Abstract
A PCR assay based on oligonucleotide primers derived from the sequence of the gene coding for the 43,000-Da (gp43) antigen was developed to detect Paracoccidioides brasiliensis DNA in sputa. In the standardized conditions, it could detect 10 cells/ml of sputum, providing sufficient accuracy to be useful for diagnosis of paracoccidioidomycosis.
Paracoccidioides brasiliensis, a thermodimorphic fungus, is the causative agent of paracoccidioidomycosis (PCM), the most prevalent systemic mycosis in Latin America (12). The yeast (tissue phase) synthesizes components that interact with the immune system and may have a role in pathogenesis (1, 18). The disease has multiple manifestations, and two progressive clinical forms (acute and chronic) are recognized (6, 12). The “gold standards” for diagnosis of PCM are the isolation of the fungus in culture and the positive identification of multibudding and birefringent yeast cells by direct examination of biologic fluids or biopsy specimens. Serological diagnosis relies on the detection of specific antibodies (2, 3, 5); methods for detecting circulating antigens may be used, although they have poor sensitivity (7, 9, 13). The main diagnostic antigen is the 43,000-Da glycoprotein (gp43); sera from over 90% of PCM patients react with gp43 in immunodiffusion assays, and virtually 100% are positive in immunoblotting assays (2); gp43 elicits delayed-type hypersensitivity reactions in patients (16) and binds to murine laminin; laminin-coated P. brasiliensis yeast cells showed a marked increase in their ability to invade and destroy the infected tissue (20); gp43 is predominantly found in circulating immunoglobulin G immunocomplexes in PCM patient sera (19). The complete sequence of the gene encoding the gp43 antigen has been given previously (4). The in vitro amplification of specific DNA sequences by PCR is a sensitive method that may be used for the detection of viruses, bacteria, and fungi in pathological samples (10, 11, 14, 17, 21). In this work, we prospectively used PCR for the detection of P. brasiliensis DNA in clinical specimens, as sputum, using primers based on the sequence of the gp43 gene.
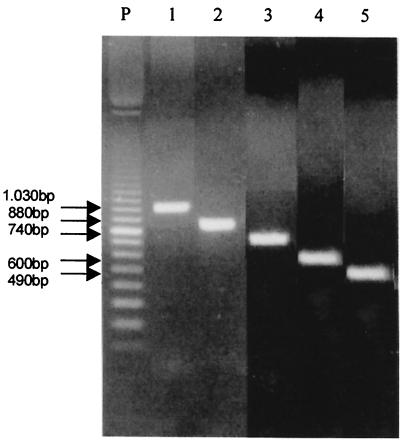
The deduced amino acid sequence of the gp43 gene (GenBank-EMBL data bank accession no. U26160) (4) showed homology (56 to 58%) among blocks of amino acids with exo-1,3-β-d-glucanases from Saccharomyces cerevisiae (vegetative EXG1 and spore-specific SPR1) and Candida albicans (CAXOG). To choose oligonucleotide primers for PCR, the nucleotide sequences of the molecules corresponding to those regions which did not show amino acid homology to the fungal exoglucanases were aligned and compared by BLAST search. The selected sequences are given in Table 1. The primer pairs PC1-PC5, PC1-PC6, PC2-PC5, PC2-PC6, and PC3-PC5 were expected to amplify fragments of 1.03, 0.88, 0.74, 0.60, and 0.49 kb, respectively. They were tested with total DNA extracted from 42 P. brasiliensis isolates, from clinical and environmental sources (data not shown).
TABLE 1.
Sequences of primers used
| Primera | Sequence |
|---|---|
| PC1 | (5′ TCA TCT CAC GTC GCA TCT CAC ATT 3′) |
| PC2 | (5′ ATA GAG GGA GAG CCA TAT GTA CAA GGT 3′) |
| PC3 | (5′ ATC AAA CAA ACC CTG ATC GGC AT 3′) |
| PC5 | (5′ AGC GCC AGA TGG TTT GCC CGC TAG GAA CGA A 3′) |
| PC6 | (5′ GGC TCC TCA AAG TCT GCC ATG AGG AAG 3′) |
PC1, PC2, and PC3 are sense; PC4 and PC5 are antisense.
