AS program is essential for HSC formation.
Abstract
Single-cell transcriptional profiling has rapidly advanced our understanding of the embryonic hematopoiesis; however, whether and what role RNA alternative splicing (AS) plays remains an enigma. This is important for understanding the mechanisms underlying splicing-associated hematopoietic diseases and for the derivation of therapeutic stem cells. Here, we used single-cell full-length transcriptome data to construct an isoform-based transcriptional atlas of the murine endothelial-to-hematopoietic stem cell (HSC) transition, which enables the identification of hemogenic signature isoforms and stage-specific AS events. We showed that the inclusion of these hemogenic-specific AS events was essential for hemogenic function in vitro. Expression data and knockout mouse studies highlighted the critical role of Srsf2: Early Srsf2 deficiency from endothelial cells affected the splicing pattern of several master hematopoietic regulators and significantly impaired HSC generation. These results redefine our understanding of the dynamic HSC developmental transcriptome and demonstrate that elaborately controlled RNA splicing governs cell fate in HSC formation.
INTRODUCTION
The first hematopoietic stem cells (HSCs) are generated in the aorta-gonad-mesonephros (AGM) region during mammalian embryonic development. In mice, this process begins on E10.5 (embryonic day 10.5) (1–4). Lineage tracing and real-time in vivo observations show that HSCs are derived from hemogenic endothelial cells (HECs), a transient population lining the ventral wall of the dorsal aorta (5). Most recently, these cells are isolated and characterized by single-cell transcriptomics, leading to the identification of a unique population of HSC-competent HECs (CD31+CD45−CD41−CD43−CD201+Kit+CD44+) (6). HSC-primed HECs pass through two sequential pre-HSC stages, CD45− type 1 (T1) and CD45+ type 2 (T2) pre-HSCs (3, 7), to become mature HSCs and colonize the fetal liver from late E11.5. This process is termed as endothelial-to-hematopoietic transition (EHT).
EHT is a central process during HSC development. The procedure of EHT requires highly coordinated gene expression that is regulated through transcriptional and posttranscriptional mechanisms. Early studies identified key roles for the transcription factors runt related transcription factor 1 (RUNX1), GATA binding protein 2 (GATA-2), and core binding factor beta (CBFβ) in HSC formation in vivo (8–10); while recent advances based on single-cell transcriptional profiling have uncovered the importance of mTOR signaling, long noncoding RNAs, and RNA modification factors (7, 11). Intriguingly, both our previous transcriptome screening of pre-HSCs (7) and the most recent decoding of HECs (6) revealed that HSC emergence is accompanied by significant changes in expression of the genes encoding RNA processing and splicing proteins. Together, these findings imply a role for alternative splicing (AS) in EHT, but, until now, this phenomenon has not been assessed in developing HSCs.
AS is a central mechanism that acts upon the pre-mRNA transcripts of more than 90% of multiexon protein-coding genes in mammals, enabling cells to generate vast transcript and protein diversity from a limited number of genes. Careful coordination of AS networks occurs during the development of various tissues, including the brain (12–14), heart (15, 16), liver (17), and skeletal muscle (18), as well as in hematopoiesis. Deep sequencing and comprehensive analysis of adult human HSCs and progenitor cells uncovered thousands of cell type–specific AS transcripts and identified a previously unknown isoform of nuclear factor Ι B (NFΙB) that regulates megakaryopoiesis (19). Moreover, several diseases of the hematopoietic system, such as myelodysplastic syndromes, are linked to mutations that affect mRNA splicing (20). At present, however, the role of AS in the endothelial-to-HSC transition, the earliest stages of HSC formation, is undefined, and the key mediators of pre-mRNA splicing in HSC ontogeny are unknown.
Here, we used single-cell full-length transcriptome data from seven HSC-associated populations to identify tens of thousands of AS events and constructed a dynamic splicing landscape during entire HSC ontogeny. We found that EHT is accompanied by a significant AS modality switch, which is mainly orchestrated by the splicing regulator serine and arginine-rich splicing factor 2 (Srsf2). These findings help to explain active RNA processing during HSC ontogeny, extend our understanding of the molecular mechanisms underlying hematopoietic specification, and provide a resource for the future elucidation of other splicing regulators pivotal for embryonic HSC development.
RESULTS
A predominant transcript diversity in T1 pre-HSCs during HSC development
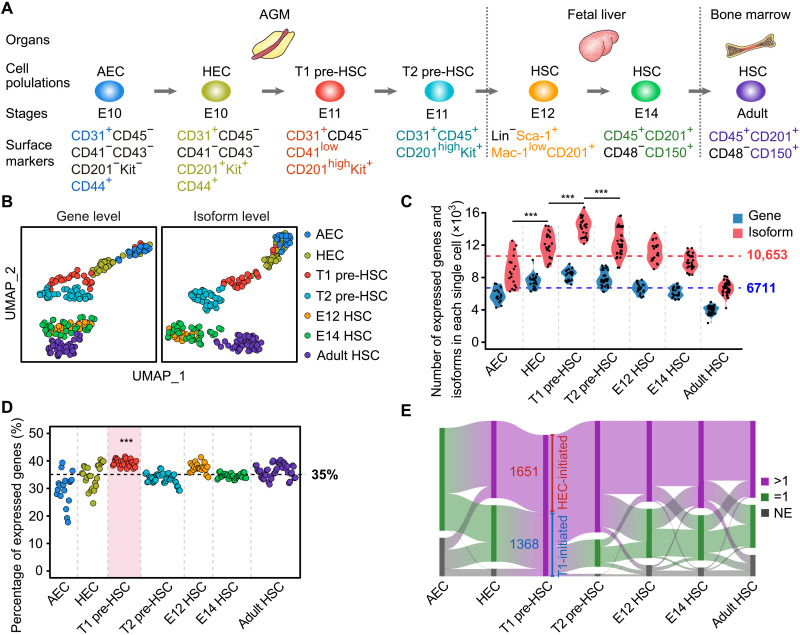
Most recently, we have deciphered multistep specification of ECs toward HSCs, with the arterial ECs (AECs) featured by CD44 expression lie upstream of the HSC-primed HECs in the AGM region (6, 21). To more accurately witness the transition process from AECs toward HSCs, we sampled AECs (CD31+CD45−CD41−CD43−CD201−Kit−CD44+) and HSC-competent HECs (CD31+CD45−CD41−CD43−CD201+Kit+CD44+) at the E10 AGM region (6) and performed full-length single-cell RNA sequencing (RNA-seq) using the same strategy as we previously used (fig. S1, A to C) (7, 22). To determine the diversity and abundance of alternative transcript expression during HSC ontogeny in mouse embryos, we used our newly acquired and previously reported (7) single-cell full-length transcriptome of totally seven sequential cell populations during HSC development (aortic ECs and HSC-primed HECs from E10 AGM, T1 and T2 pre-HSCs from E11 AGM, HSCs from E12 and E14 fetal liver, and HSCs from adult bone marrow) for systematic analyses (Fig. 1A). Uniform manifold approximation and projection (UMAP) analyses at isoform level largely recapitulated the performance at gene level, with the seven phenotypic populations basically separated except for the two fetal liver HSC populations (Fig. 1B).
Fig. 1. Global isoform expression dynamics during HSC development.
(A) Schematic representation of mouse embryonic hematopoietic development processes. (B) UMAP analysis of genes and isoforms during HSC development. (C) Number of genes and isoforms in an individual cell of each cell population. One-sided Wilcoxon rank sum test was used to determine the difference (***P < 0.001). (D) Percentage of genes expressing more than one isoform (multi-isoform genes) in an individual cell of each cell population. One-sided Wilcoxon rank sum test was used to determine the difference between T1 pre-HSC and other cell populations (***P < 0.001). (E) Sankey diagram showing the dynamic changes of HEC-initiated multi-isoform genes (HEC-initiated) and T1 pre-HSC–initiated multi-isoform genes (T1-initiated) during HSC development. The numbers on the graph indicate the number of genes. >1, multi-isoform gene; =1, single-isoform gene; NE, no expression.
Across all cell populations, we detected an average of 6711 expressed genes and 10,653 transcript isoforms per cell, with the number of isoforms gradually increased from AEC, HEC, to T1 pre-HSCs and then decreased. T1 pre-HSCs had the highest number of expressed genes and isoforms (Fig. 1C), indicating particularly high transcriptional and posttranscriptional diversity at this stage. We then calculated the constitutions in the expressed genes according to whether more than one isoform was detected. On average, 35% of genes generated multiple isoforms, while in T1 pre-HSCs, this number was significantly higher (39%) (Fig. 1D). Together, these data indicate a specific and sustained requirement for diverse transcripts during all stages of HSC ontogeny, but especially for HEC and pre-HSC fate specification.
We then looked in more detail at the T1 pre-HSC multi-isoform genes that were absent or were single-isoform genes at the AEC stage, and we saw that more than half of them (1651) began to express multiple isoforms from the intermediate HEC stage (defined as HEC-initiated multi-isoform genes), while the others (1368) did not display transcript diversity until the T1 pre-HSC stage (defined as T1 pre-HSC–initiated multi-isoform genes) (Fig. 1E and table S1). HEC multi-isoform genes were enriched in RNA processing and cell cycle (fig. S1D), with 775 (47%) of the HEC-initiated multi-isoform genes maintaining transcript diversity until the adult HSC stage (Fig. 1E, fig. S1E, and table S1), and the functions of these persistently diversely expressed genes were mainly related to RNA processing and hematopoietic cell differentiation (fig. S1F), indicating the ongoing requirement for these processes throughout HSC development. In contrast, the T1 pre-HSC–initiated multi-isoform genes gradually stopped generating multiple isoforms, with only 238 (17%) of the 1368 retaining transcript diversity until adult HSCs (Fig. 1E, fig. S1E, and table S1), which were enriched in terms related to cellular functions, including organelle assembly and localization, cellular response, and endocytosis (fig. S1G). Together, these results suggest that a stage-specific pattern of transcript diversity accompanies the stepwise EHT, while the T1 multi-isoform genes might be only responsible for its cell fate specification.
Gene expression data obscure functionally relevant differences in transcriptional isoform abundance during EHT
As traditional transcriptional analyses consider all isoforms of an expressed gene together, despite potential functional differences, we next went on to generate and compare stage-specific gene expression and transcriptional isoform signatures for HEC and T1 pre-HSC. We first identified genes that were notably more highly expressed in HECs or T1 pre-HSCs, comparing with their adjacent early developmental stages (defined as HEC or T1 pre-HSC signature genes) (fig. S2A and table S2A). In line with previous findings (6), HECs expressed relatively higher ribosomal and cell cycle genes than AECs (fig. S2B), while T1 pre-HSC signature genes were mainly enriched for RNA processing and RNA metabolism (fig. S2C).
