Abstract
Introduction
Müller glial cells typically activate to react to hypoxic tissue damage in several retinal diseases. We evaluated the in vitro response to a hypoxia-mimicking stimulus on the expression of a set of genes, known to contribute to eye morphogenesis and cell differentiation.
Materials and Methods
A MIO-M1 Müller cell line was cultured in a hypoxia-mimicking environment by the addition of cobalt chloride to the culture medium, followed by a recovery time in which we mimic restoration from the hypoxic insult. The HIF-1α protein and VEGF-A gene expression were quantified to verify the induction of a hypoxia-like state.
Results
Among the genes under study, we did not observe any difference in the expression levels of Otx1 and Otx2 during treatment; conversely, Otx1 was overexpressed during recovery steps. The VEGF-A gene was strongly upregulated at both the CoCl2 and recovery time points. The transactivated isoform (TA) of the TP73 gene showed an overexpression in long-term exposure to the hypoxic stimulus with a further increase after recovery. Discussion. Our molecular analysis is able to describe the activation of a set of genes, never before described, that can drive the response to a hypoxia-like status. The improved comprehension of these cellular events will be useful for designing new therapeutical approaches for retinal pathologies.
1. Introduction
Müller cells represent the most important glial population in the retina, with both structural and functional roles in retinal homeostasis [1]. Under physiological conditions, Müller cells provide an orientated scaffold and metabolic support to neuronal activity; they maintain the integrity of the blood-retinal barrier [2, 3] and regulate the fluid homeostasis and neovascularization (NV) processes. Several pathological alterations or injuries can activate these cells, causing morphological and/or molecular alterations [4].
In several retinal diseases, such as diabetic retinopathy, retinal vascular occlusion, or retinal detachment, the reduction of vascular perfusion is responsible for the hypoxia and/or ischemia of intraocular tissues [5]. This results in the activation of cellular mechanisms such as neovascular proliferation, intraretinal cell migration, and extravascular leakage of fluids secondary to cytokine release, affecting different retinal cell types [6]. These events, defined in terms of gliosis, may contribute to intraretinal edema formation and retinal tissue degeneration [7].
Müller cells produce a vascular endothelial growth factor (VEGF) in response to hypoxic stresses to improve vascularization and vascular permeability [8]. In this setting, it plays multiple significant roles: in the early phases of oxidative or hypoxic injury, VEGF protects neuronal cells from apoptosis and leads to ischemic preconditioning, i.e., an adaptive protective response to ischemia caused by brief ischemic events. In later stages, it instead alters the expression of several genes affecting the peroxynitration balance by inducing the expression of the inducible nitric oxide synthase (iNOS), which leads to retinal inflammation, neovascularization, vascular leakage, and key pathological changes in hypoxic tissues [9]. In addition to its role on the vasculature, VEGF is an essential neurotrophic factor for the survival of photoreceptors and Müller cells, in an autocrine process that controls the proapoptotic Bax gene and its transcriptional activator p53 [10].
The Otx1 and Otx2 genes code for two transcription factors, OTX1 and OTX2, respectively [11], which are involved in cell identity specification and differentiation and positioning of the bodily axis during the embryological development of the neuroectoderm, from which originates the adult central nervous system [12, 13]. Otx genes are essential in developing the eye, due to their expression in the optic vesicle and retinal progenitor cells (RPCs) [14], and Otx2 is especially expressed in postmitotic neuroblast, which differentiates into all retinal cell types. In the adult retina, Otx2 expression has been observed in bipolar interneurons and photoreceptors [15–17], where it controls the expression of several opsin genes [18, 19].
Otx1 and Otx2 mutant mice showed no retinal pigmented epithelium development [20], while Otx2 mutations are associated with several eye defects such as microphthalmia or the lack of a lens, cornea, and iris [14].
In a previous study on samples of hypoxic retinal tissue from patients affected by proliferative vitreoretinopathy (PVR) secondary to retinal detachment, we observed an abnormal expression of both Otx1 and Otx2 associated with an increased severity of the disease in Otx2 expressing samples [21]. More recently, these two genes have also been involved in physiological and pathological functions in adult tissues, such as sinonasal mucosae [22–25], the pituitary [26–28], and the mammary gland [29–31].
The p53, TP63, and TP73 genes belong to the p53 family and code for two isoforms, one containing the transactivation domain (TA) and the other truncated (ΔN). Either p53 or the TA isoforms of the other two genes regulate the stability and activity of HIF and are known to be involved in defensive mechanisms, especially against oxidative stresses, a typical feature of hypoxia [32–34].
The TP73 gene product is also essential for proper retinal vasculature formation [35], and, contrasting with TP53 and TP63, there are no reports on the TP73 and Otx genes.
The aim of our study is to evaluate the activation and modulation of the abovementioned set of genes involved in hypoxia response and differentiation in Müller cells treated with CoCl2in vitro.
2. Materials and Methods
2.1. Cell Line
The MIO-M1 cell line derives from the spontaneous immortalization (at least 100 divisions) of Müller cells obtained from an eye of a 68-year-old female corneal donor, 36 hours after death [36]. They grow in adhesion in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin/streptomycin (complete medium).
2.2. Analysis of the Morphology of MIO-M1 Cells
The morphology of the cells was evaluated with bright-field microscopy using an OLYMPUS IX51 inverted microscope (OLYMPUS, Shinjuku, Tokyo, Japan). The physiological morphology was compared with that described by Limb's research group [37].
2.3. Cell Culture Preparation for CoCl2 Treatment
MIO-M1 cells were cultured as described in the previous paragraph. Upon confluence, the cells were washed twice with sterile PBS (Phosphate Buffered Saline) and detached from the flask using trypsin-EDTA diluted in sterile PBS (1 : 1 ratio).
