Abstract
Objective
To investigate the relationship between the long noncoding RNA (lncRNA) Prostate cancer-associated transcription factors 14 (PCAT14) and the clinical characteristics of prostate cancer and immune cell infiltration.
Methods
The relationship between PCAT14 expression and the clinicopathological characteristics of prostate cancer was analyzed based on The Cancer Genome Atlas (TCGA) database. Receiver operating characteristic (ROC) curves were used to evaluate the value of PCAT14 as a diagnostic marker for prostate cancer. The relationship between PCAT14 and immune cell infiltration was analyzed to explore the effect of PCAT14 on the immune-related functions of prostate cancer.
Results
The ROC curve showed that PCAT14 had a significant diagnostic ability (area under curve = 0.818) for prostate cancer. A reduced expression of PCAT14 in prostate cancer was related to T stage, N stage, primary therapy outcome, residual tumor, Gleason score, and age. The expression of PCAT14 was independently associated with the progression-free interval in prostate cancer patients. The infiltration of immune cells in prostate cancer showed a significant negative correlation between the expression of PCAT14 and plasmacytoid dendritic cells, activated dendritic cells, regulatory T cells, and neutrophils.
Conclusions
PCAT14 is highly expressed in prostate cancer and is expected to be a diagnostic marker. PCAT14 might promote the development of prostate cancer through chemokines, antimicrobials, and cytokines that affect the infiltration of immune cells.
1. Introduction
Prostate cancer is the second leading cause of cancer-related deaths in men, according to the 2018 global cancer incidence and mortality statistics [1]. Prostate-specific antigen (PSA) is used as the main marker for prostate cancer screening, diagnosis, and prognosis. Nevertheless, PSA alone as a single marker still has great limitations in diagnosing and determining prostate cancer prognosis. Therefore, it is necessary to continue the search for more potentially effective markers for the diagnosis and prognosis of prostate cancer. In addition, the treatment of prostate cancer has made some progress, but it is still not satisfactory. Therefore, further studies on the pathogenesis of prostate cancer are still needed to provide more clues for the treatment of prostate cancer.
Long noncoding RNAs (lncRNAs) are long RNA transcripts of over 200 nucleotides in length, but they do not encode proteins [2]. Several studies have found that lncRNAs play important roles in various types of cancer [3–7]. At present, it is generally agreed that lncRNAs have a strong application prospect as tumor diagnostic markers and might eventually be targeted for therapy [8]. Shukla et al. [9] identified the lncRNA Prostate cancer-associated transcription factors 14 (PCAT14) in prostate cancer and that the overexpression of PCAT14 inhibited the invasion of prostate cancer cells and was associated with a good prognosis of prostate cancer. Still, there are few reports on the association between PCAT14 and prostate cancer, the relationship between PCAT14 and the clinical indicators of prostate cancer, and the relationship between PCAT14 and immune cell infiltration in prostate cancer. Indeed, the tumor microenvironment and immune cell infiltration are now recognized to play crucial roles in the development and prognosis of solid cancers [10–14].
Therefore, this study is aimed at investigating the relationship between the LncRNA PCAT14 and the clinical characteristics of prostate cancer and immune cell infiltration. This study performed a detailed comparative analysis of lncRNA PCAT14 and prostate cancer-related clinical indicators (age, race, diagnostic validity, tumor-node-metastasis (TNM) staging, initial treatment outcome, residual tumor, PSA, Gleason score, and overall survival). A detailed comparative analysis was conducted to analyze the correlation between PCAT14 and prostate cancer disease progression. In addition, we attempted to explore the correlation between PCAT14 and immune cell infiltration in the prostate cancer tumor microenvironment and analyzed the immunological pathways closely related to prostate cancer. The results might provide a scientific basis for future research on prostate cancer immunotherapy.
2. Materials and Methods
2.1. The Downloading of the RNA Sequencing (RNAseq) Data and Analysis of PCAT14 Expression in Prostate Cancer
RNAseq data and clinical information were downloaded from the PRAD project in The Cancer Genome Atlas (TCGA) database. The RNAseq data were converted into the transcripts per million reads (TPM) format, and log2 conversion was performed. The data were from 499 prostate cancer samples and 52 adjacent normal tissue samples. The significance of expression level was as follows: ns, p ≥ 0.05; ∗, p < 0.05; ∗∗, p < 0.01; and ∗∗∗, p < 0.001.
