Abstract
Chest radiography (X-ray) is the most common diagnostic method for pulmonary disorders. A trained radiologist is required for interpreting the radiographs. But sometimes, even experienced radiologists can misinterpret the findings. This leads to the need for computer-aided detection diagnosis. For decades, researchers were automatically detecting pulmonary disorders using the traditional computer vision (CV) methods. Now the availability of large annotated datasets and computing hardware has made it possible for deep learning to dominate the area. It is now the modus operandi for feature extraction, segmentation, detection, and classification tasks in medical imaging analysis. This paper focuses on the research conducted using chest X-rays for the lung segmentation and detection/classification of pulmonary disorders on publicly available datasets. The studies performed using the Generative Adversarial Network (GAN) models for segmentation and classification on chest X-rays are also included in this study. GAN has gained the interest of the CV community as it can help with medical data scarcity. In this study, we have also included the research conducted before the popularity of deep learning models to have a clear picture of the field. Many surveys have been published, but none of them is dedicated to chest X-rays. This study will help the readers to know about the existing techniques, approaches, and their significance.
Keywords: Deep convolutional neural network; Computer vision; Lung segmentation; Multiclass classification; Nodule, TB, COVID-19, Pneumothorax detection; GAN
Introduction
The medical science branch can be categorized into two classes: anatomy and physiology. Information based on visual appearance comes under anatomy, while physiological information might not be visible, for example, diet, age, a parameter from the blood test. Medical imaging comes under the anatomy class. Magnetic resonance imaging (MRI), computed tomography (CT) scan, X-rays are few clinical examinations used for probing pulmonary disorders. X-rays hold a valued position and is a primary diagnostic tool for many medical conditions because of its ease to use and low cost.
The first X-ray of small animals is reported back in 1895 [113]. It is an imaging technique that uses radiations to produce images of organ tissues and bones. It is a very common clinical examination used by the radiologist for diagnosing pulmonary diseases. The chest radiograph is captured by passing the X-ray beam through the body. It is done in two ways: in posterior–anterior (PA) the beam is passed from back to front, and anterior–posterior (AP) where the beam is passed from the front to back. It appears black and white depending on the X-ray absorption because of the different density body parts. Bones are of high density so it appears white, while muscles and fat appear gray. The air in the lungs appears black because of its low density. Chest X-ray captures the lungs, heart, rib cage, airways, and blood vessels. It is a common diagnosis for pneumonia, tuberculosis (TB), pulmonary nodule, lung tissue scarring called fibrosis, and others. It provides the thorough examination of the patient’s chest but requires interpretation by a qualified radiologist. Variation in shape-size and overlapping of organs such as lung fields with the rib cage, fuzzy intensity transitions near the boundary of heart and lung, makes the interpretation difficult even for an expert. Therefore, the discrepancy is reported in the interpretation of X-rays among radiologists and physicians in an emergency [4, 129]. Further, the rapid rise in workload and complexity increases the chances of wrong interpretation.
In the 1960s, with modern digital computers, X-rays were getting analyzed. Almost two decades later, the research community was focusing on CAD to assist the radiologist [45]. In the late 90s, using training data for the development of automated supervised system were becoming popular. From deformable models (used in segmentation) to statistical classifiers were developed for diagnosing medical conditions. There are four main components of CAD systems: image pre-processing, segmentation, extraction of the region of interest (ROI), and classification. In medical imaging, pre-processing steps like enhancement technique (histogram equalization) and rib cage suppression can help in the identification of abnormality in the chest. Lung segmentation helps in removing the non-lung part from the computation as ROI lies inside it and helps in better analysis of clinical parameters. Segmentation may decrease the false positive (FP), as any knowledge outside the lung is irrelevant. Lung segmentation is a challenging task as there is variation in shape, size (due to age), gender, and the overlapping of clavicles and rib cage. Many different segmentation methods were developed such as simple rule-based [28, 40, 94] to adapting deformable models [32, 82], but since the popularity of complex deep learning architectures [12, 141], most of the segmentation studies are using them. The convolutional neural network (CNN) is the most used technique for image analysis. The first CNN model was Neocognitron [41]. CNN was used for the first time in lung nodule detection [100]. LeNet [91] was the first real-world CNN application where it recognized the handwritten digits, but CNN gained popularity with AlexNet [87] which is a deep CNN. Many research studies for multi-class pulmonary classification [53, 185], lung nodule detection [93], TB detection [49] using deep CNN are published. Apart from the medical diagnosis, CNN can be used on handwritten text segmentation [69], facial emotion [2, 80], and face detection [138]. Further, GAN models are used for generating synthetic imaging data that can reduce the dependency for collecting medical image datasets, as it involves legal and privacy issues. In recent years, a significant research on segmentation and classification involving GAN has been published.
With the resurgence of DL in CV from 2012, most of the research work published in medical imaging is using it. DL models require a large dataset for the training. Research labs and groups working in the field have collected and annotated large medical image datasets. These groups have made datasets public for research in the deep learning field. Some publicly available chest X-ray datasets are Japanese Society of Radiological Technology (JSRT) [151], Chest X-ray-14 (CXR-14) [175], CheXpert [71]. Advancement in hardware such as graphical processing unit (GPU) and release of large annotated medical datasets has increased the pace of work in the medical imaging field. Many surveys have been published on medical image analysis and deep neural networks. One of the most recent and detailed are [98] and [50], respectively.
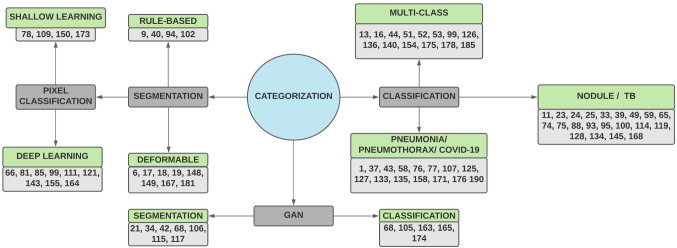
In this study, we have tried to obtain insight into the field of deep learning in medical image analysis. The intention is to collect and present the research done in the last three decades on concerning topics. An attempt has also been made to collect and discuss the publicly available chest X-ray datasets. To the best of knowledge, no survey has been published which together studied the segmentation, classification, and GAN model application involving chest X-rays. Surveys on these topics are published, but none of them is dedicated to chest X-rays only. These surveys are mostly conducted for CT scans [104, 187, 191]. This paper is categorized into eight sections. Section 2 discusses the most commonly diagnosed pulmonary disorders with X-rays. Section 3 provides a brief overview of the deep neural networks. Section 4 lists some of the publicly available datasets of chest X-rays. Section 5 discusses the work done in lung segmentation, and Sect. 6 deals with pulmonary disease detection along with multi-class classification. Section 7 followed by Sect. 6 discusses the segmentation and classification work done on X-rays using the GAN models. And Sect. 8 concludes the whole exercise. The categorization of the paper is given in Fig. 1.
Fig. 1.
Categorization of the paper
Diagnosis with chest radiographs
Chest X-rays can help in the diagnosis of different pulmonary disorders such as nodules, tuberculosis (TB), pneumothorax and many others. Pulmonary disorders can be life-threatening if not diagnosed at an early stage. In the following, we have given a brief overview of the diseases that can be diagnosed with chest X-rays.
TB is a deadly disease that is responsible for a million death according to World Health Organization Global Tuberculosis report [124]. Mantoux tuberculin skin tests (TST) or TB blood tests are conducted to detect it, but these tests are expensive. Chest X-rays are cheap and fast that can help in pulmonary TB detection. A lung nodule is another life-threatening condition that can cause cancer if not detected at an early stage. The five-year survival rate of the cancer patient is very low [153]. CT scans are most useful for early detection [57], but the low cost associated with the chest X-rays makes it popular for nodule detection.