After a cell count in Trypan Blue through a Luna Cell Counter, to evaluate their viability, the Müller cells were seeded in new flasks.
The treatment was conducted on the cells at four passages after thawing, and generally, the cells were cultured for no more than five to six passages.
2.4. Cobalt Chloride Treatment
CoCl2 at a final concentration of 100 µM, dissolved in culture-grade water, was used to mimic the hypoxic conditions. The treatment was performed in six-well plates seeded with 25000 cells/well. Before the addition of CoCl2, cells were maintained for 24 h in DMEM without FBS, to synchronize the cell cycle. Each experiment was performed in triplicate.
The cells were exposed to CoCl2 for 24 and 48 hours (samples 24 h CoCl2 and 48 h CoCl2, respectively); after each exposure, a recovery time of 24 hours followed (REC), in which the cells were cultured in the complete medium without CoCl2 (samples 24 h CoCl2 + 24 h REC and 48 h CoCl2 + 24 h REC, respectively) (Figure 1).
Figure 1.
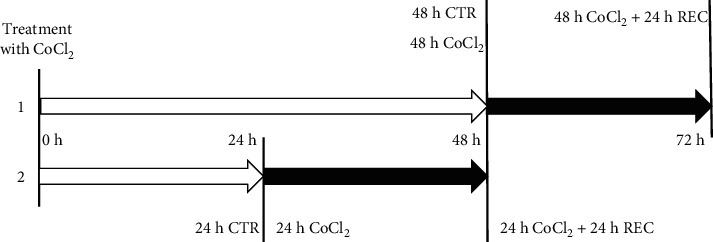
Schematic representation of two CoCl2 treatments. (1) Exposure of MIO-M1 cells to 100 µM CoCl2 for 48 h followed by 24 h of recovery in a complete medium (DMEM + 10% FBS + 1% L-Glut + 1% Pen/Strep). (2) 24 h exposure to 100 µM CoCl2 followed by 24 h of recovery. CTR: control sample. White arrow: medium added with CoCl2. Black arrow: medium without CoCl2. Black vertical lines indicate timepoints of cell collection.
In each phase, an aliquot of cells supplemented only with culture-grade water was collected in 50 mL tubes, to evaluate the effect of the solvent on the cell culture.
Bright-field images were acquired for each sample at each time point (Figure 2).
Figure 2.
Morphological analysis of MIO-M1 Müller cells in different experimental conditions. (a) CTR 24 h; (b) 24 h CoCl2; (c) 24 h CoCl2 + 24 h REC; (d) CTR 48 h; (e) 48 h CoCl2; and (f) 48 h CoCl2 + 24 h REC. Black bar: 100 μM.
The cells were centrifuged at 1200 rpm for five minutes, the supernatant was then discarded, and the resulting cellular precipitate (pellet) was resuspended in 1 mL of complete medium. The cells were then counted as previously described and split into two aliquots (500 µL each) to perform further analysis.
2.5. Protein Extraction and Quantification
An initial aliquot of treated cells was used to extract proteins and to evaluate the levels of HIF-1α (hypoxia-inducible factor 1 subunit alpha) protein, whose amount is typically increased in response to hypoxic conditions.
The cells were centrifuged at 1200 rpm for five minutes, and the obtained pellet was lysed using NP40 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) supplemented with 1 mM PMSF (phenylmethylsulphonyl fluoride) and a protease cocktail inhibitor (Sigma-Aldrich, St. Louis, Missouri, USA) for 30 min on ice and vortexed every 10 minutes. The lysed material was centrifuged at 13000 rpm for 10 minutes at 4°C, and the supernatant was collected for further analysis.
The proteins extracted were quantified using the Bradford reagent (Sigma-Aldrich, St. Louis, Missouri, USA).
2.6. SDS-PAGE
Next, 200 μg of proteins were diluted with the Laemmli buffer, boiled for 10 min, and loaded on SDS-8% polyacrylamide gel (Sodium Dodecyl Sulphate, Sigma-Aldrich, St. Louis, Missouri, USA). SDS-polyacrylamide gel electrophoresis was performed with a running buffer (Tris-HCl 25 mM, glycine 0.2 M, SDS 0.1%) at a 110 V constant voltage. Proteins were wet-transferred from the gel to a polyvinylidene fluoride membrane (PVDF Immobilon-P, Millipore) in a cold chamber (+4°C), for 1 h 30 min with a blotting buffer (glycine 0.15 M, Tris-HCl 25 mM, and methanol 10% in ultrapure water) and at a 250 mA constant current. The membrane was stained with Ponceau S solution (Sigma-Aldrich, St. Louis, Missouri, USA) to control the effective transfer of proteins, then washed five times with PBS-T (NaCl 130 mM, KCl 2.6 mM, Na2HPO4 8.4 mM, KH2PO4 1.4 mM, pH 7.4, and Tween 20 at 0.1%) and incubated with a blocking solution (5% milk in PBS-T) for 2 h, to mask aspecific sites of the membrane.
2.7. Western Blot (WB) Analysis of HIF-1α Protein Levels
The PVDF membranes were incubated with HIF-1α and β-actin primary antibodies, followed by horseradish peroxidase-conjugated secondary antisera (Table 1). The primary antibodies were diluted in a blocking solution and incubated overnight at +4°C. The membrane was then washed five times with PBS-T; secondary antibodies diluted in blocking solution were added for 1 h at room temperature and, and after five washes with PBS-T, they were ready to be analyzed.