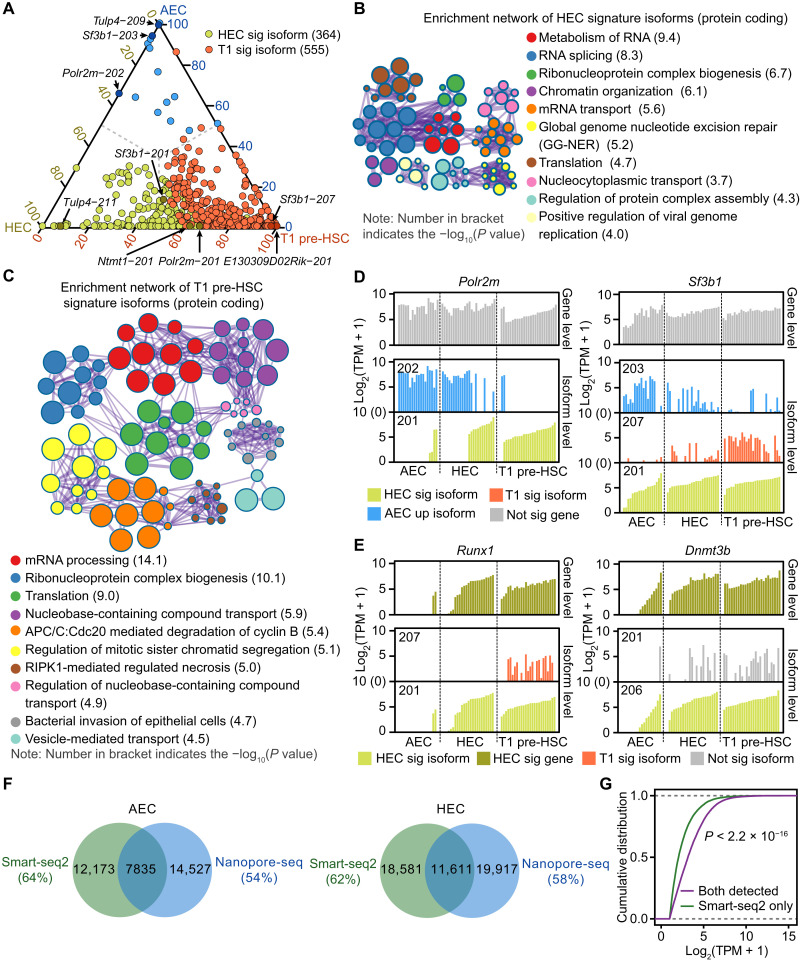
We also identified 364 HEC signature isoforms (accounting for 11% of all isoforms enriched in HEC compared to AEC) and 555 T1 pre-HSC signature isoforms (account for 20% of all isoforms enriched in T1 pre-HSC compared to HEC) that were not significantly differentially expressed at the gene level (Fig. 2A, fig. S2D, and table S2B), and whose elevated expression was masked in the gene level–based expression analysis (fig. S2A). Notably, 84% of HEC and 73% of T1 pre-HSC signature isoforms are protein-coding transcripts (fig. S2E), and their functions are mainly enriched in terms related to various RNA metabolic processes (Fig. 2, B and C), including RNA splicing, RNA transport, and ribonucleoprotein complex biogenesis, together supportive of a global activation of the posttranscriptional program during hemogenic fate specification. For example, specific isoforms of polymerase II polypeptide M (Polr2m), splicing factor 3b subunit 1 (Sf3b1), the protein N-terminal methyltransferase 1 (Ntmt1) involved in DNA damage response and cancer development (23), and tubby like protein 4 (Tulp4) associated with stress response (24) were annotated as HEC signature isoforms, while the levels of expression of these genes showed no significant changes between HECs and AECs (Fig. 2D and fig. S2F). Meanwhile, an Sf3b1 isoform Sf3b1-207 and an E130309D02Rik isoform E130309D02Rik-201 were substantially more highly expressed in T1 pre-HSCs than HECs (Fig. 2D and fig. S2F). In addition, we uncovered elevated expression of specific isoforms of some signature genes including the RUNX family transcription factor 1 (Runx1) isoform Runx1-201 (encoding the protein isoform RUNX1C) and the DNA methyltransferase 3 beta (Dnmt3b) isoform Dnmt3b-206 (representing the inactive protein variant DNMT3B4), which were both detected within the HEC signature (Fig. 2E). Collectively, the HEC and T1 pre-HSC isoform signatures clearly show the necessity of discriminating isoform-specific roles for full understanding of the transcriptional landscape of EHT.
Fig. 2. Differentially expressed genes and isoforms during EHT.
(A) Ternary phase diagram showing the relative enrichment of HEC (HEC sig) and T1 pre-HSC signature (T1 sig) isoforms. Difference at only isoform level but not gene level was defined as signature isoforms. AEC up-regulated isoforms compared to HEC were plot in blue. Indicated isoforms in (D) are also labeled. (B and C) Enrichment network representing the top 10 enriched terms of HEC (B) or T1 pre-HSC (C) signature isoforms with protein coding abilities. Enriched terms with high similarity were clustered and rendered as a network, while each node represents an enriched term and is colored according to its cluster. Node size indicates the number of enriched genes, and the line thickness indicates the similarity score shared by two enriched terms. The term with smallest P value from each cluster was labeled. RIPK1, receptor-interacting protein kinase 1. (D) Bar plot showing the expression levels of indicated HEC or T1 pre-HSC signature isoforms. Number on the upper left represents the isoform ID from Ensembl. (E) Bar plot showing the isoform expression levels of indicated HEC signature genes. Number on the upper left represents the isoform ID from Ensembl. (F) Venn diagram showing the overlap isoforms detected by Smart-seq2 and single-cell Nanopore-seq methods in AECs (left) and HECs (right). (G) Cumulative distribution showing the expression level of isoforms that were captured by both methods or Smart-seq2 only. Kolmogorov-Smirnov test was used to determine the differences.
To further validate the Smart-seq2 results, we performed single-cell Nanopore sequencing (Nanopore-seq) of full-length complementary DNA (cDNA) obtained from two cell populations: AEC and HEC. The read length from Nanopore-seq was 200 to 2000 nt, and the average read quality score was more than 7 (fig. S2G). The Venn diagram showed that more than 60% isoforms were detected by both Nanopore-seq and Smart-seq2 methods in AECs and HECs (Fig. 2F). Moreover, we found that transcripts that were specifically captured by Smart-seq2 showed relatively lower expression levels compared to the overlapped transcripts (Fig. 2G), indicating that Nanopore-seq is more likely to detect isoforms with high expression. Specifically, the expression of Sf3b1, Runx1, and Dnmt3b gene, as well as their isoforms from Nanopore-seq, showed high consistency with Smart-seq2 results (fig. S2H).
A range of AS events mediate transcript diversity during EHT
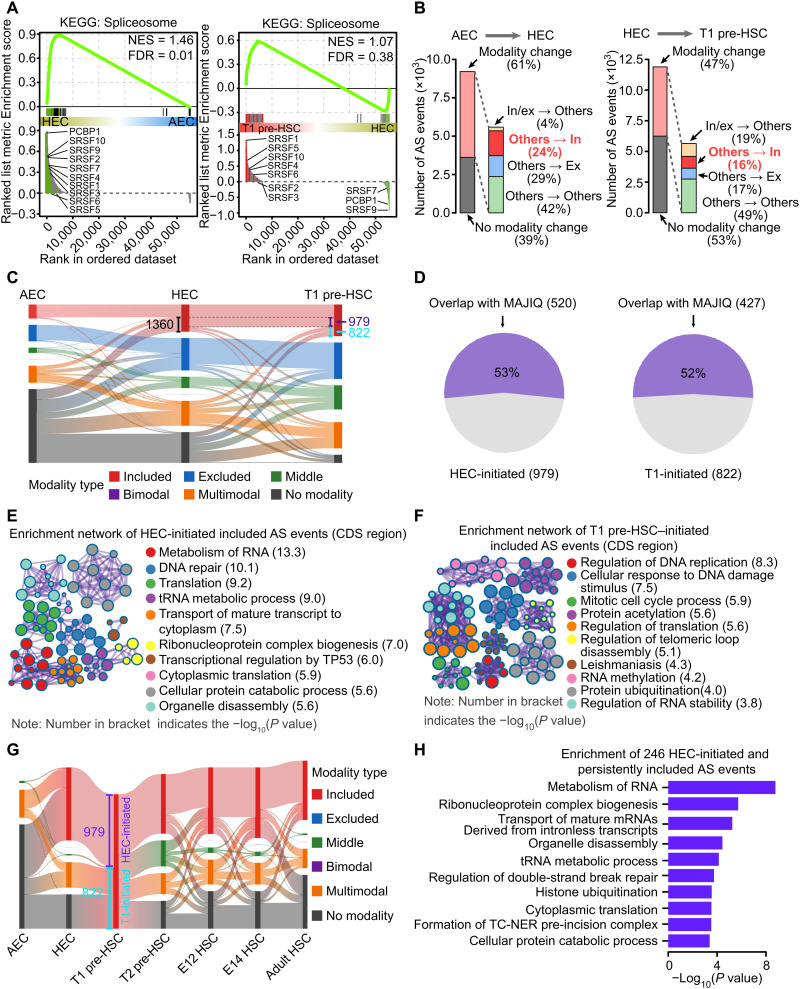
AS is one of the major mechanisms that eukaryotic cells use to generate transcript diversity from a fixed number of genes. Accordingly, alongside the increasing transcript diversity during hemogenic fate specification from AEC to HEC, gene set enrichment analysis (GSEA) demonstrated that expression of genes in the “Spliceosome” pathway was enriched in HECs compared with AECs (Fig. 3A). To quantify the contribution of AS to dynamic transcript accumulation during HSC emergence, we calculated the alternative exon usage by using the mixture-of-isoforms (MISO) statistical model (25), which assigned a “percentage spliced in” (PSI, Ψ) value to each exon by estimating its abundance compared to adjacent exons in each single cell (fig. S3A). Across all stages, we detected an average of 4309 AS events in individual cells, with considerable variation between stages and the highest number in T1 pre-HSCs, as expected (fig. S3B and S3C). We also identified five types of AS events and found that exon skipping (ES) accounted for about 51% of AS events at all stages, whereas the others included mutually exclusive exons, alternative 5′ splice sites, alternative 3′ splice sites, and intron retention (fig. S3D).
Fig. 3. HEC- and T1 pre-HSC–initiated included AS events during HSC development.
(A) GSEA showing the enrichment of Spliceosome pathway between HEC and AEC and between T1 pre-HSC and HEC. Indicated splicing factors were colored and labeled. NES, normalized enrichment score; FDR, false discovery rate. KEGG, Kyoto Encyclopedia of Genes and Genomes. (B) Bar plot showing the proportion of AS events with (modality change) or without (no modality change) modality changes between neighboring developmental stages during EHT. In/ex represents the included and excluded modality, separately. Others include the middle, bimodal, multimodal, and AS events without modality. (C) Sankey diagram showing the AS modality transition from AEC to HEC and then to T1 pre-HSC. No modality represents an AS event with less than five cells detected with PSI in a stage. The numbers on the graph indicate the number of AS events. (D) Pie chart showing the proportion of HEC-initiated (left) and T1-initiated (right) included AS events detected by both MISO and MAJIQ methods. (E and F) Enrichment network representing the top 10 enriched terms of HEC-initiated (E) or T1 pre-HSC–initiated (F) included AS events located into the CDS region. tRNA, transfer RNA. (G) Sankey diagram showing modality changes of HEC-initiated (HEC-initiated) and T1 pre-HSC–initiated (T1-initiated) included AS events at different stages. The numbers on the graph indicate the number of AS events. (H) Bar plot showing the enriched terms of 246 HEC-initiated and persistently included AS events. TC-NER, Transcription-coupled Nucleotide excision repair.
