Abstract
We present COVID-CT-Mask-Net model that predicts COVID-19 in chest CT scans. The model works in two stages: in the first stage, Mask R-CNN is trained to localize and detect two types of lesions in images. In the second stage, these detections are fused to classify the whole input image. To develop the solution for the three-class problem (COVID-19, Common Pneumonia and Control), we used the COVIDx-CT data split derived from the dataset of chest CT scans collected by China National Center for Bioinformation. We use 3000 images (about 5% of the train split of COVIDx-CT) to train the model. Without any complicated data normalization, balancing and regularization, and training only a small fraction of the model’s parameters, we achieve a 90.80% COVID-19 sensitivity, 91.62% Common Pneumonia sensitivity and 92.10% true negative rate (Control sensitivity), an overall accuracy of 91.66% and F1-score of 91.50% on the test data split with 21192 images, bringing the ratio of test to train data to 7.06. We also establish an important result that regional predictions (bounding boxes with confidence scores) detected by Mask R-CNN can be used to classify whole images. The full source code, models and pretrained weights are available on https://github.com/AlexTS1980/COVID-CT-Mask-Net.
Keywords: Chest CT scans, Image segmentation, Image classification, COVID-19
Introduction
Since the start of COVID-19 pandemic a large number of deep learning models predicting COVID-19 from chest CT scans and x-rays has been developed. One of the biggest challenges in this area is a three class problem: COVID-19 vs Common Pneumonia vs Control/Negative. Solutions for this problem include COVID Net-CT [1], that consists of a single feature extractor trained on COVIDx-CT dataset split, COVNet (augmented Res Net50) [2], ResNet18 [3] and LightCNN [4]. Some solutions use an ensemble of networks (AlexNet, GoogleNet,ResNet18) and majority voting, see [5]. In order to achieve the state-of-the-art [1] accuracy, large amounts of data are required to train (about 60K images) the model, that are often not available, which explains the need for various augmentations, both for the data and the classification model.
One approach that is used to augment the classifier, is the semantic segmentation model, e.g. in [6, 7] UNet is used as a pre-processing step: its output (mask) is concatenated with the feature maps to enhance the predictive power of the model. The advantage of using a segmentation model is that it is capable of explicitly learning and predicting areas of lesions associated with COVID-19. For a binary classification problem, COVID-19 vs non-COVID-19, COVID-CT [8] and Joint Classification and Segmentation (JCS) [7] models are publicly available. COVID-CT concatenates lung masks predicted by UNet with deep image features extracted using DenseNet169 and ResNet50 to predict the class, achieving an overall accuracy of 89% on the test data of about 350 images. JCS uses a similar approach, but with additional loss functions at deep layers (multiscale training), achieving an F1 score of 0.783 on the test data of about 120K images. Recently, in [9] a novel method was introduced that alleviates the lack of COVID-19 data by generating COVID-19 chest CT scans from lung cancer scans using CycleGAN [10]. A number of classifiers, such as ResNet50 and VGG16 are trained on the fusion of the generated and real COVID-19 images. Advanced methodology based on convnets and wavelets optimized using biogeography-based optimization was introduced in [11] to classify COVID-19 and negative images. Another approach in [12] fused convnets and Graph convolutional nets. This paper introduced a number of novelties, such as modifications of convnet operators: pooling, cropping, histogram normalization, etc. Other methodologically advanced models, such as [13] experimented with the truncation of the feature extractors and fusion of features on a small dataset with four classes; in [14] relation among different images is captured using Graph Neural Net, which is fused with a ConvNet; [15] introduced a seven-layer ConvNet with new operators like stochastic pooling and a range of data pre-processing and augmentation techniques. At least one recent publication [16] discusses the use of Mask R-CNN for predicting COVID-19 from the segmentation of CT scans.
A number of review papers compared different models directly to establish the best one for accuracy and COVID-19 sensitivity. From these papers, it appears that for the chest CT scans data, models with ResNet50, ResNeXt and DenseNet121 feature extractors produce the highest overall accuracy across a number of datasets. For further details see [17–19].
The majority of COVID-19 deep learning models use radiography (x-rays) data due to its prevalence, e.g. the state-of-the-art COVID-Net [12] that has an architecture similar to COVIDNet-CT. Also, the majority of published solutions solve two-class problems mentioned above. To the best of our knowledge, only COVIDNet-CT [1], LightCNN [4], COVNet [2] and ResNet18 in [3] use chest CT scans for a 3-class (COVID-19, Common Pneumonia, Control). This problem is more challenging due to the similarities and subtle differences between COVID-19 and Common Pneumonia (CP) on CT scans. For the discussion of these differences see [20–23].
These models have a number of drawbacks that we would like to address. COVIDNet-CT [1] requires a large training data with various augmentations and class balancing to achieve the reported accuracy and COVID-19 sensitivity, COVNet [2] was evaluated on a small dataset (about 500 images), the model using ResNet18 as a feature extractor [3] is not publicly available. Also, it reported a relatively low COVID-19 sensitivity (81.20%) and it was evaluated on a small data (90 images). Light CNN’s reported COVID-19 sensitivity is also quite low, and it was also evaluated on a small dataset. The biggest drawback though, is that these models were evaluated on the test split that was a fraction of the training split, see Table 1. For further discussion of the pitfalls and limitations of COVID-19 models see [24] and Section 5, which raises a question of overfitting and generalization to other datasets.
Table 1.