Table 1.
Antibodies used to detect HIF-1α and β-actin protein levels and primary and secondary antibodies used in WB.
| Antibody | Dilution | Host species | Source |
|---|---|---|---|
| HIF-1α | 1 : 300 | Mouse | Cell Signaling Technology (D5F3M) |
| β-Actin | 1 : 1000 | Goat | Abcam (ab8229) |
| Anti-mouse IgG-HRP | 1 : 2000 | Goat | Santa Cruz (sc2005) |
| Anti-goat IgG-HRP | 1 : 5000 | Donkey | Santa Cruz (sc2020) |
The Clarity Max™ Western ECL Substrate kit (Bio-Rad, Hercules, California, USA) was used to visualize the chemiluminescence of our proteins of interest. The membrane was exposed to a photographic film that was further processed with commercially available developer and fixer solutions (Kodak). Images were obtained by scanning the developed film, and the abundance of proteins of each band was evaluated by densitometric analysis with the ImageJ-NIH image software. The effect of CoCl2 treatment on HIF-1α protein levels was expressed as the fold change variation of the optical density (OD), expressed in arbitrary units of HIF-1α signals, normalized to the respective loading control β-actin.
2.8. RNA Isolation and Reverse Transcription
The total RNA was extracted from a second aliquot with the EuroGold Total RNA Mini Kit (Euroclone, Milan, Italy) and subsequently quantified using a NanoDrop ND 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA), following the manufacturer's instructions.
Subsequently, 2 μg of RNA was reverse-transcribed using the High-Capacity cDNA Kit (Applied Biosystems, Foster City, California, USA) following the protocol provided by the developer. Real-time PCR (qRT-Polymerase Chain Reaction) was performed by SYBR®-Green technology using the ABI Prism 7000 apparatus (Applied Biosystems, Foster City, California, USA).
2.9. Gene Expression by qRT-PCR
Gene expression analyses were performed on 20 ng cDNA in a 25 μL reaction, following this recipe: 12.5 μL 2xMasterMix, 1 μL of each primer, 20 ng cDNA, and nuclease-free water up to 25 μL.
The thermocycler program consisted of an initial hot start cycle at 50°C for two minutes and 90°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for one minute, and a final cycle at 60°C for one minute. Each reaction was performed in triplicate. Negative controls (PCR mix without the addition of cDNA) were analyzed for each reaction. Human β-actin was used as an endogenous control to normalize gene expression levels.
2.10. Molecular Characterization and Gene Expression
We firstly evaluated the expression levels of different genes in untreated MIO-M1, to characterize the expression profile of these cells. We analyzed the expression of genes of the homeobox (Otx1, Otx2, and Otx3) and p53 families (TP53, TP63, and TP73), VEGF-A and WNT1. Table 2 summarizes the primers used. Each primer pair was tested to obtain the optimal concentration and annealing temperature. The geometrical efficiency of the reactions was assessed by a visual comparison of the amplification curves of our samples.
Table 2.
List of primers used in qRT-PCR experiments. Each primer sequence is written from the 5′ end to the 3′ end.
| Gene | Fw (5′–3′) | Rv (5′–3′) |
|---|---|---|
| ACTB | CGCGAGAAGATGACCCAGAT | ACAGCCTGGATAGCAACGTACA |
| Otx1 | TGCCGACTGCTTGGATTACA | GCCATGAGGCTTGGTCCTTA |
| Otx2 | CGCAGCTAGATGTGCTGGAA | TCGACTCGGGCAAGTTGATT |
| Otx3 | CCCCAAAGCTGAGAAGAGC | GCCACAGGAGTGATGGTCA |
| TP53 | TCTTCTGTCCCTTCCCAGAA | AGAATGCAAGAAGCCCAGAC |
| TP63 (TA isoform) | TTTGAAACTTCACGGTGTGC | TGAGCTGGGGTTTCTACGA |
| TP63 (ΔN isoform) | GGTTGGCAAAATCCTGGAG | GGTTCGTGTACTGTGGCTCA |
| TP73 (TA isoform) | AACCAGACAGCACCTACTTCG | CGCCCACCACCTCATTATT |
| TP73 (ΔN isoform) | AAGCGAAAATGCCAACAAAC | AGGCTCCGCAGCTAGTGA |
| VEGF-A | TGTGTGTGTGTGAGTGGTTGA | TCTCTGTGCCTCGGGAAG |
| WNT1 | CGCTGGAACTGTCCCACT | AACGCCGTTTCTCGACAG |
Threshold cycles were used to calculate the 2−ΔCt values that give us the fold change of the expression of these genes compared to the reference actin in untreated cells.
Conversely, the ΔΔCt method was used to evaluate the effect of the treatment on the expression levels of these genes. This method allowed for obtaining the 2−ΔΔCt, a value that describes the fold change of the gene expression in treated vs. control samples, normalized to β-actin.
2.11. Statistical Analysis
Data are expressed as mean ± SD for both western blot and qRT-PCR analysis. Statistical analysis has been conducted through a one-way ANOVA test and Tukey's multiple-comparison test. Results are considered significant with p < 0.05.
3. Results
3.1. MIO-M1 Morphology
MIO-M1 cells in culture tend to spread over the entire surface of the plate, showing a bipolar morphology, cytoplasmic projections, and a rough membrane appearance. As we can see from the images in Figure 2, CoCl2 treatment does not macroscopically affect cell morphology after either 24 h (Figure 2(b)) or 48 h (Figure 2(e)).
