Summary
Studies for sepsis prediction using machine learning are developing rapidly in medical science recently. In this review, we propose a set of new evaluation criteria and reporting standards to assess 21 qualified machine learning models for quality analysis based on PRISMA. Our assessment shows that (1.) the definition of sepsis is not consistent among the studies; (2.) data sources and data preprocessing methods, machine learning models, feature engineering, and inclusion types vary widely among the studies; (3.) the closer to the onset of sepsis, the higher the value of AUROC is; (4.) the improvement in AUROC is primarily due to using machine learning as a feature engineering tool; (5.) deep neural networks coupled with Sepsis-3 diagnostic criteria tend to yield better results on the time series data collected from patients with sepsis. The new evaluation criteria and reporting standards will facilitate the development of improved machine learning models for clinical applications.
Subject areas: Clinical medicine, Machine learning
Graphical abstract
Highlights
-
•
New evaluation/reporting standard for sepsis prediction machine learning models
-
•
Major limitations in the current models for sepsis prediction have been identified
-
•
We strongly suggest using machine learning as a feature engineering tool
-
•
Recommending multilayer neural networks and Sepsis 3.0 for yield better result
Clinical medicine; Machine learning
Introduction
Sepsis is a significant threat to patients' lives. A meta-analysis estimated about 31.5 million sepsis and 19.4 million severe sepsis cases occur each year, contributing to 5.3 million deaths worldwide (Fleischmann et al., 2016). This doomed scenario is further amplified under the current COVID-19 pandemic, where most of the deceased could be traced to sepsis (Alhazzani et al.,2020).
In 2016, the Third International Consensus Definition for Sepsis and Septic Shock (Sepsis-3) defined sepsis as “life-threatening organ dysfunction resulting from dysregulated host responses to infection.” It pointed out sepsis' death risk and the necessity of early identification and intervention (Cecconi et al.,2018). Early warning and accurate prediction on sepsis, which provides opportunities for physicians to take preventative measures to alleviate its devastating consequences, is recognized by researchers. A successful early warning together with the best clinical technique provides the best chance to reduce mortality and lower the risk of the severe septic shock (Shashikumar et al.,2017; Mira et al.,2017; Singer et al.,2016). Some clinical prognostic tools, such as Sequential Organ Failure Assessment (SOFA), Modified Early Warning Score (MEWS), Systemic Inflammatory Response Syndrome (SIRS), and quick Sequential Organ Failure Assessment (qSOFA) have been developed to predict the risk of death after the onset of sepsis (Raith et al.,2017). But these are not sufficiently reliable because most values of the tested markers come from ICU admission, which can hardly be linked definitively to the onset of infection. Consequently, traditional methods have limitations to accurately identify or predict the onset of sepsis and make high a fidelity prognosis. A new methodology is clearly in need.
Two systematic reviews evaluated the performance of machine learning models used in prediction for occurrence and prognosis of sepsis in the past (Fleuren et al.,2020; Islam et al.,2019). However, the influence brought about by the evolution of diagnostic criteria has never been discussed. Compared to the old criteria, Sepsis-3 needs more clinical data to complete SOFA assessment and to confirm infection. In addition, there has not been any consensus on how to establish a reasonable dataset, an appropriate feature-treatment method, and how to obtain a prediction of sepsis development dynamically. The goal of this review is to identify the characteristics and shortcomings in the models and methods in the previous studies, and try to establish a unified standard and evaluation tool for machine learning models in order to guide the model development in medical science in the future to make reliable predictions on this deadly ailment.
Results
Studies included
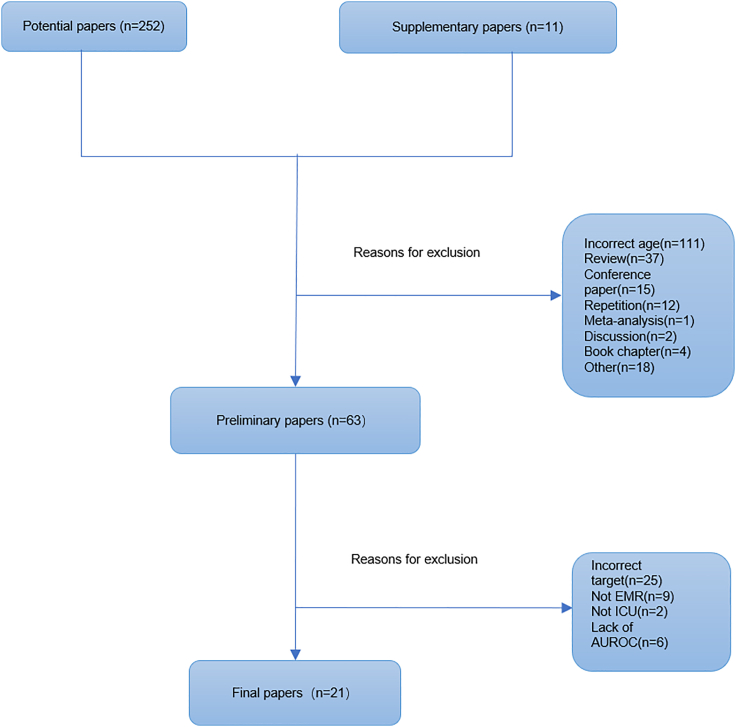
A total of twenty-one studies are included in this review from two hundred and sixty-two potentially eligible papers based on our criteria (Figure 1). Most selected studies focused on early sepsis detection, prediction, and mortality. Only two aimed at predicting severe sepsis (Table 1). We notice that seven studies used data from The Medical Information Mart for Intensive Care (MIMIC) database. Two studies used data from the University of California San Francisco Medical Center database and the Beth Israel Deaconess Medical Center database (UCSF + BIDMC database).
Figure 1.
Literature screening flowchart
Table 1.