Comparison of the models’ sizes and the sizes of the data splits used for training, validation and testing
| Model | # Total | # Trainable | Training | Validation | Test | Ratio |
|---|---|---|---|---|---|---|
| parameters | parameters | Test to Train | ||||
| Mask R-CNN (segmentation) | 31.78M | 31.78M | 650 | 100 | 0.153 | |
| COVID-CT-Mask-Net (only S) | 31.52M | 2.25M | ||||
| COVID-CT-Mask-Net (S+BN) | 2.36M | 3K | 20.6K | 21.1K | 7.060 | |
| COVID-CT-Mask-Net (full) | 31.52M | |||||
| LightCNN [4] | 1.20M | 1.20M | 1528/1768 | 118/138 | 392/203 | 0.258/0.117 |
| COVNet [2] | 25.61M | 25.61M | 3K | 370 | 438 | 0.129 |
| ResNet18 [3] | 11.69M | 11.69M | 528 | 90 | 0.170 | |
In this paper we would like to address some of these shortcomings by extending the semantic segmentation and classification solution (e.g. in [6]) to instance segmentation and COVID-19 classification using Mask R-CNN. Mask R-CNN [25] and Faster R-CNN [26] are the state-of-the-art models in instance segmentation and object detection. Mask R-CNN is an extension of Faster R-CNN with an object mask prediction branch. This is different to semantic segmentation models like Fully Convolutional Network (FCN) [27] and UNet [28], which predict objects at pixel level. Mask R-CNN localizes each object independently of others, by predicting their location (bounding box coordinates) using Region Proposal Network (RPN) and Regions of Interest (RoI). Each predicted object has therefore three properties: bounding box, class and mask. The strength of Faster and Mask R-CNN comes from the fact that the model constructs batches of data from each image to make predictions about the instances. This leverages the scarcity of the training data, and we use this strength both to obtain accurate predictions and use a small sample of images for training. We augment Mask R-CNN with a classification module and extend Mask R-CNN’s ability to detect separate objects to the classification of the whole image. The novelty of our approach to COVID-19 prediction can be summarized in the following way:
Results: we use approximately 5% of the COVIDx-CT training data, (this is approx. 3% of the whole CNCB-NCOV dataset), to train the model, and, without any data and model augmentations, e.g. class weights, background removal and batch balancing, on which COVIDNet-CT depends, achieve 90.80% COVID-19 sensitivity, and 91.66% overall accuracy on the full test split (21192 images). The ratio of the test to the training split is 7.06,
Methodology: we repurpose Mask R-CNN to predict the class of the whole image by leveraging the ability of Mask R-CNN to extract regions of interest (RoIs) from deep features and obtain spatial predictions (bounding boxes) from them to construct a batch of ranked regional predictions in each image and use it to predict the global (image) class.
Open-source solutions: We develop, train and evaluate two solutions: one for the segmentation and one for the classification problem, by training two models. Mask R-CNN segmentation model predicts and segments instances of Ground Glass Opacity and Consolidation in chest CT scans, COVID-CT-Mask-Net extends this model to predict the class of the image. Models’ code and weights are available on Github.
In short, we use much less training data than, achieve both better overall accuracy and COVID-19 sensitivity than other OS solutions, and our solution has a very good potential for generalization to other datasets, due to the ratio of test to training data. In Section 2 we discuss the datasets for both tasks, in Section 3 we introduce the segmentation and classification models, Section 4 introduces the training setup, experimental results and comparison to other models, Section 5 reports ablation studies and methodology limitations, Section 6 concludes.
Data
Segmentation data
For our segmentation model we use the publicly available dataset released by China National Center for Bioinformation (CNCB) [6], consisting of 750 scans across 150 patients with various stages of COVID-19. A total of 3 classes are segmented at pixel level: clean lungs, which we merged with the background due to its prevalence, and two types of lesions: Ground Glass Opacity (GGO) and Consolidation (C).
These two types of lesions are often associated with various stages of COVID-19 and other types of pneumonia, so we treat them as positive classes. We randomly split the provided dataset into 650 training and validation and 100 test images, maintaining the patients’ consistency. Due to the shape of the lungs, some slices of COVID-19 patients do not contain positive classes, and were therefore removed from the study.
The challenges of the data are summarized in Fig. 2: it is clear that positive scans can contain a small number of small objects of either class, and overall, the proportion of positive areas to the background is very low, making the problem of segmenting them a serious challenge. To avoid overfitting, we merged the clean lungs regions with the background. Examples of positive and negative images and their masks are presented in Fig. 1.
Fig. 2.
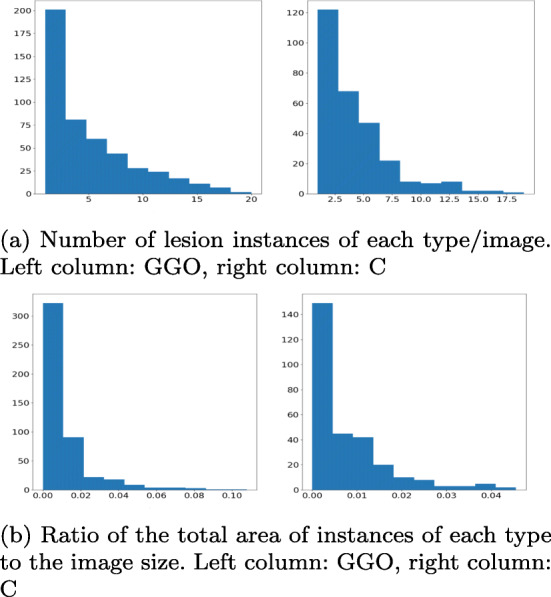
Distribution of the COVID-19 correlates in the segmentation data. The absolute majority of images have a small number (< 5 occurrences of each type) and the absolute majority of them are very small: GGO are < 2% of the image size and C are < 1%. This means that CT scans contain mostly a small number of small lesion occurrences
Fig. 1.
