Abstract
A DNA sequence was identified in isolates of Salmonella enterica serotype Typhimurium definitive type 104 (DT104). The PCR amplification of an internal segment of this sequence identified DT104 and the closely related U302 phage type among 146 isolates of S. enterica serotype Typhimurium tested, thus providing a tool for rapid identification of DT104 and related isolates.
An epidemic strain of Salmonella enterica serotype Typhimurium definitive type 104 (DT104) rose to prominence when this multidrug-resistant (R-type ACSSuT) pathogen was identified as a major cause of salmonellosis in people and farm animals in Britain (12) and in people in the United States (5). The epidemic strain of DT104 was defined by British researchers based upon Salmonella serovar Typhimurium phage type 104, the R-type ACSSuT, and a plasmid profile consisting of a single ∼60-MDa plasmid (12).
Antimicrobial susceptibility tests to determine R-type (2) and plasmid profile analyses (10) are widely available in U.S. laboratories, but identification of S. enterica serotype Typhimurium phage types requires the maintenance of a phage library and specially trained personnel. Thus, phage typing will likely always be limited to a few centralized laboratories. In addition, some isolates cannot be assigned a phage type with the available phage library. These difficulties hinder the progress of epidemiological investigations. We describe in this note the identification of a DNA sequence that is unique to the DT104 and U302 phage types among S. enterica serotype Typhimurium isolates, and the development of a PCR assay to identify isolates containing this sequence. This PCR assay can be used to rapidly screen suspect samples or isolates for further testing and identification.
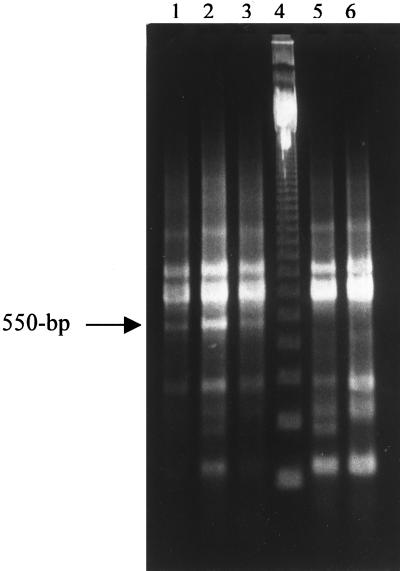
The PCR amplification of the 16S-to-23S spacer region of bacterial rRNA genes has been used to detect polymorphisms in bacterial species and to identify strains of S. enterica serotype Typhimurium associated with disease outbreaks (8, 9). Using this technique on bovine isolates of S. enterica serotype Typhimurium, the DT104 isolates demonstrated a unique band compared to other phage types (Fig. 1). This unique band was purified from the agarose gel and sequenced by the Laboratory for Bioanalysis at Washington State University. The sequence data are in boldface in Fig. 2.
FIG. 1.
PCR amplification products of isolates of S. enterica serotype Typhimurium obtained with primers to the conserved regions of the 16S and 23S ribosomal genes as described by Kostman et al. (8) and analyzed by 2% agarose gel electrophoresis and staining with ethidium bromide. Lanes: 1 to 3, DT104; 4, 123-bp DNA ladder (Life Technologies, Gaithersburg, Md.); 5, phage type 1; 6, phage type 193. The ∼550-bp polymorphism of the DT104 isolates is indicated by the arrow.
FIG. 2.
The 1,767-bp sequence identified in S. enterica serotype Typhimurium DT104. Boldface bases indicate the sequence identified as a polymorphism in Fig. 1 that was the result of the primers to the rRNA gene sequences annealing to nonribosomal sequences (identified by dotted arrows [·········>]). The positions of primers DT104F and DT104R are indicated by solid arrows (➞). The positions of additional primers designed to extend the sequence are indicated by dashed arrows (–––>).
The original sequence was extended in both the 5′ and 3′ directions by using sequence-specific primers (Fig. 2) and a kit designed to facilitate genomic walking (The Vectorette System; Genosys, The Woodlands, Tex.). A search of the nucleotide sequence databases did not produce any matches to known ribosomal sequences, demonstrating that the amplified polymorphism identified in Fig. 1 was the result of the annealing of the primers to nonribosomal sequences. The possible locations of the annealing sites are indicated on the sequence in Fig. 2.