2.2. Correlation Analysis of PCAT14 and Immune Invasion in Prostate Cancer
The single-sample Gene Set Enrichment Analysis (SSGSEA) method in the Gene Set Variation Analysis (GSVA) package (1.34.0 version) of R (3.6.3 version) was used to analyze the infiltration of immune cells in prostate cancer. The markers and classification of 24 kinds of immune cells were obtained from a published paper [11]. The correlation between PCAT14 and the level of immune cell infiltration in prostate cancer was determined using the Spearman and Wilcoxon rank-sum tests. Immlnc is a tool that can be used to study the immune function of lncRNAs in different cancer types [15]. This web tool can be used to inquire online about the relationship between lncRNAs and immune cell infiltration in specific cancers. At the same time, the tool supports queries of lncRNAs' potential immune-related biological pathways.
2.3. Statistical Methods
All analyses were performed using R (v.3.6.3). The correlations between PCAT14 and clinical indicators were analyzed by the Wilcoxon's rank-sum test, chi-square test, Fisher's exact test, and logistic regression. The Cox proportional risk model was used to assess the association between clinical indicators and progression-free interval (PFI). A P value < 0.05 was considered statistically significant.
3. Results
3.1. PCAT14 Can Be Used as a Potential Marker for Diagnosis of Prostate Cancer
We analyzed the expression of PCAT14 in 499 prostate cancer samples and 52 adjacent normal tissue samples, and the results showed that PCAT14 was highly expressed in prostate cancer tissues (Figure 1(a)). We also analyzed the expression of PCAT14 in 52 prostate cancer tissues and their matched adjacent tissues. The results also showed that PCAT14 was highly expressed in prostate cancer tissues (Figure 1(b)). In addition, receiver operating characteristic (ROC) curves were used to analyze the diagnostic validity of the PCAT14 expression level. The area under the curve (AUC) of PCAT14 was 0.818, indicating that PCAT14 can be used as an ideal biomarker for diagnosing prostate cancer (Figure 1(c)).
Figure 1.
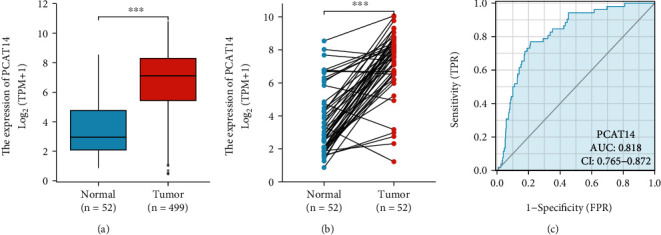
Expression and diagnostic efficacy of PCAT14 in prostate cancer. (a) Comparison of the expression of PCAT14 in 499 prostate cancer samples and 52 paracancer normal tissue samples. The results showed that PCAT14 was highly expressed in prostate cancer tissues. (b) Comparison of the expression of PCAT14 in 52 prostate cancer tissues and its matching paracancer tissues. The results showed that PCAT14 was highly expressed in prostate cancer tissues. (c) Received operating characteristics (ROC) curve analysis showed that the area under the curve (AUC) of PCAT14 was 0.818, indicating that PCAT14 has a significant diagnostic ability.
3.2. Correlation between PCAT14 and Clinical Indicators of Patients
According to the average relative expression level of PCAT14, prostate cancer patients were divided into two groups: the high PCAT14 expression group (n = 250) and the low PCAT14 expression group (n = 249). Then, the expression levels of PCAT14 and the correlations with clinical indicators were analyzed. A reduced expression of PCAT14 in prostate cancer was related to T stage (P = 0.005), N stage (P = 0.009), primary therapy outcome (P = 0.022), residual tumor (P < 0.001), Gleason score (P < 0.001), and age (P = 0.020) (Table 1, Figure 2).
Table 1.