3.2. HIF-1α Protein Level Increases after CoCl2 Exposure
To confirm the induction of hypoxic stimulus in our experimental setting, we evaluated the HIF-1α protein amount by WB at each time point of treatment (Figure 3). The HIF-1α-specific primary antibody (Ab) detected a 120 kDa band, while β-actin Ab identified a band with a molecular weight of 43 kDa (Figure 3(a)).
Figure 3.
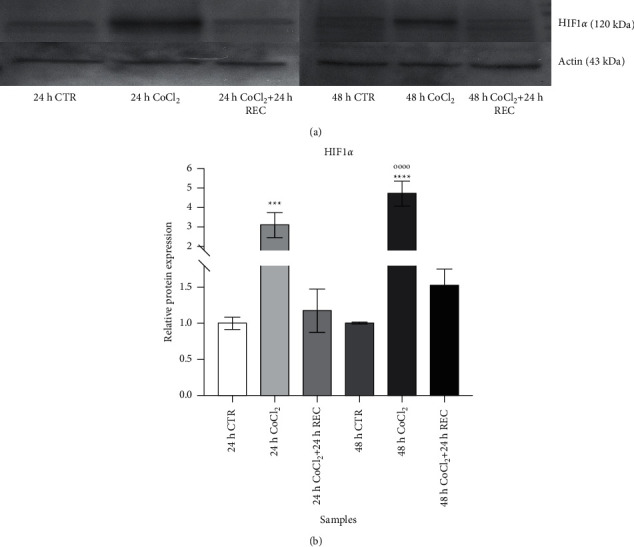
Analysis of HIF-1α and β-actin protein expression levels by western blot evaluated in MIO-M1 cells at different time points of treatment. (a) Representative immunoreactive bands for HIF-1α (top; 120 kDa) and β-actin (bottom; 43 kDa). For each sample, 200 μg of protein was loaded on SDS-8% polyacrylamide gel. (b) Relative expression of HIF-1α protein, normalized to the respective β-actin, is represented as mean ± SD of three repeated experiments. ∗∗∗p < 0.001 (24 h CoCl2 vs. 24 h CTR); ∗∗∗∗p < 0.0001 (48 h CoCl2 vs. 24 h CTR); and p < 0.0001 (48 h CoCl2 vs. 48 h CTR) by one-way ANOVA and Tukey's post hoc test.
As expected, the amount of HIF-1α protein significantly increased in response to CoCl2 treatment (24 h CoCl2 vs. 24 h CTR: p < 0.001; 48 h CoCl2 vs. 24 h CTR: p < 0.0001; and 48 h CoCl2 vs. 48 h CTR: p < 0.0001, evaluated by one-way ANOVA and Tukey's post hoc test), and the 24 h recovery phase after exposure to CoCl2 restored protein levels comparable to control samples (Figure 3(b)). Data in the graph (Figure 3(b)) represent the relative HIF-1α protein expression, normalized to the reference β-actin protein.
3.3. MIO-M1 Gene Expression Profile
We firstly evaluated the mRNA levels of a set of genes to characterize their expression profile in the untreated MIO-M1 cell lines.
Each gene expression level was normalized to β-actin, taken as a reference, applying the 2−ΔCt method to obtain a fold change value representing how much each gene of interest is expressed in untreated MIO-M1 cells (Figure 4).
Figure 4.
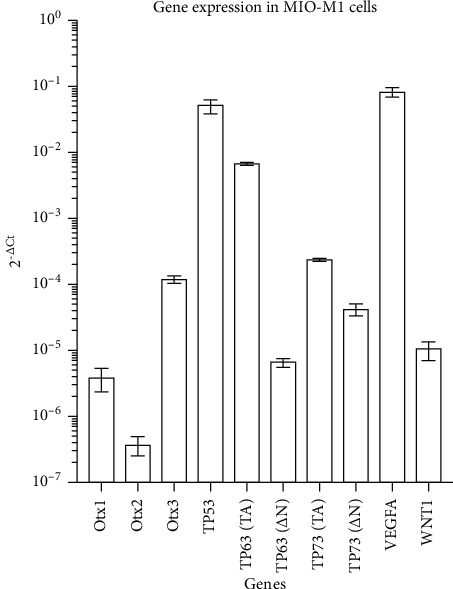
Molecular characterization of gene expression in untreated MIO-M1 cells. Each gene was normalized to β-actin expression level, to obtain a value representing the fold change between the two analyzed genes. On the Y-axis, the fold change value is reported. Each value is indicated as median ± SD.
Genes belonging to the Otx family, such as Otx1, Otx2, and Otx3, were nearly completely turned off. VEGF-A is the highest expressed gene, as expected from data observed in the literature considering the pivotal role of Müller cells in VEGF-A regulation in the retina [36, 38].
Finally, genes belonging to the p53 family showed a higher expression in the full-length TA isoforms, generally involved in tissue maintenance.
3.4. Expression of Otx1, Otx2, TP73, and VEGF-A after CoCl2 Treatment
We finally analyzed the gene expression levels of some of these genes by qRT-PCR after treatment with CoCl2. The data obtained are summarized in Figure 5, presenting fold change values for each gene, obtained through the 2−ΔΔCt method, which allows for determining the relative expression of a gene of interest, normalized toward β-actin and in treated vs. untreated samples.
Figure 5.
