1988 to 1998. This is the first evidence that cattle can be a reservoir of SF STEC O157:H− and a source of human diseases.
During the last 10 years, sorbitol-fermenting (SF) Shiga toxin (Stx)-producing Escherichia coli (STEC) strains of serotype O157:H− have emerged as important causes of hemolytic-uremic syndrome (HUS) and diarrhea in Germany (1, 8, 9). In addition to sporadic cases of human disease (8), two outbreaks of HUS caused by SF O157:H− STEC strains have been identified. The first outbreak, in 1988, lead to the discovery of the pathogen (9). The second outbreak occurred in the winter of 1995 to 1996 and included 28 HUS cases, three of which were fatal (1). Analysis of phenotypic and molecular characteristics of SF STEC O157:H− strains demonstrated that such strains represent a distinct clone within the E. coli O157 serogroup which shares virulence characteristics with non-SF (NSF) STEC O157:H7 (10). In 1995, strains belonging to the SF STEC O157:H− clone were isolated from HUS patients in the Czech Republic (2), suggesting the ability of this pathogen to spread. Despite the increasing significance of SF STEC O157:H− in the etiology of HUS and diarrhea, the epidemiology of the infection, including reservoirs and routes of transmission, remains unknown. In contrast to NSF STEC O157:H7 strains, which have cattle as their major reservoir (12), SF STEC O157 strains have not been found in cattle (9, 10).
In this study, we isolated SF STEC O157:H− strains from two patients and an epidemiologically associated cow in the Czech Republic. We compared phenotypic and genotypic characteristics of the bovine and patients' isolates to determine whether the cow was the source of human infections. Moreover, we determined the genetic relatedness of the bovine isolate to SF STEC O157:H− strains isolated previously from patients in the Czech Republic and Germany in order to investigate whether cattle can be reservoirs of SF STEC O157:H− strains.
The strains investigated are listed in Table 1. SF STEC O157:H− strains 258/98 and 269/98 were isolated from two siblings who developed HUS and diarrhea, respectively, following their visit to a dairy farm in Central Bohemia, Czech Republic. During the farm visit on 12 January 1998, both children were exposed to a herd of 32 dairy cows through touching and stroking some of the animals. The younger child (a 15-month-old boy) developed bloody diarrhea on 15 January and was hospitalized for HUS on 20 January. His 6-year-old brother experienced a 2-day period of mild watery diarrhea which began on 16 January. Investigation of fecal samples from the farm cows performed on 25 January 1998 yielded an SF STEC O157:H− strain (550/98) from 1 of the 32 animals. The SF E. coli O157 strains from the patients and the cow were isolated using the immunomagnetic separation (IMS) procedure (11), followed by slide agglutination with anti-O157 serum (ITEST, Hradec Kralove, Czech Republic) of up to 50 SF colonies from each sorbitol-MacConkey agar (SMAC) plate. The colonies that displayed agglutination were biochemically confirmed as E. coli and shown to produce Stx2 by using a commercial latex agglutination assay (Verotox-F; Denka Seiken Co., Tokyo, Japan). Serotyping by standard procedures identified serotype O157:H−.
TABLE 1.
Characteristics of SF STEC O157:H− strains isolated from a cow and patients and of control NSF STEC O157:H7 strain EDL 933
| Straina | Disease; countryb (referencec) | Phenotypic characteristicsd
|
Chromosomal characteristics
|
Plasmid-encoded genesg
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serotype | PT | SF/GUD | Stx | EHEC Hly | stxe | eae | P-gene profilef | EHEC hly | katP | espP | etp | ||
| 258/98h | HUS; CR | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 17 | −/− | −/− | −/− | −/− |
| 269/98h | WD; CR | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 17 | −/− | −/− | −/− | −/− |
| 550/98h | Cow; CR | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 17 | −/− | −/− | −/− | −/− |
| 703/88 | HUS; G (10) | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 17 | +/48.5 | −/− | −/− | +/3.9, 1.9i |
| 221/95 | HUS; CR (2) | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 9 | +/15.0 | −/− | −/− | +/3.7, 1.7 |
| 1995/96 | HUS; G (2) | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 16 | +/15.0 | −/− | −/− | +/3.9, 1.9 |
| 3573/98 | HUS; G | O157:H− | 88 | +/+ | 2 | − | stx2/18.0 | γ | 16 | +/15.0 | −/− | −/− | +/3.9, 1.9 |
| EDL 933 | HC; US (15) | O157:H7 | 21 | −/− | 1 and 2 | + | stx1/NP | γ | 2 | +/12.0 | +/9.0 | +/7.5 | +/3.9, 1.9 |
| stx2/4.7 | |||||||||||||
For the SF STEC O157:H− strains, the last two numbers indicate the year of isolation.