Other pulmonary conditions that can be diagnosed with X-rays are pneumonia, atelectasis, cardiomegaly, pneumothorax, consolidation, emphysema, fibrosis, hernia and COVID-19. Atelectasis is a collapse of complete or partial lung area because alveoli within the lung is filled with alveolar fluid or becomes deflated. The airspace between the lung and the chest wall causes pneumothorax. Cardiomegaly is due to the enlargement of the heart. It can be the result of stress on the body, abnormal heart rhythms, or kidney-related diseases. On the other hand, emphysema is a chronic obstructive pulmonary condition that results in chronic cough and difficulty in breathing. The viral/bacterial infection can be the reason of pneumonia. While COVID-19 is also a kind of pneumonia caused by coronavirus. Chest X-rays can help in early detection and isolation of patients.
Overview of deep learning
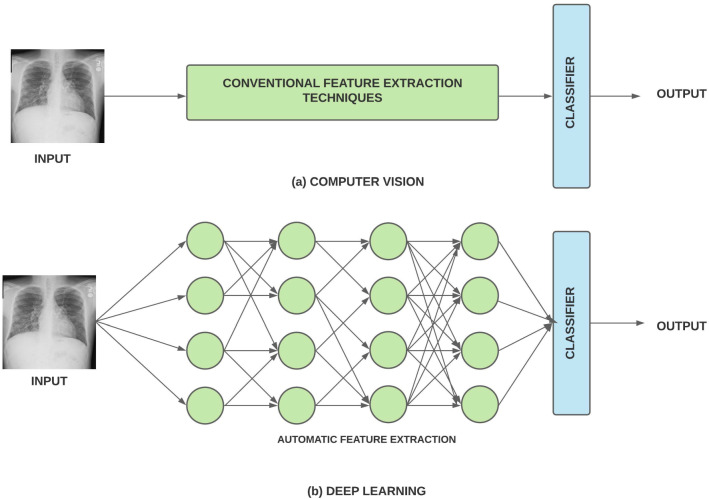
DL algorithms are a subset of machine learning that are designed to recognize or extract the patterns. It consists of artificial neurons that store the weight and help in extracting the unseen patterns. DL performs much better than traditional CV algorithms, but it is data-hungry and also requires more computational power. Improvement in computing power and availability of large annotated datasets has increased the acceptability of DL models. Unlike traditional CV algorithms that use conventional feature extraction techniques, DL models extract the feature vectors directly from the data. The workflow of CV and DL models is described in Fig. 2.
Fig. 2.
Workflow of a Computer Vision and b Deep Learning
Evolution of neural network to deep learning
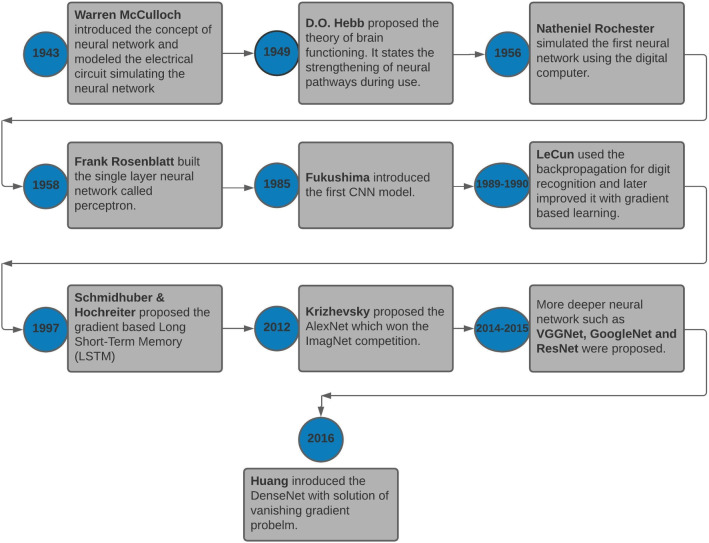
The neural networks can be traced back to 1943 when neuro-physiologist Warren McCulloch and mathematician Walter Pitts wrote about possible neuron functioning. Then, Rochester et al. [139] and Rosenblatt [142] simulated the first neural network and proposed the concept of perceptron, respectively. After the 1960s, research on neural networks partially halted, but later in the 1980s Fukushima et al. [41] proposed the first CNN model, and LeCun et al. [90] presented the first CNN application for digit recognition. Later, many advanced research models and techniques were published [56, 70, 87, 156]. A timeline of events from the proposed theory of neural network to the advanced deep learning architecture is given in Fig. 3.
Fig. 3.
Event from the theoretical neural network to Deep Learning models
Overview of deep CNN architectures
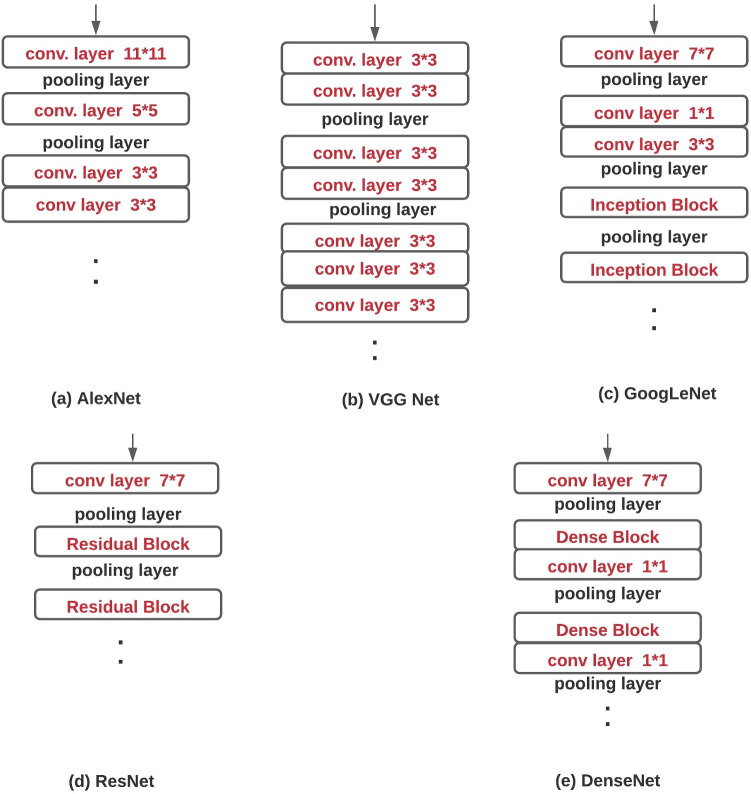
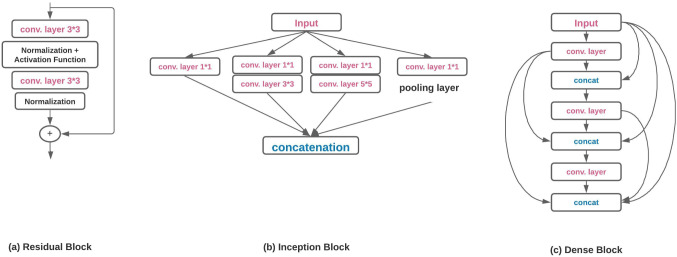
CNN was introduced with Neocognitron [41], but AlexNet revolutionizes the CV field after winning the ILSVRC-12 challenge. LeNet and AlexNet have the same design methodology but have different network depth, activation, and pooling function. Architectures proposed in later years are the AlexNet variations but are effective in classification results on the ImageNet dataset. In ILSVRC-12, AlexNet achieved 84.7% and in ILSVRC-17, NASNet [192] achieved 96.2% top-5 accuracy which is quite impressive. In this sub-section, a brief discussion of popular deep CNN architectures is presented. Figure 4 shows the basic building blocks of few popular deep learning architectures.
Fig. 4.
Basic structure of different popular deep CNN architectures
AlexNet
Krizhevsky et al. [87] proposed the eight-layer CNN for the image dataset classification. It showed that the features obtained by automatic learning could surpass the conventional methods used in the traditional CV domain. In the proposed network, there are eight stacked weighted layers out of which five are convolutional layers followed by fully connected layers. It used the dropout, data augmentation, local response normalization, and overlapped pooling for the first time. It also replaced the tanh activation function used in LeNet with the ReLU [116] activation function.