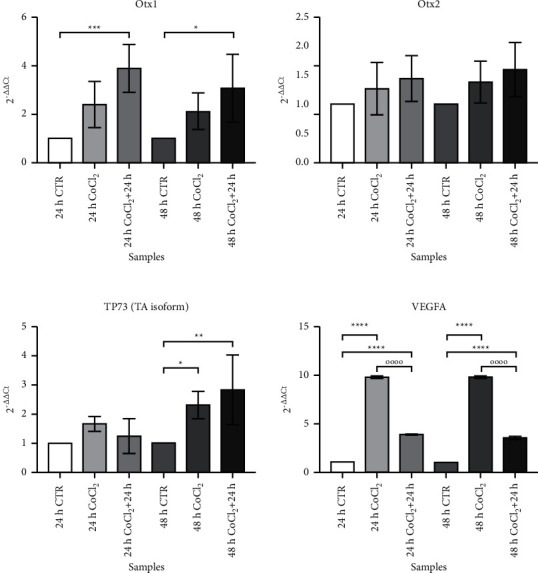
qRT-PCR on CTR- and CoCl2-treated MIO-M1 Müller cells. Each graph represents the expression levels of a single studied gene in different experimental conditions. All values are indicated as median ± SD. On the Y-axis, the fold change value is reported. Statistical analysis was performed by one-way ANOVA and Tukey's post hoc test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001.
We observed a strong upregulation, i.e., higher mRNA production, of VEGF-A in all analyzed samples, with higher fold changes in response to CoCl2 (24 h CoCl2: 9.748 ± 0.143 and 48 h CoCl2: 9.782 ± 0.191, p < 0.0001) compared to the recovery phase (3.878 ± 0.077 and 3.521 ± 0.189 for 24 h and 48 h + 24 h, with a p < 0.0001).
Otx2 expression levels did not show statistically significant modifications in response to the treatment or after the recovery time.
Interestingly, we observed an increased expression of the Otx1 gene in cells undergoing a recovery time after exposure to CoCl2 for either 24 h or 48 h (24 h CoCl2 + 24 h REC: 3.878 ± 0.9842, p < 0.001; 48 h CoCl2 + 24 h REC: 3.076 ± 1,397, p < 0.05).
The TA isoform of the TP73 gene was upregulated only in samples treated for 48 h, with a higher expression during the recovery phase compared with the treatment with only CoCl2 (48 h CoCl2: 2.301 ± 0.4676, p < 0.05; 48 h CoCl2 + 24 h REC: 2.825 ± 1.196, p < 0.01).
4. Discussion
Our research aimed to evaluate the expression modulation of genes involved in the response to stress and in differentiation, in Müller cells undergoing hypoxic stimulus, a typical condition of several retinal diseases. Cobalt chloride treatment can mimic hypoxia in two ways: cobalt inhibits the hydroxylation of a proline residue in the ODD (Oxygen-Dependent Degradation) domain of HIF-1α protein, thus preventing its recognition by the von Hippel–Lindau protein (pVHL) and the subsequent ubiquitination and degradation of HIF-1α, but it is also able to prevent the interaction between hydroxylated HIF-1α and pVHL [39, 40]. CoCl2 is also part of the vitamin B12 complex, and its accumulation causes ferroptosis-like cell death in neuronal cells. Moreover, it influences mitosis and the processes involved in preserving neuronal integrity [41].
Increased HIF-1α protein levels in response to CoCl2, but not in the recovery phase, confirmed the effectiveness of the treatment. The overexpression of VEGF-A in treated samples supported the activation of HIF-1α downstream pathways and the VEGF-A role in neuroprotection and as a survival factor [9–36].
The TP73 gene product role in hypoxia and angiogenesis is more controversial. Its activity in enhancing defense against oxidative stresses, a typical feature of hypoxia, is well documented [31–34], and it is essential for proper retinal vasculature formation [35]. Sabapathy hypothesizes a bifunctional role of the TAp73 protein: in the initial stages of hypoxia, HIF-1α activates TP73 expression and stabilizes the TAp73 protein that exerts a proangiogenic function activating downstream genes [42]. Once the vasculature is formed, TAp73 acts in a negative regulatory loop to turn off HIF-1α [42–44].
The observed upregulation of TP73 after 48 h of treatment with CoCl2 may be associated with a response to prolonged hypoxic stimulus mediated by HIF-1α accumulation.
The further increase in expression observed in the recovery phase could be due to the HIF-1α-independent protective role exerted to overcome the hypoxic insult.
To our knowledge, this is the first report that evaluates Otx1 and Otx2 expression modulation in response to hypoxia in the MIO-M1 cell line.
Both genes are involved in eye and ear tissue development and maintenance in adults [14–36, 38–46]; thus, we wanted to investigate their behavior in Müller cells during hypoxic stimuli.
We have already reported the expression and modulation of these two genes in retinal samples of patients affected by PVR, observing better surgical and clinical outcomes in patients with higher Otx1 expression compared with those with Otx2 [17].
Previous studies on two mouse models of intestinal ischemia/reperfusion and DNBS-induced (DiNitroBenzene Sulfonic acid) colitis in the enteric nervous system (ENS) showed OTX2 protein mainly expressed in neurons of the ENS in association with the neuronal isoform of nitric oxide synthase (nNOS), while OTX1 presented a glial distribution and an association with the inducible NOS [47, 48].
In accordance with these previous experiments, in our experimental models, we did not observe a variation in Otx2 expression levels during the hypoxic phase or in the recovery period, while Otx1 was upregulated during both recovery phases.