The fungi were maintained in Sabouraud dextrose agar, and DNA extraction was performed using glass beads and liquid nitrogen as previously described (11, 21). An aliquot of 60 ng of genomic DNA was used for a 25-μl PCR mixture: 12.5 pM (each) primer, 10 mM Tris-HCl, 2.0 mM MgCl2, 0.2 mM (mix) four deoxynucleotides, and 1.2 U of Taq polymerase (Amersham Pharmacia Biotech, Uppsala, Sweden). The amplification parameters included an initial denaturation at 94°C for 5 min followed by 30 cycles each of denaturation at 94°C for 1 min, annealing at 55°C for 2 min, and extension at 72°C for 2 min (Amersham Pharmacia Biotech thermal cycler). The PCR products were identified by agarose gel electrophoresis and combined ethidium bromide staining followed by observation on a UV transilluminator (Fotodyne, New Berlin, Wis.) and photographic documentation (667 Polaroid film). Under these conditions, all P. brasiliensis samples resulted in the amplification of fragments within the expected sizes, for all pairs of primers. In order to evaluate the specificity of the primers, they were used with a battery of genomic DNA templates from C. albicans, Histoplasma capsulatum, and Cryptococcus neoformans (10 isolates each) (data not shown). Figure 1 shows the amplification of P. brasiliensis genomic DNA with different pairs of primers.
FIG. 1.
Testing of the PC1-PC5 (lane 1), PC1-PC6 (lane 2), PC2-PC5 (lane 3), PC2-PC6 (lane 4), and PC3-PC5 (lane 5) primer pairs by PCR using P. brasiliensis B-339 DNA. At left are molecular size markers. (Forty-three P. brasiliensis isolates were tested with these primer pairs.)
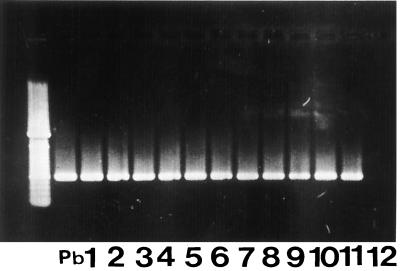
To optimize the PCR procedure for using sputum, two groups of specimens were examined. (i) We first used sputa from patients with suspected tuberculosis to which we added 105, 104, 103, 102, and 101 P. brasiliensis yeast cells per ml of sputum. Each individual seeded sample was mixed in a homogenizer; for sputum solubilization, 10 μl of 1 M dithiothreitol in 0.01 M sodium acetate (pH 5.4) and 2.0 ml of MilliQ sterile water were added to 2 ml of sample, mixed in a vortex, incubated at 37°C for 10 min, and centrifuged (1,800 × g for 5 min), after which supernatant was discarded. To the pellet was added 100 μl of lysis buffer (100 mM Tris-HCl [pH 7.5], 5% sodium dodecyl sulfate, 30 mM EDTA); this was incubated at 100°C for 15 min in a thermoblock, followed by addition of 100 μl of 2.5 M potassium acetate with vigorous agitation in a vortex and incubation at 0°C for 60 min, and centrifuged again (1,800 × g for 5 min at 4°C). Phenol-chloroform (1:1 [vol/vol]) was added to the supernatant, vigorously mixed, and centrifuged (1,800 × g for 10 min at 4°C). Isopropanol was added to the supernatant and incubated at 0°C overnight. The solution was centrifuged (1,800 × g for 5 min at 5°C), the supernatant was discarded, and the pellet was washed with 70% ethanol and resuspended in 100 μl of sterile MilliQ water. DNA was quantitated (GeneQuant apparatus). Five microliters of each solution was taken for PCR amplification. The PCR products were identified by agarose gel electrophoresis. All the primers used were able to detect at least 10 cells/ml, and combined ethidium bromide staining was followed by Southern blot hybridization to an α-32P-labeled 1.3-kb original gp43 gene insert. In order to show the specificity and sensitivity of the primers, a nested PCR was made by employing 1.0 μl of the amplification product from the pair PC1-PC5, which was added to a new reaction mixture, using as inner primers the pair PC2-PC6. The expected nested-PCR product, a 0.6-kb fragment, was detected in samples seeded with 10 cells/ml (data not shown). (ii) The second group consisted of sputa obtained from 11 proven (by serological tests and direct examination of sputa) patients with chronic PCM (male, aged 31 to 64 years) with pulmonary involvement (X ray). These specimens were processed as described above for DNA preparation and then tested by PCR. Each amplification experiment included a negative control sample without DNA and a positive control sample with 60 ng of DNA from P. brasiliensis B-339. Sputum specimens from 11 PCM patients were submitted to PCR using the primer pair PC2-PC6. All 11 sputum samples were positive and produced a band of 0.6 kb (Fig. 2).