While heterogeneity exists in AS events (as illustrated for two AS events in fig. S3, E and F), the simple comparison of PSI between two stages is informative, but it does not reflect the full level of complexity of a given AS event in a certain cell population. Even those AS events with no significant changes in average PSI between HEC and AEC showed obviously distinct distribution among the two stages when considered at the single-cell level (fig. S3G). Therefore, we applied the “modality” strategy (26), which combines the usage of alternative exons (PSI) with their distribution patterns, to provide more detailed information on the splicing profiles at the single-cell level. We began by assigning each AS event to one of five modalities: (i) included, most individual cells contained isoforms with the inclusion of this exon (Ψ ~ 1); (ii) excluded, most individual cells contained isoforms with the exclusion of this exon (Ψ ~ 0); (iii) middle, most individual cells contained isoforms with both the inclusion and exclusion of this exon (Ψ ~ 0.5); (iv) bimodal, individual cells existed as two subpopulations that contained either the included (Ψ ~ 1) or excluded (Ψ ~ 0) exon; or (v) multimodal, the distribution of the exon did not fit any of the previous categories (fig. S3H). Among all cell populations, the alternative exons within the included, excluded, and middle modalities accounted for nearly 70% of all spliced exons (fig. S3I), which is consistent with the high purity of these cell populations, because most AS events in a homogeneous cell population should exhibit homogeneity. In comparison, AS events that exhibited multimodal modalities showed relatively lower frequencies, and bimodal modality was hardly detected (fig. S3I).
We then examined the AS event modality changes alongside the stepwise specification of T1 pre-HSC from AEC. Sixty-one percent of AS events showed modality switches from AEC to HEC stages, while 47% of AS events had modality changes from HEC to T1 pre-HSC (Fig. 3B). Among these “modality change” AS events, we specifically focused on the movement to “included” (shown as “others → in”). As the specific exons were selectively and homogeneously retained by “inclusion” along with HEC or T1 pre-HSC specification, the usage of these exons might be important in HSC fate commitment. We identified 1360 AS events that changed from “other” (including middle, bimodal, multimodal, and no modality) in AECs to included modality in HECs (Fig. 3C) by following 979 of the 1360 events still remained as included modality until the T1 pre-HSC stage (defined as HEC-initiated included AS events; Fig. 3C and table S3A). In addition, another 822 included AS events were initially present in T1 pre-HSC rather than HECs and thus were annotated as T1 pre-HSC–initiated included AS events (Fig. 3C and table S3A). Overall, we uncovered a clear pattern of the accumulation of stepwise inclusion of AS events from AECs to HECs (979 events) and then to T1 pre-HSCs (822 events). In addition, to add the robustness of AS analyses, we used another algorithm, Modeling Alternative Junction Inclusion Quantification (MAJIQ), to estimate the PSI value of AS events (27). As shown in Fig. 3D, more than 50% of HEC-initiated and T1-initiated included AS events could be also detected by MAJIQ.
We then asked what types of gene products these hemogenic-specific (including HEC- and T1 pre-HSC–initiated) included AS events affected. Among the 979 HEC-initiated and 822 T1 pre-HSC–initiated included AS events, more than 60% of them are located into the Coding sequence (CDS) region, which means that these events might influence protein structures (fig. S3J). The 626 CDS-restricted HEC-initiated included AS events were enriched in the products of genes related to RNA metabolism (including transfer RNA metabolism, RNA translation, RNA transport, and ribonucleoprotein complex biogenesis) and cell cycle (Fig. 3E and table S3B), consistent with the activate RNA processing and cycling of HECs, in contrast to the quiescent state of ECs (6). The 517 CDS-restricted T1 pre-HSC–initiated included AS events mainly affected the products of genes associated with cell cycle and cellular catabolic pathways, including DNA replication, protein acetylation, and peroxisome (Fig. 3F and table S3C). Notably, the peroxisome pathway is linked with steroid hormone synthesis and cholesterol biosynthesis, which have been reported to promote the formation of HSC from HEC (28, 29). In addition, we also performed functional enrichment analysis using HEC- and T1 pre-HSC–initiated included AS events detected by MAJIQ, and the top enriched terms between MISO and MAJIQ showed high similarity, such as RNA metabolism and cell cycle (fig. S3K).
Furthermore, 246 of the HEC-initiated included AS events maintained their included modality in the subsequent mature HSC stages (T2 pre-HSCs, E12 and E14 fetal liver HSCs, and adult HSCs) (Fig. 3G, fig. S3L, and table S3A), in line with the observation that cell fate choice is initiated at the HEC stage (6). Gene Ontology (GO) enrichment analysis of these 246 HEC-initiated and persistently included AS events during HSC development revealed overrepresentation of RNA metabolic genes (Fig. 3H). In contrast with the maintenance of HEC-initiated inclusions, T1 pre-HSC–initiated inclusions lost their included modality in mature HSC populations, with most becoming undetectable (Fig. 3G, fig. S3L, and table S3A). This is consistent with the finding that a specific group of enhancers was also transiently activated at the pre-HSC stage but subsequently lost their activating markers (30). These data suggest a specific requirement of these AS events to support the transition from dual-featured transient HECs to committed T1 pre-HSCs, but not thereafter.
Hemogenic-specific included AS events are detectable in vivo and are required for hematopoietic cell generation in vitro
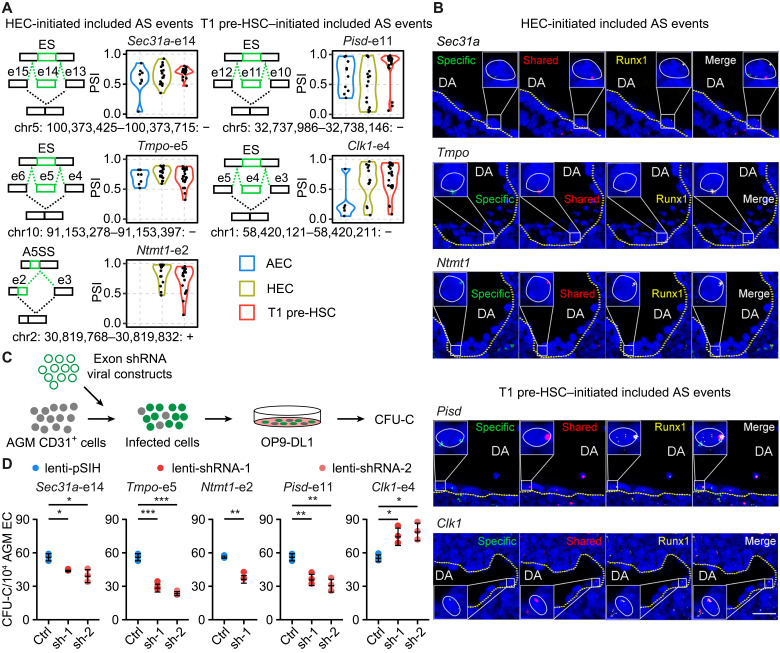
We next asked whether the HEC-initiated and T1 pre-HSC–initiated included AS events really existed in vivo. We randomly selected six HEC-initiated included AS events (from Sec31a, Tmpo, Ntmt1, Zfpl, Mpdu1, and Coro1a), which remained included in T1 pre-HSCs, and five T1 pre-HSC–initiated included AS events (from Pisd, Clk1, E130309D02Rik, Nkb1, and Ttll4) for detection by fluorescence in situ hybridization (FISH) with probes recognizing the specifically included and shared exon sequences, respectively (Fig. 4A, fig. S4A, and table S4A). We isolated the AGM region from E11 mouse embryos and used Runx1 antibodies before probing for the selected exons (Fig. 4B), as T1 pre-HSCs reside in the intra-aortic hematopoietic clusters marked by Runx1 expression (31, 32). We consistently detected the 11 inclusion events in Runx1-labeled hematopoietic cells (Fig. 4B and fig. S4B).
Fig. 4. HEC- and T1 pre-HSC–initiated included AS events were responsible for hemogenic function in vitro.
(A) The schematic illustration of AS event was plotted on the left, with the included exon shown in green. Included and excluded isoforms generated from the AS events were marked. The order of exon number is consistent with the strand direction. The genomic location of alternative spliced exon was labeled at the bottom. Violin plot on the right represents the PSI of indicated HEC- or T1 pre-HSC–initiated included AS events. ES, exon skipping; A5SS, alternative 5′ splice sites. (B) Simultaneous detection of RNA and protein by using probes against the specific and shared exons, as well as antibody against Runx1 at E11 AGM region. DA, dorsal aorta. Scale bar, 25 μm. (C) Schematic illustration of the lentivirus infection and colony-forming unit–C (CFU-C) assay strategy. (D) Relative CFU-C number of lentivirus-mediated knockdown of indicated alternative spliced exons in AGM CD31+ cells. (Data are represented as means ± SEM. Two-sided Wilcoxon rank sum test, *P < 0.05, **P < 0.01, and ***P < 0.001.)
We generated lentivirus expressing short hairpin RNAs (shRNAs) targeting the specifically included or shared exon sequences to knock down either isoforms containing the exon or all the isoforms in E11 AGM-derived CD31+ cells (Fig. 4C and table S4B), which include nearly all pre-HSCs, HSCs, and hematopoietic progenitor cells in addition to ECs. We first determined the lentivirus infection efficiency in CD31+ cells and found that it was more than 90% (fig. S4C). Consistently, the expression level of targeted isoforms was reduced by 50 to 90% (fig. S4, D and E, and table S4C). We then measured the colony-forming capacity of CD31+ cells and found that decreased expression of four of the specifically included exons (Sec31a, Pisd, Tmpo, and Ntmt1) led to significantly less colony formation compared to the vector controls (Fig. 4D), whereas repression of the shared exons from all the five genes consistently resulted in reduced colony formation capacity of CD31+ cells (fig. S4F). Notably, knockdown of the included or shared exons from the same gene has distinct effect, especially Clk1 (Fig. 4D and fig. S4F), suggesting that the included exon might play a specific role.
In addition, the specific inclusion of such exons would alter protein structures and functions to a different extent. For example, AS events from Ntmt1, Clk1, and Pisd could influence their methyltransferase, protein kinase, and phosphatidylserine decarboxylase–related domain, respectively (fig. S4G).