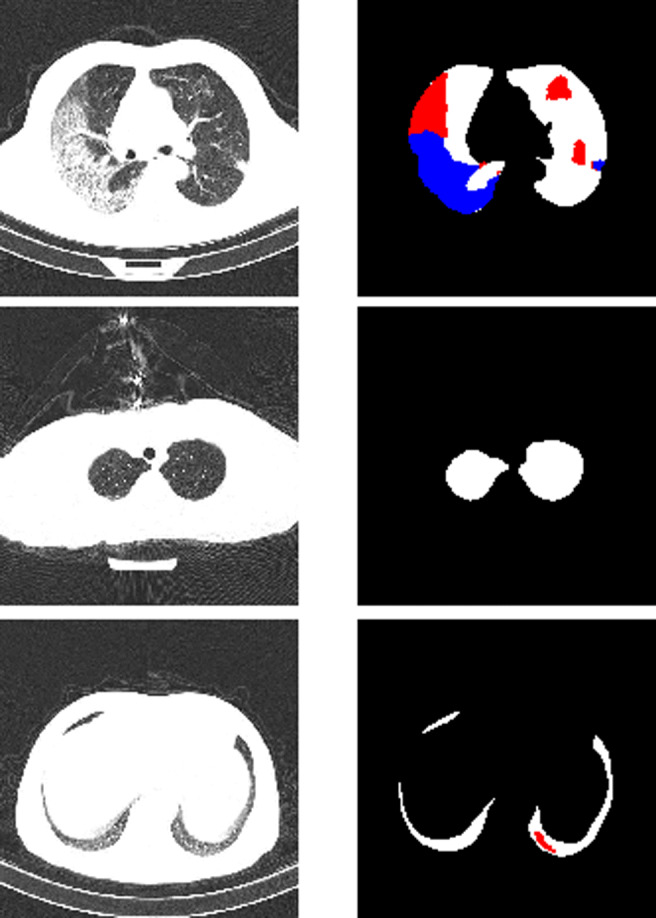
Examples of chest CT scans from the segmentation dataset with their ground truth masks. Upper row: major lungs masks, major presence of both GGO (red) and C (blue) classes, middle row: average lung mask, negative slice (no lesions), bottom row: small lung mask, small presence of GGO. In our implementation all lung masks are merged with the background
In addition to CNCB-NCOV, other open-source segmentation datasets are available, e.g. MosMedData [29], Zenodo lung and infections segmentation [30] and others. One of the key challenges in generalizing segmentation algorithms to out-of-sample data is the difference among the input images. Unlike benchmark datasets, such as Pascal VOC and MS COCO, chest CT scan datasets were collected using different methodologies and equipment. The usual approach to minimizing these differences is image normalization that we used in this study. Unfortunately, the usual normalization does not offset these differences. As a result, for all experiments, we used a single dataset. Nevertheless, development of data normalization tools and generalization across a number of datasets is one of our priorities for the future work (Fig. 2).
Classification data
To compare our model to COVIDNet-CT, we also used the second part of the dataset provided by CNCB [6], which is labelled at image level, http://ncov-ai.big.ac.cn/download and the splits from COVIDx-CT that was used to train COVIDNet-CT model (https://github.com/haydengunraj/COVIDNet-CT), both of which are publicly available. In [1] 104900 images were partitioned into 60% training, 20% validation and 20% test data. The difference between COVIDx-CT and the source data is that for COVID-19 and CP classes, only scans with observable infected regions were selected from the patients in those two classes.
One of the advantages of our approach is the size of the dataset used for training. We randomly extracted 3000 images from COVIDx-CT training split (1000/class, randomized across patients), while maintaining the full size of the validation (21036 images) and test (21192 images) splits for the direct comparison. In the validation split, the shares or Normal, CP and COVID-19 classes are 43%/35%/22%, in the test split they are 45%/35%/20%. As a result, the ratio of test to train split is 7.06, which is much higher than COVIDx-CT (0.353). These splits are also available on our Github repository.
Methodology
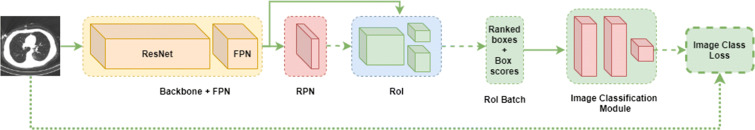
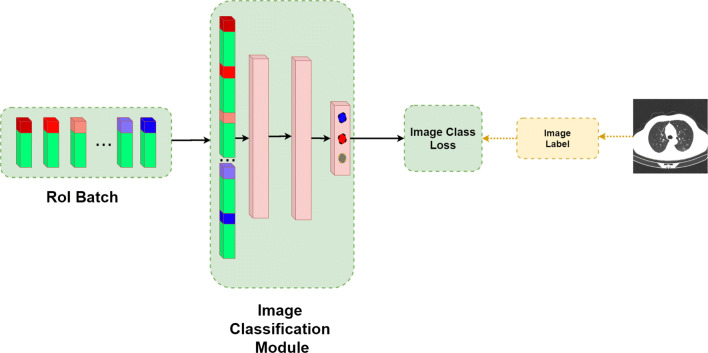
The overall flow of the algorithm is presented in Fig. 3. Our solution is split into three stages: first, we train, validate and test Mask R-CNN to predict boxes, classes and masks of GGO and C areas. After that, this model is converted to COVID-CT-Mask-Net by augmenting it with a classification module S that uses ranked bounding box predictions to classify the whole input image (Fig. 4) and the weights are copied from Mask R-CNN to COVID-CT-Mask-Net. Module S logic is presented in Fig. 5. Finally, COVID-CT-Mask-Net is tested on the test split discussed above. Overall, functionally, COVID-CT-Mask-Net extends Mask R-CNN to make global (image class) predictions (Fig. 3).
Fig. 3.
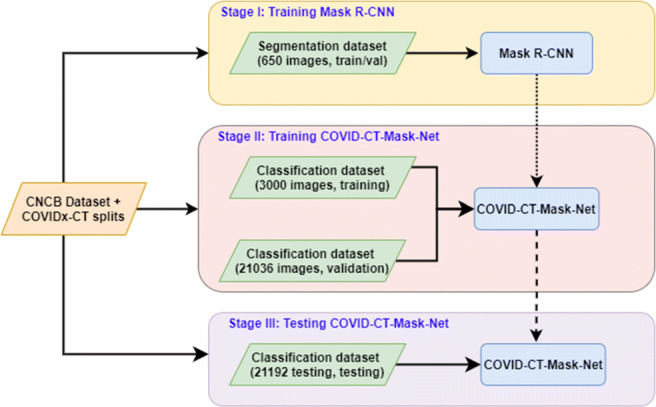
Overall flowchart of the algorithm. Normal arrows: data and labels, dotted arrow: weights copy from Mask R-CNN to COVID-CT-Mask-Net, broken arrows: copy all weights for the classifier’s evaluation
Fig. 4.