FIG. 2.
Testing of clinical specimens (sputa) from PCM patients by PCR with the PC2-PC6 primer pair. Pb, positive control (P. brasiliensis B-339 DNA). Lanes 1 to 11, PCR from clinical specimens; lane 12, negative control. At left are molecular size reference markers.
The aim of this study was to develop a PCR method for the specific and sensitive detection of P. brasiliensis DNA in clinical specimens. Although the diagnosis of PCM is relatively simple, based on the finding of multibudding yeast cells with birefringent walls in clinical materials or in biopsy specimens or by serological methods, sometimes the fungus is scarce in clinical materials or the serology is negative due to problems of anergy. For these reasons, we present here a PCR method for an early diagnosis of PCM, which is critical for a successful treatment. In this work, we devise oligonucleotide primers which do not amplify DNA from other agents of systemic mycosis, such as that of histoplasmosis, or of opportunistic mycoses such as those of cryptococcosis and candidiasis. Some investigators have reported the use of PCR to detect P. brasiliensis DNA, but not in clinical materials (8, 15). An interesting strategy based on PCR amplification of conserved regions of the multicopy ribosomal DNA genes, exclusively from fungi, and followed by specific identification of the amplicon by hybridization with species-specific oligonucleotides, also described for P. brasiliensis, is potentially useful and needs to be evaluated by clinical trials (8). To our knowledge, there is no report describing the use of PCR for detecting P. brasiliensis DNA in sputum from patients, using as primers sequences derived from the gene encoding the gp43-specific antigen. Detection of DNA specific for P. brasiliensis in sputum of PCM patients can be very useful because it is noninvasive and can be repeated several times. Although the gp43 gene is present in low copy numbers per nucleus, we should mention that P. brasiliensis yeast cells are multinucleated (four to eight nuclei per cell), and the number of targets per cell could reach at least 8 to 16 copies. Among the primers tested, we chose the PC2-PC6 pair to be used for direct amplification from clinical material (sputum) or as an inner primer on a nested-PCR assay after an initial amplification step using the pair PC1-PC5. However, all pairs of primers here tested were able to detect P. brasiliensis DNA in sputa from PCM patients. These were the procedures which presented the highest sensitivity and specificity and gave a product clearly visible (0.6 kb) on the gel. Although preliminary, our results suggest that a PCR-based test using the oligonucleotide primers described in this work can be an interesting alternative method for the fast, sensitive, and reliable detection of P. brasiliensis in clinical biological samples.
Acknowledgments
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (proc. no. 96/6635-0).
REFERENCES
- 1.Almeida S R, Unterkircher C S, Camargo Z P. The involvement of the major glycoprotein (gp43) of Paracoccidioides brasiliensis in attachment to macrophages. Med Mycol. 1998;36:405–411. doi: 10.1080/02681219880000641. [DOI] [PubMed] [Google Scholar]
- 2.Blotta M H S L, Camargo Z P. Immunological response to cell-free antigens of Paracoccidioides brasiliensis: relationship with clinical forms of paracoccidioidomycosis. J Clin Microbiol. 1993;31:671–676. doi: 10.1128/jcm.31.3.671-676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camargo Z P, Gesztesi J-L, Saraiva E C O, Taborda C P, Vicentini A P, Lopes J D. Monoclonal antibody capture enzyme immunoassay for detection of Paracoccidioides brasiliensis antibodies in paracoccidioidomycosis. J Clin Microbiol. 1994;32:2377–2381. doi: 10.1128/jcm.32.10.2377-2381.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cisalpino P S, Puccia R, Yamauchi L M, Cano M I N, Silveira J F, Travassos L R. Cloning, characterization, and epitope expression of the major diagnostic antigen of Paracoccidioides brasiliensis. J Biol Chem. 1996;271:4553–4560. doi: 10.1074/jbc.271.8.4553. [DOI] [PubMed] [Google Scholar]