Considering its role in cellular differentiation and development, we can hypothesize that the overexpression of Otx1 in Müller cells under stress conditions is involved in cell gliosis, playing a role in the induction of proliferation and morphological processes observed in retinal pathological conditions [7]. One of the most relevant factors released from Müller cells under hypoxic conditions is the vascular endothelial growth factor (VEGF-A). VEGF-A supports the survival of endothelial cells and retinal neurons and may restrict glucose- and oxidative stress-induced damage in retinal vessels [49]. The effects of VEGF-A include vasodilation, revascularization, inflammation, and glial cell proliferation, which turn into a pathological pathway when VEGF-A levels exceed a threshold level. Retinal tissue degenerates due to tissue folds, edema, and atrophy, with a decrease in visual acuity and finally a loss of function of the eye. On the other side, according to ischemic stimuli, the Otx1 and other gene pathways may control the neuroregenerative and neuroprotective VEGF-A function. Understanding how to modulate these events may allow us to direct Müller cell gliosis toward reparation of the injury, with visual function recovery, and surgical or medical therapy will show a better prognosis.
It is well known that teleost and chicken retinae maintain regenerative properties, mainly associated with the Müller cell ability to dedifferentiate and redifferentiate into any cell type of retinal tissue [50]. This ability is lost in mammals, even if Müller cells cultured in vitro exhibit neural stem cell characteristics [51] and can differentiate if properly stimulated [52].
However, most of the retinal degenerative conditions in humans are associated with an altered proliferation of Müller cells that lead to the formation of glial scars, as in proliferative vitreoretinopathy. Reactive gliosis has been described in different retinal pathologies, including age-related macular degeneration (AMD), diabetes, glaucoma, and retinal detachment.
The identification of genes conferring differentiation capacity, such as Otx1, and how they behave during the hypoxic response can lead us to a better understanding of the dual mechanism, neuroprotective and/or cytotoxic, of microglial Müller cells.
This knowledge can be helpful in modulating and treating retinal pathologies pointing to Müller cells as key targets for new drug therapies.
The main limitation of this study is the fact that we considered only Müller cells and no other subtypes of retinal cells or pigmented epithelium cells. All retinal layers are involved in maintaining the correct retinal physiology. However, our preliminary study in vitro on Müller cell genes in hypoxic conditions could represent a good first step for further studies.
5. Conclusions
In conclusion, our experimental model of mimicked hypoxia showed an interesting mechanism in Müller cells: Otx1, Otx2, and TP73 gene expressions were altered after CoCl2 treatment of MIO-M1. The significant increase in Otx1 levels in the recovery phase pointed to its involvement in processes activated after a hypoxic stimulus: we need to verify in which pathway Otx1 plays a role and its association with vasoactive dynamics, as well as in inflammation and on its pathological or protective role in tissues. TP73 upregulation may suggest an activation of mechanisms of defense usually caused by the accumulation of oxidative stresses in hypoxia.
Data Availability
The data presented in this study are available on request from the corresponding author.
Ethical Approval
The study was conducted according to the guidelines of the Declaration of Helsinki.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Claudio Azzolini, Francesca Simonelli, and Giovanni Porta conceptualized the study; Giovanni Porta formulated the methodology; Giovanni Porta and Roberto Valli performed validation; Giovanni Micheloni, Vittoria Moretti, Francesco Acquati, Costantino Lucy, and Ferrara Fulvio conducted investigation; Claudio Azzolini and Giovanni Porta obtained resources; Vittoria Moretti, Francesco Acquati, Costantino Lucy, and Ferrara Fulvio curated data; Claudio Azzolini, Simone Donati, Elias Premi, and Giovanni Micheloni prepared the original draft of the manuscript; Claudio Azzolini, Simone Donati, and Giovanni Micheloni reviewed and edited the manuscript; and Claudio Azzolini, Giovanni Porta, Davide Borroni, Francesco Testa, and Francesca Simonelli supervised the work. The authors have read and agreed to the published version of the manuscript.
References
- 1.Willbold E., Layer P. G. Müller glia cells and their possible roles during retina differentiation in vivo and in vitro. Histology & Histopathology . 1998;13(2):531–552. doi: 10.14670/HH-13.531. [DOI] [PubMed] [Google Scholar]
- 2.Huxlin K. R., Sefton A. J., Furby J. H. The origin and development of retinal astrocytes in the mouse. Journal of Neurocytology . 1992;21(7):530–544. doi: 10.1007/bf01186955. [DOI] [PubMed] [Google Scholar]
- 3.Todd L., Suarez L., Quinn C., Fischer A. J. Retinoic acid-signaling regulates the proliferative and neurogenic capacity of Müller glia-derived progenitor cells in the avian retina. Stem Cells . 2018;36(3):392–405. doi: 10.1002/stem.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bringmann A., Pannicke T., Grosche J., et al. Müller cells in the healthy and diseased retina. Progress in Retinal and Eye Research . 2006;25(4):397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Pournaras C. J., Rungger-Brändle E., Riva C. E., Hardarson S. H., Stefansson E. Regulation of retinal blood flow in health and disease. Progress in Retinal and Eye Research . 2008;27(3):284–330. doi: 10.1016/j.preteyeres.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Kermorvant-Duchemin E., Sapieha P., Sirinyan M., et al. Understanding ischemic retinopathies: emerging concepts from oxygen-induced retinopathy. Documenta Ophthalmologica . 2010;120(1):51–60. doi: 10.1007/s10633-009-9201-x. [DOI] [PubMed] [Google Scholar]