To determine the association of this sequence with S. enterica serotype Typhimurium DT104, primers were designed for PCR amplification of an internal segment of the sequence. Figure 2 illustrates the locations of the primers DT104F (5′-GTCAGCAGTGTATGGAGCGA-3′) and DT104R (5′-AGTAGCGCCAGGACTCGTTA-3′), which were designed to amplify a 162-bp segment. In addition, primers INVA-1 (5′-ACAGTGCTCGTTTACGACCTGAAT-3′) and INVA-2 (5′-AGACGACTGGTACTGATCGATAAT-3′), which amplify a 243-bp segment of the Salmonella invA gene (4, 11), were included in the PCR assay as a positive control for sample preparation and the amplification reaction.
To evaluate the multiplex PCR, 239 Salmonella isolates representing a wide range of serotypes and sources were selected from a bank of Salmonella isolates maintained by the Field Disease Investigation Unit (FDIU), College of Veterinary Medicine, Washington State University, Pullman, Wash. These isolates were associated with clinical cases of salmonellosis and were collected by the FDIU or submitted to the Washington Animal Diseases Diagnostic Laboratory between 1986 and 1997. All isolates were serogrouped and tested for antimicrobial susceptibility (2) by the FDIU. Serotypes were determined by the National Veterinary Services Laboratory (NVSL), U.S. Department of Agriculture, Ames, Iowa. Phage typing was performed with 57 of the 146 isolates of S. enterica serotype Typhimurium by the National Laboratory for Enteric Pathogens, Health Canada, Ottawa, Canada (6), or the NVSL. Plasmid profiles (10) for 21 isolates of S. typhimurium phage type 104 were determined by the FDIU to confirm these isolates were of the epidemic strain.
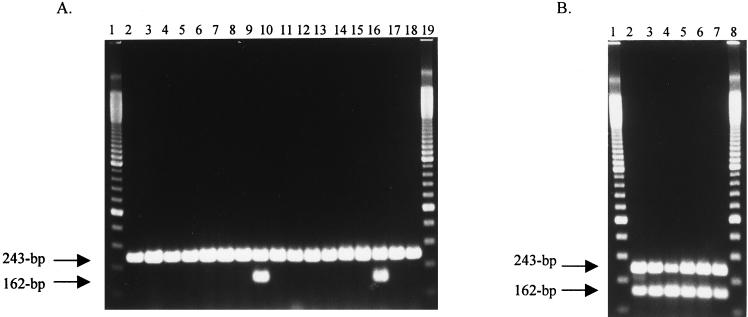
Cell lysates for each isolate were prepared in duplicate by suspending a single bacterial colony in 300 μl of sterile distilled water in a microcentrifuge tube and boiling for 20 min. Cell lysates were stored at −20°C until amplified. Aliquots (5 μl) of cell lysates were each amplified in a 25-μl reaction mixture with 1 μM (each) primers INVA-1, INVA-2, DT104F, and DT104R; 200 μM (each) deoxynucleoside triphosphates (dNTP), 2 mM MgCl2, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, and 1.25 U of Taq DNA polymerase (Life Technologies, Gaithersburg, Md.). Amplification was performed in 0.2-ml microreaction tubes in a RapidCycler (Idaho Technology, Idaho Falls, Idaho) as follows: denaturation at 96°C for 1 min; 30 cycles of 96°C for 30 s, 60°C for 30 s, and 72°C for 35 s; and 1 final extension cycle at 72°C for 30 s. The PCR products were visualized on ethidium bromide-stained agarose gels (Fig. 3).
FIG. 3.
Multiplex PCR amplification of isolates of Salmonella analyzed by 2% agarose gel electrophoresis. (A) Lanes: 1 and 19, 100-bp DNA Step Ladder (Promega Corp., Madison, Wis.); 2 to 18, isolates of S. enterica serotype Typhimurium with phage types 1, 2, 3, 10, 51, 66, 80, 104, 121, 132, 160, 193, 208, 274, U302, 771, and 811, respectively. (B) Lanes: 1 and 8, 100-bp DNA Step ladder; 2 to 7, isolates of S. enterica serotypes B (Brandenburg), C (Lille), C2 (Muenchen), D (Dublin), D (Enteritidis), and B (Typhimurium DT104), respectively. The 243-bp amplification product of the Salmonella invA gene and the 162-bp amplification product of the newly identified sequence are indicated.
Table 1 illustrates the variety of serogroups, serotypes, and sample sources tested by the multiplex PCR assay. In addition to the S. typhimurium DT104 isolates, 15 isolates of other serotypes produced the 162-bp product after PCR amplification, including S. enterica serotype Brandenburg (1 of 1), S. enterica serotype Lille (3 of 3), S. enterica serotype Muenchen (1 of 2), S. enterica serotype Dublin (9 of 9), and S. enterica serotype Enteritidis (1 of 3). The amplification product from one representative isolate of each positive serotype was purified and sequenced. The sequence data in Fig. 4 demonstrate the PCR assay amplifies a similar 162-bp segment of DNA in these diverse serotypes.