Basic information of the included studies
| Study | Sepsis definition | Target | Data sources | Missing data processing | Training data | Testing data | Validation data |
|---|---|---|---|---|---|---|---|
| Delahanty et al. (2019) | sepsis3.0 | Early prediction of sepsis | 49 urban community hospitals operated by Tenet Healthcare | NR | 1,839,503 | 920,026 | NR |
| Barton et al. (2019) | sepsis3.0 | Detection and early prediction of sepsis | UCSF data+BIDMC data | Carry-forward and replacing by mean | NR | NR | NR |
| Taylor et al. (2016) | Infection + SIRS | Mortality prediction of sepsis | Four emergency departments | K-means | 4222 | NR | 1056 |
| Kam and Kim (2017) | ICD-9 | Detection and early prediction of sepsis | MIMIC-II | Replacing by nearest measured value | 252 | 72 | 36 |
| Mao et al. (2018) | SIRS | Sepsis detection | UCSF data+BIDMC data | Carry-forward and replacing by mean | 80% | 20% | NR |
| Taneja et al. (2017) | Clinical adjudication label | Early prediction of sepsis | Carle Foundation Hospital | NR | NR | NR | NR |
| Saqib et al. (2018) | Angus | Early prediction of sepsis | MIMIC-III | Forward-filling | 81% | 10% | 9% |
| Perng et al. (2019) | SIRS + qSOFA | Mortality prediction of sepsis | Chang Gung Research Database | Replacing by medium number of the column | 70% | 30% | NR |
| Thottakkara et al. (2016) | the criteria of the Agency for Healthcare Research and Quality | Severe sepsis prediction | DECLARE data | Replacing by mean value | 70% | NR | 30% |
| Bloch et al. (2019) | Infection +SIRS | Early prediction of sepsis | Israel Rabin Medical Center | NR | 75% | 25% | NR |
| Kwon and Baek, (2020) | Infection + qSOFA | Mortality prediction of sepsis | Four hospitals of Korea | NR | 74% | 18% | 8% |
| Nemati et al. (2018) | sepsis3.0 | Early prediction of sepsis | two hospitals within the Emory Healthcare system and an ICU database | NR | 80% | 20% | NR |
| Lauritsen et al. (2020) | Infection +SIRS | Early detection and prediction of sepsis | Four Danish municipalities data | NR | 80% | 10% | 10% |
| Scherpf et al. (2019) | ICD9+SIRS | Early prediction of sepsis | MIMIC-III | Liner interpolation and “carry forward/backward” extrapolation | NR | NR | NR |
| Hou et al. (2020) | Sepsis3.0 | Mortality prediction of sepsis | MIMIC III v1.4 | Remove the variables with more than 20% observations missing + multiple imputation method | NR | NR | NR |
| Kong et al. (2020) | Sepsis3.0 | Mortality prediction of sepsis | MIMIC III | Remove the patients with more than 30% predictor variable missing + Replace by mean value | NR | NR | NR |
| Bedoya et al.(2020) | SIRS + infection + end organ failure | Early detection of sepsis | ED of a quaternary academic hospital | NR | NR | NR | NR |
| van Doorn et al. (2021) | Infection + SIRS/SOFA | Mortality prediction of sepsis | ED at the Maastricht University Medical Center+ | NR | 1244 | NR | 100 |
| Li et al. (2021) | ICD-9 | Mortality prediction of sepsis | MIMIC-III V1.4 | Remove the patients with data missing more than 30% + Replace by mean value | NR | NR | NR |
| Burdick et al. (2020) | SIRS | Early severe sepsis prediction | The Dascena Analysis Dataset and the Cabell Huntington Hospital Dataset | last-one carry forward | NR | NR | NR |
| Qi et al. (2021) | Sepsis3.0 | Mortality prediction of sepsis | MIMIC-III | Remove the patients with data missing more than 40% + Replace by 21% and mean value | NR | NR | NR |
Abbreviation:SIRS: Systemic Inflammatory Response Syndrome; ICD9:international classification of diseases 9; NR: not reported.
Thirteen studies described preprocessing methods for the clinical data with various methods, including filling missing data by mean, median or nearest measured values, K-means clustering, forward-filling, linear interpolations, and carry forward/backward extrapolations. Twelve studies provided detailed descriptions of sample sizes or proportions between training groups and test/verification groups. However, not a single study discussed the rationale for adopting their methods. Only six studies adopted the latest Sepsis-3 definition, and the others used old criteria (SIRS/ICD [the international classification of diseases definition]/Angus/the criteria of the Agency for Healthcare Research and Quality).
-
•
SIRS: Heart rate >90 beats/min; Body temperature >38°C or <36°C; Respiration rate >20 times/min or PaCO2<32mm Hg; White blood cell count >12 × 109/L or < 4 × 109/L
-
•
Sepsis-3: infection + SOFA≥2
Quality evaluation
Using the Joanna Briggs Institute Critical Appraisal (JBI) tool, Kwong et al. proposed a method to evaluate the quality of machine learning research. JBI is a checklist for cross-sectional research, which has been adopted by Islam et al. and Kwong et al. to evaluate quality of machine learning studies (Kwong et al.,2019; Islam et al.,2018). It consists of eight items. We first applied their tool to evaluate the included studies, and the results are shown in Table 2.
Table 2.
Quality evaluation of including studies
| Study | Inclusioncriteria | Data preprocessed | Data source and collection | The source of the feature | Ethical issue | Detail discussion | Measurement of models' performance | Cross-validation/evaluation method |
|---|---|---|---|---|---|---|---|---|
| Delahanty et al. (2019) | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| Barton et al. (2019) | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Taylro, 2015 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| Kam and Kim (2017) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| Mao et al. (2018) | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Taneja et al. (2017) | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| Saqib et al. (2018) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| Perng et al. (2019) | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Thottakkara et al. (2016) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Bloch et al. (2019) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Kwon and Baek (2020) | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| Nemati et al. (2018) | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| Lauritsen et al. (2020) | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| Scherpf et al. (2019) | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 |
| Hou et al. (2020) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| Kong et al. (2020) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bedoya et al. (2020) | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| van Doorn et al. (2021) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Li et al. (2021) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Burdick et al. (2020) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Qi et al. (2021) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
Annotation: The contents of have been tweaked to better fit machine learning research.
Prediction in time
Considering that sepsis progression is time-sensitive, a good predictive model should be able to verify the accuracy at different times. However, we find only seven studies provided the information (see Table 3).
Table 3.
Prediction (AUROC) of each model at different hours in the sepsis studies
| Study | Model | Algorithm | Different hours |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −48 | −24 | −12 | −10 | −8 | −6 | −5 | −4 | −3 | −2 | −1 | −0.25 | 0 | |||
| Delahanty et al. (2019) | RoS | Gradient boosting | 0.97 | 0.93 | |||||||||||
| Barton et al. (2019) | MLA | Gradient boosted trees | 0.83 | 0.84 | 0.88 | ||||||||||
| Kam and Kim (2017) | SepLSTM | long short-term memory | 0.93 | 0.94 | 0.96 | 0.99 | |||||||||
| Bloch et al. (2019) | SVM-RBF | SVM-RBF | 0.8141 | 0.8879 | 0.8807 | 0.8639 | 0.8675 | ||||||||
| Nemati et al. (2018) | Weilbull-Cox proportional hazards | Weilbull-Cox proportional hazards | 0.79 | 0.8 | 0.81 | 0.82 | |||||||||
| Lauritsen et al. (2020) | CNN-LSTM | CNN-LSTM | 0.752 | 0.792 | 0.842 | 0.879 | |||||||||
| Scherpf et al. (2019) | RNN | RNN | 0.76 | 0.79 | 0.81 | ||||||||||
Abbreviation:RoS: Risk of Sepsis; MLA: machine learning algorithm; LSTM: long short-term memory; SVM-RBF: support vector machines with radial basis function; CNN-LSTM: convolutional neural network-long short-term memory; RNN: recurrent neural network.