GoogLeNet
GoogLeNet [159] won the ILSVRC-14 challenge with 6.67% top-5 error. It introduced a new block called Inception (Fig. 5(b)). It uses padding to give the same height and width to the input/output. There are nine inception blocks in the architecture and at the end; fully connected (FC) layers are replaced by Global Average Pooling(GAP)[97]. The replacement of FC layers with GAP reduces the trainable parameters.
Fig. 5.
Basic diagram of a Residual Block b Inception Block c Dense Block
VGGNet
Visual Geometry Group from Oxford University introduced the VGG [152] in the ILSVRC-14 challenge. It became popular for the introduction of the repetitive blocks consisting of two/three convolution layers with ReLU activation function, followed by max-pooling layers. Unlike the AlexNet, VGGNet has only 33 convolutional layers. It achieved significant improvement in the results by increasing the network depth to 16-19 layers.
ResNet
He et al. [56] introduced the ResNet model in the ILSVRC-15 challenge and achieved 3.57% top-5 error. The plain network introduced in the original paper was inspired by the VGG nets where all convolutional layers were stacked. However, residual learning (shortcut connections) was introduced which turns the plain network into a residual version (Fig. 5(a)). The introduction of shortcut connection improved the performance of the network.
DenseNet
Huang et al. [61] proposed the Dense Convolutional Network (DenseNet) where the layers are connected in a feed-forward manner. The proposed network ensures the maximum information flow between all the connected layers (Fig. 5(c)). Except the first layer, all the layers take input from the preceding layers and pass the feature maps to successive layers.
EfficientNet
Most of the proposed CNN architectures are scaled by depth, width, or dimension. VGG [152], ResNet [56], and DenseNet [61] are few architectures that are scaled by depth. Gpipe [63] uses 557 million parameters for the image classification task. Due to large number of parameters, it needs a parallel pipeline library and more than one accelerator for training. To provide the solution for compound scaling of the network, Tan et al. [162] introduced the EfficientNet. It is the first work in the DL, to empirically quantify the relationship among scaling dimensions. With compound scaling, they have proposed the eight variants of EfficientNet (B0-B7).
Chest X-ray datasets
Deep learning models require a large dataset for the training. Collecting such a large medical dataset is a hectic process as there are legal and privacy issues. However, some research groups have collected and annotated the chest X-rays and released them for the research studies. Labeling can be done in two ways: manual or automatic. Manual labeling requires experience and expertise which is not a feasible or practical solution for a large dataset. Automatic annotation is fast but error-prone. Many researchers rely on these datasets for their work. A brief details about these datasets are given in Table 1.
Table 1.
Publicly available datasets
| S. no. | Dataset | Year | Modality | Instances | Resolution | Projection | Publicly available |
|---|---|---|---|---|---|---|---|
| 1 | JSRT | 2000 | X-Ray | 247 | 2048 2048 | PA | Yes |
| 2 | MC | 2014 | X-Ray | 138 | 4020 4892 | PA | Yes |
| 3 | Shenzhen | 2014 | X-Ray | 762 | 3000 3000 | AP | Yes |
| 4 | Indiana | 2016 | X-Ray | 7470 | – | PA | Yes |
| 5 | CXR-14 | 2017 | X-Ray | 112,120 | 1024 1024 | PA/AP | Yes |
| 6 | CheXpert | 2019 | X-Ray | 224,316 | – | – | Yes |
| 7 | MIMIC | 2019 | X-Ray | 371,920 | – | – | No |
| 8 | COVID-19 | 2020 | X-ray | 679 | 224224 | PA/AP | Yes |
JSRT [151] is the most used dataset for nodule detection and lung segmentation. It has 247 posterior X-rays collected from fourteen institutions in Japan and the USA. Montgomery County (MC) dataset [73] is available on the National library of medicine. It consists of 138 PA radiographs of which 80 are normal and 58 are infected with TB. Another such dataset is Shenzhen Hospital X-ray [73] which consists of 326 normal and 336 abnormal radiographs showing TB. A COVID-19 dataset [31] comprising more than 600 frontal X-rays collected from 412 people in 26 countries is released recently.
In 2017, CXR-14 [175] was made public for the research community by the National Library of Medicine, National Institutes of Health (NIH), USA. Initially, it was labeled with eight pulmonary diseases, but later, updated to 14 diseases. CheXpert dataset released by the Stanford ML Group is another large dataset. It is labeled with fourteen diseases namely pneumonia, atelectasis, lung nodule, lung opacity, edema, pleural effusion, and others. Figure 6 shows the common eight pulmonary conditions that can be examined in chest X-rays.
Fig. 6.
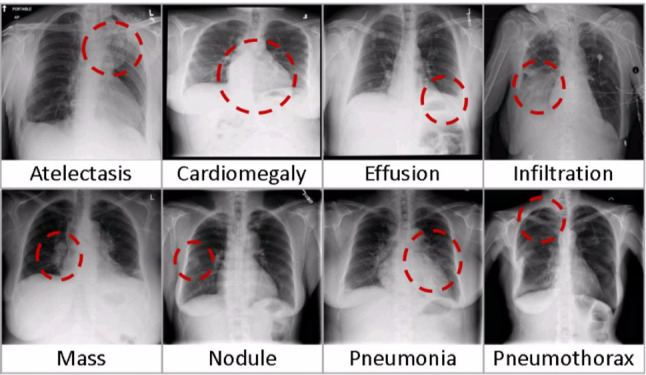
Diseases diagnosis with chest X-rays [175]
MIMIC-CXR dataset [79] contains 371,920 x-ray images from 60,000 patients. However, this dataset is not publicly available. Indiana dataset [35] contains 7470 images with 3955 radiology reports. The dataset is text-mined with eight disease labels (each image can have multi-labels). It is collected from the various hospital associated with Indiana University.
ImageNet dataset [36] is important while discussing deep learning in medical image analysis. DL models require a large dataset and enormous computation power for training [89]. If the model is not trained enough, then there are chances of overfitting. Therefore, many researchers have released pre-trained deep learning models. These pre-trained models are used for further fine-tuning the models. This technique is known as transfer learning. With no other medical dataset available of ImageNet size, transfer learning is extensively applied in the medical image domain.
Segmentation
Extracting the information from an image with a computer can be termed image processing. Dissecting the image, depending on the objects or regions present in it is termed image segmentation. It helps in locating and identifying the boundaries of different objects. Object detection helps to differentiate among objects but does not help in identifying the boundaries of objects. It simply puts the square box around the object to identify it. In chest X-rays, accurate and automatic segmentation is a challenging problem due to variation in size of the lung, edges at the rib cage, and clavicle. Segmentation in chest X-rays is performed for segregation or separation of lungs, heart, and clavicles. Many methods are proposed for lung segmentation to determine the ROI for specific abnormalities such as lung nodules, pulmonary TB, cavities, pulmonary opacity, and consolidations. Usually, the whole X-ray is used for training purposes, but the presence of unnecessary organs can contribute to noise and false positive (FP). Ginneken et al. [169, 170] classified the segmentation methods into four categories (i) rule-based, (ii) pixel classification-based, (iii) deformable-based, and (iv) hybrid methods. This categorization is generally followed in the academia for writing research or survey papers. Table 2 lists the below-mentioned rule-based and deformable methods, and Table 3 lists the pixel classification methods for lung segmentation. In the tables, if the author had calculated the result for both lungs separately, then the average score is mentioned. Further, if the author has used more than one dataset for the performance evaluation, then the best score is reported.
Table 2.
