Abstract
A competitive PCR (cPCR) assay was developed for monitoring porcine circovirus (PCV) DNA in serum samples from piglets. The cPCR was based on competitive coamplification of a 502- or 506-bp region of the PCV type 1 (PCV1) or PCV2 ORF2, respectively, with a known concentration of competitor DNA, which produced a 761- or 765-bp fragment, respectively. The cPCR was validated by quantification of a known amount of PCV wild-type plasmids. We also used this technique to determine PCV genome copy numbers in infected cells. Furthermore, we measured PCV DNA loads in clinical samples. More than 50% of clinically healthy piglets could harbor both types of PCV. While PCV1 was detected in only 3 of 16 pigs with postweaning multisystemic wasting syndrome (PMWS), all the sick piglets contained PCV2. A comparison of the PCV2 DNA loads of healthy and sick animals revealed a significant difference, indicating that the development of PMWS may require a certain amount of PCV2.
Postweaning multisystemic wasting syndrome (PMWS) was first observed in western Canada in 1991 and in many other countries later on (2, 3, 6, 8, 11, 14, 16–18). Clinical signs of the disease include progressive weight loss, dyspnea, diarrhea, pallor, and jaundice. It was histologically recognized by interstitial pneumonia, lymphoid depletion, and histiocytosis in lymphoid tissues and, less frequently, hepatitis and nephritis. Although the causal agent has not been identified explicitly, a new strain of porcine circovirus (PCV), PCV type 2 (PCV2), has been isolated from a range of tissues of piglets affected by PMWS. PCV2 is distinguishable antigenically and genomically from nonpathogenic PCV (PCV1) from permanent pig kidney (PK-15) cells (24). Preliminary studies on the pathogenesis of PMWS have shown that PCV2 alone could not reproduce the severe forms of PMWS and that a coinfection with other disease-producing agents was required (4, 7, 10, 12). At present, PMWS is diagnosed by histopathology coupled with the identification of PCV infection by multiplex or type-specific PCR, in situ hybridization, immunohistochemistry, and enzyme-linked immunosorbent assay (1, 2, 5–7, 9, 13, 15, 17, 19, 20, 22, 26, 27). However, measurement of the PCV load and the dynamics of PCV in the pathogenesis of PMWS have not been described. Bearing this goal in mind, we believe that the availability of a quantitative assay for PCV DNA would be especially useful for addressing whether there is any correlation between the viral DNA load and PMWS development. In this paper, we report on a new competitive PCR (cPCR) assay that allows the quantification of PCV DNA in the sera of piglets with PMWS.
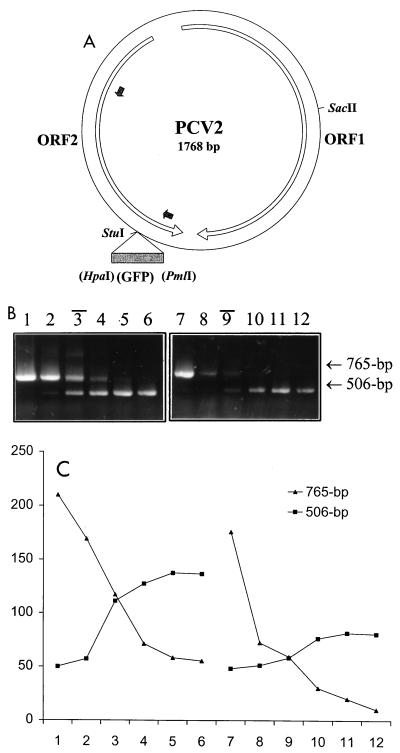
To obtain a PCV type-specific signal in the cPCR assay, a fragment of the ORF2 open reading frame was chosen as the target sequence because of the high degree of diversity of this region between the two types of PCVs (positions 1066 to 1568 in the PCV1 genome and positions 1081 to 1587 in the PCV2 genome). Two pairs of type-specific primers were designed accordingly. The primers for PCV1, I-FD (5′-GGGGGG GGATCC CATAAA TAGTCA GCCTTA CCACA-3′) and I-BD (5′-GGGGGG GGATCC TTCTAC AGAATT TGTACT CACCA-3′), were designed on the basis of the sequence of the prototype ATCC CCL-33 strain of PCV1 (GenBank accession no. U49186), whereas the primers for PCV2, II-FD (5′-GGGGGG GGATCC CATACA TGGTTA CACGGC TATTG-3′) and II-BD (5′-GGGGGG GGATCC CCGCAC CTTCGG ATATAC TG-3′), were established in reference to the sequence of the PCV2 strain isolated in our laboratory (GenBank accession no. AF086834). A BamHI site (underlined) was added at the 5′ end of each primer for the cloning of the PCR fragment. For the construction of plasmids, total DNA was extracted from PK-15 cells or the liver tissue of a piglet with PMWS by using a QIAamp kit (Qiagen Inc., Mississauga, Ontario, Canada), and PCV DNAs were amplified by PCR with virus-specific primers covering the whole genome and cloned into the pBKSII(+) vector (Stratagene). The PCV1 or PCV2 full-length genomic DNA could be released from the resulting constructs with the SapI (the unique recognition site at position 574 in the PCV1 genome) or SacII (the unique recognition site at position 491 in the PCV2 genome) restriction enzyme. To construct the competitive templates for cPCR, the HpaI-PmlI fragment from pQBI25 (Quantum Biotechnologies Inc.) containing a portion of the coding sequence for green fluorescent protein (GFP) was inserted into the unique StuI site (recognition site at position 1128 of the PCV1 genome or position 1143 of the PCV2 genome), generating two new plasmids with a 259-bp insertion. The resulting PCV2 plasmids are demonstrated in Fig. 1A. Thus, the PCR with wild-type PCV1 or PCV2 template would produce a 502- or 506-bp fragment, respectively, whereas a 761- or 765-bp fragment was synthesized when the respective competitor plasmid was used as the template.
FIG. 1.
Construction of wild-type and competitor plasmids and validation of cPCR. (A) Diagrammatic representation of PCV2 wild-type and competitor constructs. The whole genome (1,768 nucleotides) of PCV2 was cloned with a SacII restriction enzyme into the pBKSII(+) vector. The largest two open reading frames (ORFs) in the PCV2 genome are indicated by open arrows. Two small filled arrows point out the primer binding sites. A portion of the GFP coding sequence as the insert for the construction of the competitor plasmid is demonstrated by a filled box. (B and C) Quantification of a known amount of PCV2 wild-type DNA. (B) cPCR assay performed with two series of 10-fold dilutions of competitor DNA (lanes 1 to 6, 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, and 1 pg, respectively; lanes 7 to 12, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg, respectively) to compete against two fixed amount of PCV2 wild-type DNA per reaction (lanes 1 to 6, 1 ng; lanes 7 to 12, 1 pg). Black bars above the lane numbers indicate the equivalent point. (C) The densities of DNA bands were measured with SPOT DENSO software by using an AlphaImager 2200 instrument, and values representing the average density per area unit were plotted. The numbers on the x axis correspond to the respective lane in panel B. The value determined for the wild-type DNA was in excellent agreement with the actual input amount.
The type specificity of the PCR assay was tested with each pair of primers in a noncompetitive PCR. It turned out that both pairs of primers were type specific, as expected, and no cross-reaction was observed (data not shown). To examine whether there was competition between these two pairs of plasmids, when they were coamplified in one reaction tube, cPCR was carried out with a 50-μl reaction mixture containing 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate, 500 nM (each) primer, and 2.5 U of platinum Taq polymerase (Gibco-BRL) with a fixed amount of wild-type DNA and various quantities of competitor DNA. Following incubation at 94°C for 2 min, PCR was performed for 30 cycles, with each cycle consisting of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 7 min in a GeneAmp PCR System 2400 instrument (Perkin-Elmer). Ten microliters of the amplification products was analyzed on a 1% agarose gel (Ultrapure agarose; Gibco-BRL) in 1× Tris-acetate-EDTA electrophoresis buffer and 0.2 μg of ethidium bromide per ml. The equivalence points, at which the 502- and 506-bp bands and the 761- and 765-bp bands had equal intensities, were determined by visual inspection and densitometric scanning in an AlphaImager 2200 instrument (Alpha Innotech Corporation). Figure 1B shows a progressive competition between PCV2 wild-type DNA at two fixed amounts (1 ng or 1.8 × 108 molecules and 1 pg or 1.8 × 105 molecules) and 10-fold dilutions ranging from 100 ng to 1 pg or 100 pg to 1 fg of competitor DNA, indicating a broad range of sensitivity of the cPCR. A similar competition was also observed between PCV1 wild-type and competitor DNAs (data not shown).