Performance in predictions
Compared with traditional predictive tools in single studies, AUROC of machine learning models mostly scored more than 0.8, with some studies even over 0.9, which was significantly higher than the traditional predictive tools where the results were around 0.7 (Table S1). Meanwhile, two studies also detected sepsis. Their predictive models showed AUROC value around 0.9 (Table 3), demonstrating strong ability to distinguish sepsis from no-sepsis patients at 0 h. We are therefore confident that machine learning algorithms can effectively predict sepsis.
Time sensitivity
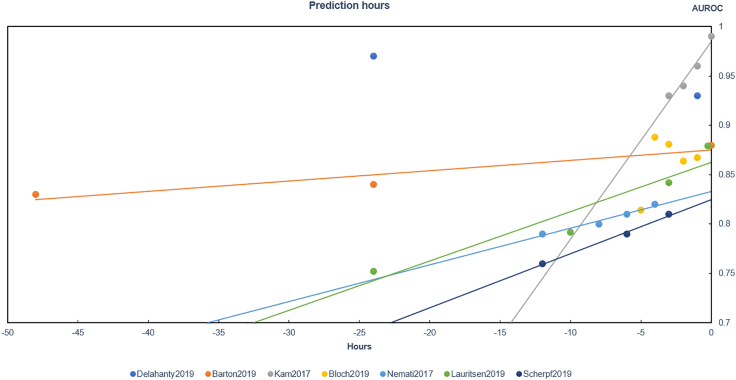
The predictions can be divided into three categories: (1.) using only one model, (2.) using more than one model, and (3.) using the best model among several for the prediction. Among the 21 included studies, most belong to the third category and focused on the prediction of an early occurrence of sepsis. With the completion of information collection, the prediction performance of the third category at different hours is shown in Figure 2. Here, we make trend lines of AUROC in five studies, and find that the model's performance increased notably as the time gets closer to the onset of sepsis. The ideal time period for early sepsis prediction ranges from 0 to 24 h.
Figure 2.
Predicting performance of multi-time points, related to Table 3
Mortality prediction
There are eight studies targeted at predicting sepsis mortality in emergency departments or ICUs, and we list seven studies' models, algorithm, AUROC, and prediction time in Table 4. These researchers tried many algorithms to build their predictive models. In a study of 28-days mortality prediction by Perng et al., the use of convolutional neural networks (CNN) + SoftMax resulted in AUROC =0.92, which is the highest among all the models in the study. Meanwhile, it predicted 72-h mortality, proving that CNN + SoftMax was the best model (AUROC = 0.94). And we find Ke Li et al. used Gradient Boosting Decision Tree (GBDT) and random forest (RF) to predict in-hospital mortality. They attained remarkably high AUROC scores (0.992, 0.980) and demonstrated excellent predictive ability of ensemble learning and traditional machine learning algorithm in sepsis.
Table 4.
AUROC and time points of mortality prediction studies
| Study | Model | Algorithm | AUROC | Time |
|---|---|---|---|---|
| Taylro, 2015 | Logistic regression | Logistic regression | 0.755 | 28 days |
| CART | Classification and regression tree | 0.693 | ||
| Random forest | Random forest | 0.860 | ||
| MEDS score | NR | 0.705 | ||
| CURB-65 score | NR | 0.734 | ||
| REMS score | NR | 0.717 | ||
| Perng et al. (2019) | KNN | KNN | 0.84 | 28 days |
| SoftMax | SoftMax | 0.88 | ||
| PCA + SoftMax | PCA + SoftMax | 0.91 | ||
| AE + SoftMax | AE + SoftMax | 0.90 | ||
| CNN + SoftMax | CNN + SoftMax | 0.92 | ||
| Kwon and Baek (2020) | qSOFA scores | NR | 0.78 | 3 days |
| qSOFA-based machine-learning models | Extreme gradient boosting, light gradient boosting machine, and random forest | 0.86 | ||
| Hou et al. (2020) | XGBoost | eXtreme Gradient Boosting | 0.857 | 30 days |
| logistic regression | logistic regression | 0.819 | ||
| SAPS-II scores | Simplified acute physiology score-II | 0.797 | ||
| Kong et al. (2020) | LASSO | least absolute shrinkage and selection operator | 0.829 | In hospital |
| RF | random forest | 0.829 | ||
| GBM | gradient boosting machine | 0.845 | ||
| LR | logistic regression | 0.833 | ||
| SAPS II | Simplified acute physiology score-II | 0.77 | ||
| Li et al. (2021) | GBDT | GBDT | 0.992 | In hospital |
| LR | Logistic regression | 0.876 | ||
| KNN | k-nearest neighbor | 0.877 | ||
| RF | Random forest | 0.980 | ||
| SVM | Support vector machine | 0.898 | ||
| Qi et al. (2021) | XGBoost | Extreme gradient boosting | 0.848 | In hospital |
| SAPSII | The simplified acute physiology score | 0.777 | ||
| SOFA | Sequential organ failure assessment score | 0.704 | ||
| SIRS | Systemic inflammatory response syndrome | 0.609 | ||
| qSOFA | Quick sequential organ failure assessment | 0.580 |
Abbreviation: CART: classification and regression tree; MEDS: mortality in emergency department sepsis score; KNN: K nearest neighbor; REMS: rapid emergency medicine score; CURB-65 score: the confusion, urea nitrogen, respiratory rate, blood pressure, 65 years of age and older; PCA: principal component analysis; AE: Autoencoder; CNN: Convolutional Neural Network; qSOFA: quick Sequential Organ Failure Assessment.
Feature engineering
All studies collected vital signs and laboratory data. For vital signs, researchers collected body temperature, heart rate, blood pressure, and respiratory rate. For laboratory data, researchers collected white blood cell count, lactic acid etc. Furthermore, demographic characteristics, clinical scores, and other features were also included in a few studies. We show representative ten studies and list their results in Table 5.
Table 5.