- 7.Bringmann A., Iandiev I., Pannicke T., et al. Cellular signaling and factors involved in Müller cell gliosis: neuroprotective and detrimental effects. Progress in Retinal and Eye Research . 2009;28(6):423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y. J., Park S. J., Kim N. R., Chin H. S. Effects of histone deacetylase inhibitor (valproic acid) on the expression of hypoxia-inducible factor-1 alpha in human retinal müller cells. Korean Journal of Ophthalmology . 2017;31(1):80–85. doi: 10.3341/kjo.2017.31.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishijima K., Ng Y.-S., Zhong L., et al. Vascular endothelial growth factor-A is a survival factor for retinal neurons and a critical neuroprotectant during the adaptive response to ischemic injury. American Journal Of Pathology . 2007;171(1):53–67. doi: 10.2353/ajpath.2007.061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saint-Geniez M., Maharaj A. S. R., Walshe T. E., et al. Endogenous VEGF is required for visual function: evidence for a survival role on müller cells and photoreceptors. PLoS One . 2008;3(11) doi: 10.1371/journal.pone.0003554.e3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kastury K., Druck T., Huebner K., et al. Chromosome locations of human EMX and OTX genes. Genomics . 1994;22(1):41–45. doi: 10.1006/geno.1994.1343. [DOI] [PubMed] [Google Scholar]
- 12.Boncinelli E., Simeone A., Acampora D., Gulisano M. Homeobox genes in the developing central nervous system. Annales de Genetique . 1993;36(1):30–37. [PubMed] [Google Scholar]
- 13.Klein F., Mahr P., Galova M., et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell . 1999;98(1):91–103. doi: 10.1016/s0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 14.Bovolenta P., Mallamaci A., Briata P., Corte G., Boncinelli E. Implication of OTX2 in pigment epithelium determination and neural retina differentiation. Journal of Neuroscience . 1997;17(11):4243–4252. doi: 10.1523/jneurosci.17-11-04243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida A., Furukawa A., Koike C., et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nature Neuroscience . 2003;6(12):1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 16.Koike C., Nishida A., Ueno S., et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Molecular and Cellular Biology . 2007;27(23):8318–8329. doi: 10.1128/mcb.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fossat N., Le Greneur C., Béby F., et al. A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors. BMC Developmental Biology . 2007;7(10):p. 122. doi: 10.1186/1471-213X-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa T., Morrow E. M., Cepko C. L. Crx, a novel Otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell . 1997;91(4):531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 19.Ba-Charvet K. T., von Boxberg Y., Guazzi S., Boncinelli E., Godement P. A potential role for the OTX2 homeoprotein in creating early “highways” for axon extension in the rostral brain. Development . 1998;125(21):4273–4282. doi: 10.1242/dev.125.21.4273. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Morales J. R., Signore M., Acampora D., Simeone A., Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development . 2001;128(11):2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- 21.Azzolini C., Pagani I. S., Pirrone C., et al. Expression of VEGF-A, Otx homeobox and p53 family genes in proliferative vitreoretinopathy. Mediators of Inflammation . 2013;2013:8. doi: 10.1155/2013/857380.857380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuo I., Kuratani S., Kimura C., Takeda N., Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes & Development . 1995;9(21):2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 23.Mallamaci A., Di Blas E., Briata P., Boncinelli E., Corte G. OTX2 homeoprotein in the developing central nervous system and migratory cells of the olfactory area. Mechanisms of Development . 1996;58(1-2):165–178. doi: 10.1016/s0925-4773(96)00571-0. [DOI] [PubMed] [Google Scholar]
- 24.Pirrone C., Chiaravalli A. M., Marando A., et al. OTX1 and OTX2 as possible molecular markers of sinonasal carcinomas and olfactory neuroblastomas. European Journal of Histochemistry . 2017;61:p. 2730. doi: 10.4081/ejh.2017.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micheloni G., Millefanti G., Conti A., et al. Identification of OTX1 and OTX2 as two possible molecular markers for sinonasal carcinomas and olfactory neuroblastomas. Journal of Visualized Experiments . 2019;144 doi: 10.3791/56880. [DOI] [PubMed] [Google Scholar]
- 26.Acampora D., Mazan S., Tuorto F., et al. Transient dwarfism and hypogonadism in mice lacking Otx1 reveal prepubescent stage-specific control of pituitary levels of GH, FSH and LH. Development . 1998;125(7):1229–1239. doi: 10.1242/dev.125.7.1229. [DOI] [PubMed] [Google Scholar]
- 27.Diaczok D., Romero C., Zunich J., Marshall I., Radovick S. A novel dominant negative mutation of OTX2 associated with combined pituitary hormone deficiency. Journal of Clinical Endocrinology & Metabolism . 2008;93(11):4351–4359. doi: 10.1210/jc.2008-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.del Blanco D. G., de Graaff L. C. G., Visser T. J., Hokken-Koelega A. C. S. Growth hormone insensitivity syndrome caused by a heterozygous GHR mutation: phenotypic variability owing to moderation by nonsense-mediated decay. Clinical Endocrinology . 2012;76(5):706–712. doi: 10.1111/j.1365-2265.2011.04304.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen H., Sukumar S. Role of homeobox genes in normal mammary gland development and breast tumorigenesis. Journal of Mammary Gland Biology and Neoplasia . 2003;8(2):159–175. doi: 10.1023/a:1025996707117. [DOI] [PubMed] [Google Scholar]
- 30.Pagani I. S., Terrinoni A., Marenghi L., et al. The mammary gland and the homeobox gene Otx1. Breast Journal . 2010;16(1):S53–S56. doi: 10.1111/j.1524-4741.2010.01006.x. [DOI] [PubMed] [Google Scholar]