WD, watery diarrhea; HC, hemorrhagic colitis; CR, Czech Republic; G, Germany; US, United States.
Strains for which no references are given are from this study.
PT, phage type; SF/GUD, sorbitol fermentation/β-d-glucuronidase activity; Stx, Stx phenotype; EHEC Hly, production of EHEC hemolysin; +, positive result; −, negative result.
stx PCR result/size (in kilobases) of EcoRI restriction fragment of the genomic DNA hybridizing to the stx2 probe. NP, not performed.
P-gene profile 17 is characterized by three fragments of 20, 6.4, and 3.7 kb; the related P-gene profile 16 is characterized by two fragments of 20 and 3.7 kb, and the other related P-gene profile 9 is characterized by two fragments of 6.4 and 3.7 kb. P-gene profile 2 consists of two fragments of 21.2 and 16.2 kb and is unrelated to P-gene profiles 17, 16, and 9.
Detection of the gene by PCR/size (in kilobases) of the plasmid DNA fragment hybridizing to the respective probe. +, positive result; −, no signal obtained.
Epidemiologically related strains; the other four SF STEC O157:H− strains are representative isolates from 1988 to 1998 and are epidemiologically unrelated.
Fragments of two different sizes hybridized with the probe.
All of the isolates were tested for sorbitol fermentation, β-d-glucuronidase activity, and the production of enterohemorrhagic E. coli (EHEC) hemolysin according to procedures described by Gunzer et al. (8) and Schmidt et al. (20). Phage typing was performed according to the procedure described by Khakhria et al. (13). Since the isolates were nonmotile and their H antigens could not be determined by serotyping, the gene encoding the flagellin subunit (fliC) was detected and characterized using the fliC restriction fragment length polymorphism (fliC-RFLP) method described by Fields et al. (7). The presence of stx1, stx2, and stx2c genes was investigated by PCR procedures described previously (8, 19). The location of stx2 was determined by hybridization of EcoRI-digested genomic DNA with digoxigenin-labeled stx2 probe (6). The intimin-encoding eae gene was detected using primers SK1 and SK2 (19) and further characterized using primer pairs SK1-LP2, SK1-LP3, SK1-LP4, and SK1-LP5 (16), which are specific to eae types α, γ, β, and ɛ, respectively (16). The P-gene profile, which reflects the number and positions of lambdoid phages in the genome, was determined as described by Datz et al. (6). Pulsed-field gel electrophoresis (PFGE) was performed according to the PulseNet protocol of the Centers for Disease Control and Prevention (17), except that the gel running time was increased to 40 h. PFGE patterns were analyzed using the RFLPscan software (Scanalytics; CSP Inc.). Plasmid profiles were determined as described previously (22). The presence of plasmid-encoded putative virulence genes (EHEC hly, katP, espP, and etp) was investigated by PCR (4, 5, 20, 21) and by hybridization of plasmid DNA digested with BamHI or SmaI with digoxigenin-labeled probes (22).
Four representative SF STEC O157:H− strains isolated from patients in Germany and the Czech Republic during 1988 to 1998 (Table 1) and a control STEC O157:H7 strain, EDL 933 (15), were investigated by the same procedures. Stx-negative SF E. coli O157 strains 1083-36/91 (O157:H45) and 693/91 (O157:H19) (2), isolated from infants with diarrhea, were used as controls in fliC-RFLP and PFGE.