Features engineering and included features of each study
| Study | Number of initial features | Number of final features | Including features |
|---|---|---|---|
| Delahanty et al. (2019) | 217 | 13 | Lactic acid (max), Shock index age (last), WBC count(max), Lactic acid(change), Neutrophils(max), Glucose(max), Blood urea nitrogen(max), Shock index age (first), Respiratory rate (max), Albumin (last), Systolic blood pressure (min), Serum creatinine (max), Temperature (max) |
| Barton et al. (2019) | 6 | 6 | SpO2, heart rate, respiratory rate, temperature, systolic blood pressure, diastolic blood pressure |
| Taylro, 2015 | 566 | 20 | Oxygen saturation, Respiratory rate, Blood pressure, BUN, Albumin, Intubation, Procedures (in ED), Need for vasopressors, Age, RN resp care, RDW, Potassium, AST, Heart rate, Acuity level(triage), ED impression (Dx), CO2 (Lab), ECG performed, Beta-blocker (Home Med), Cardiac dysrhythmia (PMHx) |
| Kam and Kim (2017) | 9 | 9 | systolic pressure, pulse pressure, heart rate, body temperature, respiratory rate, WBC count, pH, blood oxygen saturation,age |
| Mao et al. (2018) | 6 | 6 | systolic blood pressure, diastolic blood pressure, heart rate, respiratory rate, temperature, peripheral capillary oxygen saturation |
| Taneja et al. (2017) | 31 | NR | TNF-α, IL-1β, GCSF, IL-6, PCT, sTREM1, IL18, MMP9, TNFR1, TNFR2, IP10, MCP1, IL-1ra, NA, CD64, WBC, Lactic Acid, Systolic Blood Pressure, Diastolic Blood Pressure, Pulse, Temperature, Respirations, PCO2, Age, Gender, Bilirubin, Glasgow Coma Scale, Creatinine, Platelet, SOFA score, qSOFA score |
| Saqib et al. (2018) | 47 | 34 | White blood cell count, Heart rate, Diastolic blood pressure, Systolic blood pressure, Mean blood pressure, Weight, Anion gap, Bicarbonate, Oxygen saturation, Height, Temperature, pH |
| Bloch et al. (2019) | 20 | 4 | the number of trend changes in respiratory rate and arterial pressure, the minimal change in respiratory rate, and the median change in heart rate |
| Kwon and Baek (2020) | 14 | NR | Age, sex, diagnoses at the ED, systolic blood pressure, respiration rate, mental status, body temperature, heart rate, arterial partial pressure of carbon dioxide, white blood cell count, duration of hospitalization, ICU admission, mechanical ventilation, mortality. |
| Nemati et al.(2018) | 65 | 65 | RRSTD, MAPSTD, HRV1, BPV1, HRV2, BPV2, MAP, HR, O2Sat, SBP, DBP, RESP, Temp, GCS, PaO2, FIO2, WBC, Hemoglobin, Hematocrit, Creatinine, Bilirubin and Bilirubin direct, Platelets, INR, PTT, AST, Alkaline Phosphatase, Lactate, Glucose, Potassium, Calcium, BUN, Phosphorus, Magnesium, Chloride, B-type BNP, Troponin, Fibrinogen, CRP, Sedimentation Rate, Ammonia, pH, pCO2, HCO3, Base Excess, SaO2, Care Unit (Surgical, Cardiac Care, or Neuro intensive care), Surgery in the past 12 h, Wound Class (clean, contaminated, dirty, or infected), Surgical Specialty (Cardiovascular, Neuro, Ortho-Spine, Oncology, Urology, etc.), Number of antibiotics in the past 12, 24, and 48 h, Age, CCI, Mechanical Ventilation, maximum change in SOFA score over the past 6 h. |
| Hou et al. (2020) | 22 | 11 | urine output, lactate, Bun, sysbp, INR, age, cancer, SpO2, sodium, AG, creatinine |
Annotation: The study of Tanejia2017 and YS2020 has established a variety of different models with different numbers of included features, so all features are provided. The Saqib et al. (2018) study provides only partial features.
Abbreviation: WBC count: white blood cell count; BUN: blood urea nitrogen; RDW: Red blood cell distribution width; AST: aspartate transaminase; ED: emergency department; ECG: electrocardiogram; SOFA: Sequential Organ Failure Assessment; qSOFA: quick Sequential Organ Failure Assessment; RRSTD: standard deviation of respiratory rate intervals; MAPSTD: standard deviation of mean arterial pressure; HRV1: average multiscale entropy of respiratory rate; BPV1:averagemultiscale entropy of mean arterial pressure; HRV2:average multiscale conditional entropy of respiratory rate; HRV2:average multiscale conditional entropy of respiratory rate; MAP: Mean Arterial Blood Pressure; HR: Heart Rate; O2Sat: Oxygen Saturation; SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure; RESP: Respiratory Rate; Temp: Temperature; GCS: Glasgow Coma Scale; PaO2: Partial Pressure of Arterial Oxygen; FIO2: Fraction of Inspired O2; INR: International Normalized Ratio, PTT: Partial Prothrombin Time, AST: Aspartate Aminotransferase, BNP:B-type Natriuretic Peptide; CCI: Charleston Comorbidity Index; sysbp: systolic blood pressure; AG: anion gap.
In general, feature preprocessing can also be divided into two categories. The studies in category one used feature engineering methods to identify the key factors/features that can be used for machine learning processes. For example, Bloch et al. recorded four vital signs of data at the frequency of 6 times an hour, found median, and calculated mean values. They obtained 20 features and selected the most important 4 in their machine learning models (Bloch et al.,2019). The studies in category two rely on researchers' expertise to choose what factors/variables should be used to devise models. For example, Barton et al. used six factors, including heart rate and respiratory rate to develop their models to predict sepsis occurrence (Barton et al.,2019). Mao et al. chose the data that are easily available in intensive care unit and emergency department as features (Mao et al.,2018).
Discussion
As the first attempt to systematically review methodologies of sepsis prediction studies, we find that most studies focused on early prediction and detection of sepsis and mortality. Except for the results mentioned above, there are nine issues that we would like to address in this review.
Diagnostic criteria
Those studies, which adopt old sepsis definition or improper inclusion criteria, performed adequately. They were however viewed as too lax in sample inclusion and lacking enough specificity and sensitivity. For example, Mao et al. and Bloch et al. considered patients over 18 years old with a slight limitation and selected SIRS as diagnostic criteria (Mao et al.,2018; Bloch et al.,2019). They all used large datasets and had enough patients meeting the SIRS criteria, which led to high AUROC values. Compared to older diagnostic criteria, the latest Sepsis-3 includes more stringent clinical features and describes sepsis more accurately.
Large disparities are found among sepsis definitions, making it impossible to compare AUROC of each study to find the best machine learning model. However, it should be noted that Kam et al. used the long short-term memory (LSTM) model to predict sepsis occurrence and obtained high AUROC value (Kamand Kim, 2017). In addition, 1D CNN combined with SoftMax model was selected by Perng et al., which significantly improved the performance of mortality prediction compared to the traditional predictive models in the single study. The CNN model reached AUROC 0.92 while the traditional models, such as KNN, got AUROC only 0.84 (Perng et al., 2019). This is because deep learning algorithms can remove many redundant dimensions by self-learning (Kamand Kim,2017; Mücke et al.,2021), and multiclass classification problem can be resolved with SoftMax. We also noted that Ke Li et al. reported that GBDT and RF predicted sepsis mortality well. GBDT is an ensemble learning method and may correct the training results and reduce the degree of overfitting by a regularization function (Chen et al.,2020). However, there are conflicting l studies which reported not good performance of random forest (Table 4); thus, further studies are needed to determine the robustness of RF for sepsis prediction.
Prediction time
Prediction times of the studies were different and AUROC changed with time (Figure 2). These characteristics corroborates with clinical experience. The closer to the onset of sepsis, the more accurately the model predicts. We find a study by Delahanty et al. where the results were inconsistent with other conclusions (Delahanty et al.,2019). This study used inappropriate inclusion criteria and concluded that neutrophil count had a negative effect on the RoS model, which was contrary to the pathophysiological mechanism. Therefore, we think the robustness of RoS should be further discussed. Because there is huge heterogeneity between studies predicting sepsis mortality, we cannot compare them reasonably to obtain similar rules.