Correlation between PCAT14 and clinical indicators of patients.
| Low expression of PCAT14 | High expression of PCAT14 | P | |
|---|---|---|---|
| n | 249 | 250 | |
| T stage, n (%) | 0.005 | ||
| T2 | 76 (15.4%) | 113 (23%) | |
| T3 | 160 (32.5%) | 132 (26.8%) | |
| T4 | 7 (1.4%) | 4 (0.8%) | |
| N stage, n (%) | 0.009 | ||
| N0 | 169 (39.7%) | 178 (41.8%) | |
| N1 | 52 (12.2%) | 27 (6.3%) | |
| M stage, n (%) | 0.249 | ||
| M0 | 230 (50.2%) | 225 (49.1%) | |
| M1 | 3 (0.7%) | 0 (0%) | |
| Primary therapy outcome, n (%) | 0.022 | ||
| PD | 16 (3.7%) | 12 (2.7%) | |
| SD | 18 (4.1%) | 11 (2.5%) | |
| PR | 27 (6.2%) | 13 (3%) | |
| CR | 156 (35.6%) | 185 (42.2%) | |
| Race, n (%) | 0.195 | ||
| Asian | 9 (1.9%) | 3 (0.6%) | |
| Black or African American | 30 (6.2%) | 27 (5.6%) | |
| White | 204 (42.1%) | 211 (43.6%) | |
| Age, n (%) | 0.342 | ||
| ≤60 | 106 (21.2%) | 118 (23.6%) | |
| >60 | 143 (28.7%) | 132 (26.5%) | |
| Residual tumor, n (%) | <0.001 | ||
| R0 | 133 (28.4%) | 182 (38.9%) | |
| R1 | 100 (21.4%) | 48 (10.3%) | |
| R2 | 1 (0.2%) | 4 (0.9%) | |
| PSA (ng/ml), n (%) | 0.347 | ||
| <4 | 199 (45%) | 216 (48.9%) | |
| ≥4 | 16 (3.6%) | 11 (2.5%) | |
| Gleason score, n (%) | < 0.001 | ||
| 6 | 16 (3.2%) | 30 (6%) | |
| 7 | 103 (20.6%) | 144 (28.9%) | |
| 8 | 34 (6.8%) | 30 (6%) | |
| 9 | 92 (18.4%) | 46 (9.2%) | |
| 10 | 4 (0.8%) | 0 (0%) | |
| OS event, n (%) | 0.339 | ||
| Alive | 246 (49.3%) | 243 (48.7%) | |
| Dead | 3 (0.6%) | 7 (1.4%) | |
| Age, mean ± SD | 61.74 ± 6.48 | 60.32 ± 7.08 | 0.020 |
| PSA (ng/ml), mean ± SD | 2.79 ± 22.41 | 0.76 ± 3.34 | 0.179 |
PD: progressive disease; SD: stable disease; PR: partial response; CR: complete response; PSA: prostate-specific antigen; OS: overall survival.
Figure 2.
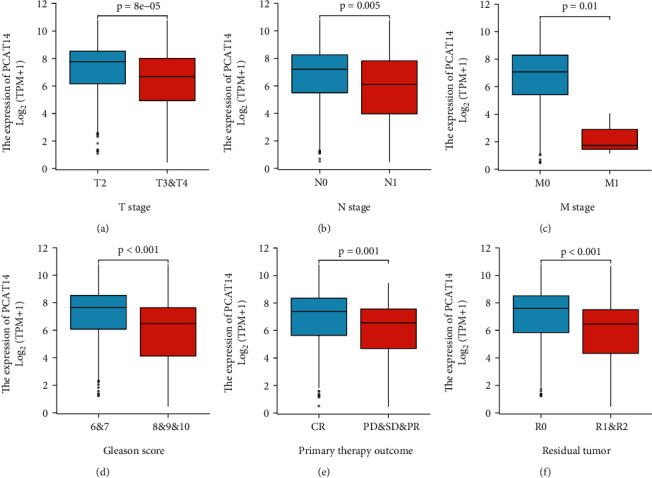
Relationship between PCAT14 expression and clinically relevant indicators in prostate cancer. (a) Compared with T2 stage prostate cancer, PCAT14 was lowly expressed in T3-4 stage prostate cancer tissues. (b) Compared with N0 stage prostate cancer, PCAT14 was lowly expressed in N1 stage prostate cancer tissues. (c) Compared with M0 stage prostate cancer, PCAT14 was lowly expressed in M1 stage prostate cancer tissue. (d) Compared with the prostate cancer group with Gleason score = 6-7, PCAT14 was lowly expressed in prostate cancer tissue with Gleason score = 8-10. (e) PCAT14 was lowly expressed in prostate cancer tissues with the initial treatment outcome of progressive disease, stable disease, and partial response compared with prostate cancer tissues with initial treatment outcome of complete response. (f) PCAT14 was lowly expressed in R1&2 prostate cancer tissue compared with prostate cancer in R0 prostate cancer tissue.