Mask R-CNN (Backbone+FPN, RPN, RoI) and COVID-CT-Mask-Net architectures. The architecture of Mask R-CNN at training and test time is the same, except that at training time LSEG is computed for RPN and RoI. At test time, RPN and RoI do not compute any losses. See Section 3.1 for a detailed discussion of its functionality. The new classification module S (Fig. 5) takes the batch size N of the ranked encoded boxes with their confidence scores as an input and predicts the class of the input image. Normal arrows: tensors or data, broken arrows: boxes, dotted arrow: image class label. Best viewed in color
Fig. 5.
Batch output from RoI layer and image classification module S of COVID-CT-Mask-Net: RoI batch is reshaped from N × 5 to a feature vector size 1 × N ⋅ 5 by concatenating the encoded boxes (green) and their scores(red, blue), followed by two fully connected layers, and the last prediction layer outputting 3 logits (scores), 1 per image class. The colors in each element reflect the normalized (sigmoid) confidence score (red:high, blue:low). Best viewed in color
Mask R-CNN
We start with a brief overview of the functionality of the segmentation model that is at the core of our approach.
Mask R-CNN can be in one of the two stages: training and testing. At training stage, ground truth data (class labels, box coordinates and masks) are used to compute the loss and update weights. At test time, the model outputs the predicted boxes, masks and class confidence. One of its strength is the construction of batches of predictions from each image, which to some extent alleviates the demand for more data.
At training time, the backbone, which consists of a ResNet feature extractor and Feature Pyramid Net (FPN, [31]) extracts features from the input image and outputs the final image-level feature map. Backbone passes this map to the Region Proposal Net (RPN) module that uses a large number of anchors (predefined rectangles) and these features, to construct a batch of candidate bounding boxes and their objectness (object vs background scores) and compute losses by matching anchors to the ground truth.
Next, Region of Interest (RoI) module maps these candidates to the backbone’s feature map and extracts regional feature maps (also known as regions of interest) of the predefined dimensionality. This is done in three steps: 1) align the box coordinates predicted by RPN to the feature map, 2) crop the local features to match the coordinates of the object’s box, 3) resize the cropped features to the predefined size using RoIAlign functionality. As a result, each region of interest has the same dimensionality, C × H × W (C: number of channels, H,W: height and width of the region).
First, RoIs output encoded box coordinates, that are used to compute the box loss. For each box, its class and mask losses are computed too. In total, 5 loss functions are computed: objectness loss, , box coordinates in the RPN module, class loss, box coordinates in RoI and pixel-wise loss for masks, LMask (1). Mask loss is class-aware, i.e. its loss is calculated only for the correct class. Mask and bounding box losses are calculated only for positive predictions.
| 1 |
All loss terms in (1) are taken from the respective publications ( from Faster R-CNN and LMask from Mask R-CNN) and their out-of-the-box implementation from Torchvision library v0.8.0.
At test time, the model outputs predictions that consist of decoded boxes, masks and class confidence scores. Those that have confidence score below a certain threshold are discarded. Also, NMS threshold is used to discard overlapping predictions with higher confidence scores. For the details of NMS threshold, see Section 3.2.3. Also, some important bits of RoI functionality are discussed in Section 3.2.1. Final box and mask predictions are resized to the object’s and image dimensions.
COVID-CT-Mask-Net
The main motivation for the development of the classifier is to explore the idea of fusing local (object) information to make a global (image) prediction (image class). In this study, we select a set of RoI encoded boxes and class confidence scores, as they can explicitly detect both lesions and the background. In this section we introduce our model and explain its functionality.
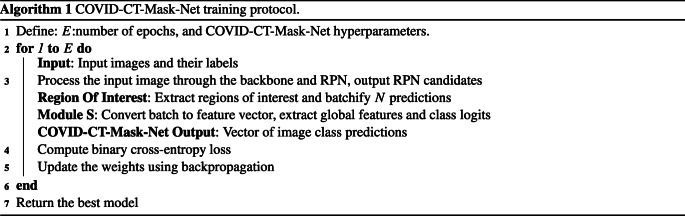
The training of COVID-CT-Mask-Net algorithm is formalized in Algorithm 1 and visualized in Fig. 4. The architecture of the image classification layer S is presented in Fig. 5. The image class loss is computed using (2), where is the vector of class logits (COVID-19, CP, Control) output by the model, σ is a sigmoid function and C∗ is the correct class. We chose to use binary cross-entropy loss (each class is either 0 or 1) instead of the multilabel cross-entropy (softmax) to improve the total loss computation.
| 2 |
Detection of regions of interest
The backbone, anchor scales and sizes, architecture of RPN and RoI layers in COVID-CT-Mask-Net are identical to Mask R-CNN, but RoI hyperparameters and functionality is quite different and needs to be put in the context of the classification problem.
As discussed previously, at test time, each region predicts objects’ classes and box coordinates (including the background class). RoI collects all of these predictions, filters out backgrounds, and outputs positive object predictions with class confidence score exceeding a predefined threshold (RoI confidence score𝜃). The maximum number of predictions is also capped at a predefined number N. We adapt this functionality for the image classification problem.
Our objective is to extract a N (fixed number, defined as a hyperparameters) of predictions from each image: obviously, in Negative images there are no lesions at all, and our objective is to address this fact. We do this by accepting all N predictions, regardless of their confidence scores. This is achieved by setting the threshold that we call RoI classification score𝜃 to a value that guarantees acceptance of exactly the predefined number of predictions.
We discard the decoded, object and image-adjusted box coordinates predicted by RoI, and, instead, use the encoded ones (confidence scores are kept the same). Next, all of these predictions are ranked in the decreasing order of their confidence scores. This ranking is essential for the next step. At this stage we are ready to extract the output batch of fixed size N from RoI (N = 256 in Table 3) of top-ranking predictions from this set, which is used as an input in the image classification module. The challenge of the classification problem is that RoI box scores for lesions in Control images are very low, barely above 0. Additionally, in order to keep the batch size fixed at N, we need a sufficient number of proposals after discarding highly overlapping and empty boxes. For this reason, RoI classification score𝜃 is set to − 0.01, which ensures both of these condition. As a result, we extract the same number of predictions from each type of image. All predictions from Normal/Control images are in fact background, for the obvious reason, but they are still ranked in the same decreasing order of confidence scores, however low.