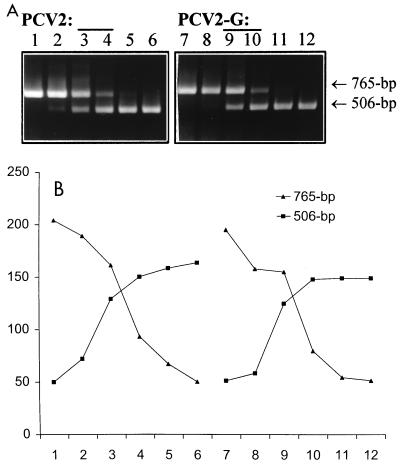
We then applied this cPCR method to evaluate whether there were any changes in the amount of viral DNA after d-glucosamine treatment of PK-15 cells or PCV2-infected cells. About 106 PK-15 cells or PCV2-infected PCV-free PK-15 cells (at a multiplicity of infection of ∼1) were incubated with 300 mM d-glucosamine for 30 min, and 24 h later total DNA was extracted with a QIAamp kit and eluted into 200 μl of H2O. Five microliters of the 10-fold-diluted (PCV1) or undiluted (PCV2) DNA was subjected to cPCR with various known amounts of competitor DNA. It turned out that the PCV1 DNA content in PK-15 cells was about 103-fold the PCV2 DNA content in PCV2-infected cells under our experimental conditions. As illustrated in Fig. 2, d-glucosamine did not cause any increase in viral DNA levels in PCV-infected cells. These findings were compatible with those of previous studies in which d-glucosamine promoted the entry of viral DNA into the nucleus prior to mitosis but the number of viral inclusion bodies per infected cell was not increased after d-glucosamine induction (23, 25).
FIG. 2.
Application of cPCR for detection of PCV2 DNA in cell culture. (A) cPCR assay of PCV2 DNA in infected PCV-free PK-15 cells with or without d-glucosamine treatment. About 106 cells were infected with PCV2 at a multiplicity of infection of ∼1. One flask was incubated with 300 mM d-glucosamine for 30 min at 24 h postinfection. Both treated and untreated cells were harvested 24 h later. DNAs extracted from PCV2-infected cells (designated PCV2; lanes 1 to 6) and PCV2-infected, d-glucosamine-treated cells (designated PCV2-G; lanes 7 to 12) were coamplified with a selected range of 10-fold dilutions of competitor DNA (lanes 1 to 6 or 7 to 12, 1 ng, 100 pg, 10 pg, 100 fg, 10 fg, and 1 fg, respectively). (B) Quantification after densitometric scanning of the DNA bands in panel A. In both cases (PCV2 and PCV2-G) the equivalent point corresponds to 10 to 1 pg of competitor DNA.
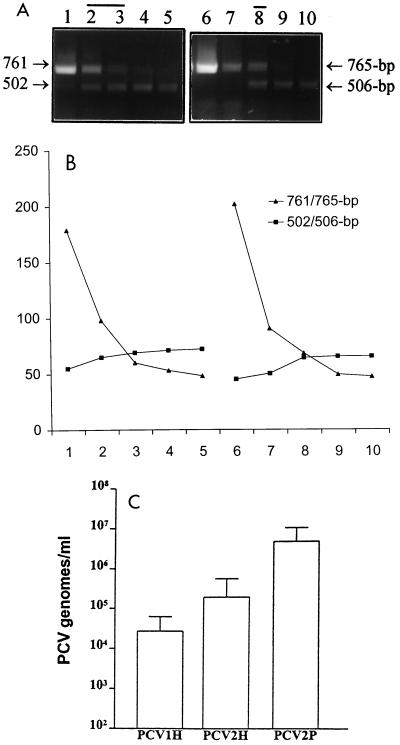
Subsequently, we measured the viral DNA loads in serum samples from piglets with various clinical signs and lesions. Serum samples were collected from two herds in Canada. Total DNA was extracted from 200 μl of each serum sample and was eluted into 200 μl of H2O. A standard, noncompetitive PCR was carried out with type-specific primers to detect PCV before the quantitative cPCR was performed. Figure 3A and B show two examples of the results of the cPCR experiments. The specificity of the cPCR assay was evaluated by sequencing several randomly selected PCR products (two for PCV1 and five for PCV2). For this purpose, the samples were subjected to a standard PCR and were cloned into a pBKSII(+) vector via BamHI, which was incorporated into the 5′ ends of the primers. The inserts were sequenced with an automated DNA sequencer.