3.3. Cox Univariable and Multivariable Analysis of Prognostic Factors in Prostate Cancer
Table 2 shows the results of the Cox univariable and multivariable analyses of PFI in prostate cancer patients. In the Cox univariable regression model, the variables with P < 0.01 were T stage (P < 0.001), N stage (P = 0.007), primary treatment outcome (P < 0.001), PSA (P < 0.001), and the Gleason score (P < 0.001). The multivariable analysis showed that the primary treatment outcome (P < 0.001) and Gleason score (P < 0.001) were independent prognostic factors for PFS in prostate cancer patients.
Table 2.
Cox univariable and multivariable analysis of PFI in prostate cancer patients.
| Total (n) | Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| T stage (T3&T4 vs. T2) | 492 | 3.785 (2.140-6.693) | <0.001 | 1.473 (0.710-3.059) | 0.299 |
| N stage (N1 vs. N0) | 426 | 1.946 (1.202-3.150) | 0.007 | 0.861 (0.500-1.483) | 0.589 |
| M stage (M1 vs. M0) | 458 | 3.566 (0.494-25.753) | 0.208 | ||
| Primary therapy outcome (PD&SD&PR vs. CR) | 438 | 6.627 (4.337-10.126) | <0.001 | 3.576 (2.054-6.226) | <0.001 |
| Race (Asian & Black or African American vs. White) | 484 | 0.751 (0.417-1.352) | 0.339 | ||
| Age (>60 vs. ≤60) | 499 | 1.302 (0.863-1.963) | 0.208 | ||
| Residual tumor (R1&R2 vs. R0) | 468 | 2.365 (1.566-3.570) | <0.001 | 1.017 (0.598-1.728) | 0.951 |
| PSA (ng/ml) (≥4 vs. <4) | 442 | 4.196 (2.095-8.405) | <0.001 | 1.650 (0.743-3.667) | 0.219 |
| Gleason score (8-10 vs. 6-7) | 499 | 4.675 (2.957-7.391) | <0.001 | 2.901 (1.603-5.253) | <0.001 |
CI: confidence interval; PSA: prostate-specific antigen.
3.4. Relationship between PCAT14 Expression and Immune Cell Infiltration in Prostate Cancer
We analyzed the relationship between PCAT14 expression and immune cell infiltration in prostate cancer using SSGSEA. The results showed that PCAT14 expression was negatively correlated with the infiltration of plasmacytoid dendritic cells (pDC), activated dendritic cells (aDC), regulatory T cells (Tregs), and neutrophils (Figures 3 and 4).
Figure 3.
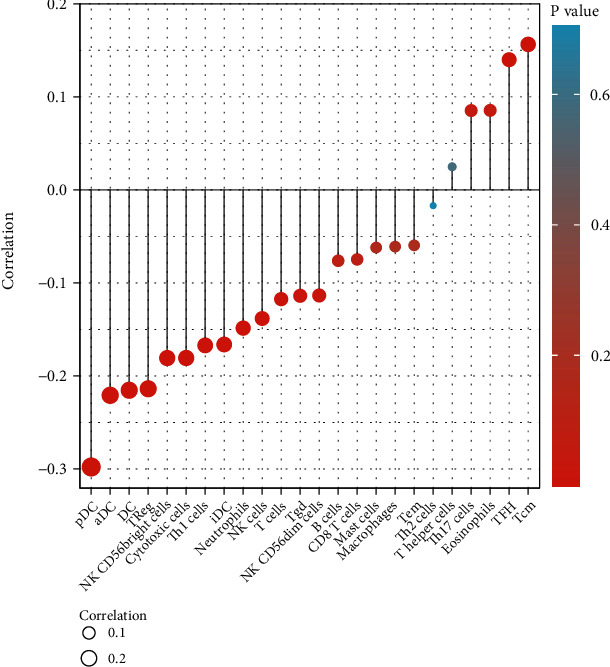
Forest plot showed that the expression of PCAT14 in the tumor microenvironment of prostate cancer was negatively correlated with the infiltration of plasmacytoid dendritic cells, activated dendritic cells, regulatory T cells, and neutrophils.