Srsf2 orchestrates the inclusion of hemogenic-specific included AS events
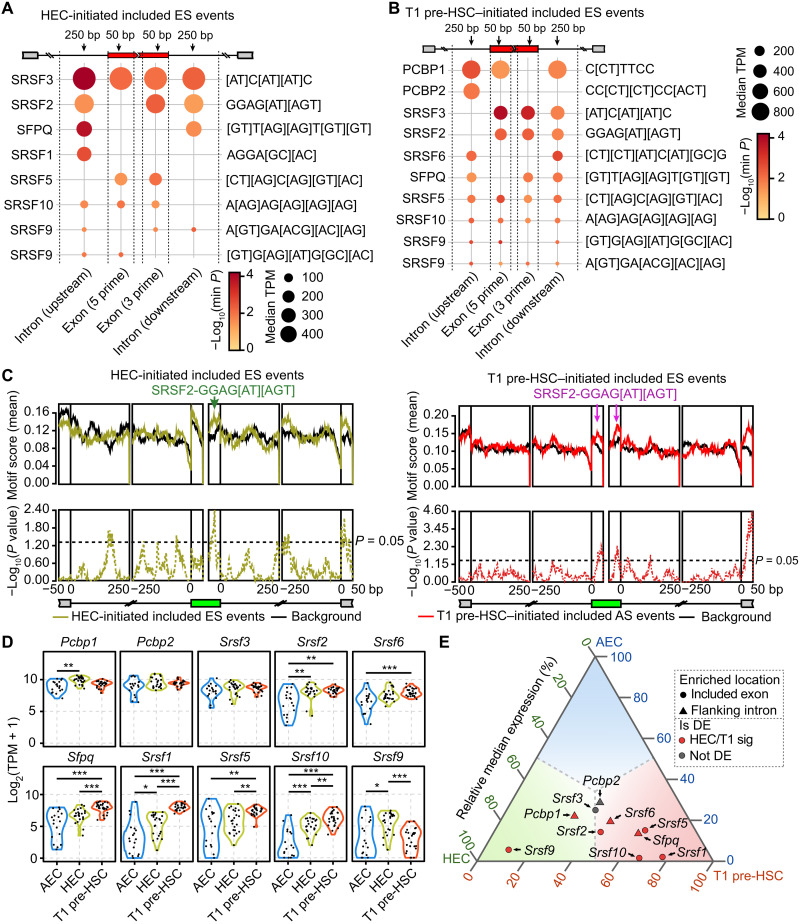
Having demonstrated the in vivo presence and in vitro functional importance of the selected hemogenic-specific included AS events, we next aimed to identify the splicing regulators responsible for them. We used an online web server named rMAPS2 (RNA Map Analysis and Plotting Server 2) (33) to identify discriminative cis-regulatory sequence motifs enriched in the cassette exons that are preferentially included in HECs or T1 pre-HSCs, as well as motifs in their flanking introns. These identified motifs corresponded to seven predicted RNA binding proteins (RBPs) for the HEC-specific included events: SRSF3, SRSF2, SFPQ, SRSF1, SRSF5, SRSF10, and SRSF9 (Fig. 5A and table S5A). Another nine RBPs were overrepresented for the T1 pre-HSC–specific included events: PCBP1, PCBP2, SRSF3, SRSF2, SRSF6, SFPQ, SRSF5, SRSF10, and SRSF9 (Fig. 5B and table S5A). The result that six of the RBPs (SRSF3, SRSF2, SFPQ, SRSF5, SRSF10, and SRSF9) were predicted from both HEC and T1 pre-HSC stages suggested a common regulatory effect of these splicing factors throughout the entire EHT process. In line with the inclusion of these AS events accompanied by hematopoietic specification, the accumulated expression of splicing factors Srsf2 (Fig. 5C), Sfpq (fig. S5A), Srsf5 (fig. S5B), and Srsf10 (fig. S5C) was greater in HEC and T1 pre-HSC stages compared with AEC (Fig. 5, D and E, and table S5B). Notably, the relative expression level of Srsf2, a splicing factor with a known pivotal role for HSC survival in fetal liver and bone marrow (34), as well as for T cell development (35), was higher in both HECs and T1 pre-HSCs compared with AECs (Fig. 5D). Moreover, Srsf2 is one of the most frequently mutated genes in myelodysplasia/myeloproliferative neoplasm overlap syndromes (36, 37). Thus, we selected Srsf2 for further investigation of its role in HSC emergence.
Fig. 5. Srsf2 regulated the inclusion of HEC- and T1 pre-HSC–initiated included AS events.
(A and B) Bubble plot showing the predicted splicing factors from unbiased motif analysis of HEC-initiated (A) and T1 pre-HSC–initiated (B) included ES events. Motif sequences of corresponding splicing factors were labeled on the right. The dot color represents the smallest P value in each enriched region, while the dot size indicates the median expression level of the splicing factors in HEC (A) and T1 pre-HSC (B). TPM, transcripts per million reads. (C) Positional distribution of Srsf2-binding motifs of HEC-initiated (left) and T1 pre-HSC–initiated (right) included AS events. Motif enrichment scores (top, solid line) and P values (bottom, dashed line) were plotted according to AS event positions. Arrows indicate peaks of enrichment for exons. (D) Violin plot representing the expression of indicated splicing factors during EHT. (Two-sided Wilcoxon rank sum test, *P < 0.05, **P < 0.01, and ***P < 0.001). (E) Ternary phase diagram showing the relative expression of splicing factors predicted from HEC- and T1 pre-HSC–initiated included ES events. HEC/T1 sig, HEC or T1 pre-HSC signature gene; Not DE, not differentially expressed.
Loss of Srsf2 from the endothelial stage disrupts EHT
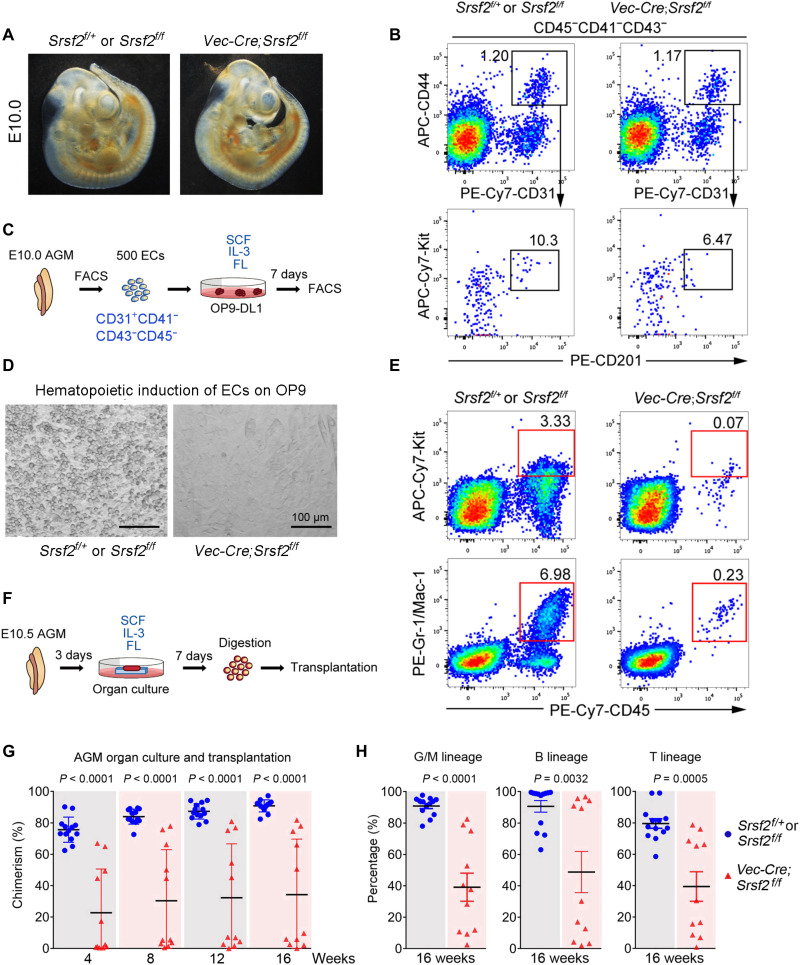
To determine whether Srsf2 is required for the process of EHT, we disrupted Srsf2 expression from the embryonic endothelial stage using Vec-Cre transgenic mice (fig. S6, A and B). As most of the mutant (Vec-cre;Srsf2f/f) embryos died from E11, we used morphologically normal E10.0 or E10.5 embryos for the subsequent experiments (Fig. 6A). By fluorescence-activated cell sorting (FACS) analysis, we witnessed a comparable constitution of CD44+ ECs at the E10.0 AGM region between mutant and control embryos. Notably, the immunohistochemistry on the AGM region from mutant embryos showed that the number of small intra-aortic clusters (less than five cells) was significantly reduced upon Srsf2 deficiency (fig. S6, C and D). In addition, the frequency of the immunophenotypic HECs (CD31+CD45−CD41−CD43−CD201+Kit+CD44+) in the mutant embryos was clearly decreased, featured by the obvious decrease in the expression of CD201 (Fig. 6B). Then, we assessed the influence of Srsf2 on the hemogenic capacity of ECs (CD31+CD41−CD43−CD45−) sorted from the E10 AGM region of these mice by measuring the number and type of hematopoietic cells generated following coculture with the stromal cells OP9-DL1 in vitro (Fig. 6C). We found that Srsf2-deficient ECs generated remarkably fewer hematopoietic cells than the ones from the littermate control embryos, evidenced by both morphological observation and FACS analysis (Fig. 6, D and E). These results together suggested that the hemogenic specification of ECs in the AGM region was at least partially blocked in the absence of Srsf2.
Fig. 6. Srsf2 was required for HSC emergence in AGM.
(A) Morphology of E10 Srsf2f/+ or Srsf2f/f (33 somite pairs) and Vec-Cre;Srsf2f/f (33 somite pairs) embryos. (B) Representative flow cytometry analysis of the frequencies of CD44+ ECs and immunophenotypic defined HECs in the E10 AGM of Srsf2f/+ or Srsf2f/f and Vec-Cre;Srsf2f/f embryos. APC, allophycocyanin; PE, phycoerythrin. (C) Schematic illustration of hematopoietic induction from ECs. IL-3, interleukin-3. FL, FLT-3 ligand. (D) Representative view of the hematopoietic cells (round) generated from 500 ECs after coculture with OP9-DL1 cells for 7 days. (E) Representative flow cytometry analysis of the frequencies of CD45+Kit+ hematopoietic progenitors and Gr1/Mac1+myeloid cells after OP9-DL1 coculture. (F) Schematic illustration of E10.5 AGM organ culture. (G) Donor chimerism in peripheral blood of recipients after transplantation of E10.5 AGM organ culture. Data are collected from 13 (Srsf2f/+ or Srsf2f/f) and 11 (Vec-Cre; Srsf2f/f) recipients. (Data are represented as means ± SEM.) (H) Donor chimerism of myeloid (Gr1+/Mac1+), B lymphoid (B220+), and T lymphoid (CD3+) cells repopulated by E10.5 AGM organ culture after 16 weeks of transplantation. (Data are represented as means ± SEM.)