FIG. 3.
Quantification of PCV DNA in serum samples from piglets with different clinical conditions. (A and B) Two representative samples of PCV DNA in serum samples from piglets with different clinical signs. Total DNA was purified from 200 μl of serum, and 10 μl was used per reaction mixture in a cPCR with 10-fold dilutions of competitor DNA. (A) cPCR assay of the PCV1 DNA load present in a clinically healthy piglet (lanes 1 to 5) and the PCV2 DNA burden present in a piglet with PMWS (lanes 6 to 10) with 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg of PCV1 or PCV2 competitor DNA, respectively. The equivalent zone was determined by measuring and plotting the densities of DNA bands (B). (C) PCV DNA loads of healthy and PMWS piglets. The viral loads of PCV1 (n = 11) and PCV2 (n = 25) in healthy piglets (designated PCV1H and PCV2H, respectively) and PCV2 DNA content (n = 16) in piglets with PMWS (designated PCV2P) were demonstrated. Error bars represent the standard deviation from the mean.
The majority of clinically healthy piglets had both PCV1 and PCV2. The DNA loads ranged from 1.8 × 107 to 1 × 105 copies/ml of serum for PCV1 (n = 11) or 1.8 × 109 to 1 × 104 copies/ml of serum for PCV2 (n = 25), with PCV2 DNA loads (mean, 1.89 × 105) being approximately 1 log higher than the PCV1 DNA loads (mean, 2.68 × 104) (Fig. 3C). In contrast, 3 of 16 piglets with PMWS had detectable PCV1, whereas all 16 had PCV2, suggesting that PCV2 might play a role in the development of PMWS and PCV1 might not be involved. It was noteworthy that the mean PCV2 DNA content (4.61 × 106 copies/ml) obtained for the 16 piglets with PMWS was significantly higher than that (1.89 × 105 copies/ml) obtained for 25 healthy animals (P = 0.0273 in a two-tailed t test) (Fig. 3C), indicating that there might be a threshold of the PCV2 amount which was needed to trigger the disease.
Furthermore, we wanted to examine the relationship between the PCV2 DNA content and the clinical expression of disease in different phases of PMWS. Two piglets with poor growth conditions suggestive of possible early signs of PMWS were sampled at 10-day intervals until they were diagnosed with clinical acute PMWS. One animal showed a continuous increase in PCV2 content from 105 to 107 copies/ml of serum during the development of PMWS, whereas the PCV2 load of the other piglet remained at a stable (and high) level (106) during this process. In another series of experiment, a follow-up sample was collected from six piglets 10 days after the onset of the acute phase of PMWS since they were found to be recovering from PMWS, as judged clinically. At the acute phase of PMWS, the piglets had 106 to 107 copies/ml of serum. The cPCR assay with the follow-up samples did not reveal any change in the viral DNA content, indicating that PCV2 had not been effectively eliminated in this 10-day period, even though our preliminary data demonstrated that the animals mounted an antibody response (data not shown). The observation of the apparent irrelevance of the PCV2 DNA content (at a certain level, though) to the alleviation and aggravation of clinical PMWS was in line with the findings that PCV2 is not the only infectious agent required for the production of PMWS (7, 10, 12).