Figure 4.
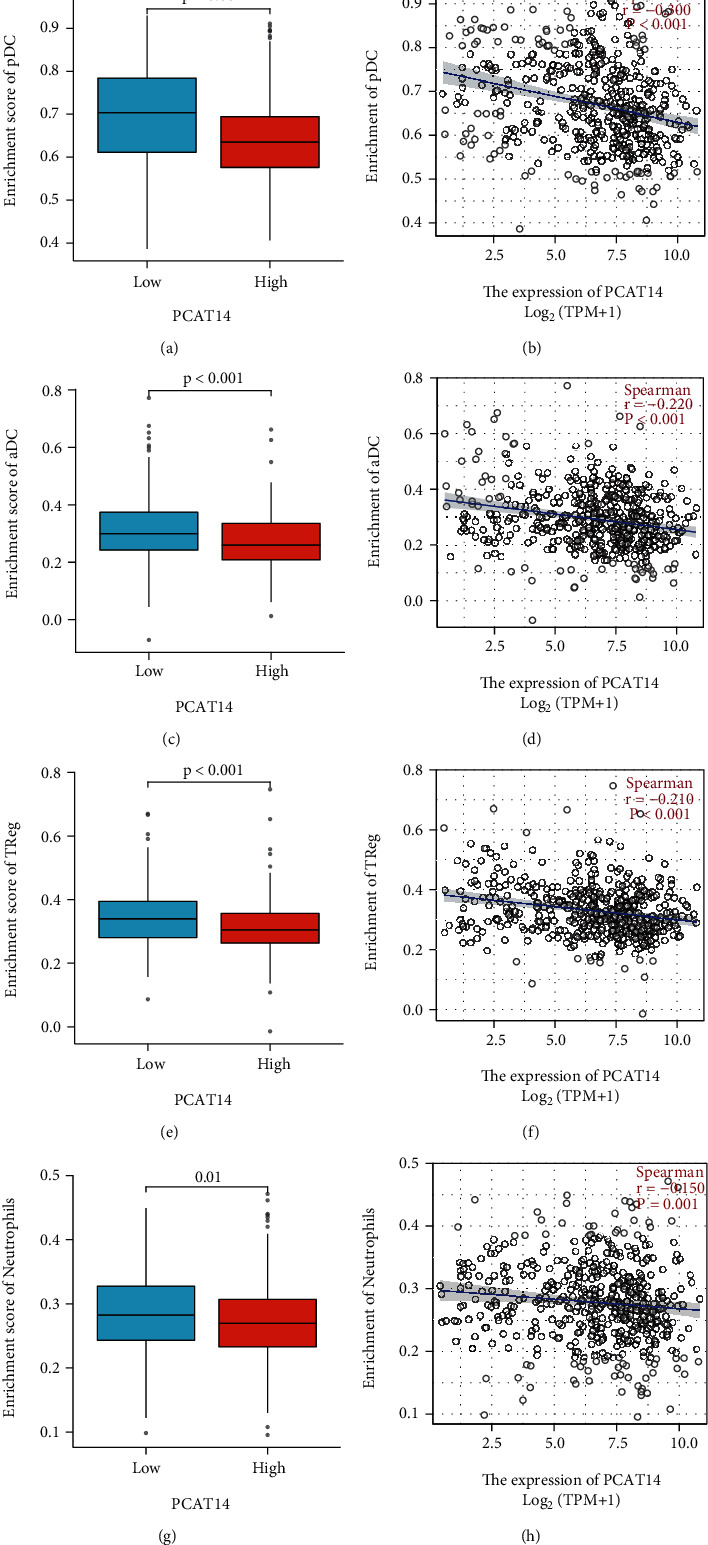
Correlation analysis of PCAT14 expression level with pDC, aDC, Treg, and neutrophil infiltration. (a) The Wilcoxon rank-sum test analysis showed that the plasmacytoid dendritic cell (pDC) infiltration level of the high PCAT14 expression group was low, and the pDC infiltration level of the low PCAT14 expression group was high, and the difference was statistically significant. (b) The Spearman correlation analysis showed that the expression level of PCAT14 was negatively correlated with the infiltration of pDC. (c) The Wilcoxon rank-sum test analysis showed that the activated dendritic cells (aDC) infiltration level in the high PCAT14 expression group was low, and the aDC infiltration level in the low PCAT14 expression group was high, and the difference was statistically significant. (d) The Spearman correlation analysis showed that the expression level of PCAT14 was negatively correlated with the infiltration of aDC. (e) The Wilcoxon rank-sum test analysis showed that the level of regulatory T cell (Treg) infiltration in the high PCAT14 expression group was low, the Treg infiltration level in the low PCAT14 expression group was high, and the difference was statistically significant. (f) The Spearman correlation analysis showed that the expression level of PCAT14 was negatively correlated with Treg infiltration. (g) The Wilcoxon rank-sum test analysis showed that the level of neutrophil infiltration in the high PCAT14 expression group was low, the neutrophil infiltration level in the low PCAT14 expression group was high, and the difference was statistically significant. (h) The Spearman correlation analysis showed that the expression level of PCAT14 was negatively correlated with the infiltration of neutrophils.