We next used an organ culture and transplantation strategy to determine whether Srsf2 deletion would affect the ontogeny of HSCs in vivo. E10.5 AGM regions from Srsf2 mutant and control embryos (CD45.2/2) were isolated and cultured for 3 days in a growth factor cocktail to promote HSC formation. Then, the tissues were digested into single cells that were subsequently injected into the tail veins of lethally irradiated CD45.1/1 adult recipients (Fig. 6F). After 16 weeks, we euthanized the recipient mice and measured the degree of donor cell (CD45.2/2) chimerism and the relative frequencies of key hematopoietic cell populations. We found that the recipients of cells from littermate control embryos exhibited an average of 83.96% chimerism in the peripheral blood, while those from Srsf2 mutant embryos showed an average of only 30.93% chimerism (Fig. 6G), with this decrease in donor cell frequency affecting myeloid, B, and T cell lineages (Fig. 6H). Together, the splicing factor Srsf2 is required for the normal generation of HSCs in the AGM region, probably acting on the initial specification of HECs.
Srsf2 coordinates hemogenic-specific AS events
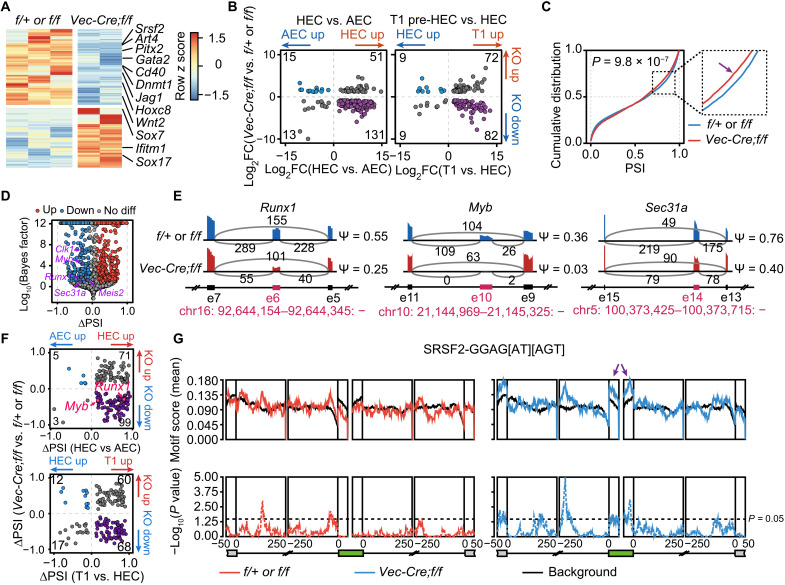
To understand the role of Srsf2 in hemogenic potential, we compared the transcriptomes of CD44+ ECs, involving both phenotypic AECs and HECs, from E10 AGM regions of Srsf2 mutant and control embryos. We identified 2026 genes with significantly different expression between Srsf2 mutant and control ECs, of which 1036 were decreased and 990 were increased in the mutants (Fig. 7A and table S6A). In ECs lacking Srsf2, the expression of several hematopoietic regulators was down-regulated including Gata2 (38), Dnmt1 (39), Cd40 (40), Jag1 (41), and Wnt2 (42) (Fig. 7A and table S6A). In contrast, endothelium-associated factors such as Sox7 (43) and Sox17 (44) were up-regulated (Fig. 7A and table S6A). Overall, the expression of many of the HEC or T1 pre-HSC signature genes (Fig. 2A) was lower under Srsf2 deficiency (Fig. 7B and table S6B). These results reflect a profound disruption to the gene expression programs required for EHT, accounting for the hematopoietic deficiency observed in Srsf2 mutant embryos and strongly evidencing the key role of this splicing regulator in embryonic hematopoiesis.
Fig. 7. Loss of Srsf2 disrupts gene programs required for HSC formation.
(A) Heatmap showing the differential expressed genes between Srsf2f/+ or Srsf2f/f (f/+ or f/f) and Vec-Cre;Srsf2f/f (Vec-Cre;f/f) cells. (B) Dot plot showing the expression comparison between HEC (left) or T1 pre-HSC (right) signature genes (x axis) and differentially expressed genes upon loss of Srsf2 (y axis). Number of genes in each quadrant was marked in the corner. Divergent expressed genes were labeled in purple or blue. KO, knockout; FC, fold change. (C) Cumulative distribution showing the distribution of PSI in f/+ or f/f and Vec-Cre;f/f cells. Kolmogorov-Smirnov test was used to determine the differences. (D) Volcano plot representing the differential AS events upon loss of Srsf2. Several indicated AS events were labeled in purple and marked with gene names. (E) Sashimi plots showing indicated ES AS events in f/+ or f/f and Vec-Cre;f/f cells. The alternative spliced exons were colored in red and plotted according to genomic locations. Numbers of exon-exon junction reads were marked above gray arcs. PSI of AS events was labeled on the right. The genomic location of alternative exon was labeled in red at the bottom. (F) Dot plot showing the PSI change of AS events during EHT (x axis) upon loss of Srsf2 (y axis). Number of AS events in each quadrant was marked in the corner. Divergent AS events were labeled in purple or blue. (G) Positional distribution of Srsf2-binding motifs of the down-regulated AS events in f/+ or f/f and Vec-Cre;f/f cells. Motif enrichment scores (top, solid line) and P values (bottom, dashed line) were plotted according to AS event positions. Arrows indicate peaks of enrichment for exons. The red and blue lines indicated the enrichment of up-regulated and down-regulated AS events upon loss of Srsf2, respectively.
We next went on to determine the molecular changes underpinning the profound effects of Srsf2 deficiency on the transcriptomes of ECs. We first quantified global changes in the inclusion of exons in Srsf2 mutant and control ECs and found that the PSI of certain AS events was slightly lower in Srsf2 mutants than controls, indicating reduced exon inclusion (Fig. 7C). This is consistent with the bias of SRSF2 toward exon recognition and inclusion (45, 46). Notably, among the down-regulated AS events, several were annotated to essential EHT regulators, including the transcription factors Runx1 and Myb and the vesicular transport COPII (coat protein complex II) component Sec31a (Fig. 7, D and E, and table S6C) whose alternative exon 14 affected hemogenic function in vitro (Fig. 4D). Moreover, the PSI of changed AS events during EHT was more decreased after Srsf2 knockout (KO) (Fig. 7F), including AS events from Runx1 and Myb. RNA motif enrichment analyses showed that the down-regulated AS events were significantly enriched with Srsf2 recognition sequences (GGAG[AU][AGU]), while the up-regulated AS events were not (Fig. 7G), suggesting a specific requirement for Srsf2 on the inclusion of its target AS events. Together, these results indicate that Srsf2 regulates the emergence of HSC during embryonic hematopoiesis, which is likely through defining the splicing patterns of essential EHT regulators.
DISCUSSION
The current interest in stem cell–based therapies has emphasized the importance of understanding how tissue-specific stem cells are specified during development. The hematopoietic system may be a paradigm for this. Research over recent decades has begun to clarify the cellular and molecular processes underlining the initial emergence of HSCs (47, 48). However, our current knowledge of gene expression and function is mainly based on the observations at the gene level. This is problematic, because it is well established that transcribed mRNAs commonly undergo AS to generate distinct transcript isoforms and ultimately yield protein products that differ in their functional properties; therefore, transcriptome analyses that do not consider transcriptional isoforms will miss important information and paint an incomplete or misleading picture of the complex process of hematopoietic development. In the current study, we described the genome-wide isoform-based transcriptome landscape at single-cell resolution, which enabled the identification of cell type–specific isoforms, AS events, and a previously unidentified regulatory mechanism of HSC formation.
Our analysis of transcriptional isoform expression during EHT identified numerous previously undetected AS switches that occurred in the absence of appreciable changes at the gene expression level, providing evidence of an additional layer of regulation of HSC formation in the embryo. For example, we illustrated that AS events in the products of genes Sec31a, Pisd, Tmpo, and Ntmt1 had a positive hemogenic effect in in vitro functional assays. Among these, Sec31a encodes a core component of the COPII complex, trafficking newly synthesized membrane and secretory proteins from the endoplasmic reticulum to Golgi (49). Because the impaired hematopoiesis seen in congenital dyserythropoietic anemia type II and combined deficiency of coagulation factors V and VIII (F5F8D) results from defects in the COPII transport system (50), the involvement of Sec31a, and especially the Sec31a-202 isoform, in embryonic hematopoiesis should be further explored. Moreover, genes involved in other cellular processes, including Pisd, which is associated with mitochondrial functions (51), and Tmpo, which is related to cell proliferation and tumorigenesis (52), might also contribute to the embryonic hematopoiesis. Although the precise expression and roles of these genes and their specific isoforms need to be further defined in hematopoietic context, there is already evidence of the isoform-specific functions in different systems. As to the essential regulator of HSC formation, Runx1, we detected two isoforms, Runx1-201 and Runx1-207, respectively, corresponding to protein isoform Runx1c and a noncoding transcript, in HECs and T1 pre-HSCs, indicating the importance of Runx1c for pre-HSC emergence. Although data from an in vitro mouse pluripotent stem cell system indicate that the protein levels of the Runx1b isoform need to be restricted for the successful completion of EHT (53), the exact roles of Runx1c and Runx1b isoforms during AGM EHT need to be further investigated in vivo. There is already evidence from other systems that specific AS isoforms can have key regulatory roles: the Rbfox1 isoforms, which have different cellular localizations and are associated with distinct gene expression programs in neuronal differentiation (54); the exon 2–skipping MKK7 isoform, which is specifically required for T cell activation as it includes an additional site for c-Jun N-terminal kinase docking (55); and the titin isoforms, whose precise ratios determine the passive tension of cardiomyocytes and the stiffness of the myocardium wall (56). Together, with our own findings, these studies provide further evidence of the need for a paradigm shift from the current “gene centric” to an “isoform centric” approach to transcriptional analyses, making the study of gene isoforms an important aspect of biological networks. Accordingly, acquiring knowledge of transcript diversity during EHT is a necessary first step for understanding the isoform-specific functions of key proteins and their regulators during embryonic hematopoiesis.
The role of AS in expanding transcriptomic diversity is already known. Using full-length single-cell transcriptome data, we were able to explore the distribution and variation of AS events at the single-cell level using the modality strategy. The representation of AS modalities across different cell populations was remarkably consistent, with nearly 70% of AS events being unimodal (~26% inclusion, ~31% exclusion, and ~17% middle). A distinct property of included AS events is high evolutionary conservation, possibly to endow cells with specific functions. As to the HEC- or T1 pre-HSC–specific included events, almost half were switched from other modalities in their adjacent early developmental stages. This gradual modality switch occurred concomitantly with the stepwise cell fate commitment from AEC to HEC and then to T1 pre-HSC. A major cluster of the hemogenic-specific included exons were enriched for genes involved in the posttranscriptional regulation of gene expression, indicating a previously unrecognized importance of RNA regulation in this process.