Table 3.
Key hyperparameters of COVID-CT-Mask-Net
| Backbone | Anchor | Anchor | RPN | RoI | RPN | RoI | RoI | Classifier |
|---|---|---|---|---|---|---|---|---|
| sizes | scales | NMS𝜃 | NMS𝜃 | output | batch | class. score𝜃 | Module S | |
| ResNet50 | 22:5 | 0.1, 0.25, 0.5, | 0.75 | 0.75 | 1000 | 256 | -0.01 | 2.26M |
| +FPN | 1, 1.5, 2 |
Conversion of the RoI batch to a feature vector
The ranked RoI predictions from the previous step are concatenated into a batch with dimensions N × 5 (N predictions × 4 encoded box coordinates + 1 confidence score), which is illustrated in Fig. 5. As a result of this operation, this batch has three important properties that the image classification module S can learn:
Object’s location (encoded box coordinates),
Object’s confidence score (actual predicted class is discarded),
Object’s area (box size, boxes below a threshold are discarded),
These properties are important factors in determining the difference between the classes:
COVID-19 vs Control, CP vs Control: higher box scores, different box coordinates,
COVID-19 vs CP: different box coordinates, higher number of high-scoring boxes, larger box area
Finally, image classification module S accepts the batch and reshapes it into a single feature vector with dimensionality 1 × (N ⋅ 5), by vertically concatenating the predictions in the batch while maintaining their ranking order explained above. This feature vector is passed through two fully connected layers in S that outputs three class logits, predicting the class of the image. Finally, the loss, (2) is computed and backpropagated through the model (including RoI and RPN layers), updating the weights.
NMS threshold
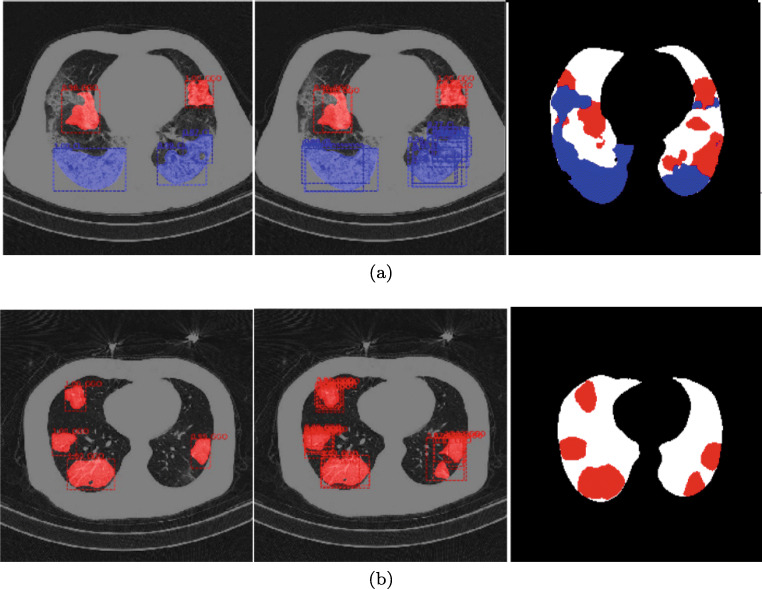
As discussed in Section 3.1, this hyperparameter is used to filter out overlapping predictions, which is essential to the detection/segmentation problem, both at training and test stages to avoid multiple predictions for the same object. For the classification problem, its role is different. As shown in [23, 32], the frequently observed difference between COVID-19 and other types of pneumonia is the distribution of the location of lesions in the lungs, e.g. COVID-19 lesions tend to be bilateral in comparison to other types of pneumonia, therefore the presence of a larger number of high-scoring overlapping box predictions can be learnt by S to indicate the presence of COVID-19 rather than CP. This is illustrated in Fig. 6: left column is the output with RoI NMS𝜃 = 0.25, central column is the output with RoI NMS𝜃 = 0.75. This motivated our choice of selecting RPN and RoI NMS𝜃 for Mask R-CNN, set out in Table 2 and COVID-CT-Mask-Net, set out in Table 3. This threshold ensures a higher number of high scoring predictions, which are an important factor in distinguishing between COVID-19 and CP.
Fig. 6.
GGO and C (a) and only GGO (b) segmentation by the same model with RoI NMS𝜃 = 0.25 in the left column and RoI NMS𝜃 = 0.75 in the central column. The right column is the ground-truth mask for each scan slice. Predictions with scores above RoI confidence score𝜃 = 0.05 for each detection and all pixels in mask logits> 0 are considered positive
Table 2.
Key hyperparameters of Mask R-CNN. Hyperparameters marked with ∗ are used only at test time
| Backbone | Anchor | Anchor | RPN | RoI | RPN | RoI | RPN | RoI | RPN | RoI | RoI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| sizes | scales | NMS𝜃 | NMS𝜃 | batch | batch | output | output∗ | IoU𝜃 | IoU𝜃 | conf. score | |
| ResNet50 | 22:5 | 0.1, 0.25, 0.5, | 0.75 | 0.25 | 256 | 256 | 1000 | 100 | 0.75 | 0.75 | 0.05 |
| +FPN | 1, 1.5, 2 |
Experiments and results
Mask R-CNN
We design hyperparameters of Mask R-CNN to maximize its capacity to detect and segment a number of small objects of varying shapes, which are widespread in chest CT scans of patients with COVID-19, see Table 2, Figs. 1 and 2. Most anchor sizes are small (< 32 × 32 pixels) and have a large number of scales (6 in total between 0.1 and 2), allowing for accurate detection of various shapes of GGO and C. Examples of Mask R-CNN’s outputs are presented in Fig. 6.
Mask R-CNN model was trained for 100 epochs on the train split using Adam optimizer [34] with a learning rate of 1e − 5 and regularization factor if 1e − 3. Tables 1 and 2 report the key hyperparameters of Mask R-CNN. At training time, RPN/RoI IoU𝜃 are thresholds for determining whether the prediction is positive. RPN and RoI batches are the number of candidates selected for training. RPN output is the set of positive predictions passed from RPN to RoI at both stages. At test stage, RoI output is the cap on the number of predictions, and RoI confidence score𝜃 is the cut-off value for positive predictions. RPN/RoI NMS𝜃 are as described in Section 3.2.3, and are also the same in both stages.