We verified the correlation between PCAT14 and immune cell infiltration using the Immlnc database. The results also showed that PCAT14 was negatively correlated with neutrophils and dendritic cells (P < 0.05) (Table 3). In addition, we analyzed the relationship between PCAT14 and immune-related pathways in prostate cancer by GSEA. The results showed that the immune pathways related to PCAT14 included chemokines, antimicrobials, and cytokines (P < 0.05) (Table 4).
Table 3.
Correlation between PCAT14 and immune cells in prostate cancer.
| Cancer | lncRNA ID | lncRNA symbol | Immune cell | P value |
|---|---|---|---|---|
| PRAD | ENSG00000280623 | PCAT14 | Dendritic | <0.001 |
| PRAD | ENSG00000280623 | PCAT14 | Neutrophil | < 0.001 |
lncRNA: long noncoding RNA; PRAD: prostate adenocarcinoma.
Table 4.
Correlation between PCAT14 and immune-related pathways in prostate cancer.
| Cancer | lncRNA ID | lncRNA symbol | Immune pathway | P valuea | ESb |
|---|---|---|---|---|---|
| PRAD | ENSG00000280623 | PCAT14 | Chemokines | 0.003384 | 0.488026 |
| PRAD | ENSG00000280623 | PCAT14 | Antimicrobials | 0.025140 | 0.327294 |
| PRAD | ENSG00000280623 | PCAT14 | Cytokines | 0.028011 | 0.334931 |
| PRAD | ENSG00000280623 | PCAT14 | TNF family members | 0.090566 | 0.658357 |
| PRAD | ENSG00000280623 | PCAT14 | Interferon receptor | 0.206693 | 0.791169 |
| PRAD | ENSG00000280623 | PCAT14 | Interleukins receptor | 0.164966 | 0.440330 |
a P value: the P values for GSEA analysis; bES: enrichment scores; lncRNA: long noncoding RNA; ES: enrichment score; PRAD: prostate adenocarcinoma.
4. Discussion
At present, scholars have explored the relationship between lncRNAs and prostate cancer. He et al. [16] identified regulatory relationships among lncRNAs, miRNAs, and mRNAs that might be involved in the pathogenesis of prostate cancer. Still, few studies have reported the association between PCAT14 and prostate cancer. The research on the tumor microenvironment is also a hot topic at present. Immune cells, as important members of the tumor microenvironment, are closely related to the tumor [17]. At present, the relationship between PCAT14 and immune cell infiltration in prostate cancer is still unclear.