Importance of feature engineering
There are commonly two major steps in machine learning studies. The first is to extract features from input samples and the second is to feed feature vectors into the machine learning algorithm for training and making the prediction. It is especially important to select key features and reduce the data dimension, which is known as feature engineering or order reduction. Feature engineering can not only significantly reduce redundant information and improve computational efficiency but also keep lowest negative influence of complex data dimensions on model robustness (Dai et al., 2020). There are two categories of feature engineering among the studies: one is designed by the domain expertise while the other is designed using machine learning methods (Miotto et al.,2018).
Designed by the domain expertise
Three studies chose features that were common and easily obtained in hospitals or based on the researchers' clinical experience. For example, several features that were common in the intensive care units (ICU) or emergency departments (systolic blood pressure, diastolic blood pressure, heart rate, respiratory rate, and body temperature) were selected in studies by Barton et al. and Mao et al. (Barton et al.,2019; Mao et al.,2018). However, only relying on clinical expertise could lead to strong subjectivity and may overlook some key features (Garcia et al.,2014). Even though the models performed adequately, the outcomes were difficult to be validated by external data; therefore, the applicability of these models is limited.
Designed by machine learning methods
Traditional reduced-order or feature-extraction algorithms, represented by principal component analysis (PCA) or auto decoder-encoder methods, can reduce data dimensions and significantly improve model performance. Perng et al. increased the accuracy of support vector machine (SVM) from 74.33% to 78.91% using PCA to preprocess the data. Meanwhile, AUROC of SoftMax increased from 0.88 to 0.91 (Perng et al., 2019). We can see the same situation here again, where Thottakkara et al. succeeded in improving the accuracy and AUROC after using PCA to preprocess their data (Thottakkara et al.,2016).
In addition, the deep feedforward neural network (DFN) can independently learn and obtain the most crucial features. For instance, Kam et al. used DFN to detect early sepsis (Kamand Kim,2017). In addition, LSTM, a deep recurrent neural network, was adopted to learn long-range dependencies and handle vanishing gradient. As a result, the accuracy, sensitivity, and AUROC of LSTM are the highest and the number of the final features is the smallest.
Data granularity
Before processing data, researchers should first establish the definition for data granularity. The degree of data refinement and predictive performance can be improved by changing the granularity level (Dormosh et al.,2020). Here, we screened studies that refined data. In the 21 studies we included in this review, only Bloch et al. reported and discussed the issue of data granularity in detail, and the others did not mention this essential information at all (Bloch et al.,2019). Bloch et al. selected four features at the early stage and then expanded them to 20 features by calculating mean, median, minimum, maximum, and standard deviation. As a result, they obtained four satisfying features by ranking the features' importance. To some extent, it is another type of feature engineering used to explore intrinsic regularities of the clinical data.
Missing data
It is inadequate to build predictive models when there are missing data (Beaulieu-Jones et al.,2018). There were 12 studies that reported their methods on how to deal with missing data; six studies filled the missing data with mean value or median value. Three studies used methods such as filling in missing data by liner interpolation or “carry forward/backward” extrapolation methods (Table 1). It is a consensus that missing data should be processed before conducting any analysis (Mehrabani-Zeinabad et al., 2020), but obviously in the machine learning research field on sepsis, this standard operation has not been widely followed.
Machine learning algorithms
Various algorithms were applied, and their predictive performance is summarized in Tables 3 and 4, respectively. These consisted of popular current machine learning algorithms, including logistic regression, decision tree, support vector machine, random forest, and deep learning algorithms in supervised learning. The other category is unsupervised learning, including principal component analysis, K-means clustering, and autoencoder. Regardless of the influence of diagnostic criteria, the neural network-based algorithm performed better on average in sepsis detection, mortality, and early prediction. GBDT, which is a kind of classical and popular ensemble algorithm, may also have a broad prospect in sepsis prediction.
Continuous dataset
The studies contained in this review established outcome prediction models based on sectional data. To the contrary, some researchers predicted sepsis by using continuous data (time series). Kamaleswaran et al., Mohammed et al., and Wyk et al. constructed models for predicting sepsis onset with continuous physiologic data streams (Mohammed et al., 2021; Kamaleswaran et al., 2021a, 2021b, van Wyk et al., 2019). After preprocessing, they put data into selected algorithms and obtain the best predictive model. In addition, Kamaleswaran et al. also studied the significance of continuous data in predicting sepsis patients' response for volume treatment. We noticed this study reported better performance based on continuous data than EMR (Kamaleswaran et al., 2021a, 2021b), high-frequency data containing more patients' information, which may account for this result. This was an interesting finding. In fact, our team is conducting a similar research currently, which will be reported later.
Data heterogeneity
Predictive models of sepsis were mostly based on large databases. However, every sepsis patient is unique, therefore significant differences among the patients. For example, sepsis detection is to distinguish the confirmed sepsis patients from the non-sepsis patients, but it is difficult to carry out the classification in clinical settings because the symptoms and therapeutic medicine of every patient are different. There are large discrepancies among each individual patient so that it could be misleading to put all patients' data into a single dataset for training or testing when conducting machine learning. Therefore, all models mentioned above lack certain universality in machine learning protocols and cannot be used widely to assist any clinical decision-making (Fohner et al.,2019).
Combined with clinical experience, researchers can collect necessary higher frequency clinical data every day to observe dynamical evolution of sepsis. Meanwhile, sepsis progression can be simulated by the machine learning model based on neural networks so that patients' prognosis will be predicted. Although we have discussed that we cannot solely rely on physician's experiences to select feature, it is necessary to integrate physician's experiences when transferring the model to new patient. It will mitigate the inherent heterogeneity. In a recent study, we have developed a deep learning method to integrate the clinical knowledge with clinical data to make successful short-term predictions (up to 48 h) for clinical practitioners (Lei et al.,2020).
Based on the above discussions, we recommend strongly that future model development should incorporate clinical experience into data preprocessing instead of relying solely on the routinely collected data. Certain objective-oriented data preprocessing standard must be established so that the preprocessed data will be AI-ready for machine learning use.
Quality reevaluation and reporting standards
Through comparative study, we believe the JBI is a crude tool for evaluating machine learning methods. Based on the analysis above, we propose a new quality evaluation tool for machine learning methods. (1.) The evaluation methodology should include an appropriate and accurate disease definition, a data preprocessing protocol, and reasonable inclusion criteria. For example, we think that only Sepsis-3 can describe patient conditions accurately and be a basis for patient inclusion. (2.) For common problems in clinical data, such as missing data, data redundancy, data collected in different forms, noisy data etc., one should develop a protocol to produce standardized or normalized datasets, making sparse data non-sparse and “smooth” and improve data granularity. (3.) To avoid data redundancy and improve computational efficiency, feature engineering should include how many types of the original features are included, how many key features are selected, and how many types are classified. (4.) The process of sample removing, and grouping should be provided in the flowchart, and the rationales clearly explained. (5.) One needs to introduce algorithms, including the rationale for their choices based on relevant mathematical and statistical principles. (6.) Every model needs to have a set of corresponding evaluation criteria. We suggest adopting AUROC as an evaluation standard for the model performance. (7.) Finally, a prospective validation process is needed to ensure the predictive model developed can be adapted to clinical settings.