TABLE 1.
Serogroup, serotype, antibiotic susceptibility, PCR assay, and source of 239 isolates of Salmonella used in this study
| Serogroup | Serotype | n | ACSSuTa | 162-bp ampliconb
|
Source | |
|---|---|---|---|---|---|---|
| + | − | |||||
| A | Blockley | 1 | S | 1 | Frog | |
| Senftenberg | 2 | S | 2 | Bovine, feed | ||
| B | 4,12 Monophasic | 1 | S | 1 | Bovine | |
| Agona | 4 | S | 4 | Bovine, canine | ||
| Arizona | 2 | S | 2 | Bovine, ovine | ||
| Brandenburg | 1 | S | 1 | Porcine | ||
| Bredeney | 4 | S | 4 | Bovine, feed | ||
| California | 1 | S | 1 | Bovine | ||
| Derby | 2 | S | 2 | Cheetah | ||
| Heidelberg | 23 | S | 22 | Bovine | ||
| R | 1 | Cheetah | ||||
| Infantis | 1 | S | 1 | Bovine | ||
| Java | 2 | S | 2 | Cheetah | ||
| Reading | 2 | S | 2 | Canine, mink | ||
| Saint Paul | 1 | S | 1 | Bovine | ||
| Typhimurium | 146 | S | 117 | Bovine, emu, feline, human | ||
| R | 24 | 5 | Bovine, coyote, elk, human | |||
| C | Blockley | 2 | S | 2 | Bovine | |
| Cholerasuis | 1 | S | 1 | Bovine | ||
| Hadar | 2 | S | 2 | Bovine, canine | ||
| Istanbul | 2 | S | 2 | Avian | ||
| Johannesburg | 1 | S | 1 | Canine | ||
| Lille | 3 | S | 3 | Bovine | ||
| Tennessee | 1 | S | 1 | Canine | ||
| C1 | Infantis | 2 | S | 2 | Bovine, porcine | |
| Mbandka | 2 | S | 2 | Bovine | ||
| Montevideo | 1 | S | 1 | Bovine | ||
| Newport | 1 | S | 1 | Bovine | ||
| Thompson | 2 | S | 2 | Bovine | ||
| C2 | Blockley | 2 | S | 2 | Bovine | |
| Hadar | 2 | S | 2 | Canine, mink | ||
| Muenchen | 2 | S | 1 | 1 | Equine, iguana | |
| Newport | 2 | R | 2 | Bovine | ||
| D | Berta | 1 | S | 1 | Bovine | |
| Dublin | 9 | S | 8 | Bovine | ||
| R | 1 | Bovine | ||||
| Enteritidis | 3 | S | 1 | 1 | Hedgehog, canine | |
| R | 1 | Equine | ||||
| Loma Linda | 1 | S | 1 | Squirrel | ||
| E | Anatam | 2 | S | 1 | Equine | |
| R | 1 | Equine | ||||
| Given | 2 | S | 2 | Bovine, opossum | ||
R and S, resistance (R) to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline or susceptibility (S) to one or more of these antibiotics.
+ and −, presence (+) or absence (−) of a 162-bp product following amplification with primers DT104F and DT104R.
FIG. 4.
Comparison of the sequences of the 162-bp product generated by the PCR amplification of an isolate of each positive serotype by using primers DT104F and DT104R. Nucleotides that differ from the consensus sequence are highlighted.
Table 2 illustrates that among the 54 isolates of S. enterica serotype Typhimurium with phage typing data, all 21 DT104 isolates tested by this multiplex PCR produced the 162-bp amplification product. In addition, three isolates with phage type U302 produced the 162-bp amplification product, and U302 is considered an offspring of the DT104 strain (R. D. Khakhria, personal communication). None of the 15 other defined phage types of S. enterica serotype Typhimurium tested produced the 162-bp product following PCR. Three isolates of multidrug-resistant S. enterica serotype Typhimurium submitted for phage typing could not be assigned a defined phage type, and these isolates also did not produce the 162-bp product following PCR.
TABLE 2.