This study first analyzed the expression of the lncRNA PCAT14 in prostate cancer and its relationship with prostate cancer prognosis. The results showed that PCAT14 was significantly overexpressed in prostate cancer. In addition, the ROC curve analysis showed that PCAT14 could be a biomarker for diagnosing prostate cancer. PCAT14, a new potential diagnostic marker for prostate cancer, could make up for the nonincreased PSA levels in diagnosing prostate cancer.
At present, several studies have shown that the infiltration of immune cells in prostate cancer promotes the occurrence and development of prostate cancer [18]. Jiang et al. [19] found that infiltrating immune cells could promote the proliferation, invasion, and distant metastasis of tumor stem cells in prostate and breast cancers. Gannon et al. [20] found that androgen deprivation therapy could regulate the infiltration of immune cells and proved that NK cells had a protective effect on tumor progression, while macrophages were beneficial to the progression of advanced prostate cancer. The highlight of our study was to reveal the relationship between PCAT14 expression and a variety of immune cells infiltrating prostate cancer. Our results showed that the expression of PCAT14 was negatively correlated with the infiltration level of pDC, aDC, Tregs, and neutrophils in prostate cancer. Since dendritic cells (DC) [12] and neutrophils [13] mainly play an antitumor role in the tumor microenvironment, PCAT14 might promote the progression of prostate cancer by inhibiting the antitumor effect of DC and neutrophils. We also observed a negative correlation between PCAT14 expression and the infiltration level of Tregs in prostate cancer. Currently, it is widely believed that Tregs generally inhibit the immune response of tumors [21]. Still, some authors have proposed that Treg cells play a double-edged role in solid tumors [22]. Further experiments are needed to confirm the association and analyze the specific biological significance of the various immune cell subtypes in prostate cancer progression. In addition, we analyzed the relationship between PCAT14 and immune-related pathways in prostate cancer by GSEA. The results showed that the immune pathways related to PCAT14 included chemokines, antimicrobials, and cytokines. Several studies suggested that cytokines and chemokines, along with their receptors and signaling axes, are important factors driving prostate cancer metastasis [10–14].
This study provides a scientific basis for further research on the pathogenesis of the lncRNA PCAT14 in prostate cancer. This study showed that PCAT14 has significant diagnostic ability in prostate cancer and might be expected to become an effective screening and diagnostic marker for prostate cancer. This research also found that the expression of PCAT14 is negatively correlated with the infiltration of pDC, aDC, Tregs, and neutrophils in the tumor microenvironment of prostate cancer. PCAT14 might affect the infiltration of immune cells through chemokines, antibacterial agents, and cytokines. Such immunological pathways promote the pathogenesis and progression of prostate cancer [10–14], providing a theoretical basis for future research on prostate cancer immunotherapy. In vitro and in vivo studies are necessary to confirm these roles and explore potential interventions based on PCAT14.
In conclusion, we observed an increased expression of PCAT14 in prostate cancer tissue. PCAT14 might be promising as a diagnostic marker for prostate cancer. In addition, PCAT14 probably influences the infiltration of immune cells through chemokines, antimicrobials, and cytokines, thus promoting prostate cancer development.
Acknowledgments
This research was funded by the Zhanjiang Unfunded Science and Technology Project (No. 2018B01134).
Data Availability
We did not create any data, and all the results we analyzed in our paper were based data from public database. So we do not have any data that could be uploaded to a repository.
Consent
Consent is not applicable.