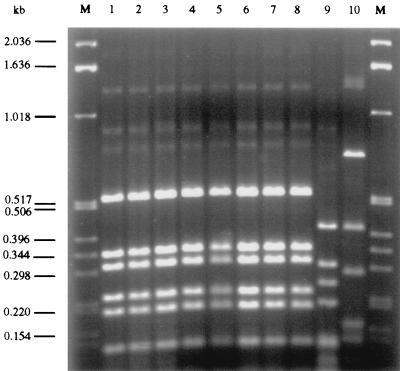
As shown in Fig. 1, the SF STEC O157:H− isolates from the cow (lane 1) and from the two epidemiologically related patients (lanes 2 and 3) and the four representative German and Czech SF STEC O157:H− isolates from 1988 to 1998 (lanes 4 to 7) shared a fliC-RFLP pattern which was identical to that of E. coli O157:H7 strain EDL 933 (lane 8) but clearly differed from fliC-RFLP patterns of E. coli O157 strains possessing H45 (lane 9) or H19 (lane 10). This demonstrated the presence of the H7-encoding fliC gene in all SF STEC O157:H− strains. The other characteristics of the strains are summarized in Table 1. These results show that the epidemiologically related isolates from the cow (550/98) and the patients with HUS (258/98) and diarrhea (269/98) had identical phenotypic and genotypic characteristics. Moreover, these three isolates shared phenotypic features and chromosomal characteristics, including the presence of stx2 only, the presence of eae type γ, and closely related P-gene profiles, with the four representative SF STEC O157:H− strains isolated from HUS patients in Germany and the Czech Republic during 1988 to 1998 (Table 1); in all strains, stx2 was localized to the same 18-kb EcoRI restriction fragment of the genomic DNA (Table 1). In addition, all seven STEC O157:H− isolates had identical plasmid profiles, possessing a single large plasmid of 90 to 100 kb. However, while the large plasmids of the bovine and two epidemiologically related human isolates did not contain any of the putative virulence genes (Table 1), the other four SF STEC O157:H− strains possessed a combination of EHEC hly and etp (Table 1). This suggests the presence of two different large plasmids in SF STEC O157:H− strains. None of the SF STEC O157:H− strains harbored the full spectrum of the plasmid-encoded genes found in a prototype STEC O157:H7 strain, EDL 933 (Table 1).
FIG. 1.
Agarose gel electrophoresis of fliC PCR products of SF STEC O157:H− strains and of control E. coli O157 strains after restriction with RsaI. Lanes M, molecular size marker (1-kb DNA ladder; Gibco BRL). In lanes 1 to 7, the following SF STEC O157:H− strains are shown: lane 1, 550/98; lane 2, 258/98; lane 3, 269/98; lane 4, 703/88; lane 5, 221/95; lane 6, 1995/96; and lane 7, 3573/98. Lanes 8 to 10 contain control E. coli O157 strains as follows: lane 8, EDL 933 (O157:H7); lane 9, 1083-36/91 (O157:H45); and lane 10, 693/91 (O157:H19).
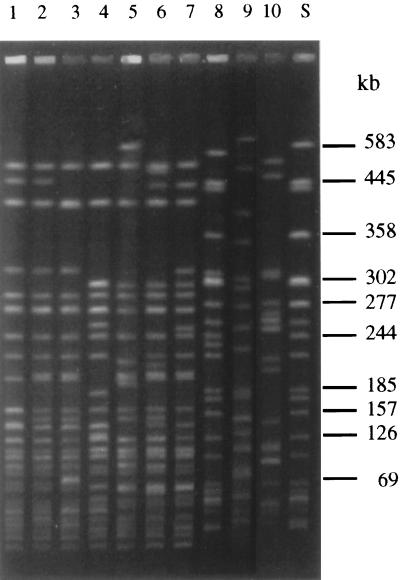
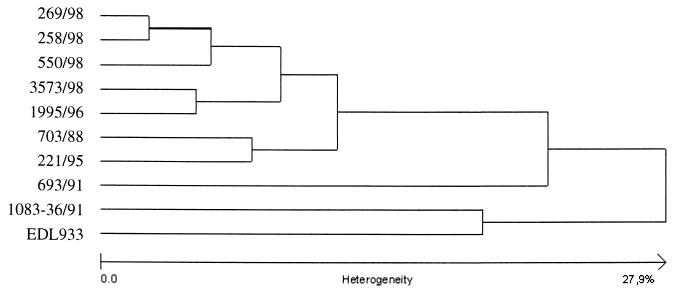
The clonal relatedness of SF STEC O157:H− isolates was investigated using PFGE. As shown in Fig. 2, the PFGE pattern of the bovine isolate (lane 3) was closely related to the patterns of the two epidemiologically related human isolates (lanes 1 and 2) as well as to the patterns of the four representative, epidemiologically unrelated, human SF STEC O157:H− isolates (lanes 4 to 7). In contrast, the PFGE patterns of the bovine and all human SF STEC O157:H− isolates differed markedly from the patterns of all of the control E. coli O157 strains, including NSF STEC O157:H7 strain EDL 933 (Fig. 2, lane 8) and two SF Stx-negative strains of serotypes O157:H45 (Fig. 2, lane 9) and O157:H19 (Fig. 2, lane 10). Analysis of the PFGE patterns by the RFLPscan system (Fig. 3) demonstrated that all seven SF STEC O157:H− strains, including the bovine and human isolates, belonged to one cluster but were only distantly related to NSF STEC O157:H7 strain EDL933 and to the SF Stx-negative E. coli O157:H19 and O157:H45 strains. Taking these findings together with the other chromosomal characteristics, it can be concluded that the SF STEC O157:H− isolates of bovine and human origin belong to one clone complex. The observed differences in the gene composition of large plasmids in strains of the same clone can be due to the loss, acquisition, or exchange of plasmid DNA during lateral transfer of these mobile elements.