Based on the new criteria alluded to above, we reevaluate the models in the 21 studies. We score 1 for any item meeting a criterion above and 0 otherwise. Total score more than or equal to 8 is considered high quality, 5–8 (including 5) average quality, and less than 5 low qualities. The quality reevaluation results of this review are shown in Tables 2, 10 studies are ranked low and 11 average. There are no high-quality models based on the score table.
It is obvious that there are significant differences in data sources, data preprocessing, and feature engineering among sepsis prediction models. In addition, using diverse types of evaluation indices and predicted sepsis occurrence in various times, could result in distinctive model performance. Naturally, we believe there should be a unified standard to be a guideline of machine learning models in clinical research and applications. Referring to Standards for Reporting Diagnostic Accuracy (STARD) and combining with the above quality evaluation table, we propose a new report list of machine learning models in clinical medicine (Table 6).
Table 6.
Report standards list of machine learning in clinical medication
| Section and topic | Item | Description |
|---|---|---|
| Title/Abstract/Keywords | 1 | Can be judged as a machine learning predictive research. (Keywords,such as machine learning,prediction) |
| Introduction | 2 | Introduce background, existing problems, and study targets,such as evaluating machine learning models to predict prognoses and probability of disease occurrence |
| Method research subject | 3 | Inclusion and exclusion criteria, locations where data is collected and time range |
| 4 | Describe reasons of patients' selection, including symptoms, laboratorial results, or disease golden standard. | |
| 5 | Describe golden standard and provide references | |
| Research data | 6 | Describe whether study is based on past datasets (retrospective study) or latest collection data (prospective study). |
| 7 | Describe the data collection process. | |
| 8 | Describe the process of feature engineering. At least explain why choose this way to select features. | |
| Results Building model | 9 | Provide flowchart of the including and excluding process, describe demographic and clinical characteristics (such as age, sex, height, and weight) |
| 10 | Describe data preprocessing methods, including missing data processing, and smoothly processing sparse data. | |
| 11 | Describe the mathematical theory of the algorithm and its advantages. | |
| 12 | Describe numbers and names of finally including features | |
| Research results | 13 | Describe models performance at different time points (provide at least one evaluation indicator, such as AUROC, accuracy). |
| Discussion | 14 | Discuss clinical universality of predictive models, including heterogeneity discussion and clinical prospective validation. |
Conclusions
Through a systematical review, we find that the number of studies using machine learning to predict the occurrence and mortality of sepsis grows rapidly in recent years, and the accuracy of predictions has improved considerably. However, there is no model that can be widely adopted in the real world yet, because of the lack of unified validation standard and procedure and the heterogeneity in a cohort of patients. In addition, the data collected from patients with sepsis are normally high-dimensional, highly heterogeneous, including both structured and unstructured data that evolve in a time-sensitive fashion and static data. Compared to traditional tools, deep neural networks are more suitable for this type of data. The traditional SIRS criteria cannot describe sepsis comprehensively due to the lack of sufficient features, and cannot be included for sepsis machine learning study. We note that studies based on Sepsis-3 just begin so that further studies are necessary. Hence, the new quality evaluation tool and reporting standard list suggested in this review would help improve the effective use of machine learning methods in clinical medicine.
Limitations of the study
We do not have access to enough medical information on the treatment process of sepsis and therefore cannot evaluate its significance in the model development. Moreover, limited to very few open-source databases, it is difficult to compare them and have a meaningful discussion. We do not find any study that described the specific influence of data preprocessing and have not come to the conclusion on which method is the best.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Studies' methodologies and AUROC of prediction | Contained in the article | N/A |
| Other | ||
| MIMIC database | MIMIC database | https://mimic.mit.edu/ |
Resource availability
Lead contact
Further requests for resources and materials should be directed to and will be fulfilled by the Lead Contact, Hua Jiang (cdjianghua@qq.com).
Materials availability
This study did not yield new unique reagents.
Methods details
Eligibility criteria
There should be a consensus that eligible studies should provide clear data source based on Electronic Medical Record (EMR) or Electronic Healthy Record (EHR) from Emergency Department (ED) or Intensive Care Unit (ICU), so we can obtain disease and demographic information from the patients. In addition, we need AUROC and a clearly delineated detail of predictive models to compare and determine which model is the best. Considering the definition of sepsis has changed several times in the past, we require each study to provide at least one acceptable definition based on the current standard. We only study the prospection of machine learning algorithms in adults' sepsis and the target conditions include early detection and mortality of sepsis and severe sepsis.
Search strategy
A comprehensive literature retrieval is conducted in PubMed, ScienceDirect, Engineering Index (EI), Web of Science, China National Knowledge Infrastructure (CNKI) and WANFANG DATA for papers published between January 2010 and November 2021. Keywords like sepsis/machine learning/prediction are used for the search
A literature retrieval strategy for sepsis prediction
| Databases | Search strategy |
|---|---|
| PubMed | ((sepsis [Title/Abstract]) and (machine learning [Title/Abstract])) and (prediction [Title/Abstract]) |
| ScienceDirect | Title, abstract, keywords: sepsis, machine learning, predict |
| The engineering index | (((sepsis) and (machine learning) and (prediction) and (mortality) and (onset)) WN KY) |
| Web of science | Title:(sepsis) and Title:(machine learning) and Title:(prediction) |
| CNKI | ky = 'sepsis' and ky = 'machine learning' and ky= 'prediction' |
| WANFANG DATA | Title or keywords: “sepsis” and “machine learning” and “prediction” |
.
All the included papers are perused by two independent reviewers (TL and DHL), including title-abstract and full text. All disagreements between the two authors are resolved by a third author (TY) and principal investigators (HFD, HJ). The chosen papers are limited to languages in Chinese and English.
Evaluating tool and reporting standard
After rules of evaluating machine learning models on sepsis prediction are established, we realize that, like clinical medicine, there is the need for specialized tools for quality evaluation and reporting standard to guide research analogous to those used in evidence-based medicine. Therefore, based on the above analysis, we propose a new quality evaluation tool and a new reporting standard from the aspects of data acquisition, algorithm selection, feature engineering, and model building with reference to the Standards for Reporting Diagnostic Accuracy (STARD). These are more comprehensive than existing tools and standards, and more appropriate for machine learning research in medicine.
Quantification and statistical analysis
This work systematically evaluate the method of statistical and quantification analysis of published researches. The authors of this work did not do further quantification analysis, eg, meta-analysis.