Quantitative comparison of rule and deformable model-based lung segmentation methods. Accuracy, dice similarity coefficient (DSC), jaccard index (JI), and inference time are reported for comparison
| Methods | Dataset | Technique | Results(%) | Time(s) | ||
|---|---|---|---|---|---|---|
| Accuracy | DSC | JI | ||||
| Intensity based [9] | Private | Thresholding, Filtering, Contour smoothing | 79.0 | – | – | – |
| Edge based [40] | Private | Edge tracing algorithm | 95.85 | – | – | 0.84 |
| Edge based [94] | Private | Derivative function, Iterative contour smoothing | 95.6 | – | – | 0.775 |
| Knowledge based [102] | Private | Robust snake model, Textual, Spatial and Shape characteristics | 80.0 | – | – | – |
| Parametric model [167] | Private | ASM, Adaptive gray-level appearance model, kNN | – | – | 95.5 | 4.1 |
| Parametric model [149] | JSRT | SIFT, Optimization algorithm | – | – | 94.9 | 75 |
| Geometric model [6] | Private | Level set energy, Equalized histogram, Canny edge detector, Otsu thresholding | – | 88.0 | – | 7 |
| Geometric model [17] | JSRT | Lung model calculation, Graph cut segmentation | 91.0 | 91.0 | – | 8 |
| Geometric model [19] | JSRT | Adaptive parameter learning, Graph cut segmentation | – | – | 91.1 | 8 |
| Geometric model [18] | JSRT, MC | Image retrieval, SIFT flow, Graph cut optimization | 95.4 | – | – | 20–25 |
| Hybrid model [148] | JSRT | Robust shape initialization, Local sparse shape composition, Local appearance model, Hierarchical deformable segmentation framework | – | 97.2 | 94.6 | 35.2 |
| Hybrid method [181] | JSRT | Modified gradient vector flow-ASM | – | – | 89.1 | – |
Table 3.
Quantitative comparison of shallow and deep learning models. Accuracy (Acc.), dice similarity coefficient (DSC), jaccard index (JI), and time are reported for comparison. If the time is in second(s), then it is the inference time; otherwise, it is the model training time. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Methods | Optimizer | AF | LR scheduling | Images size | Pre-processing step | Dataset | Technique | Acc. | DSC | JI | Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SL [150] | – | – | – | 440 440 | Image resizing | JSRT | Gaussian kernel distance matrix, FCM | 97.8 | – | – | – |
| SL [78] | – | – | – | 2048 2048, 4020 4892 | No pre-processing is performed | JSRT, MC, CXR-14 | FCM, Level set algorithm | – | 97.6 | 95.6 | 25–30(s) |
| SL [109] | – | – | – | – | No pre-processing is performed | Private | Linear discriminant, kNN, Neural Network, gray level thresholding | 76.0 | – | – | – |
| SL [173] | – | – | – | 1024 1024 | Images are resized using the bilinear interpolation | Private | Markov random field classifier, Iterated conditional modes | 94.8 | – | – | – |
| DL [66] | SGD | – | lr = 0.1 and it decreased to 0.01 after training 70 epochs | 256 256 | Image resizing | JSRT; MC | Residual learning, atrous convolution layers, network wise training | – | 98.0 | 96.1 | – |
| DL [121] | Adam | ELU, Sigmoid | lr = 0.00001 with = 0.9 and = 0.999 | 128 128, 256 256 | Image resizing | JSRT | UNet, ELU, Highly restrictive regularization | – | 97.4 | 95.0 | 33.0(hr) |
| DL [99] | Adam | ReLu | lr = 0.001 | 256 256 | Image resizing and data augmentation by affine transformations | JSRT | UNet, cross-validation | – | 98.0 | 97.0 | – |
| DL [155] | SGD | ReLu, Softmax | lr = 0.01 | 512 512, 128 128 | Image resizing and scaling | MC | AlexNet, ResNet-18, Patch classification, Reconstruction of lungs | 96.9 | 94.0 | 88.07 | – |
| DL [81] | SGD | ReLu, Softmax | – | 2048 2048 | Histogram equalization and local contrast normalization is applied | JSRT | Modified SegNet | 96.2 | 95.0 | – | 3.0(hr) |
| DL [143] | – | ReLu, Softmax | – | 2048 2048 | No pre-processing is performed | JSRT | SegNet | – | 95.9 | – | – |
| DL [111] | Adam | ReLu, Softmax | lr = 0.0001 with = 0.9 and = 0.999 | 224 224 | Image resizing and data augmentation by flipping, rotating, and cropping | JSRT, MC | Modified SegNet (Lf-SegNet) | 98.73 | – | 95.10 | – |
| DL [164] | SGD | – | lr = 0.02 with poly learning rate policy | 512 512 | Image resizing and data augmentation by image to image translation | JSRT, MC, NIH | ResNet-101, dilated convolution, CCAM, MUNIT | – | 97.6 | – | – |
| DL [85] | SGD | ReLu, Sigmoid | lr = 0.01 with decrease in by factor 10 when validation accuracy is not improved | 512 512 | Image resizing | JSRT, MC, Shenzhen | ResNet-101, UNet, self-attention modules | – | 97.2 | – | 1.4(s) |
Rule-based segmentation
Rule-based segmentation uses prior information such as shape, position, texture, lung intensity, and imaging features to formulate the rules. Rule-based segmentation is done using thresholding, edge detection, and knowledge-based rules.
Intensity thresholding identifies the lung or non-lung region based on the threshold value, but the challenge is to find the suitable value. A low value can result in insufficient lung contour and a high value can merge the contour between the lung and non-lung region. Using a single gray-level threshold value based on knowledge of X-ray, Cheng et al. [28] segmented the left and right lung separately, while Armato et al. [9] proposed the method based on the combination of iterative gray-level thresholding (global and local), contour smoothing using the morphological, and rolling-ball techniques. Iterative thresholding improves the segmentation but makes the process computationally insufficient and slow, while the use of static threshold values can result in poor segmentation performance with different contrast.
Edge detection depends on the pixel intensity difference in the lung and non-lung regions. There are sharp changes in intensity near the lung edges. Edge detection in lung segmentation does not require any prior knowledge of the X-ray. Duryea et al. [40] used a heuristic edge-tracing approach with validation against hand-drawn lung fields. The proposed algorithm is capable of segmenting both lungs separately. The proposed lung detection algorithm took 673 seconds to determine both lungs for 802 images (0.84 second/image). Powell et al. [131] first used the derivative for rib cage boundary detection that was later improved by Xu et al. [182, 183]. Both have used the second derivative for contour detection. The first-order derivative produces the thicker edges, while the second-order derivative generates fine details such as thin lines, isolated points, and noise.
Later, Li et al.[94] used the first derivative which includes costal and mediastinal edges. The first derivative is sensitive to noise, thus is unreliable for edge detection, but Li et al. [94] successfully used it for lung segmentation. Rib structure hindrance and false contour problem are successfully eliminated which remained in Duryea et al. [40]. It took 0.775 second per image to determine the lung contours.
In intensity and edge-based segmentation, it is possible, not to get the desired results for a deformed anatomical structure. Prior knowledge of the lung’s spatial, texture, and shape characteristics can help with this issue. Brown et al. [15] proposed a knowledge-based approach where anatomical structure information is used for extracting the edges. Another study which used the knowledge-based approach is Luo et al. [102]. It used the lung characteristics for constructing an object-oriented hierarchical model for lung regions. After defining the initial lung contour with knowledge-based rules, it is further improved with a robust snake model [103].
Deformable methods
The deformable models define the geometric shape of the object. The object’s shape can change under internal forces, external forces, or user-defined constraints. These models have been used extensively in motion tracking, edge detection, segmentation, and shape modeling. Initially, they were used in geometric applications but later found scope in medical imaging. These models can be categorized into two types: parametric and geometric deformable models. Lung segmentation applications using the parametric and geometric deformable models are discussed in the following sub-sections.