To evaluate the model on the test split in Table 1 we use the main criteria from MS COCO dataset introduced in [35]: average precision at IoU thresholds, 0.5 and 0.75, and a mean average precision across 10 IoU thresholds, 0.5 : 0.95 with a step of 0.05. For each image in the test split, the model’s predictions are compared to the ground truth masks (GGO, C, see Fig. 1). If the IoU between the predicted and gt masks exceeds the IoU threshold, and the class prediction is correct, it is considered a True Positive. Other predictions are False Positives. Gt objects without positive predictions are False Negatives. Average precision across all images is similar to a Precision-Recall curve. For further details see [35].
Segmentation results are reported in Table 6. Backbones in both networks were initialized from the weights of the model trained on ImageNet. We trained two Mask R-CNN models, which is a common practice in the literature: only RPN and RoI modules (‘heads’), and the ‘full’ model: backbone, RPN and RoI. The ‘full’ model strongly outperforms the ‘heads’ across all IoU thresholds. We explain it by the fact that both coarse and semantic features in the pretrained backbone do not immediately translate from the general-purpose ImageNet model to a specific chest CT scans dataset. Although both results appear strong, we could not compare them to any benchmark, as we did not find another Mask R-CNN model trained on a chest CT scans dataset that uses the same precision metrics, and our results cannot be directly compared to MS COCO leaderboard. Examples of lesion instance prediction by Mask R-CNN are presented in Fig. 6.
Table 6.
Average Precision on the segmentation data test split (100 images). Best results in bold
| Model | AP@0.5 IoU | AP@0.75 IoU | AP@[0.5:0.95] IoU |
|---|---|---|---|
| Mask R-CNN (head only) | 0.511 | 0.301 | 0.298 |
| Mask R-CNN (full) | 0.565 | 0.413 | 0.352 |
COVID-CT-Mask-Net
The weights from the best Mask R-CNN model were used to initialize COVID-CT-Mask-Net. As explained in Section 3, mask branch in RoI is not used in our implementation, which is reflected in the model sizes (# Total parameters in Table 1).
Key hyperparameters for the training of COVID-CT-Mask-Net are presented in Table 3 (the number of trainable parameters in S is 2.26M). We reimplement Torchvision’s Mask R-CNN library for the necessary augmentation and hacking. During the training of the classifier, RPN and RoI do not compute any loss. RoI classification score𝜃, as mentioned above, is set to − 0.01 to accept all box predictions, however low-scoring, to guarantee the RoI batch size is equal to 256, this is particularly important for Negative images without lesions. In their case all predictions are very low-scoring (e.g. 0.001), which is a pattern that S can learn. RPN and RoI NMS𝜃 defined in Section 3.2.3 are set to 0.75. RPN output is the same as in Mask R-CNN.
We train COVID-CT-Mask-Net in three different ways, which determines the total number of trainable parameters, see Table 1: 1) only classification module S, 2) S+batch normalization (BN) layers in the backbone, 3) all weights.
To train the full model, a large hack was applied: all layers, including the backbone, were set to test mode (no targets for object detection and segmentation, batch normalization layers’ tracking of means and standard deviations switched off), while the gradients were computed for all layers. Therefore, although formally, RPN and RoI were in the test mode, in fact their weights were updated using image class loss. We use a small fraction of the dataset of COVIDx-CT for training, while maintaining the full size of the test and validation sets. We use Adam optimizer [34], the learning rate of 1e − 5, weight regularization parameter of 1e − 3, and train each algorithm for 50 epochs.
As pointed out in Sections 1 and 2, the share of test to training split in our experiments is very high compared to other solutions. We explain high accuracy of the models trained on this small split by Mask R-CNN’s innovative functionality to construct batches of candidates from each image in RPN and RoI modules. This functionality greatly augments the classifier’s ability to learn from a single image and reduces its demand for larger dataset.
To evaluate each model, we compute the sensitivity (recall) and precision (positive predictive value) for each class Cl, overall accuracy and class-adjusted F1 score, see (3)-(6) (TP: true positive, FP: false positive, FN: false negative). In (6) wCl is the share of class Cl in the test split.
| 3 |
| 4 |
| 5 |
| 6 |
Best results for each version of COVID-CT-Mask-Net are presented in Table 4. The variant where we train S and batch normalization layers achieved the highest COVID-19 sensitivity, while keeping the sensitivity to other classes, overall accuracy and F1-score above 90%. The model training all parameters achieves the highest overall accuracy and F1-score and the second best COVID-19 sensitivity. Comparison of the models’ sizes and main results for other COVID-19 classifiers for 3 classes are presented in Tables 1 and 5.
Table 4.
Results on COVIDx-CT test split (21192 images). Sensitivity (PPV) per class. Best results in bold
| Model | COVID | Pneumonia | Normal | Overall | F1-score |
|---|---|---|---|---|---|
| COVID-CT-Mask-Net (only S) | 76.30% (81.13%) | 71.13% (67.70%) | 82.37% (83.38%) | 77.20% | 77.30% |
| COVID-CT-Mask-Net (S+BN) | 90.80% (94.75%) | 91.62% (87.08%) | 91.10% (94.33%) | 91.66% | 91.50% |
| COVID-CT-Mask-Net (full) | 82.26% (87.01%) | 91.70% (95.22%) | 97.21% (95.33%) | 92.22% | 92.93% |
Table 5.