Acknowledgments
This work is support by Funding to Sichuan Provincial Research Center for Emergency Medicine and Critical Illness from Sichuan Department of Science and Technology through award #2019YFS0534, 2019YFS0303 and 2020YFS0392, NSFCaward #11971051, # 72074222 and NSAF-U1930402. The authors thank Dr. Charles Damien Lu and Mr. Chao Zhang for their significant help in the manuscript writing and English proofing.
Author contributions
HFD, MWS, and HJ conducted the protocol and drafting of the systematic review. TY, DHL, and TL performed literature search and data retrieval. YW and QW provided guidance and made suggestions on machine learning algorithms. HFD, HJ, JZ, MWS, PZ, and QW interpret the analysis result. WC, QW, and HJ contributed to the subsequent revisions of the manuscripts. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103651.
Contributor Information
Qi Wang, Email: qwang@csrc.ac.cn.
Hua Jiang, Email: cdjianghua@qq.com.
Supplemental information
Data and code availability
•This paper analyzes existing, publicly available data, they can be shared by the lead contact upon request.
•This paper does not report original code.
•Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alhazzani W., Møller M.H., Arabi Y.M., Loeb M., Gong M.N., Fan E., Oczkowski S., Levy M.M., Derde L., Dzierba A., et al. Surviving sepsis campaign: guidelines on the management of critically Ill adults with coronavirus disease 2019 (COVID-19) Crit. Care Med. 2020;48:e440–e469. doi: 10.1097/ccm.0000000000004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton C., Chettipally U., Zhou Y., Jiang Z., Lynn-Palevsky A., Le S., Calvert J., Das R. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput. Biol. Med. 2019;109:79–84. doi: 10.1016/j.compbiomed.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu-Jones B.K., Lavage D.R., Snyder J.W., Moore J.H., Pendergrass S.A., Bauer C.R. Characterizing and managing missing structured data in electronic health records: data analysis. JMIR Med. Inform. 2018;6:e11. doi: 10.2196/medinform.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoya A.D., Futoma J., Clement M.E., Corey K., Brajer N., Lin A., Simons M.G., Gao M., Nichols M., Balu S., et al. Machine learning for early detection of sepsis: an internal and temporal validation study. JAMIAOpen. 2020;3:252–260. doi: 10.1093/jamiaopen/ooaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch E., Rotem T., Cohen J., Singer P., Aperstein Y. Machine learning models for analysis of vital signs dynamics: a case for sepsis onset prediction. J. Healthc. Eng. 2019;2019:5930379. doi: 10.1155/2019/5930379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick H., Pino E., Gabel-Comeau D., Gu C., Roberts J., Le S., Slote J., Saber N., Pellegrini E., Green-Saxena A., et al. Validation of a machine learning algorithm for early severe sepsis prediction: a retrospective study predicting severe sepsis up to 48 h in advance using a diverse dataset from 461 US hospitals. BMC Med. Inform. Decis.Mak. 2020;20:276. doi: 10.1186/s12911-020-01284-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi M., Evans L., Levy M., Rhodes A. Sepsis and septic shock. Lancet. 2018;392:75–87. doi: 10.1016/s0140-6736(18)30696-2. [DOI] [PubMed] [Google Scholar]
- Chen T., Zhu L., Niu R.-q., Trinder C.J., Peng L., Lei T. Mapping landslide susceptibility at the Three Gorges Reservoir, China, using gradient boosting decision tree, random forest and information value models. J. Mountain Sci. 2020;17:670–685. doi: 10.1007/s11629-019-5839-3. [DOI] [Google Scholar]
- Dai D., Xu T., Wei X., Ding G., Xu Y., Zhang J., Zhang H. Using machine learning and feature engineering to characterize limited material datasets of high-entropy alloys. Comput.Mater.Sci. 2020;175:109618. doi: 10.1016/j.commatsci.2020.109618. [DOI] [Google Scholar]
- Delahanty R.J., Alvarez J., Flynn L.M., Sherwin R.L., Jones S.S. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann. Emerg. Med. 2019;73:334–344. doi: 10.1016/j.annemergmed.2018.11.036. [DOI] [PubMed] [Google Scholar]
- Dormosh N., Abu-Hanna A., van der Velde N., Schut M. Impact of altering data granularity levels on predictive modelling: a case study of fall risk prediction in older persons. Stud. Health Technol. Inform. 2020;270:257–261. doi: 10.3233/shti200162. [DOI] [PubMed] [Google Scholar]
- Fleischmann C., Scherag A., Adhikari N.K., Hartog C.S., Tsaganos T., Schlattmann P., Angus D.C., Reinhart K. Assessment of global incidence and mortality of hospital-treated sepsis.Current estimates and limitations. Am. J. Respir. Crit. Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- Fleuren L.M., Klausch T.L.T., Zwager C.L., Schoonmade L.J., Guo T., Roggeveen L.F., Swart E.L., Girbes A.R.J., Thoral P., Ercole A., et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46:383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fohner A.E., Greene J.D., Lawson B.L., Chen J.H., Kipnis P., Escobar G.J., Liu V.X. Assessing clinical heterogeneity in sepsis through treatment patterns and machine learning. J. Am. Med. Inform. Assoc. 2019;26:1466–1477. doi: 10.1093/jamia/ocz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E.V., Klein J.L., Taylor A.T. Clinical decision support systems in myocardial perfusion imaging. J. Nucl. Cardiol. 2014;21:427–439. doi: 10.1007/s12350-014-9857-9. [DOI] [PubMed] [Google Scholar]
- Hou N., Li M., He L., Xie B., Wang L., Zhang R., Yu Y., Sun X., Pan Z., Wang K. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J. Transl. Med. 2020;18:462. doi: 10.1186/s12967-020-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.M., Nasrin T., Walther B.A., Wu C.C., Yang H.C., Li Y.C. Prediction of sepsis patients using machine learning approach: a meta-analysis. Comput.Methods Programs Biomed. 2019;170:1–9. doi: 10.1016/j.cmpb.2018.12.027. [DOI] [PubMed] [Google Scholar]
- Islam M.S., Hasan M.M., Wang X., Germack H.D., Noor E.A.M. A systematic review on healthcare analytics: application and theoretical perspective of data mining. Healthcare (Basel, Switzerland) 2018;6:54. doi: 10.3390/healthcare6020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam H.J., Kim H.Y. Learning representations for the early detection of sepsis with deep neural networks. Comput. Biol. Med. 2017;89:248–255. doi: 10.1016/j.compbiomed.2017.08.015. [DOI] [PubMed] [Google Scholar]
- Kamaleswaran R., Lian J., Lin D.L., Molakapuri H., Nunna S., Shah P., Dua S., Padman R. Predicting volume responsiveness among sepsis patients using clinical data and continuous physiological waveforms. AMIAAnnu.Symp.Proc. 2021;2020:619–628. PMC8075451. [PMC free article] [PubMed] [Google Scholar]