Parametric deformable
Cootes et al. [32] introduced the Active Shape Model (ASM) in 1995. The method is similar to the Active Contour Model (ACM) [82] but different in global shape constraint. Many researchers improved it later [167, 189]. Yuan et al. [189] proposed the gradient vector flow driven active shape model (GV-ASM). However, this algorithm is not applied to medical datasets. Ginneken et al. [167] extended the ASM and test the method for lung segmentation in chest X-ray using the gray-level appearance model. It moves the points along the lung contour to a better location during optimization. Pixel classification inside or outside of the lung is determined through feature selection and a kNN classifier. One advantage that the original ASM has over the proposed scheme is the speed. The total time for segmentation is 4.1 seconds for the proposed method, while with original ASM is 0.17 seconds. Gleason et al. [46] proposed the deformable model that optimizes the objective function generated by global shape and local gray-level characteristics, simultaneously. The local shape characteristics that are obtained in the training are used for boundary detection. Shi et al. [149] proposed the deformable model using the population-based and patient-specific shape (PBPS) statistics trained using hierarchical principal component analysis (PCA). Further, they used the scale-invariant feature transform (SIFT) to improve the segmentation accuracy. The proposed method took 75 seconds, while the standard ASM took 30 seconds for one chest radiograph segmentation. The limitation of the ASM is that it only considers the shape of an object, not texture or gray-level appearance. This limitation is removed by the active appearance model (AAM) that is an extension of ASM. It took both shape and appearance into account for segmentation but is computationally expensive.
Geometric deformable
Caselles et al. [20] proposed the geometric deformable model to overcome the parametric model limitations. These models are based on the level set method and curve evolution theory [86, 146]. Geometric measures evolve the curve and surfaces that are independent of the parameterization. This results in the smooth handling of the automatic topology changes. In parametric methods, evolution is mapped with the image data for recovering the contour.
Annangi et al. [6] used the active contour with the prior shape knowledge for the lung segmentation. Lungs are segmented with otsu thresholding and level set minimal energy that uses extracted edge/corner feature with region-based data. Extracted features along with region-based data help in the handling of local minima issues. The proposed algorithm took seven second per image for segmenting both lungs.
Graph cut is another segmentation method where the objective function is similarly minimized as the level set method. Candemir et al. [17] proposed the segmentation model based on graph cut and later expanded the work using multi-class regularization to detect lung boundary [19] in chest X-rays. The work done in [17, 19] was extended in [18]. Candemir et al. [17] proposed the graph-cut-based lung segmentation model. It consists of two stages: in first, the average lung shape model is calculated, and in second, the lung boundary is calculated using graph-cut. It took eight second for image segmentation of 1024 1024 resolution. The drawback of graph cut segmentation is that it produces the static shape model and the chest X-rays have variable lung shape [17]. Another drawback of the graph cut method is high computation time. In [19], adaptive parameter learning methodology is described to improve the segmentation process using a multi-class classifier approach. The proposed method takes eight second for segmentation using graph cut. Candemir et al. [18] improved the lung segmentation method by using the anatomical atlases with non-registration. In this work, lung contour is detected using non-rigid registration by applying an image retrieval-based patient-specific adaptive lung model. The execution time for the proposed method is 20–25 seconds on 256 256 resolution.
Segmentation methods (rule-based, parametric) reported good results for the lung segmentation, but the hybrid approach can do better. Shao et al. [148] combined the active shape and active appearance model. In the proposed method, three models are used for robust and accurate segmentation. First, the shape initialization model is used to detect the salient landmarks on the lung boundary, and then the local sparse appearance model is used to learn local sparse shapes. Finally, a local appearance model is built for capturing the local appearance characteristics. The computation time for the proposed method is 35.2 seconds. Xu et al.[181] proposed a modified gradient vector flow-based ASM (GVF-ASM) for lung segmentation in X-rays. The experimental results are 3-5% better than the ASM technique.
Pixel classification-based segmentation
Lung segmentation in radiographs allows quantitative analysis of clinical parameters. It is the first step of the CAD diagnosis. Classifiers such as kNN or support vector machine (SVM) can extract pixel spatial and texture information from gray-scale values, and according to it, the pixel is assigned to the corresponding anatomical structure. Segmentation is one of the most common topic for researchers applying deep learning on medical images [98]. With the best of knowledge, the earliest work for lung segmentation on chest radiography using convolutional neural networks and neural networks is Hasegawa et al. [55] and McNittGray et al. [109], respectively. Hasegawa et al.[55] used downsampled chest X-rays as input for shift-invariant convolutional neural network, and then post-processing methods, such as adaptive thresholding, noise reduction, were applied for lung field segmentation.
Pixel classification can be sub-divided into shallow and deep learning (DL). DL models outperform the shallow learning models as it extracts the features automatically from raw data, while later uses the conventional methods. The disadvantage with DL models is that they need large annotated datasets and have high computational complexity. Few deep learning architectures have been proposed for the segmentation of medical images only. One such architecture is UNet proposed by Ronneberger et al. [141] for biomedical image segmentation. The main highlight of the architecture is an equal number of upsampling and downsampling layers along with skip connections. Milletari et al. [110] proposed the variant of UNet called VNet. It performed the 3D medical image segmentation using the 3D convolution layers. Another encoder–decoder-based architecture that is quite popular for image segmentation is SegNet [12]. The encoder architecture is similar to that of VGG-16 except fully connected layers were replaced by the decoder. One major difference between UNet and SegNet is that U-Net does not use the pooling indices and transfers the whole feature map to the corresponding decoding layer [111]. In the following, lung segmentation in chest X-rays using shallow and deep learning-based segmentation models is discussed.
Shallow learning
In shallow learning (SL), the features is extracted with conventional methods and the challenge is to find a suitable class for the extracted features robustly. McNittGray et al. [109] were first to propose lung segmentation using a neural network. With kNN, linear discriminant analysis (LDA), and feed-forward neural network classifiers (FFNNC), chest X-ray pixels are classified into anatomical classes. Each classifier is trained using local texture, gray-level-based features. Among all three classifiers, the neural network outperformed the other two and predicted more than 76% pixels correctly on the test set. Vittitoe et al. [173] identified lung region in X-rays using spatial and texture characteristics with Markov random field modeling (MRFM). The proposed method classified 94.8% pixels correctly in comparison with 76% pixels in [109]. For medical image segmentation, the fuzzy c-means clustering algorithm (FCM) is often used. It permits a single data point to belong to many clusters. It is used in pattern recognition problems. Many research studies have used the FCM for the MRI image segmentation [3, 26, 27]. The basic difference between MRI and chest X-ray is that in X-ray there is the possibility of ribs and nodule overlap, but in MRI both are separated. Shi et al. [150] proposed an algorithm for lung segmentation in X-rays using FCM. The proposed algorithm is based on Gaussian kernel-based fuzzy clustering. The original c-means algorithm is modified by altering the objective function using the Gaussian kernel-induced distance metric. Jangam et al. [78] proposed an algorithm based on a firefly optimized fuzzy c-means clustering algorithm. The model combines spatial fuzzy clustering and a level set algorithm for lung segmentation. Lung region pixels are classified using a fuzzy clustering algorithm, and then the outcome is applied to the level set algorithm for segmentation. The computation time for the proposed method is 25–30 seconds.
Deep learning
Loss of a lung region while segmentation can be disastrous. So, the need for an algorithm for accurate segmentation is very important. The shallow learning-based method relies on conventional feature extraction methods, while deep learning models extract the features automatically. So, the deep learning feature extraction method has a huge advantage over shallow learning-based methods. To the best of knowledge, Long et al. [101] was the first study to use the deep fully convolutional neural network for pixel classification in images. The encoder–decoder architecture is extensively used for semantic segmentation [120] in which an image is processed for multiple segmentation. The encoder works as CNN for feature extraction, while the decoder is used for upsampling operations to get the final segmentation results. The studies on lung segmentation can be classified according to whether the network uses transfer learning or is trained from scratch. The pre-trained network has the advantage of knowledge transfer from a large image classification dataset.