Notably, a large subset of HEC- and pre-HSC–specific included AS events were predicted to be under the control of Srsf2, one of the founding members of the serine/arginine-rich protein family of splicing factors that is involved in both constitutive and AS. The potential close relationship between Srsf2 and the hematopoietic system is evidenced by the frequent mutation in Srsf2 gene in myelodysplastic syndromes and chronic myelomonocytic leukemia (36, 37). Mechanicistic investigation of its role in leukemia indicated that mutations altered the RNA-binding characteristics of Srsf2 and caused extensive mis-splicing of hundreds of genes (57). Here, we showed that Srsf2 was essential for the onset of hematopoiesis, as Srsf2 deficiency in endothelial cells resulted in failure to generate HSCs from endothelial precursors. By comparison, Srsf2 deletion from mature HSCs led to enhanced apoptosis and decreased HSPCs (34), suggesting the sustainable requirement of Srsf2 from HSC emergence to maintenance. Loss of Srsf2 changed the splicing pattern of two master EHT regulators, Runx1 and Myb, in HECs, the transient population between endothelial cells and specified HSCs. As to Runx1, the Srsf2-orcherasted alternative usage of exon 6 led to the generation of Runx1 isoforms with (canonical Runx1c) or without exon 6 (Runx1cEx6−), which have been demonstrated to have distinct roles in the maintenance of HSC pool size (58). Moreover, the expression of the two best known Myb isoforms, p75 and p89, of which the larger isoform is encoded by a transcript containing the additional exon 10 (59), was also disturbed by loss of Srsf2. The inclusion of exon 10 equips Myb p89 with additional amino acids involved in protein-protein interaction (59), possibly rendering it specifically required for HSC emergence. Yet, the exact function of these isoforms needs to be further investigated in the context of embryonic hematopoiesis.
Characterizing the landscape of AS profiles during hematopoietic ontogeny allowed us to explore previously unidentified regulatory mechanisms that complement the conventional epigenetic and transcriptional processes at work in EHT. These findings also contribute to the functional annotation of the genome by indicating the necessity for the study of alternatively spliced isoforms during transcriptional analysis. Regulators that govern the selection of cell type–specific AS events may have dominant functions in driving the completion of these cellular processes. This knowledge will be essential for the continued effort to validate the existence and molecular significance of transcript diversity across all biological systems.
MATERIALS AND METHODS
Mice
Mice were bred in specific pathogen–free condition, and mice experiments were approved by the ethics committee of the affiliated hospital of Academy of Military Medical Sciences. Vec-Cre mice were purchased from the Jackson Laboratory. Srsr2fl/fl (B6.129S4-SRsf2tm1Xdfu/J, the Jackson Laboratory) mice on c57BL/6 background (CD45.2/2) were provided by X.-D. Fu. Vec-Cre male mice were crossed with female Srsr2fl/fl mice to obtain conditional Srsf2 KO embryos and littermate controls at E10.0 (31 to 35 somite pairs) or E10.5 (36 to 40 somite pairs).
Flow cytometry
Cells were sorted and analyzed by flow cytometers FACSAria 2 and Calibur (BD Biosciences), and the data were analyzed using FlowJo software (Tree Star). 7-aminoactinomycin D (eBioscience) was used to exclude dead cells. For index sorting, the FACSDiva 8 “index sorting” function was activated, and sorting was performed in single-cell mode.
Preparation of single-cell RNA-seq library
FACS-enriched cells were kept on ice until lysed and reverse-transcribed. Morphologically deformed cells were discarded. Single cells were rinsed in phosphate-buffered saline–bovine serum albumin and manually transferred into cell lysis buffer with a mouth pipette; 0.05 μl of 1:200,000 dilution of External RNA Controls Consortium (ERCC) RNA spike-in Mix1 (Ambion) was added to lysis buffer per reaction. The cDNA libraries from single cells were generated as described previously (7). Fifty to 200 ng of amplified single-cell or 10-cell-pool cDNAs were sonicated to ~250–base pair fragments by the Covaris S2 system, and libraries were generated using a NEBNext Ultra DNA Prep Kit for Illumina (New England Biolabs, E7370) following the manufacturer’s protocol.
Single-cell Nanopore-seq library preparation and sequencing
After digestion, single cells were placed into 1.35-μl lysis buffer by mouth pipette. Then, the cDNA libraries from single cells were generated and amplified as described using a method named SCAN-seq (60).
Colony-forming unit assay
Lentivirus-infected CD31+ cells were seeded onto OP9-DL1 stromal cells and cocultured 3 days for subsequent colony-forming unit–C (CFU-C) assay. These cells were collected and plated in a 35-mm petri dish containing 2 ml of methylcellulose-based medium with recombinant cytokines (MethoCult). The cells were incubated at 37°C with 5% CO2 for 7 days for colony quantification.
Hematopoietic induction on OP9
E10.0 AGM was dissected into single-cell suspensions as routine aseptic operation. After antibody staining, 500 FACS-sorted CD31+CD41−CD43−CD45− endothelial cells were cultured on mouse OP9 stromal cells and supplemented with hematopoietic cytokines [stem cell factor (SCF), 50 ng/ml; interleukin-3 (IL-3), 10 ng/ml; and FLT-3 ligand (FL), 10 ng/ml]. After culture for 7 days, cells were harvested by mechanical pipetting for flow cytometry analysis.
Organ culture and transplantation
AGMs were isolated from the caudal of E10.5 embryos (four female mice) and cultured using an ex vivo organ culture system as previously described (61). The medium used for the AGM culture contained M5300 (STEMCELL Technologies), 1% penicillin/streptomycin (Gibco), 10−6 M hydrocortisone (STEMCELL Technologies), and cytokines cocktail (SCF, 50 ng/ml; IL-3, 10 ng/ml; and Flt3 ligand, 10 ng/ml). After being cultured for 3 days, AGMs were dissociated in collagenase for transplantation. Either 0.5 or 1 embryo equivalent (ee) of donor mice was transplanted into one recipient mouse (one AGM from one donor mouse for one or two recipient mice equally). The ee of KO and control mice in each experiment was consistent. Eight- to 12-week-old female C57BL/6 (CD45.1/1) mice were exposed to a dose of 4.5-gray γ-irradiation (60Co) twice with a 2-hour interval. AGM cells (CD45.2/2), together with 2 × 104 nucleated bone marrow cells (CD45.1/2), were injected into irradiated adult recipients via the tail vein. Peripheral blood samples from recipients were analyzed to assess donor chimerism every 4 weeks until 16 weeks after transplantation. Peripheral blood samples from recipients were analyzed to assess donor chimerism every 4 weeks until 16 weeks after transplantation. Red blood cells (RBCs) were lysed with RBC lysis buffer before antibody staining. The recipients demonstrating ≥5% donor-derived chimerism in peripheral blood were counted as successfully reconstituted (11).
DNA extraction and genotyping
For adult mouse or mouse embryo genotyping, the mouse tails or embryonic tails were separated and digested. DNA was briefly centrifuged and isolated as a template for polymerase chain reaction (PCR). PCR genotyping for Srsf2 mice was performed on the mouse tail DNA using a cocktail of two primers (forward: GGTTATTTGGCCAAGAATCAC; reverse: CCATGGACCGATGGACTGAG). Thermal cycling was carried out as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s; primer annealing at 60°C for 30 s, chain elongation at 72°C for 90 s, and final extension at 72°C for 10 min. PCR genotyping for Vec-Cre mice was performed using a cocktail of two primers (forward: CCAGGCTGACCAAGCTGAG; reverse: CCTGGCGATCCCTGAACA). Thermal cycling was carried out as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, primer annealing at 57°C for 30s, chain elongation at 72°C for 90 s, and final extension at 72°C for 10 min.
RNA FISH
E11 mouse embryos were isolated, fixed with 10% neutral buffered formalin for 24 hours at 4°C, embedded in paraffin, and sectioned at 5 to 6 μm with Leica RM2235. Slides were baked at 60°C for 1 hour and deparaffinized with xylene followed by 100% alcohol for two rounds and then subjected to the manufacturer’s instruction provided with the BaseScope Duplex Detection Reagent Kit (ACD, a Bio-Techne brand). Briefly, slides were incubated with hydrogen peroxide for 10 min before boiled in RNAscope 1× Target Retrieval Reagent at 95 to ~99°C for 15 min. After target retrieval, the sections were treated with RNAscope Protease IV and incubated at 40°C for 30 min. The prepared sections were then hybridized with corresponding probes (each specific probe was mixed with Runx1 probe, and the specific probes and Runx1 probe were labeled by different dyes), and 12 steps of signal amplification treatment with BaseScope Duplex Amp 1 to 12 reagents and 2 steps of dyeing. After all the hybridization and sequential amplification, the slides were stained with 4′,6-diamidino-2-phenylindole. Images were collected with a fluorescence microscope (Zeiss Axio Imager M2).
Lentivirus infection
For lentivirus production, recombination lentiviruses expressing shRNAs against T1 pre-HSC specific isoforms or empty vector were produced using pSIH-based constructs, and the lentivirus packaging was performed using the pPACKH1 Lentiviral Vector Packaging Kit (LV500A-1, SBI System Biosciences) according to the manufacturer’s instructions. The virus particles were collected and condensed using the PEG-it Virus Precipitation Solution (SBI System Biosciences).
Magnetic-activated cell sorting–sorted AGM CD31+ cells were first cultured in 24-well plate for 12 hours before lentivirus infection and then were infected with lentivirus at a multiplicity of infection of 100 in the presence of polybrene (8 μg/ml) for 6 hours.
Quantitative PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. The RNA was quantified by absorbance at 260 nm. cDNA was synthesized by Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Invitrogen) from total RNA. Oligo(dT) 18 primers were used as the reverse transcription (RT) primers for reverse transcription of mRNAs. Quantitative real-time PCR was carried out using LightCycler 480 Instrument II (Roche) in triplicate. The data were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA expression.
RNA-seq data processing
All single-cell RNA-seq data were firstly trimmed for adaptor sequences and low-quality bases using trimmomatic (version 0.36) (62) with parameters “LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 HEADCROP:20 CROP:101 MINLEN:101.” Trimmed reads were mapped to GENCODE (version M22) mouse genome (mm10) with STAR (version 2.7.3a) (63) using parameters “--outSAMattributes All --outSAMtype BAM SortedByCoordinate --quantMode TranscriptomeSAM GeneCounts --twopassMode Basic.” Then, SAMtools (version 1.9) (64) was used to sort and index aligned BAM files. Isoform-level expression was quantified as transcripts per million reads (TPM) with Salmon (version 1.1.0) (65) using aligned transcriptome BAM files from STAR output. R package “tximport” (version 1.16.1) (66) was used to map transcripts to gene-level expression.
Single-cell Nanopore-seq processing
Guppy (version 3.5.2) was used for basecalling and for generating the FASTQ data from electric signals. Then, the nanopack software (https://github.com/wdecoster/nanopack) were used to perform quality control. After that, all sequenced reads were mapped to GENCODE (version M22) mouse genome (mm10) with minimap2 (version 2.18-r1015) (67) with default parameters. Isoform-level expression was firstly quantified as counts with Salmon (version 1.1.0) (65) and then normalized as reads per transcript per 10,000 reads.
Transcriptional diversity analysis
Genes and isoforms with TPM > 1 in at least one cell were kept for further analysis. The UMAP analysis was applied to isoform and gene levels for dimensional reduction and clustering with R package “Seurat” (version 3.1.4) (68) using normalized value TPM as input. Top 10,000 features with high variation were kept for dimensional reduction and clustering analysis. Genes with more than one isoform expressed (TPM > 1) in one cell were consider as “multi-isoform gene.” In Fig. 1E, a multi-isoform gene detected in at least five cells in a stage was regarded as a multiple type in this stage. HEC-initiated multi-isoform genes were defined as genes that do not express any or express only one isoform at AEC stage but express multiple isoforms in both the HEC and T1 pre-HSC stages. T1 pre-HSC–initiated multi-isoform genes were defined as genes that do not express any or express only one isoform at the AEC and HEC stages but express multiple isoforms in the T1 pre-HSC stage. R package “ggalluvial” (version 0.11.3) was used to draw a Sankey diagram.