FIG. 2.
PFGE patterns of XbaI-digested genomic DNAs of SF STEC O157:H− strains and control E. coli O157 strains. Lane S, molecular size standard (DNA from E. coli strain G5244 restricted with XbaI). In lanes 1 to 7, the following SF STEC O157:H− strains are shown: lane 1, 258/98; lane 2, 269/98; lane 3, 550/98; lane 4, 221/95; lane 5, 703/88; lane 6, 1995/96; and lane 7, 3573/98. In lanes 8 to 10, control E. coli O157 strains are shown as follows: lane 8, EDL933 (NSF STEC O157:H7); lane 9, 1083-36/91 (SF, Stx-negative E. coli O157:H45); and lane 10, 693/91 (SF, Stx-negative E. coli O157:H19).
FIG. 3.
Cluster analysis, derived from PFGE data, of SF STEC O157:H− isolates from the cow and patients, NSF STEC O157:H7 strain EDL 933, and SF Stx-negative strains O157:H45 and O157:H19 with the RFLPscan software.
By demonstrating the clonal relatedness between SF STEC O157:H− strains isolated from a cow and patients, we provide the first evidence that cattle can be a reservoir of SF STEC O157:H− strains and a source of the infection for humans. Importantly, the bovine SF STEC O157:H− isolate contained both stx2 and eae type γ genes, which are the virulence characteristics possessed by NSF and SF STEC O157 strains isolated from patients (3, 8, 16) (Table 1). This strongly supports the pathogenic potential of the bovine isolate for humans. On the other hand, the bovine isolate and both epidemiologically related SF STEC O157:H− strains isolated from patients lacked the plasmid-encoded putative virulence genes (Table 1), suggesting that these genes may not be significant in the genesis of human disease or that they have been lost during infection or storage.
Both patients in this study were likely to be infected through direct contact with the cow that shed the SF STEC O157:H− strain in its feces. This observation is in agreement with previous reports on direct transmission of STEC O157:H7 from cattle to humans (12, 18) and contributes to the increasing body of evidence that contact with farm animals, especially with cattle, is an important risk factor for acquiring STEC infection (14).
The failure to isolate SF STEC O157 strains from cattle in previous studies that used molecular techniques (9, 10) was probably due to the fact that such techniques are 100- to 1,000-fold less sensitive than the IMS procedure (11) that was used for the detection of SF E. coli O157 strains in this study. However, despite the fact that we introduced the IMS enrichment step, subsequent identification of SF O157 colonies on SMAC plates was a laborious procedure that required slide agglutination of almost 50 colonies per plate. The widespread distribution of the SF STEC O157:H− clone in central Europe (2, 8) and the emergence of cattle as a reservoir of such strains thus place significant limitations on using SMAC as the sole method for detecting STEC O157. This, combined with the fact that SF STEC O157 strains do not express EHEC hemolysin (2, 8) (Table 1) and thus cannot be detected on enterohemolysin agar (20), accentuates the need to develop a selective medium for such strains. Additional studies using appropriate diagnostic methods are necessary to determine the significance of cattle as a reservoir of SF STEC O157:H− and to further investigate the epidemiology of the infection.
Acknowledgments
This study was supported by grant 4563-3 from the Ministry of Health of the Czech Republic, by grant 525/97/0373 from the Czech Grant Agency, and by grants 01 KI 9903 and 1368/343 from Bundesministerium für Bildung und Forschung (BMBF), Germany.