In the following, the studies that have used transfer learning are discussed. Tang et al. [164] use the pre-trained ResNet-101 [56] along with the criss-cross attention module (CCAM) [64]. CCAM captures the contextual information to improve the pixel-wise representation. Further, to deal with the insufficient data, the image-to-image translation is proposed using the MUNIT [62]. This is the only study that has performed cross-dataset generalization. The proposed model has trained on JSRT and MC dataset and tested it on NIH dataset [164]. Kim et al. [85] proposed the self-attention module to capture the global features in input X-ray images. The proposed attention module is applied to the UNet for lung segmentation. They have used the pre-trained ResNet-101 [56] as UNet backbone. The training time of UNet is 4 hours approximately, but when attention modules are added in the architecture, the training time dropped to 2.5 hours. While the inference time is 1.4 second per chest X-ray image.
In the following, we have discussed the studies that have used the training from scratch strategy. One such study is Hwang et al. [66] that proposed the model for chest X-ray segmentation based on residual learning [56] with atrous convolution layers. The global receptive field of the network can be increased by stacking the convolutional layer for a larger context. But this can increase the computational complexity, so the atrous convolution is used with a dilation rate of 3 in the last two residual layers. The proposed method uses the multi-stage training strategy called network-wise training. In network-wise training, the output of the pre-stage network is used as input. The proposed model has 120,672 parameters which are less than the UNet model. Novikov et al. [121] performed the clavicles, lungs, and heart segmentation. The proposed modified UNet (InvertedNet) architecture outperformed the radiologist for lungs and heart segmentation. In the modified architecture, Gaussian dropout is added after every convolutional layer. The delayed sub-sampling has been introduced in the contraction part of the network, and the exponential linear units (ELU) [30] are used instead of ReLu to speed up training. The method performed better even after with ten times fewer parameters compared to the state of art methods. The proposed InvertedNet took 33 hours to train and have 3,140,771 parameters. Other studies have also used the UNet architecture, and one such is Liu et al. [99] that modified the architecture for the lung segmentation. The false-positive (FP) region is dealt with using post-processing techniques after segmentation. External air or intestinal gas can cause the FP area to appear. The actual segmented lung is found by measuring the distance between the picture center and the segmented area centroid.
SegNet [12] is another popular encoder–decoder architecture for the image segmentation. Kalinovsky et al. [81] modified the SegNet and segmented the lungs on 354 radiographs with 96.2% accuracy. They used the max-pool index from the corresponding encoder layer to upsample feature map in the decoding layer which is not clearly defined in the SegNet. The proposed network took nearly three hours for the training. Another study that uses the SegNet for lung segmentation is Saidy et al. [143]. The model is tested on 35 unseen images and a dice coefficient (DC) or DSC value of 96.0% is achieved. Similarly, Mittal et al. [111] performed the lung segmentation using the encode-decoder network, named as LF-SegNet. They performed the segmentation on the JSRT and MC dataset and achieved 98.73% accuracy and 95.10% Jaccard Index value.
In Souza et al. [155] the weights are initialized using normal initialization [47]. The proposed method performs lung segmentation using the reconstruction technique. Initial segmentation is done using AlexNet [87] on lung patches to generate a segmentation mask. Many false positives generated in the resultant segmentation mask during the plotting of lung patches are removed with morphological operations. In healthy radiographs where no abnormality is present, the initial segmentation can produce good results, but the lung regions severely affected by opacities or consolidations can result in poor segmentation. To provide a general solution, a second CNN (ResNet-18) is trained for lung reconstruction. The initial segmentation and reconstruction outputs are combined with binary OR operation for the final segmentation.
Most of the lung segmentation models in chest X-rays have used the UNet and SegNet architecture or have proposed their modified variants, but in the following, we have mentioned some recurrent neural network (RNN) models that have been used for medical image segmentation. The RNN models have not been exploited for lung segmentation in chest X-rays. One such model is Xie et al. [180] that uses the spatial clockwork RNN for the segmentation of the histopathology images, while Stollenga et al. [157] use the 3D LSTM-RNN for membrane and brain segmentation. Another model that has used RNN is Andermatt et al. [5]. It has used the 3D RNN for brain segmentation, while Chen et al. [22] used the bi-directional LSTM-RNN and U-Net architecture together for tubular fungus and neuronal structures segmentation in 3D images.
Discussion
A clear boundary between the lung and non-lung parts is needed for lung segmentation. Variation in lung shape and ambiguity in boundary makes it difficult for the rule-based methods. Later many other methods such as deformable methods were proposed, and they have shown better results in comparison with rule-based segmentation. The problem with deformable models is that they are knowledge-based and do not work accurately on different datasets. Deep learning methods have given the state of the art results in several image domains. In deep learning models, the features are automatically extracted, they do not use the handcraft features or conventional methods. After the introduction of UNet, SegNet models, many research studies have proposed their variant architectures for medical image segmentation. In the above-discussed paper, we can analyze that transfer learning and cross-dataset generalization has not been exploited for lung segmentation in comparison with image classification. Further, to get the insight of the field, Ginneken et al.[170] have conducted a comparative study between ASM, AAM, and pixel classification methods for lung segmentation and Terzopoulos et al.[108] have done a detailed survey on deformable models in medical image analysis. Recently, Tajbakhsh et al. [161] have conducted a detailed survey addressing the techniques to handle the scarcity of segmentation datasets for deep learning models.
Classification
Lung diseases are the primary cause of death in most countries. The chest X-rays, CT scans, and MRI scans are few available diagnosis methods for detecting and analyzing the disease severity. Among the diagnoses stated, X-ray is the most commonly used modality. The use of medical imaging with deep learning for classification is the current academic research direction. After segmentation, classification is the most common topic for research in medical image analysis [98]. The deep learning model requires a large dataset for training. It is hard to acquire a large dataset due to privacy issues. Labeling or annotation is another issue related to the medical datasets. Many research groups have collected datasets for their study and made them available publicly. The size of these datasets is small, ranging from a few hundred to thousands. CXR-14 [175] is the largest X-ray dataset that is released publicly in 2017. They have collected more than 100,000 chest X-rays from nearly 30,000 patients. Most of the multi-class classification studies using deep learning have used this dataset. In recent years, some new datasets have been made publicly available. For example, the CheXpert dataset is publicly available, whereas MIMIC-CXR is available only for the credential authors. Other chest X-ray datasets (e.g., JSRT, Indiana, etc.) are publicly available, and many studies have evaluated their models on these datasets. In the below sub-sections, we have discussed the studies that have used automatic models to detect pulmonary conditions.
Disease detection
Nodule detection
Commonly referred to as a ‘spot on the lung’, a nodule is a dense round area in comparison with normal tissue. They appear as white shadows in the chest X-rays. A lung nodule is an early stage of lung cancer. According to WHO, 9.6 million people have died from cancer in 2018. The nodule of size 5-10 mm is visible in X-rays, and less than that is not likely to be detected. Nodule detection is also difficult sometimes because it may be hidden by strong ribs or clavicle [10, 147]. There are no symptoms of the pulmonary nodule. It can be diagnosed only through screening. In Table 4, we have enlisted the nodule detection work reported on X-rays.
Table 4.