- Kamaleswaran R., Sataphaty S.K., Mas V.R., Eason J.D., Maluf D.G. Artificial intelligence may predict early sepsis after liver transplantation. Front. Physiol. 2021;12:692667. doi: 10.3389/fphys.2021.692667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong G., Lin K., Hu Y. Using machine learning methods to predict in-hospital mortality of sepsis patients in the ICU. BMC Med. Inform. Decis.Mak. 2020;20:251. doi: 10.1186/s12911-020-01271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.S., Baek M.S. Development and validation of a quick sepsis-related organ failure assessment-based machine-learning model for mortality prediction in patients with suspected infection in the emergency department. J. Clin. Med. 2020;9:875. doi: 10.3390/jcm9030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong M.T., Colopy G.W., Weber A.M., Ercole A., Bergmann J.H.M. The efficacy and effectiveness of machine learning for weaning in mechanically ventilated patients at the intensive care unit: a systematic review. Bio-Design and Manufacturing. 2019;2:31–40. doi: 10.1007/s42242-018-0030-1. [DOI] [Google Scholar]
- Lauritsen S.M., Kalør M.E., Kongsgaard E.L., Lauritsen K.M., Jørgensen M.J., Lange J., Thiesson B. Early detection of sepsis utilizing deep learning on electronic health record event sequences. Artif.Intell.Med. 2020;104:101820. doi: 10.1016/j.artmed.2020.101820. [DOI] [PubMed] [Google Scholar]
- Lei C., Wang Y., Zhao J., Li K., Jiang H., Wang Q. A patient specific forecasting model for human albumin based on deep neural networks. Comput.Methods Programs Biomed. 2020;196:105555. doi: 10.1016/j.cmpb.2020.105555. [DOI] [PubMed] [Google Scholar]
- Li K., Shi Q., Liu S., Xie Y., Liu J. Predicting in-hospital mortality in ICU patients with sepsis using gradient boosting decision tree. Medicine (Baltimore). 2021;100:e25813. doi: 10.1097/md.0000000000025813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q., Jay M., Hoffman J.L., Calvert J., Barton C., Shimabukuro D., Shieh L., Chettipally U., Fletcher G., Kerem Y., et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJOpen. 2018;8:e017833. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabani-Zeinabad K., Doostfatemeh M., Ayatollahi S.M.T. An efficient and effective model to handle missing data in classification. Biomed.Res Int. 2020;2020:8810143. doi: 10.1155/2020/8810143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto R., Wang F., Wang S., Jiang X., Dudley J.T. Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinform. 2018;19:1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira J.C., Gentile L.F., Mathias B.J., Efron P.A., Brakenridge S.C., Mohr A.M., Moore F.A., Moldawer L.L. Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immunosuppression and catabolism syndrome. Crit. Care Med. 2017;45:253–262. doi: 10.1097/ccm.0000000000002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed A., Van Wyk F., Chinthala L.K., Khojandi A., Davis R.L., Coopersmith C.M., Kamaleswaran R. Temporal differential expression of physiomarkers predicts sepsis in critically Ill adults. Shock. 2021;56:58–64. doi: 10.1097/SHK.0000000000001670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mücke N.T., Bohté S.M., Oosterlee C.W. Reduced order modeling for parameterized time-dependent PDEs using spatially and memory aware deep learning. J. Comput. Sci. 2021;53:101408. doi: 10.1016/j.jocs.2021.101408. [DOI] [Google Scholar]
- Nemati S., Holder A., Razmi F., Stanley M.D., Clifford G.D., Buchman T.G. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit. Care Med. 2018;46:547–553. doi: 10.1097/ccm.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng J.W., Kao I.H., Kung C.T., Hung S.C., Lai Y.H., Su C.M. Mortality prediction of septic patients in the emergency department based on machine learning. J. Clin. Med. 2019;8:1906. doi: 10.3390/jcm8111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Xu H., Hu J., Mao Z., Hu X., Zhou F.H. Early mortality risk prediction model for sepsis patients in intensive care unit based on machine learning. Acad. J.Chin.PLA Med.Sch. 2021;42:150–155. doi: 10.3969/j.issn.2095-5227.2021.02.006. [DOI] [Google Scholar]
- Raith E.P., Udy A.A., Bailey M., McGloughlin S., MacIsaac C., Bellomo R., Pilcher D.V. Prognostic accuracy of the SOFA score, SIRS criteria, and qSOFAscore for in-hospital mortality among adults with suspected infection admitted to the intensive care unit. JAMA. 2017;317:290–300. doi: 10.1001/jama.2016.20328. [DOI] [PubMed] [Google Scholar]
- Saqib M., Sha Y., Wang M.D. Early prediction of sepsis in EMR records using traditional ML techniques and deep learning LSTM networks. Annual International Conference of the IEEE Engineering in Medicine and Biology Society.IEEE Engineering in Medicine and Biology Society. Annual International Conference. 2018;2018:4038–4041. doi: 10.1109/embc.2018.8513254. [DOI] [PubMed] [Google Scholar]
- Scherpf M., Gräßer F., Malberg H., Zaunseder S. Predicting sepsis with a recurrent neural network using the MIMIC III database. Comput.Biol. Med. 2019;113:103395. doi: 10.1016/j.compbiomed.2019.103395. [DOI] [PubMed] [Google Scholar]
- Shashikumar S.P., Stanley M.D., Sadiq I., Li Q., Holder A., Clifford G.D., Nemati S. Early sepsis detection in critical care patients using multiscale blood pressure and heart rate dynamics. J. Electrocardiol. 2017;50:739–743. doi: 10.1016/j.jelectrocard.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja I., Reddy B., Damhorst G., Dave Zhao S., Hassan U., Price Z., Jensen T., Ghonge T., Patel M., Wachspress S., et al. Combining biomarkers with EMRdata to identify patients in different phases of sepsis. Sci. Rep. 2017;7:10800. doi: 10.1038/s41598-017-09766-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.A., Pare J.R., Venkatesh A.K., Mowafi H., Melnick E.R., Fleischman W., Hall M.K. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad. Emerg. Med. 2016;23:269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thottakkara P., Ozrazgat-Baslanti T., Hupf B.B., Rashidi P., Pardalos P., Momcilovic P., Bihorac A. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoSONE. 2016;11:e0155705. doi: 10.1371/journal.pone.0155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W., Stassen P.M., Borggreve H.F., Schalkwijk M.J., Stoffers J., Bekers O., Meex S.J.R. A comparison of machine learning models versus clinical evaluation for mortality prediction in patients with sepsis. PLoSONE. 2021;16:e0245157. doi: 10.1371/journal.pone.0245157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk F., Khojandi A., Mohammed A., Begoli E., Davis R.L., Kamaleswaran R. A minimal set of physiomarkers in continuous high frequency data streams predict adult sepsis onset earlier. Int. J. Med. Inform. 2019;122:55–62. doi: 10.1016/j.ijmedinf.2018.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
•This paper analyzes existing, publicly available data, they can be shared by the lead contact upon request.
•This paper does not report original code.
•Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.