We thank R. Ahmed and W. Demczuk (Health Canada, Winnipeg, Canada) for providing phages and reference strains for phage typing and L. Durso (University of Nebraska, Lincoln) for critical reading of the manuscript. The excellent technical assistance of B. Plaschke (Würzburg), A. Reischelova (Prague), and G. Bartel, U. Siewert, and B. Tannert (Wernigerode) is highly appreciated.
REFERENCES
- 1.Ammon A, Peterson L R, Karch H. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain E. coli O157:H−. J Infect Dis. 1999;179:1274–1277. doi: 10.1086/314715. [DOI] [PubMed] [Google Scholar]
- 2.Bielaszewska M, Schmidt H, Karmali M A, Khakhria R, Janda J, Blahova K, Karch H. Isolation and characterization of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157H− strains in the Czech Republic. J Clin Microbiol. 1998;36:2135–2137. doi: 10.1128/jcm.36.7.2135-2137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bockemühl J, Karch H, Tschäpe H. Zur Situation der Infektionen des Menschen durch enterohämorrhagische Escherichia coli (EHEC) in Deutschland, 1997. Bundesgesundhbl. 1998;Suppl.(October):2–5. doi: 10.1007/s00103-002-0458-4. [DOI] [PubMed] [Google Scholar]
- 4.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 5.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7, cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 6.Datz M, Janetzki-Mittmann C, Franke S, Gunzer F, Schmidt H, Karch H. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl Environ Microbiol. 1996;62:791–797. doi: 10.1128/aem.62.3.791-797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunzer F, Böhm H, Rüssmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karch H, Wiss R, Glonning H, Emmrich P, Aleksic S, Bockemühl J. Hämolytisch-urämisches Syndrom bei Kleinkindern durch Verotoxin-produzierende Escherichia coli. Dtsch Med Wochenschr. 1990;115:485–495. doi: 10.1055/s-2008-1065036. [DOI] [PubMed] [Google Scholar]
- 10.Karch H, Böhm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1200–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karch H, Janetzki-Mittmann C, Aleksic S, Datz M. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods, and direct culture. J Clin Microbiol. 1996;34:516–519. doi: 10.1128/jcm.34.3.516-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karch H, Bielaszewska M, Bitzan M, Schmidt H. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn Microbiol Infect Dis. 1999;34:229–243. doi: 10.1016/s0732-8893(99)00031-0. [DOI] [PubMed] [Google Scholar]
- 13.Khakhria R, Duck D, Lior H. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol Infect. 1990;105:511–520. doi: 10.1017/s0950268800048135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michel P, Wilson J B, Martin S W, Clarke R C, McEven S A, Gyles C L. Temporal and geographical distribution of reported cases of Escherichia coli O157:H7 infection in Ontario. Epidemiol Infect. 1999;122:193–200. doi: 10.1017/s0950268899002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien A D, Lively T A, Chen M E, Rothman S W, Formal S B. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet. 1983;i:702. doi: 10.1016/s0140-6736(83)91987-6. [DOI] [PubMed] [Google Scholar]
- 16.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun. 2000;68:64–71. doi: 10.1128/iai.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PulseNet. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Atlanta, Ga: Centers for Disease Control and Prevention; 1998. [Google Scholar]
- 18.Renwick S A, Wilson J B, Clarke R C, Lior H, Borczyk A A, Spika J, Rahn K, McFadden K, Brouwer A, Copps A, Anderson N G, Alves D, Karmali M A. Evidence of direct transmission of Escherichia coli O157:H7 infection between calves and a human. J Infect Dis. 1993;168:792–793. doi: 10.1093/infdis/168.3.792. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H, Rüssmann H, Schwarzkopf A, Aleksic S, Heesemann J, Karch H. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Zentbl Bakteriol. 1994;281:201–213. doi: 10.1016/s0934-8840(11)80571-2. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt H, Henkel B, Karch H. A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohaemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett. 1997;148:265–272. doi: 10.1111/j.1574-6968.1997.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W-L, Bielaszewska M, Liesegang A, Tschäpe H, Schmidt H, Bitzan M, Karch H. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J Clin Microbiol. 2000;38:2134–2140. doi: 10.1128/jcm.38.6.2134-2140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]