Comparison to OS models trained on the 3-class problem (COVID-19 vs CP vs Control). Due to the difference in sample size/COVID-19 prevalence, in fact, models are not directly comparable
| Model | COVID Sensitivity | Overall accuracy | COVID prevalence | #Test images |
|---|---|---|---|---|
| Ours (best) | 90.80% | 91.66% | 20.00% | 21191 |
| ResNet50 [8] | 85.90% | 88.10% | 46.84% | 746 |
| LightCNN [4] | 88.23% | 84.56% | 25.39% | 392 |
| COVNet [2] | 90.00% | 89.04% | 30.00% | 434 |
| ResNet18 [3] | 81.30% | 86.70% | 35.79% | 210 |
| DarkCOVIDNet [33] | 85.25% | 87.02% | 50.00% | 1000 |
| DreNet [17] | 93.00% | 87.00% | 47.38% | 57 |
| WRE [11] | 86.40% | 86.12% | 50.00% | 29 |
Although S adds only a small overhead in terms of weights, the results are quite strong compared to other models with a feature extractor + classification head architecture, that are mostly much larger. To obtain results for LightCNN and COVIDNet-CT we used the best reported models (resp. Model1 and COVIDNet-CT-A), COVNet and ResNet18 in [3] report only one model. The results for COVIDNet-CT were obtained by running the publicly available model on the test split. Results for the other models are taken from the respective publication. Ours(best) is the model with the highest COVID-19 sensitivity (Table 6).
Although OS models in Table 5 report very high accuracy and COVID-19 sensitivity, they are not directly comparable for a number of reasons. These reasons are discussed in-depth in [24] and include the size of the datasets, reproducibility of the solutions, lack of the details of the training and test protocols, and many other. These methodological flaws prevent their comparison, generalization, and application in-the-wild, i.e. in radiological departments. In this study, to address the issue of the size of the datasets, we used CNCB-NCOV, the largest open-source dataset to adapt and evaluate our models.
Ablation studies
We perform additional testing of the introduced model. First, we use the remaining 58782 images from the training dataset of CNCB-NCOV that were left after the random the sampling of 3000 training images. Results in Table 7 are consistent with the test results in Table 4.
Table 7.
Results on COVIDx-CT left-out train split (58782 images). Sensitivity (PPV) per class. Best results in bold
| Model | COVID | Pneumonia | Normal | Overall | F1-score |
|---|---|---|---|---|---|
| COVID-CT-Mask-Net (only S) | 79.21% (86.01%) | 70.12% (68.34%) | 85.73% (82.28%) | 78.56% | 78.30% |
| COVID-CT-Mask-Net (S+BN) | 93.18% (96.51%) | 90.68% (88.04%) | 94.22% (97.31%) | 93.89% | 93.55% |
| COVID-CT-Mask-Net (full) | 81.14% (85.91%) | 90.01% (95.22%) | 94.00% (91.32%) | 90.22% | 90.93% |
To address the issue of the ability of the model to generalize to out-of-sample data, we use the 2-class publicly available iCTCF [36, 37] dataset (Table 8) to finetune the models for 10 epochs. Only 600 images from the training data were used to finetune each model that took about 15 minutes on a single GPU. Each model was evaluated on 12976 images in the test split. Results in Table 9 confirm the ability of our approach to quickly and successfully adapt to the new data.
Table 8.
| Split | COVID-19 | Negative | Total |
|---|---|---|---|
| Train/Val | 300 | 300 | 600 |
| Test | 3701 | 9275 | 12976 |
Table 9.
Accuracy results on the iCTCF-CT test split (12976 images). Best results in bold
| Model | COVID-19 | Negative | F1score |
|---|---|---|---|
| Ours (only S) | 85.45% | 81.27% | 82.01% |
| Ours (S+BN) | 93.91% | 91.46% | 92.20% |
| Ours (full) | 91.31% | 87.27% | 88.91% |
Although in this ablation study we had to turn to an additional finetuning on the new dataset, we kept the ratio of the train to test splits low, following the setup of the base dataset. Fast and simple finetuning protocol demonstrate the potential of our model to generalize to new data. Nevertheless, we see this as the major limitation of our solution, that we share, to the best of our knowledge, with all other COVID-19 solutions, both that report a very high accuracy one dataset, and those that claim out-of-the box generalization to other datasets, because in the latter case the reported accuracy on the other dataset is low.
We also addressed other limitations in the COVID-19 literature discussed in [24]: we provided the OS dataset details, such as test splits and class distribution; critical hyperparameters for both models, results on the test splits for both problems; comparison to a set of OS models; other details, including all remaining hyperparameters, can be found in the source code in a Github repository that we made available. Therefore, we minimized the list of methodological flaws discussed in [24], and our solution can be both verified and used in other studies.
Conclusions
It is often a challenge to find a sufficiently large dataset to train models for accurate predictions of COVID-19. This means that the model must either be trained using various augmentation tricks, or it is evaluated on a relatively small dataset, and therefore may not generalize well to the new data. One of the strongest features of COVID-CT-Mask-Net’s methodology is the ability to train on very small training split relative to the test split, without any balancing and augmentation tweaks due to the functionality of Mask R-CNN.
We trained our model on 3000 images from COVIDx-CT training split, and evaluated it on more than 21192 test images achieving a 91.66% overall accuracy and 90.80% COVID-19 sensitivity. The model can be easily and quickly adapt to new chest CT scans data to achieve a high sensitivity to COVID-19. Mask R-CNN achieved a 0.352 average precision of the segmentation of instances of Ground Glass Opacity and Consolidation lesions in chest CT scans. The source code with all models and weights are on https://github.com/AlexTS1980/COVID-CT-Mask-Net.