Quantitative comparison of nodule detection methods. Accuracy, sensitivity for false positive per image (FPPI), and inference time are reported for comparison. For [33] training time is reported. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Method | Optimizer | AF | LR scheduling | Images size | Pre-processing step | Dataset | Technique | Accuracy | Sensitivity | Time(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| ANN [100] | – | – | – | 32 32 | Background removal for nodule enhancement and contrast enhancement | Private | CNN with fuzzy training, circular background subtraction technique | – | – | 15 |
| ANN [33] | Gradient-descent | – | lr = 0.1 | 128 128, 512 512 | Lung field segmentation using deformable models (snakes) | JSRT, Private | Neural network, LoG, gabor kernel | 95.7-98.0(4-10) | 60.0-75.0(4-10) | 500 |
| CAD [145] | – | – | – | 256 256 | Deformable model (ASM) is used for lung field segmentation and local normalization is performed to achieve the global contrast equalization | JSRT | ASM, local normalization (LN) filtering, Lindeberg detector, Gaussian filter bank | 51.0(2), 67.0(4) | – | – |
| CAD [25] | – | – | – | 2048 2048 | Lung segmentation using multi-segment ASM, gray-level morphological operators for enhancing nodules and rib suppression | JSRT, Private | ASM, watershed algorithm, leave-one out cross-validation, SVM with a Gaussian kernel | – | 77.1(2), 83.3(5) | 70 |
| ANN [24] | – | – | – | 2048 2048 | Lung segmentation using multi-segment ASM, gray-level morphological operators for enhancing nodules and rib suppression using MTANNs | JSRT, Private | VDE, MTANN, morphological filtering, support vector classifier | – | 85.0(5) | 115 |
| DL [93] | SGD | ReLu | – | 229 229 | Unsharp mask sharpening technique | JSRT | Unsharp mask sharpening, E-CNN, five-fold cross-validation | – | 84.0(2), 94.0(5) | – |
| DL [95] | SGD | ReLu | lr = 0.001 which drops 0.00001 after every iteration | 224 224 | Random rotation and mirroring, image enhancement with gray-level stretching and histogram matching, lung field segmentation and rib suppression using ASM and PCA, respectively | JSRT, Shenzhen | ASM, PCA, dense blocks, fivefold cross-validation | 99.0(0.2) | – | – |
| DL [23] | SGD | ReLu, Softmax | lr = 0.001 | 229 229 | Horizontal inversion, angle rotation and flipping, lung field segmentation using multi-segment ASM | JSRT | ASM, watershed algorithm, GoogLeNet | – | 91.4(2), 97.1(5) | – |
Lo et al. [100] is the first study to apply CNN for nodule detection. Here, the proposed CNN is the simplified version of Neocognitron [41]. The results on the test set demonstrated the effective use of CNN in nodule detection. Further, the fuzzy training method is used with CNN to improve the obtained results. The processing time taken by the proposed model for nodule detection is 15 seconds on a DEC Alpha workstation. Coppini et al. [33] proposed an approach based on multi-scale processing and artificial neural network (ANN) for detecting lung nodules. The feed-forward ANN uses prior knowledge such as the shape and size of lung nodules. The proposed method works in twofold: first, the network narrows down the regions of possibly having the nodule with high true positive. Second, evaluating the region for the abnormality presence to detect the nodule. It helps in reducing the false positive and increasing the detection rate. The Laplacian of Gaussian (LoG) and Gabor kernels are used to amplify the features of X-rays. The average training time for the proposed method is 500 seconds.
Schilham et al. [145] proposed a method for nodule detection and selection to reduce the FP for candidate classification. To avoid nodules candidates outside the lungs, lung segmentation is performed using ASM before nodule detection. The detection of lung nodules is a multi-scale problem since nodule size varies. The bright spots help in blob detection, so the authors use the local maxima to find the candidate nodule locations with the corresponding radius. The detected blob is then segmented to separate the nodule from the background with ray casting procedure [54]. After the candidate blob detection, features from a multi-scale Gaussian filter bank are used to classify the nodule with a kNN classifier. It is employed to locate the k closest candidates in the database.
The detection of lung nodules is challenging since their size varies, and if they’re under the ribs, they are easily overlooked. So, Chen et al. [25] proposed the multi-stage method to improve the nodule detection and to reduce the false positive. First, the lungs are segmented using the multi-segment ASM. Then, a gray-level morphological operation is applied to enhance the nodule, and a line-structure suppression technique to detect the nodules overlapping the rib cage and clavicles. At the third stage, in a nodule likelihood map obtained from the second stage, noise is reduced by using the Gaussian filter to further improve the performance. After, the candidate nodule detection from the images, the improved watershed algorithm is applied for nodule segmentation. The features such as texture, shape, gray-level, etc are extracted, and then a nonlinear SVM with a Gaussian kernel is used lung nodule classification. The proposed method took 70 seconds to detect the nodule in a image.
In their other work, Chen et al. [24] also suppress ribs to reduce the FP. The same method is applied for the lung segmentation, but for the ribs and clavicle suppression, they have used the virtual dual-energy (VDE) X-ray images with massive training artificial neural network (MTANNs) instead of subtracting the rib pattern from the enhanced image. The ribs and clavicles in X-rays can be significantly repressed with this approach, while soft tissues like lung nodules and vessels remain preserved. The candidate nodules are detected by using morphological filtering techniques and the nonlinear support vector classifier. The processing time of the proposed method for each image is 115 seconds. In comparison with [25], sensitivity is improved, but processing time is increased.
All the above-mentioned work has produced good sensitivity with decreasing false positive per image (FPPI), but none of the above-mentioned research has used deep learning. Li et al. [93] in their proposed work have used the deep ensemble network for nodule detection and FPPI reduction. The proposed method uses an ensemble convolutional neural network (E-CNN) and does not employ the lung segmentation procedure as in the above-mentioned methods. The nodule is amplified using the unsharp mask image sharpening technique. Patches are cut from the processed image with the slide-window method before feeding to the CNN. Three different CNN models with dissimilar input sizes and layers are trained, and the results of all models are fused using the logical AND operator. The proposed ensemble network has outperformed the other state-of-the-art methods. In their another study, Li et al. [95] have done segmentation and rib suppression using the modified ASM and PCA, respectively. Image enhancement is performed using the efficient gray-level stretching operation and histogram matching. Then, three CNN with dense blocks is fused with four varying strategies to find the probability of the selected candidate nodule. Further, the FP is removed using morphological operations in post-processing steps.
Chen et al. [23] proposed the CAD scheme using the CNN for nodule detection. They have used the classical method for candidate detection and the CNN for FP reduction. Authors termed the methodology as balanced CNN with classic candidate detection. First, pulmonary parenchyma is segmented by using the multi-segment ASM. Then, the gray-scale morphological enhancement technique is applied to improve the visibility of the nodule. After selecting the nodule from enhanced images, the watershed algorithm is applied to segment the candidate nodule. At last, a pre-trained CNN model (GoogLeNet) is used for the classification of nodule candidates. On analyzing Table 4, it can be seen that the deep learning models provide better sensitivity and accuracy in comparison with shallow learning methods.
Tuberculosis detection
TB is the main reason for morbidity and mortality worldwide. It is the second disease after HIV that has resulted in million death alone in 2010 [124]. Chest X-rays are the most common diagnosis for TB examination [92]. With an increasing number of cases and a handful of trained professionals to interpret the radiographs, there is a need for automatic detection. Many research studies are available that has exploited the deep learning models for tuberculosis detection. These models need minimum human intervention and give accurate results. We have also discussed few shallow methods for result comparison with deep models. Table 5 enlists the work done for TB detection on chest X-rays.
Table 5.