- 5.de Camargo Z P, Unterkircher C, Campoy S, Travassos L R. Production of Paracoccidioides brasiliensis exoantigens for immunodiffusion tests. J Clin Microbiol. 1988;26:2147–2151. doi: 10.1128/jcm.26.10.2147-2151.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco M F, Montenegro M R, Mendes R P, Marques S A, Dillon N L, Mota N G S. Paracoccidioidomycosis: a recent proposed classification of clinical forms. Rev Soc Bras Med Trop. 1987;20:129–131. doi: 10.1590/s0037-86821987000200012. [DOI] [PubMed] [Google Scholar]
- 7.Freitas da Silva G, Roque-Barreira M C. Antigenemia in paracoccidioidomycosis. J Clin Microbiol. 1992;30:381–385. doi: 10.1128/jcm.30.2.381-385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldani L Z, Maia A L, Sugar A M. Cloning and nucleotide sequence of a specific DNA fragment from Paracoccidioides brasiliensis. J Clin Microbiol. 1995;33:1652–1654. doi: 10.1128/jcm.33.6.1652-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez B L, Figueroa J I, Hamilton A J, Ortiz B, Robledo M A, Hay R J, Restrepo A. Use of monoclonal antibodies in diagnosis of paracoccidioidomycosis: new strategies for detection of circulating antigens. J Clin Microbiol. 1997;35:3278–3283. doi: 10.1128/jcm.35.12.3278-3283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guatelli J C, Gingeras T R, Richman D D. Nucleic acid amplification in vitro: detection of sequences with low copy numbers and application to diagnosis of human immunodeficiency virus type 1 infection. Clin Microbiol Rev. 1989;2:217–226. doi: 10.1128/cmr.2.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kersulyte D, Woods J P, Keath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacaz C S. Paracoccidioides brasiliensis: morphology, evolutionary cycle, maintenance during saprophytic life, biology, virulence, taxonomy. In: Franco M, Lacaz C S, Restrepo-Moreno A, Del Negro G, editors. Paracoccidioidomycosis. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 13–22. [Google Scholar]
- 13.Mendes-Giannini M J S, Bueno J P, Shikanai-Yassuda M A, Ferreira A W, Masuda A. Detection of 43,000-molecular-weight glycoprotein in sera of patients with paracoccidioidomycosis. J Clin Microbiol. 1989;27:2842–2845. doi: 10.1128/jcm.27.12.2842-2845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niesters H G M, Goessens W H F, Meis J F M G, Quint W G V. Rapid, polymerase chain reaction-based identification assays for Candida species. J Clin Microbiol. 1993;31:904–910. doi: 10.1128/jcm.31.4.904-910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandhu G S, Alleff R A, Kline B C, da Silva Lacaz C. Molecular detection and identification of Paracoccidioides brasiliensis. J Clin Microbiol. 1997;35:1894–1896. doi: 10.1128/jcm.35.7.1894-1896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saraiva E C O, Altemani A, Franc M F, Unterkircher C S, Camargo Z P. Paracoccidioides brasiliensis-gp43 used as paracoccidioidin. J Med Vet Mycol. 1996;34:155–161. doi: 10.1080/02681219680000261. [DOI] [PubMed] [Google Scholar]
- 17.Thierry D, Brisson-Noel A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon J-L, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travassos L R. Immunochemistry of Paracoccidioides brasiliensis. In: Franco M, Lacaz C S, Restrepo-Moreno A, Del Negro G, editors. Paracoccidioidomycosis. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 67–86. [Google Scholar]
- 19.Unterkircher C S, Yazaki S C, Shimizu M T, Jorge A O C, Camargo Z P. Specific components found in circulating immune complexes (CIC) in paracoccidioidomycosis. J Med Vet Mycol. 1996;34:273–277. doi: 10.1080/02681219680000461. [DOI] [PubMed] [Google Scholar]
- 20.Vicentini A P, Gesztesi J-L, Franco M F, de Souza W, de Moraes J Z, Travassos L R, Lopes J D. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect Immun. 1994;62:1465–1469. doi: 10.1128/iai.62.4.1465-1469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J Clin Microbiol. 1996;34:2464–2468. doi: 10.1128/jcm.34.10.2464-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]