Differential expression analysis
A gene/isoform expressed in at least five cells was remained for differential analysis. Two-tailed Wilcoxon rank sum test was used to estimate the statistical significance of differential expressed isoforms and genes between neighboring stages. Log2-transformed fold change of a gene was calculated as follows
where Ex and Ey represent the expression values of stages x and y, while c is a minimum constant (0.01) set to avoid infinite value.
Differential expressed genes and isoforms between two stages were identified with Benjamini-Hochberg correction P value < 0.05 as the cutoff, while nondifferential expressed genes were calculated with false discovery rate > 0.1 as the cutoff. Differential expressed genes or isoforms with median TPM > 1 in at least one of the two stages were kept for further analysis.
HEC and T1 pre-HSC signature genes in fig. S2A were defined as up-regulated genes in HECs and T1 pre-HSCs compared with adjacent early developmental stages. Overlap genes between HEC and T1 pre-HSC signature genes were regarded as HEC signature genes. Signature isoforms in Fig. 2A were obtained in a similar way as signature genes expect that the differentially expressed isoforms at only the isoform level but not the gene level were kept. R package “ggtern” (version 3.3.0) was used to draw a ternary phase diagram.
AS analysis
PSI (Ψ) of alternative spliced events were quantified with filtered and sorted BAM files using MISO (version 0.5.4) (25) and MAJIQ (version 2.0) (27) with default parameters. The minimum counts of junction reads were set to 5. We developed Perl scripts to map AS events to genes with exon location. Meanwhile, we developed an R script to map AS event IDs from MISO and MAJIQ together on the basis of the alternative spliced exon location. Anchor (version 1.1.1) (26) was used to estimate the modalities of AS events per stage. Only the AS events observed in at least five cells per stage were considered. The modalities of AS events were assigned according to the maximum values of log2 Bayes factor with a cutoff of 1.
Two-tailed Wilcoxon rank sum test was used to estimate the statistical significance of differential AS events between AEC and HEC stages. If the PSI of AS events was detected within less than five cells in one of the two compared stages, then the PSI of the AS events in the stage was set to 0 before testing.
AS events with modality change from nonincluded to included modality from AEC to HEC and remained included in T1 pre-HSC were defined as HEC-initiated included AS events. AS events with nonincluded modalities in AEC and HEC but changed to included modality in T1 pre-HSC were defined as T1 pre-HSC–initiated included events.
Motif enrichment, GSEA, and functional enrichment analysis
Motif enrichment analysis of alternative spliced exons was performed with ES events using rMAPS2 (version 2.2.0) (33) with default parameters. Eighty-one human RBPs in rMAPS2 were transformed into mouse homolog genes and then used for motif enrichment (table S5C). Motif enrichment in Fig. 5 (A and B) was performed using HEC- and T1 pre-HSC–initiated included ES events against with background ES events. The nonmultimodal ES events with no modality change during EHT and the absolute value of δPSI (differences between the median PSI of two stages) less than 0.05 were selected as background events. The significant enriched RBPs of HEC- and T1 pre-HSC–initiated included ES events were determined with following strategies: The minimum P value in at least one of the four regions should be less than 0.05, and the maximum mean motif enrichment score in at least one of the four regions should be more than 0.1. Only the smallest P value in an individual enriched region of each RBP was used to plot the bubble plot in Fig. 5 (A and B).
GSEA was performed with neighboring stages during the EHT process by using GSEA software (http://software.broadinstitute.org/gsea/index.jsp) (69) against Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
The Metascape online web server (70) was used to perform GO and pathway enrichment analysis with the “Express Analysis” option. Top 10 enriched terms were selected for enrichment network visualization. Cytoscape (version 3.7.1) (71) was used to modify and visualize the network of enriched terms. To perform enrichment analysis of isoform with protein coding abilities, we first mapped ensembl isoform IDs to ensembl protein IDs and then input them into Metascape. At the AS event level, we only selected the AS events that fall into the CDS region and then mapped AS event IDs to ensembl gene IDs before functional enrichment analysis.
Srsf2 KO analysis
All bulk RNA-seq data were processed and aligned with the same strategy as single-cell RNA-seq data. Then, gatk MarkDuplicates (72) was used to remove PCR duplicates. Gene level expression was quantified as raw counts and TPM with salmon. R package “edgeR” (version 3.26.8) (73) was applied to determine the statistical significance between f/+ or f/f and Vec-Cre;f/f samples. Genes with P value < 0.05 and absolute value of logFC more than 1 were identified as differential expressed gene. R package “pheatmap” (version 1.0.12) was used to draw a heatmap.
MISO (version 0.5.4) (25) was used to identify differential AS events between f/+ or f/f and Vec-Cre;f/f samples. AS events with Bayes factors >5 and absolute value of δPSI more than 0.1 were identified as differential. Kolmogorov-Smirnov test was used to determine the significance of PSI distribution between f/+ or f/f and Vec-Cre;f/f samples. Motif enrichment analysis of Srsf2 was performed using rMAPS2 with up-regulated and down-regulated AS events between f/+ or f/f and Vec-Cre;f/f samples against background AS events. AS events with Bayes factors <2 and absolute value of δPSI less than 0.05 were selected as background events when performing motif enrichment analysis. Sashimi plots and peak distribution of indicated AS events were visualized with Integrative Genomics Viewer (version 2.3.83) (74).
Statistics
No statistical methods were used to predetermine sample size. In general, chimerism comparisons and CFU-C number comparisons were performed using two-sided Wilcoxon rank sum test. Statistical analysis was performed using R.
Acknowledgments
We thank Y. Si for preparing laboratory consumables.
Funding: This work was supported by the National Key Research and Development Program of China (2019YFA0801800, 2019YFA0111700, 2017YFA0103401, 2016YFA0100601, 2020YFA0112402, 2019YFA0110201, and 2017YFA0106000); the National Science Foundation of China (82022001, 81530007, 81970103, 31725013, 81890991, 31871173, 31930054, 81600077, 82070109 and 81770104); CAMS Innovation Fund for Medical Sciences (2021-I2M-1-019 and 2021-I2M-1-040); the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S347); the Key Research and Development Program of Guangdong Province (2019B020234002); the CAMS Young Talents Award Program (2018RC310013); the CAMS (2017-I2M-B&R-04); grant from Medical Epigenetics Research Center, CAMS (2017PT31035); and Basic and Applied Basic Research Fund of Guangdong Province (2019A1515010784 and 2019A1515110701).
Author contributions: J.Y., B.L., D.W., and Y.Lan conceived and supervised the project. J.Z. performed single-cell RNA-seq with help from S.H., L.Z., and C.W.; P.Z. and Y.Li performed the pre-HSC– and HSC-related experiments with help from W.T.; P.T. performed the bioinformatics analysis with help from Y.Huo, Y. Hu, and T.C.; F.W. and Y.R. performed FISH and shRNA construct with help from Y.M. and X.W.; S.L. performed IHC experiment with the help from C.N.; and F.W., P.T., and Y.Lan wrote the manuscript with help from J.Y. and B.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The accession number for the RNA-seq data reported in this paper is GEO: GSE185555. All the data used in this study are available at Zenodo (https://doi.org/10.5281/zenodo.5463924). The codes for data processing and visualization were uploaded to Zenodo (https://doi.org/10.5281/zenodo.5706760).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Medvinsky A., Dzierzak E., Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906 (1996). [DOI] [PubMed] [Google Scholar]
- 2.de Bruijn M. F. T. R., Ma X., Robin C., Ottersbach K., Sanchez M.-J., Dzierzak E., Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 16, 673–683 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Rybtsov S., Sobiesiak M., Taoudi S., Souilhol C., Senserrich J., Liakhovitskaia A., Ivanovs A., Frampton J., Zhao S., Medvinsky A., Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 208, 1305–1315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanovs A., Rybtsov S., Welch L., Anderson R. A., Turner M. L., Medvinsky A., Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J. Exp. Med. 208, 2417–2427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zovein A. C., Hofmann J. J., Lynch M., French W. J., Turlo K. A., Yang Y., Becker M. S., Zanetta L., Dejana E., Gasson J. C., Tallquist M. D., Iruela-Arispe M. L., Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3, 625–636 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou S., Li Z., Zheng X., Gao Y., Dong J., Ni Y., Wang X., Li Y., Ding X., Chang Z., Li S., Hu Y., Fan X., Hou Y., Wen L., Liu B., Tang F., Lan Y., Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 30, 376–392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou F., Li X., Wang W., Zhu P., Zhou J., He W., Ding M., Xiong F., Zheng X., Li Z., Ni Y., Mu X., Wen L., Cheng T., Lan Y., Yuan W., Tang F., Liu B., Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487–492 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A., Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R., AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Tsai F. Y., Keller G., Kuo F. C., Weiss M., Chen J., Rosenblatt M., Alt F. W., Orkin S. H., An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Zhou J., Xu J., Zhang L., Liu S., Ma Y., Wen X., Hao J., Li Z., Ni Y., Li X., Zhou F., Li Q., Wang F., Wang X., Si Y., Zhang P., Liu C., Bartolomei M., Tang F., Liu B., Yu J., Lan Y., Combined single-cell profiling of lncRNAs and functional screening reveals that H19 is pivotal for embryonic hematopoietic stem cell development. Cell Stem Cell 24, 285–298.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Zhang X., Chen M. H., Wu X., Kodani A., Fan J., Doan R., Ozawa M., Ma J., Yoshida N., Reiter J. F., Black D. L., Kharchenko P. V., Sharp P. A., Walsh C. A., Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166, 1147–1162.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M., Ergin V., Lin L., Stork C., Chen L., Zheng S., Axonogenesis is coordinated by neuron-specific alternative splicing programming and splicing regulator PTBP2. Neuron 101, 690–706.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weyn-Vanhentenryck S. M., Feng H., Ustianenko D., Duffié R., Yan Q., Jacko M., Martinez J. C., Goodwin M., Zhang X., Hengst U., Lomvardas S., Swanson M. S., Zhang C., Precise temporal regulation of alternative splicing during neural development. Nat. Commun. 9, 2189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labeit S., Kolmerer B., Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science 270, 293–296 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Guo W., Schafer S., Greaser M. L., Radke M. H., Liss M., Govindarajan T., Maatz H., Schulz H., Li S., Parrish A. M., Dauksaite V., Vakeel P., Klaassen S., Gerull B., Thierfelder L., Regitz-Zagrosek V., Hacker T. A., Saupe K. W., Dec G. W., Ellinor P. T., MacRae C. A., Spallek B., Fischer R., Perrot A., Özcelik C., Saar K., Hubner N., Gotthardt M., RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 18, 766–773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elizalde M., Urtasun R., Azkona M., Latasa M. U., Goñi S., García-Irigoyen O., Uriarte I., Segura V., Collantes M., di Scala M., Lujambio A., Prieto J., Ávila M. A., Berasain C., Splicing regulator SLU7 is essential for maintaining liver homeostasis. J. Clin. Invest. 