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Yunkun Yan and Jianjun Liu are responsible for the conception. Yunkun Yan is assigned to the interpretation or analysis of data and the preparation of the manuscript. Mushi Ye and Jianchang Li revised the important intellectual content. Zhijian Xu and Jianjun Liu did the supervision.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians . 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Quinn J. J., Chang H. Y. Unique features of long non-coding RNA biogenesis and function. Nature Reviews. Genetics . 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 3.Han T., Hu H., Zhuo M., et al. Long non-coding RNA: an emerging paradigm of pancreatic cancer. Current Molecular Medicine . 2016;16(8):702–709. doi: 10.2174/1566524016666160927095812. [DOI] [PubMed] [Google Scholar]
- 4.Khandelwal A., Bacolla A., Vasquez K. M., Jain A. Long non-coding RNA: a new paradigm for lung cancer. Molecular Carcinogenesis . 2015;54(11):1235–1251. doi: 10.1002/mc.22362. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L., Chen S., Wang B., et al. An eight-long noncoding RNA expression signature for colorectal cancer patients' prognosis. Journal of Cellular Biochemistry . 2019;120(4):5636–5643. doi: 10.1002/jcb.27847. [DOI] [PubMed] [Google Scholar]
- 6.Li C., Hu J., Hu X., et al. LncRNA SNHG9 is a prognostic biomarker and correlated with immune infiltrates in prostate cancer. Translational Andrology and Urology . 2021;10(1):215–226. doi: 10.21037/tau-20-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luan C., Li Y., Liu Z., Zhao C. Long noncoding RNA MALAT1 promotes the development of colon cancer by regulating miR-101-3p/STC1 Axis. Oncotargets and Therapy . 2020;Volume 13:3653–3665. doi: 10.2147/OTT.S242300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Bhan A., Soleimani M., Mandal S. S. Long noncoding RNA and cancer: a new paradigm. Cancer Research . 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla S., Zhang X., Niknafs Y. S., et al. Identification and Validation of _PCAT14_ as Prognostic Biomarker in Prostate Cancer. Neoplasia . 2016;18(8):489–499. doi: 10.1016/j.neo.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adekoya T. O., Richardson R. M. Cytokines and chemokines as mediators of prostate cancer metastasis. International Journal of Molecular Sciences . 2020;21(12):p. 4449. doi: 10.3390/ijms21124449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bindea G., Mlecnik B., Tosolini M., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity . 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Gardner A., Ruffell B. Dendritic cells and cancer immunity. Trends in Immunology . 2016;37(12):855–865. doi: 10.1016/j.it.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giese M. A., Hind L. E., Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood . 2019;133(20):2159–2167. doi: 10.1182/blood-2018-11-844548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rani A., Dasgupta P., Murphy J. J. Prostate cancer: the role of inflammation and chemokines. The American Journal of Pathology . 2019;189(11):2119–2137. doi: 10.1016/j.ajpath.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Jiang T., Zhou W., et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nature Communications . 2020;11(1):p. 1000. doi: 10.1038/s41467-020-14802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J. H., Han Z. P., Zou M. X., et al. Analyzing the LncRNA, miRNA, and mRNA regulatory network in prostate cancer with bioinformatics software. Journal of Computational Biology . 2018;25(2):146–157. doi: 10.1089/cmb.2016.0093. [DOI] [PubMed] [Google Scholar]
- 17.Wu X. L., Ma H., Li Y. Y. Infiltration patterns of immune cells in gastric cancer and their clinical correlation with prognosis . Research Square; 2020. [Google Scholar]
- 18.Yan Y., Mao X., Zhang Q., et al. Molecular mechanisms, immune cell infiltration, and potential drugs for prostate cancer. Cancer Biomarkers . 2021;31(1):87–96. doi: 10.3233/CBM-200939. [DOI] [PubMed] [Google Scholar]
- 19.Jiang B., Mason J., Jewett A., et al. Tumor-infiltrating immune cells: triggers for tumor capsule disruption and tumor progression. International Journal of Medical Sciences . 2013;10(5):475–497. doi: 10.7150/ijms.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gannon P. O., Poisson A. O., Delvoye N., Lapointe R., Mes-Masson A. M., Saad F. Characterization of the intra-prostatic immune cell infiltration in androgen- deprived prostate cancer patients. Journal of Immunological Methods . 2009;348(1-2):9–17. doi: 10.1016/j.jim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka A., Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Research . 2017;27(1):109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf D., Sopper S., Pircher A., Gastl G., Wolf A. M. Treg(s) in cancer: friends or foe. Journal of Cellular Physiology . 2015;230(11):2598–2605. doi: 10.1002/jcp.25016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We did not create any data, and all the results we analyzed in our paper were based data from public database. So we do not have any data that could be uploaded to a repository.