- 31.Terrinoni A., Pagani I. S., Zucchi I., et al. OTX1 expression in breast cancer is regulated by p53. Oncogene . 2011;30(27):3096–3103. doi: 10.1038/onc.2011.31. [DOI] [PubMed] [Google Scholar]
- 32.Amelio I., Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends in Biochemical Sciences . 2015;40(8):425–434. doi: 10.1016/j.tibs.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Velletri T., Romeo F., Tucci P., et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle . 2013;12(22):3564–3573. doi: 10.4161/cc.26771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Alessandro A., Amelio I., Berkers C. R., et al. Metabolic effect of TAp63alpha: enhanced glycolysis and pentose phosphate pathway, resulting in increase dantioxidant defense. Oncotarget . 2014;5:7722–7733. doi: 10.18632/oncotarget.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Alonso R., Martin-Lopez M., Gonzalez-Cano L., et al. p73 is required for endothelial cell differentiation, migration and the formation of vascular networks regulating VEGF and TGFbeta signaling. Cell Death & Differentiation . 2015;22(8):1287–1299. doi: 10.1038/cdd.2014.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Y.-Z. VEGF production and signaling in Müller glia are critical to modulating vascular function and neuronal integrity in diabetic retinopathy and hypoxic retinal vascular diseases. Vision Research . 2017;139:108–114. doi: 10.1016/j.visres.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limb G. A., Salt T. E., Munro P. M., Moss S. E., Khaw P. T. In vitro characterization of a spontaneously immortalized human müller cell line (MIO-M1) Investigative Ophthalmology & Visual Science . 2002;43(3):864–869. [PubMed] [Google Scholar]
- 38.Fu S., Dong S., Zhu M., et al. Müller glia are a major cellular source of survival signals for retinal neurons in diabetes. Diabetes . 2015;64(10):3554–3563. doi: 10.2337/db15-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sears J. E., Hoppe G. Triamcinolone acetonide destabilizes VEGF mRNA in Müller cells under continuous cobalt stimulation. Investigative Opthalmology & Visual Science . 2005;46(11):4336–4341. doi: 10.1167/iovs.05-0565. [DOI] [PubMed] [Google Scholar]
- 40.Yuan Y., Hilliard G., Ferguson T., Millhorn D. E. Cobalt inhibits the interaction between hypoxia-inducible factor-α and von Hippel-lindau protein by direct binding to hypoxia-inducible factor-α. Journal of Biological Chemistry . 2003;278(18):15911–15916. doi: 10.1074/jbc.M300463200. [DOI] [PubMed] [Google Scholar]
- 41.Gupta G., Gliga A., Hedberg J., et al. Cobalt nanoparticles trigger ferroptosis‐like cell death (oxytosis) in neuronal cells: potential implications for neurodegenerative disease. The FASEB Journal . 2020;34(4):5262–5281. doi: 10.1096/fj.201902191RR. [DOI] [PubMed] [Google Scholar]
- 42.Sabapathy K. p73: a positive or negative regulator of angiogenesis, or both? Molecular and Cellular Biology . 2015;36:848–854. doi: 10.1128/MCB.00929-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amelio I., Inoue S., Markert E. K., et al. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1α degradation. Proceedings of the National Academy of Sciences . 2015;112(1):226–231. doi: 10.1073/pnas.1410609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stantic M., Sakil H. A. M., Zirath H., et al. TAp73 suppresses tumor angiogenesis through repression of proangiogenic cytokines and HIF-1α activity. Proceedings of the National Academy of Sciences . 2015;112(1):220–225. doi: 10.1073/pnas.1421697112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu B., Zhang H., Zhu M., Le Y.-Z. Critical role of trophic factors in protecting müller glia: implications to neuroprotection in age-related macular degeneration, diabetic retinopathy, and anti-VEGF therapies. Retinal Degenerative Diseases . 2019;1185:469–473. doi: 10.1007/978-3-030-27378-1_77. [DOI] [PubMed] [Google Scholar]
- 46.Palombo R., Porta G., Bruno E., et al. OTX2 regulates the expression of TAp63 leading to macular and cochlear neuroepithelium development. Aging . 2015;7:928–36. doi: 10.18632/aging.100839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filpa V., Carpanese E., Marchet S., et al. Nitric oxide regulates homeoprotein OTX1 and OTX2 expression in the rat myenteric plexus after intestinal ischemia-reperfusion injury. American Journal of Physiology-Gastrointestinal and Liver Physiology . 2017;312(4):G374–G389. doi: 10.1152/ajpgi.00386.2016. [DOI] [PubMed] [Google Scholar]
- 48.Bistoletti M., Micheloni G., Barazzini N., et al. Homeoprotein OTX1 and OTX2 involvement in rat myenteric neuron adaptation after DNBS-induced colitis. PeerJ . 2020;8 doi: 10.7717/peerj.8442.e8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada H., Yamada E., Hackett S. F., Ozaki H., Okamoto N., Campochiaro P. A. Hyperoxia causes decreased expression of vascular endothelial growth factor and endothelial cell apoptosis in adult retina. Journal of Cellular Physiology . 1999;179(2):149–156. doi: 10.1002/(sici)1097-4652(199905)179:2<149::aid-jcp5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Lenkowski J. R., Raymond P. A. Müller glia: stem cells for generation and regeneration of retinal neurons in teleost fish. Progress in Retinal and Eye Research . 2014;40:94–123. doi: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence J. M., Singhal S., Bhatia B., et al. MIO‐M1 cells and similar müller glial cell lines derived from adult human retina exhibit neural stem cell characteristics. Stem Cells . 2007;25(8):2033–2043. doi: 10.1634/stemcells.2006-0724. [DOI] [PubMed] [Google Scholar]
- 52.Giannelli S. G., Demontis G. C., Pertile G., Rama P., Broccoli V. Adult human Müller glia cells are a highly efficient source of rod photoreceptors. Stem Cells . 2011;29(2):344–356. doi: 10.1002/stem.579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.