Despite these achievements, unlike Faster and Mask R-CNN, that were trained on large benchmark datasets, our model at present does not generalize out-of-the-box. In our future work we will focus on developing models that, without any additional finetuning, will generalize to other datasets, and could be introduced in radiology departments. Very likely, this will include new architectural solutions, pre-processing algorithms and loss functions.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gunraj H, Wang L, Wong A (2020) Covidnet-ct: A tailored deep convolutional neural network design for detection of covid-19 cases from chest ct images, arXiv:2009.05383 [DOI] [PMC free article] [PubMed]
- 2.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al (2020) Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology [DOI] [PMC free article] [PubMed]
- 3.Butt C, Gill J, Chun D, Babu BA (2020) Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell:1–7 [DOI] [PMC free article] [PubMed]
- 4.Polsinelli M, Cinque L, Placidi G (2020) A light cnn for detecting covid-19 from ct scans of the chest, arXiv:2004.12837 [DOI] [PMC free article] [PubMed]
- 5.Zhou T, Lu H, Yang Z, Qiu S, Huo B, Dong Y (2021) The ensemble deep learning model for novel covid-19 on ct images. Appl Soft Comput 98:106885 [DOI] [PMC free article] [PubMed]
- 6.Zhang K, Liu X, Shen J, Li Z, Sang Y, Wu X, Zha Y, Liang W, Wang C, Wang K et al (2020) Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell [DOI] [PMC free article] [PubMed]
- 7.Wu Y-H, Gao S-H, Mei J, Xu J, Fan D-P, Zhao C-W, Cheng M-M (2020) Jcs: An explainable covid-19 diagnosis system by joint classification and segmentation. arXiv:2004.07054 [DOI] [PubMed]
- 8.Zhao J, Zhang Y, He X, Xie P (2020) Covid-ct-dataset: a ct scan dataset about covid-19, arXiv:2003.13865
- 9.Jiang H, Tang S, Liu W, Zhang Y. Deep learning for covid-19 chest ct (computed tomography) image analysis: a lesson from lung cancer. Comput Struct Biotechnol J. 2021;19:1391–1399. doi: 10.1016/j.csbj.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J-Y, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision, pp 2223–2232
- 11.Wang S-H, Wu X, Zhang Y-D, Tang C, Zhang X. Diagnosis of covid-19 by wavelet renyi entropy and three-segment biogeography-based optimization. Int J Comput Intell Syst. 2020;13(1):1332–1344. doi: 10.2991/ijcis.d.200828.001. [DOI] [Google Scholar]
- 12.Wang L, Wong A (2020) Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. arXiv:2003.09871 [DOI] [PMC free article] [PubMed]
- 13.Wang S-H, Nayak DR, Guttery DS, Zhang X, Zhang Y-D. Covid-19 classification by ccshnet with deep fusion using transfer learning and discriminant correlation analysis. Inf Fusion. 2021;68:131–148. doi: 10.1016/j.inffus.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S-H, Govindaraj VV, Górriz JM, Zhang X, Zhang Y-D. Covid-19 classification by fgcnet with deep feature fusion from graph convolutional network and convolutional neural network. Inf Fusion. 2021;67:208–229. doi: 10.1016/j.inffus.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y-D, Satapathy SC, Zhu L-Y, Górriz JM, Wang S-H (2020) A seven-layer convolutional neural network for chest ct based covid-19 diagnosis using stochastic pooling. IEEE Sensors Journal [DOI] [PMC free article] [PubMed]
- 16.Aleem M, Raj R, Khan A (2020) Comparative performance analysis of the resnet backbones of mask rcnn to segment the signs of covid-19 in chest ct scans. arXiv:2008.09713
- 17.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, Chong Y, Shen J, Zha Y, Yang Y Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images, medRxiv [DOI] [PMC free article] [PubMed]
- 18.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D (2020) Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE reviews in biomedical engineering [DOI] [PubMed]
- 19.Liu B, Yan B, Zhou Y, Yang Y, Zhang Y (2020) Experiments of federated learning for covid-19 chest x-ray images. arXiv:2007.05592
- 20.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Ct scans of patients with 2019 novel coronavirus (covid-19) pneumonia. Theranostics. 2020;10(10):4606. doi: 10.7150/thno.45016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao W. Relation between chest ct findings and clinical conditions of coronavirus disease (covid-19) pneumonia: a multicenter study. Am J Roentgenol. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 22.Yan T, Wong PK, Ren H, Wang H, Wang J, Li Y (2020) Automatic distinction between covid-19 and common pneumonia using multi-scale convolutional neural network on chest ct scans. Chaos Solitons Fractals 140:110153 [DOI] [PMC free article] [PubMed]
- 23.Zhao D, Yao F, Wang L, Zheng L, Gao Y, Ye J, Guo F, Zhao H, Gao R (2020) A comparative study on the clinical features of covid-19 pneumonia to other pneumonias. Clinical Infectious Diseases [DOI] [PMC free article] [PubMed]
- 24.Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, Aviles-Rivero AI, Etmann C, McCague C, Beer L, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for covid-19 using chest radiographs and ct scans. Nat Mach Intell. 2021;3(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 25.He K, Gkioxari G, Dollár P, Girshick R (2017) Mask r-cnn. In: Proceedings of the IEEE international conference on computer vision, pp 2961–2969
- 26.Ren S, He K, Girshick R, Sun J (2015) Faster r-cnn: Towards real-time object detection with region proposal networks. In: Advances in neural information processing systems, pp 91–99 [DOI] [PubMed]
- 27.Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440 [DOI] [PubMed]
- 28.Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234–241
- 29.Morozov S, Andreychenko A, Pavlov N, Vladzymyrskyy A, Ledikhova N, Gombolevskiy V, Blokhin IA, Gelezhe P, Gonchar A, Chernina VY (2020) Mosmeddata: Chest ct scans with covid-19 related findings dataset. arXiv:2005.06465
- 30.Ma J, Wang Y, An X, Ge C, Yu Z, Chen J, Zhu Q, Dong G, He J, He Z et al (2020) Towards efficient covid-19 ct annotation: A benchmark for lung and infection segmentation. arXiv:2004.12537
- 31.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 32.Li X, Fang X, Bian Y, Lu J (2020) Comparison of chest ct findings between covid-19 pneumonia and other types of viral pneumonia: a two-center retrospective study. Eur Radiol:1–9 [DOI] [PMC free article] [PubMed]
- 33.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput Biol Med 121:103792 [DOI] [PMC free article] [PubMed]
- 34.Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv:1412.6980
- 35.Lin T-Y, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, Zitnick CL (2014) Microsoft coco: Common objects in context. In: European conference on computer vision. Springer, pp 740–755
- 36.Ning W, Lei S, Yang J, Cao Y, Jiang P, Yang Q, Zhang J, Wang X, Chen F, Geng Z et al (2020) Open resource of clinical data from patients with pneumonia for the prediction of covid-19 outcomes via deep learning. Nat Biomed Eng:1–11 [DOI] [PMC free article] [PubMed]
- 37.Ning W (2020) ictcf: an integrative resource of chest computed tomography images and clinical features of patients with covid-19 pneumonia