In conclusion, the viral DNA levels in serum carried by the piglets in the acute (and recovering) phases of PMWS were significantly higher than those in clinically healthy piglets. This observation suggested that the development of PMWS might require a certain amount of PCV2. Although all the piglets with PMWS tested in this study had high PCV2 genome contents (above the threshold), some animals carrying the same amount of PCV2 remained healthy, leading us to speculate that PCV2 is required, but may not be sufficient, to cause PMWS. The ultimate occurrence of PMWS may probably rely on a secondary pathogen and/or the conditions of individual piglets. It is also intriguing that the clinically healthy piglets could harbor both types of PCV, whereas the majority of the clinically PMWS-positive piglets tested in this study contained only PCV2. The fairly high incidence of PCV1 and PCV2 in healthy piglets and the rather low incidence of PCV1 in piglets with PMWS are in agreement with several recent serological surveys (9, 13, 19). The biological significance of the coexistence of the nonpathogenic and apparently pathogenic strains of PCV in healthy piglets and the almost entire exclusion of the nonpathogenic PCV strain from the piglets with PMWS require further studies. Also compatible with a previous study (13) is the observation of the apparent predominance of PCV2 in all the samples tested, suggesting that PCV2 might have developed a much more efficient way of transmission.
The type-specific primers used in this study were designed on the basis of the sequences of one PCV1 strain and one PCV2 strain, respectively. Compared to all four complete PCV1 sequences in GenBank, the forward primer for detecting PCV1 had an entire match to three isolates and a one-nucleotide mismatch to the French isolate, whereas the backward primer had an one-nucleotide mismatch to the French and the U.S. isolates. In the case of PCV2-specific primers, the backward primer had full homology to all 26 PCV2 sequences in GenBank, while the forward primer had an one-nucleotide mismatch to the two isolates reported from Taiwan. Because of the mismatches of the primers, we could not exclude the possibility of missing certain genetically divergent PCV strains, albeit it seemed somewhat unlikely since the one-nucleotide mismatches were located internally and not at the extreme 3′ ends of the primers, which would not affect the primer-template binding dramatically (21).
In summary, we have described the development of a cPCR and the monitoring of PCV DNA levels in serum samples, which makes it possible to analyze the correlation between the amount of circulating viruses and clinical parameters in different groups of piglets. Moreover, the cPCR method will also benefit studies of PMWS as well as PCV, e.g., the spread of virus within tissues of swine.
Acknowledgments
This research was supported by Boehringer-Ingelheim and the Natural Science and Research Council of Canada.
The technical assistance of Betty Chow, Elaine Van Moolehem, and Tammy Karkut is greatly appreciated.
REFERENCES
- 1.Allan G M, Mackie D P, McNair J, Adair B M, McNulty M S. Production, preliminary characterisation and applications of monoclonal antibodies to porcine circovirus. Vet Immunol Immunopathol. 1994;43:357–371. doi: 10.1016/0165-2427(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 2.Allan G M, McNeilly F, Kennedy S, Daft B, Clarke E G, Ellis J A, Haines D M, Meehan B M, Adair B M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Investig. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 3.Allan G M, McNeilly F, Meehan B M, Kennedy S, Mackie D P, Ellis J A, Clark E G, Espuna E, Saubi N, Riera P, Bøtner A, Charreyre C E. Isolation and characterization of circoviruses from pigs with wasting syndromes in Spain, Denmark and North Ireland. Vet Microbiol. 1999;66:115–123. doi: 10.1016/s0378-1135(99)00004-8. [DOI] [PubMed] [Google Scholar]
- 4.Balasch M, Segales J, Rosell C, Domingo M, Mankertz A, Urniza A, Plana-Duran J. Experimental inoculation of conventional pigs with tissue homogenates from pigs with post-weaning multisystemic wasting syndrome. J Comp Pathol. 1999;121:139–148. doi: 10.1053/jcpa.1999.0310. [DOI] [PubMed] [Google Scholar]
- 5.Choi C, Chae C. In-situ hybridization for the detection of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Comp Pathol. 1999;121:265–270. doi: 10.1053/jcpa.1999.0315. [DOI] [PubMed] [Google Scholar]