124, 2909–2920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R. K., Xia Z., Bland C. S., Kalsotra A., Scavuzzo M. A., Curk T., Ule J., Li W., Cooper T. A., Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol. Cell 55, 592–603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Kostadima M., Martens J. H. A., Canu G., Garcia S. P., Turro E., Downes K., Macaulay I. C., Bielczyk-Maczynska E., Coe S., Farrow S., Poudel P., Burden F., Jansen S. B. G., Astle W. J., Attwood A., Bariana T., de Bono B., Breschi A., Chambers J. C.; BRIDGE Consortium, Choudry F. A., Clarke L., Coupland P., van der Ent M., Erber W. N., Jansen J. H., Favier R., Fenech M. E., Foad N., Freson K., van Geet C., Gomez K., Guigo R., Hampshire D., Kelly A. M., Kerstens H. H. D., Kooner J. S., Laffan M., Lentaigne C., Labalette C., Martin T., Meacham S., Mumford A., Nürnberg S. T., Palumbo E., van der Reijden B. A., Richardson D., Sammut S. J., Slodkowicz G., Tamuri A. U., Vasquez L., Voss K., Watt S., Westbury S., Flicek P., Loos R., Goldman N., Bertone P., Read R. J., Richardson S., Cvejic A., Soranzo N., Ouwehand W. H., Stunnenberg H. G., Frontini M., Rendon A., Transcriptional diversity during lineage commitment of human blood progenitors. Science 345, 1251033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scotti M. M., Swanson M. S., RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Y., He J., Bai Z., Li Z., Gong Y., Liu C., Ni Y., du J., Ma C., Bian L., Lan Y., Liu B., Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 29, 881–894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B. B., Siddiqui A., Lao K., Surani M. A., mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Dong C., Mao Y., Tempel W., Qin S., Li L., Loppnau P., Huang R., Min J., Structural basis for substrate recognition by the human N-terminal methyltransferase 1. Genes Dev. 29, 2343–2348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., Xu Z., Kong Y., The tubby-like proteins kingdom in animals and plants. Gene 642, 16–25 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Katz Y., Wang E. T., Airoldi E. M., Burge C. B., Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Botvinnik O. B., Lovci M. T., Kakaradov B., Liu P., Xu J. L., Yeo G. W., Single-cell alternative splicing analysis with expedition reveals splicing dynamics during neuron differentiation. Mol. Cell 67, 148–161.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaquero-Garcia J., Barrera A., Gazzara M. R., González-Vallinas J., Lahens N. F., Hogenesch J. B., Lynch K. W., Barash Y., A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife 5, e11752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortes M., Chen M. J., Stachura D. L., Liu S. Y., Kwan W., Wright F., Vo L. T., Theodore L. N., Esain V., Frost I. M., Schlaeger T. M., Goessling W., Daley G. Q., North T. E., Developmental vitamin D availability impacts hematopoietic stem cell production. Cell Rep. 17, 458–468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Q., Yang X., Lv J., Zhang J., Xia B., Kim J.-D., Wang R., Xiong F., Meng S., Clements T. P., Tandon B., Wagner D. S., Diaz M. F., Wenzel P. L., Miller Y. I., Traver D., Cooke J. P., Li W., Zon L. I., Chen K., Bai Y., Fang L., AIBP-mediated cholesterol efflux instructs hematopoietic stem and progenitor cell fate. Science 363, 1085–1088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao P., Chen C., Howell E. D., Li Y., Tober J., Uzun Y., He B., Gao L., Zhu Q., Siekmann A. F., Speck N. A., Tan K., Transcriptional regulatory network controlling the ontogeny of hematopoietic stem cells. Genes Dev. 34, 950–964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boisset J. C., Clapes T., Klaus A., Papazian N., Onderwater J., Mommaas-Kienhuis M., Cupedo T., Robin C., Progressive maturation toward hematopoietic stem cells in the mouse embryo aorta. Blood 125, 465–469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron C. S., Kester L., Klaus A., Boisset J. C., Thambyrajah R., Yvernogeau L., Kouskoff V., Lacaud G., van Oudenaarden A., Robin C., Single-cell transcriptomics reveal the dynamic of haematopoietic stem cell production in the aorta. Nat. Commun. 9, 2517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang J. Y., Jung S., Kook T. L., Rouchka E. C., Bok J., Park J. W., rMAPS2: An update of the RNA map analysis and plotting server for alternative splicing regulation. Nucleic Acids Res. 48, W300–W306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komeno Y., Huang Y.-J., Qiu J., Lin L., Xu Y.-J., Zhou Y., Chen L., Monterroza D. D., Li H., DeKelver R. C., Yan M., Fu X.-D., Zhang D.-E., SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol. Cell. Biol. 35, 3071–3082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H. Y., Xu X., Ding J. H., Bermingham J. R. Jr., Fu X. D., SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7, 331–342 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Arbab Jafari P., Ayatollahi H., Sadeghi R., Sheikhi M., Asghari A., Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: A meta-analysis. Hematology 23, 778–784 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Rahman M. A., Lin K. T., Bradley R. K., Abdel-Wahab O., Krainer A. R., Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes Dev. 34, 413–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim K. C., Hosoya T., Brandt W., Ku C.-J., Hosoya-Ohmura S., Camper S. A., Yamamoto M., Engel J. D., Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J. Clin. Invest. 122, 3705–3717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trowbridge J. J., Snow J. W., Kim J., Orkin S. H., DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5, 442–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solanilla A., Déchanet J., el Andaloussi A., Dupouy M., Godard F.ṃ., Chabrol J., Charbord P., Reiffers J., Nurden A. T., Weksler B., Moreau J.-F., Ripoche J., CD40-ligand stimulates myelopoiesis by regulating flt3-ligand and thrombopoietin production in bone marrow stromal cells. Blood 95, 3758–3764 (2000). [PubMed] [Google Scholar]
- 41.Gama-Norton L., Ferrando E., Ruiz-Herguido C., Liu Z., Guiu J., Islam A. B. M. M. K., Lee S.-U., Yan M., Guidos C. J., López-Bigas N., Maeda T., Espinosa L., Kopan R., Bigas A., Notch signal strength controls cell fate in the haemogenic endothelium. Nat. Commun. 6, 8510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Gilner J. B., Bautch V. L., Wang D.-Z., Wainwright B. J., Kirby S. L., Patterson C., Wnt2 coordinates the commitment of mesoderm to hematopoietic, endothelial, and cardiac lineages in embryoid bodies. J. Biol. Chem. 282, 782–791 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Costa G., Mazan A., Gandillet A., Pearson S., Lacaud G., Kouskoff V., SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development 139, 1587–1598 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Serrano A. G., Gandillet A., Pearson S., Lacaud G., Kouskoff V., Contrasting effects of Sox17- and Sox18-sustained expression at the onset of blood specification. Blood 115, 3895–3898 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Liu H. X., Chew S. L., Cartegni L., Zhang M. Q., Krainer A. R., Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20, 1063–1071 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zahler A. M., Damgaard C. K., Kjems J., Caputi M., SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279, 10077–10084 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Gritz E., Hirschi K. K., Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 73, 1547–1567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swiers G., Rode C., Azzoni E., de Bruijn M. F. T. R., A short history of hemogenic endothelium. Blood Cells Mol. Dis. 51, 206–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khoriaty R., Vasievich M. P., Ginsburg D., The COPII pathway and hematologic disease. Blood 120, 31–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarz K., Iolascon A., Verissimo F., Trede N. S., Horsley W., Chen W., Paw B. H., Hopfner K.-P., Holzmann K., Russo R., Esposito M. R., Spano D., de Falco L., Heinrich K., Joggerst B., Rojewski M. T., Perrotta S., Denecke J., Pannicke U., Delaunay J., Pepperkok R., Heimpel H., Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat. Genet. 41, 936–940 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Zhao T., Goedhart C. M., Sam P. N., Sabouny R., Lingrell S., Cornish A. J., Lamont R. E., Bernier F. P., Sinasac D., Parboosingh J. S.; Care4Rare Canada Consortium, Vance J. E., Claypool S. M., Innes A. M., Shutt T. E., PISDis a mitochondrial disease gene causing skeletal dysplasia, cataracts, and white matter changes. Life Sci. Alliance 2, e201900353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L., Wang G., Chen S., Ding J., Ju S., Cao H., Tian H., Depletion of thymopoietin inhibits proliferation and induces cell cycle arrest/apoptosis in glioblastoma cells. World J. Surg. Oncol. 14, 267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sroczynska P., Lancrin C., Kouskoff V., Lacaud G., The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood 114, 5279–5289 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Lee J. A., Damianov A., Lin C. H., Fontes M., Parikshak N. N., Anderson E. S., Geschwind D. H., Black D. L., Martin K. C., Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron 89, 113–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez N. M., Agosto L., Qiu J., Mallory M. J., Gazzara M. R., Barash Y., Fu X.-D., Lynch K. W., Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev. 29, 2054–2066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutchinson K. R., Saripalli C., Chung C. S., Granzier H., Increased myocardial stiffness due to cardiac titin isoform switching in a mouse model of volume overload limits eccentric remodeling. J. Mol. Cell. Cardiol. 79, 104–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim E., Ilagan J. O., Liang Y., Daubner G. M., Lee S. C.-W., Ramakrishnan A., Li Y., Chung Y. R., Micol J.-B., Murphy M. E., Cho H., Kim M.-K., Zebari A. S., Aumann S., Park C. Y., Buonamici S., Smith P. G., Deeg H. J., Lobry C., Aifantis I., Modis Y., Allain F. H.-T., Halene S., Bradley R. K., Abdel-Wahab O., SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komeno Y., Yan M., Matsuura S., Lam K., Lo M.-C., Huang Y.-J., Tenen D. G., Downing J. R., Zhang D.-E., Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood 123, 3760–3769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manzotti G., Mariani S. A., Corradini F., Bussolari R., Cesi V., Vergalli J., Ferrari-Amorotti G., Fragliasso V., Soliera A. R., Cattelani S., Raschellà G., Holyoake T. L., Calabretta B., Expression of p89(c-Mybex9b), an alternatively spliced form of c-Myb, is required for proliferation and survival of p210BCR/ABL-expressing cells. Blood Cancer J. 2, e71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan X., Tang D., Liao Y., Li P., Zhang Y., Wang M., Liang F., Wang X., Gao Y., Wen L., Wang D., Wang Y., Tang F., Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing. PLoS Biol. 18, e3001017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ganuza M., Chabot A., Tang X., Bi W., Natarajan S., Carter R., Gawad C., Kang G., Cheng Y., McKinney-Freeman S., Murine hematopoietic stem cell activity is derived from pre-circulation embryos but not yolk sacs. Nat. Commun. 9, 5405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., Benner C., Chanda S. K., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for tables S1 to S6
Tables S1 to S6