Phage type, antibiotic susceptibility, and PCR assay of 57 isolates of S. enterica serotype Typhimurium
| Phage type | ACSSuTa | 162-bp ampliconb
|
|
|---|---|---|---|
| + | − | ||
| 1 | S | 1 | |
| 2 | S | 1 | |
| 3 | S | 1 | |
| 10 | S | 5 | |
| R | 1 | ||
| 51 | S | 3 | |
| 66 | S | 1 | |
| 80 | S | 1 | |
| 104 | R | 21 | |
| 121 | S | 1 | |
| 132 | S | 1 | |
| 160 | S | 1 | |
| 193 | S | 6 | |
| 208 | S | 2 | |
| 274 | S | 1 | |
| U302 | R | 3 | |
| 771 | S | 2 | |
| R | 1 | ||
| 811 | S | 1 | |
| Unknownc | R | 3 | |
R and S, resistance (R) to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline or susceptibility (S) to one or more of these antibiotics.
+ and −, presence (+) or absence (−) of a 162-bp product following amplification with primers DT104F and DT104R.
Three isolates could not be typed with the available phage library.
A sequence identity search (1) produced a closely matched alignment (98% identity) between bases 1645 and 1716 of the newly identified sequence in S. enterica serotype Typhimurium DT104 and 71 of 72 bases of an unfinished fragment of S. enterica serotype Typhi (microbial genome BLAST database accession no. Sanger_601). An additional alignment (80% identity) was identified for 382 bp of the newly identified sequence with a cDNA clone from a mouse blastocyst that is described as being similar to replication protein 14 from bacteriophage Φ80 (GenBank accession no. AA574821).
Recent studies have described PCR assays to detect S. enterica serotype Typhimurium DT104 based upon amplification of regions of antibiotic resistance genes (3, 7). Our data indicate that the sequence we have identified in DT104 is not associated with the R-type ACSSuT. Fifteen isolates among other serotypes yield an amplification product similar to DT104, but these isolates were predominantly sensitive to one or more of the antibiotics that distinguish multidrug-resistant DT104. There was no common resistance pattern among these non-Typhimurium-positive isolates, and 5 of the 15 isolates were sensitive to all five antibiotics. Also, five isolates of S. enterica serotype Typhimurium—one phage type 10, one phage type 771, and three isolates that could not be phage typed—all have the R-type ACSSuT and do not yield an amplification product in our PCR assay (Table 2).
In conclusion, we have identified a 1,767-bp sequence in S. enterica serotype Typhimurium DT104 and developed a PCR assay to amplify an internal segment of this sequence. This multiplex PCR assay, in combination with serotype data, identifies the DT104 and U302 phage types. Furthermore, the similarity of the 162-bp amplification products among different serotypes of Salmonella and the alignment of a portion of the extended sequence with an unfinished fragment of S. enterica serotype Typhi suggest the possibility of the horizontal transfer of DNA.
Nucleotide sequence accession number.
The sequence of DT104 has been submitted to GenBank under accession no. AF275268.
Acknowledgments
This work was funded by a grant from the American Veterinary Medical Foundation and the USDA Fund for Rural America.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;36:493–496. [PubMed] [Google Scholar]
- 3.Carlson S A, Bolton L F, Briggs C E, Hurd H S, Sharma V K, Fedorka-Cray P J, Jones B D. Detection of multiresistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol Cell Probes. 1999;13:213–222. doi: 10.1006/mcpr.1999.0240. [DOI] [PubMed] [Google Scholar]
- 4.Chiu C-H, Ou J T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol. 1996;34:2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 6.Khakhria R D, Woodward D, Johnson W M, Poppe C. Salmonella isolated from humans, animals and other sources in Canada, 1983–92. Epidemiol Infect. 1997;119:15–23. doi: 10.1017/s0950268897007577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan A A, Nawaz M S, Khan S A, Cerniglia C E. Detection of multidrug-resistant Salmonella typhimurium DT104 by multiplex polymerase chain reaction. FEMS Microbiol Lett. 2000;182:355–360. doi: 10.1111/j.1574-6968.2000.tb08921.x. [DOI] [PubMed] [Google Scholar]
- 8.Kostman J R, Edlind T D, LiPuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natasi A, Mammima C. Epidemiological evaluation by PCR ribotyping of sporadic and outbreak-associated strains of Salmonella enterica serotype typhimurium. Res Microbiol. 1995;146:99–106. doi: 10.1016/0923-2508(96)80274-9. [DOI] [PubMed] [Google Scholar]
- 10.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahn K, DeGrandis S A, Clarke R C, McEwen S A, Gal'an J E, Ginochhio C, Curtiss R, Gyles C L. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of salmonella. Mol Cell Probes. 1992;6:271–290. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 12.Wall P G, Morgan D, Lamden K, Ryan M, Griffin M, Threlfall E J, Ward L R, Rowe B. A case control study of infection with an epidemic strain of multiresistant Salmonella typhimurium DT 104 in England and Wales. Commun Dis Rep CDR Rev. 1994;4:R130–R135. [PubMed] [Google Scholar]