- 6.Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strokappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998;39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Investig. 1999;11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- 8.Hamel A L, Lin L L, Nayar G P S. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72:5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamel A L, Lin L L, Sachvie C, Grudeski E, Nayar G P S. PCR detection and characterization of type-2 porcine circovirus. Can J Vet Res. 2000;64:44–52. [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy S, Moffett D, McNeilly F, Meehan B, Ellis J, Krakowka S, Allan G M. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J Comp Pathol. 2000;122:9–24. doi: 10.1053/jcpa.1999.0337. [DOI] [PubMed] [Google Scholar]
- 11.Kiupel M, Stevenson G W, Mittal S K, Clark E G, Haines D M. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet Pathol. 1998;35:303–307. doi: 10.1177/030098589803500411. [DOI] [PubMed] [Google Scholar]
- 12.Krakowak S, Ellis J A, Meehan B, Kennedy S, McNeilly F, Allan G. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet Pathol. 2000;37:254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- 13.Larochelle R, Antaya M, Morin M, Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods. 1999;80:69–75. doi: 10.1016/s0166-0934(99)00032-4. [DOI] [PubMed] [Google Scholar]
- 14.Mankertz A, Domingo M, Folch J M, LeCann P, Jestin A, Segales J, Chmielewicz B, Plana-Duran J, Soike D. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res. 2000;66:65–77. doi: 10.1016/s0168-1702(99)00122-7. [DOI] [PubMed] [Google Scholar]
- 15.McNeilly F, Kennedy S, Moffett D, Meehan B M, Foster J C, Clarke E G, Ellis J A, Haines D M, Adair B M, Allan G M. A comparison of in situ hybridization and immunohistochemistry for the detection of a new porcine circovirus in formalin-fixed tissues from pigs with post-weaning multisystemic wasting syndrome (PMWS) J Virol Methods. 1999;80:123–128. doi: 10.1016/s0166-0934(99)00043-9. [DOI] [PubMed] [Google Scholar]
- 16.Meehan B M, McNeilly F, Todd D, Kennedy S, Jewhurst V A, Ellis J A, Hassard L E, Clark E G, Haines D M, Allan G M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 17.Morozov I, Sirinarumitr T, Sorden S D, Halbur P G, Morgan M K, Yoon K J, Paul P S. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. 1998;36:2535–2541. doi: 10.1128/jcm.36.9.2535-2541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onuki A, Abe K, Togashi K, Kawashima K, Taneichi A, Tsunemitsu H. Detection of porcine circovirus from lesions of a pig with wasting disease in Japan. J Vet Med Sci. 1999;61:1119–1123. doi: 10.1292/jvms.61.1119. [DOI] [PubMed] [Google Scholar]
- 19.Ouardani M, Wilson L, Jette R, Montpetit C, Dea S. Multiplex PCR for detection and typing of porcine circoviruses. J Clin Microbiol. 1999;37:3917–3924. doi: 10.1128/jcm.37.12.3917-3924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosell C, Segales J, Plana-Duran J, Balasch M, Rodriguez-Arrioja G M, Kennedy S, Allan G M, McNeilly F, Latimer K S, Domingo M. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol. 1999;120:59–78. doi: 10.1053/jcpa.1998.0258. [DOI] [PubMed] [Google Scholar]
- 21.Sommer R, Tautz D. Minimal homology requirements for PCR primers. Nucleic Acids Res. 1989;17:6749. doi: 10.1093/nar/17.16.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorden S D, Harms P A, Nawagitgul P, Cavanaugh D, Paul P S. Development of a polyclonal-antibody-based immunohistochemistry method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J Vet Diagn Investig. 1999;11:528–530. doi: 10.1177/104063879901100607. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson G W, Kiupel M, Mittal S K, Kanitz C L. Ultrastructure of porcine circovirus in persistently infected PK-15 cells. Vet Pathol. 1999;36:368–378. doi: 10.1354/vp.36-5-368. [DOI] [PubMed] [Google Scholar]
- 24.Tischer I, Gelderblom H, Vettermann W, Koch M A. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 25.Tischer I, Peters D, Rash R, Pociuli S. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch Virol. 1987;96:39–57. doi: 10.1007/BF01310989. [DOI] [PubMed] [Google Scholar]
- 26.Tischer I, Bode L, Peters D, Pociuli S, Germann B. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Arch Virol. 1995;140:737–743. doi: 10.1007/BF01309961. [DOI] [PubMed] [Google Scholar]
- 27.Tischer I, Bode L, Apodaca J, Timm H, Peters D, Rasch R, Pociuli S, Gerike E. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch Virol. 1995;140:1427–1439. doi: 10.1007/BF01322669. [DOI] [PubMed] [Google Scholar]