Quantitative comparison of TB detection methods. Accuracy, sensitivity, and area under curve (AUC) are reported for comparison. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Method | Optimizer | AF | LR scheduling | Images size | Pre-processing step | Dataset | Technique | Accuracy | Sensitivity | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| CAD [168] | – | – | – | 932 932 | Lung field segmentation using ASM | Private | ASM, Gaussian derivative filter, kNN algorithm | – | 86.0 | 82.0 |
| CAD [74] | – | – | – | 2048 2048, 1023 1005 | Lung field segmentation using intensity, lung model and Log Gabor masks | JSRT, MC | lung model, intensity and LoG mask, SVM, leave-one out evaluation | 75.0 | – | 83.12 |
| CAD [75] | – | – | – | 2048 2048 | Lung field segmentation using graph cut method | JSRT, MC, Shenzhen | Graph cut segmentation, ODI, CBIR, SVM | 84.0 | – | 90.0 |
| DL [65] | SGD | ReLu, Softmax | lr = 0.01 and it is decreased by a factor of 2 for every 30 epochs | 520 520 | Images resizing and data augmentation | MC, Shenzhen | AlexNet, transfer learning, dataset augmentation, threefold cross-validation | 90.3 | – | 96.4 |
| DL [88] | SGD | – | Training from scratch, lr = 0.01, pre-trained network, lr = 0.001 | 256 256 | Data augmentation and histogram equalization | MC, Shenzhen | GoogLeNet, AlexNet, transfer learning, data augmentation | – | – | 99.0 |
| DL [59] | Adam | ReLu, Softmax | lr = 0.0001 | 224 224 | No pre-processing is performed | MC, Shenzhen | CNN | 94.73 | – | – |
| DL [49] | Nesterov ADAM | ReLu | lr = 0.001 and decreased by factor of 10 when validation loss stops improving | 224 224 | Image resizing, normalization, and data augmentation | MC, Shenzhen, CXR-14 | DenseNet121, transfer learning, meta data | – | – | 93.7 |
| DL [128] | Adam | ReLu, Softmax | lr = 0.001 | 512 512 | Image cropping, resizing, and normalization | MC, Shenzhen | CNN, grad-CAM, cross-validation | 86.2 | – | 92.5 |
| DL [134] | SGD | ReLu, Softmax | lr = 0.001 | 224 224 | Image normalization using Z-score, lung segmentation and data augmentation | MC, Shenzhen | ResNet18, ResNet-101, VGG-19, InceptionV3, UNet, transfer learning, score-CAM | 98.6 | 98.56 | – |
| DL [114] | SGD | ReLu | lr = 0.01 with rate decay of 0.5 | 224 224 | Unsharp Masking, High-Frequency Emphasis Filtering, and Contrast Limited Adaptive Histogram Equalization, cropping, image normalization | Shenzhen | ResNet-18, ResNet-50, EfficientNet-B4, UM, HEF, CLAHE, transfer learning | 89.92 | – | 94.8 |
| DL [11] | – | ReLu | – | 300 300, 229 229 | Image normalization and resizing | MC, Shenzhen | Inception-v3, MobileNet, ResNet50, Gabor filter, cross-validation | 97.59 | – | 99.0 |
| DL [39] | – | ReLu, Softmax | – | 224 224, 320 320 | Image resizing and data augmentation | MC, Shenzhen, COVID-19 | Transformer model, EfficientNetB0, EfficientNetB1, transfer learning | 97.72 | – | 100 |
| DL [119] | – | – | – | 512 512 | Image resizing, normalization, and data augmentation | MC, Shenzhen, Private | ResNext, UNet | 91.0 | 85.7 | 91.0 |
Ginneken et al. [168] were the first to propose an automatic method for TB detection. The lungs are initially segmented using ASM in the suggested method. Then, the segmented lung fields are divided into several regions. The texture features are extracted from each region using the multi-scale filter bank and the analysis of these texture features results in feature vectors. At last, kNN is applied to classify the feature vector. The proposed method obtained an AUC of 0.82 and a sensitivity of 0.86. Jaegar et al. [74] proposed the automatic method for TB detection. Initially, they used three different masks, namely intensity mask, lung model mask, and Log Gabor mask for lung segmentation. Then, the shape and texture feature descriptors are used for finding normal/abnormal patterns. At last, the SVM classifier is trained to classify the images into normal and abnormal. The overall accuracy reached 75.0% when all three masks are used together. In their other work [75], they used the graph cut method for lung segmentation. The object detection inspired (ODI) and content-based image retrieval (CBIR) based texture and shape features are applied to the segmented lung fields for describing normal and abnormal patterns. At last, the SVM classifier categorizes the computed features into abnormal or normal classification.
Deep learning is not employed for feature extraction in any of the above-mentioned three papers. The features are extracted using conventional image processing methods in these research papers. These strategies have the drawback of making the model domain-specific, while deep learning models are not domain-specific since they can automatically extract significant characteristics. Hwang et al. [65] were the first to propose TB detection using the deep learning model. The pre-trained AlexNet is used for the feature extraction, and at the end, Softmax is used for the classification. The method is validated with the cross-dataset generalization technique. Lakhani et al. [88] detected the TB using the pre-trained CNN with data augmentation techniques such as rotation and histogram equalization. They evaluated the three training strategy, i.e., pre-trained and untrained networks, pre-trained and untrained networks with data augmentation, and ensemble pre-trained networks with augmented data. Among all the different training methods, an ensemble pre-trained network reported the best AUC of 99.0%.
Gozes et al. [49] employed DenseNet-121 network trained on CXR-14 dataset for classification. The proposed model (MetaChexnet) is trained in two phases. In the first phase, a pre-trained DenseNet-121 learns the dataset-specific features with the CXR-14 dataset and meta-data. Then, the network is fine-tuned for TB detection with Shenzhen and MC dataset. The proposed model is compared with the ChexNet and pre-trained DenseNet-121 for the classification. The proposed model gives a comparable result with the ChexNet [135] and outperforms the pre-trained DenseNet-121. Rahman et al. [134] evaluated the nine different pre-trained CNN models for TB detection. They used two strategies for training the model. In the first strategy, the lung region is extracted with UNet architecture [141] and then it is fed as input to the CNN models. In the second strategy, full X-rays are used as input to CNN models. Among all the pre-trained models, DenseNet-201 achieved the highest accuracy for TB detection on the segmented lungs.
Hooda et al. [59] proposed the CNN with 7 convolutional layers, 7 activation layers, 2 dropout layers, and 3 fully connected layers for the TB detection. They used the MC and Shenzhen datasets to validate the proposed model. The proposed CNN does not use transfer learning. Similar to Hooda et al. [59], Pasa et al. [128] also proposed the model that does not use transfer learning. The proposed model is faster and efficient for TB detection compared to the state-of-the-art deep learning models. With a depth of 5 blocks, the proposed method reported good classification performance. The proposed network has only 230,000 parameters and took approximately one hour for the training. The inference time of the proposed model is five-six milliseconds.
The deep learning models require high-quality images for training, and the low contrast chest X-ray may impact the results. Munadi et al. [114] evaluated the effect of image enhancement for TB detection in chest X-rays. They used the Unsharp Masking (UM) and High-Frequency Emphasis Filtering (HEF) technique for image enhancement. They used enhanced images to fine-tune ResNet-18, ResNet-50, and EfficientNet-B4. EfficientNet-B4 outperformed the other models on UM enhanced images. Training time for the proposed models was 14 minutes approximately. Ayaz et al. [11] proposed the CAD for early TB detection using chest X-rays. They have applied hybrid learning approach that uses the handcraft feature extraction (Gabor filter) and deep feature learning (CNN) technique. They ensembles several classifiers through logistic regression to achieve the best classification accuracy. The proposed methodology was evaluated on Shenzhen and MC datasets with K-fold cross-validation.
Transformer models are mostly used in natural language processing and their application in images are very limited. Duong et al. [39] proposed the hybrid deep learning model consisting of EfficientNet [162] and Transformer model [38] for TB detection. Moreover, three different learning strategies are considered for training the proposed model to prevent overfitting. The proposed model outperformed the other state-of-the-art models with 97.72% accuracy and 100% AUC. The proposed model has 94,814,915 parameters. All the research papers discussed above performed the image classification for TB detection, while Nijiati et al. [119] proposed the model to detect the TB affected regions in the lungs to assist the radiologist. For the same, TB-UNet model is proposed that uses the ResNext [179] as the encoder for UNet architecture [141]. Further, they have compared the performance of radiologists with and without the proposed model assistance for TB diagnosis in patients. The obtained results show that the radiologist performance is improved with the help of the proposed model.
Many research studies have been proposed for detecting respiratory illnesses such as pneumonia, COVID-19, and pneumothorax. Table 6 and Table 7 enlist the few works for these pulmonary conditions on chest X-rays. In the following, we have discussed the deep learning models proposed for pneumothorax, COVID-19, and pneumonia.
Table 6.
Quantitative comparison of pneumonia and pneumothorax detection methods. Dice similarity coefficient (DSC), F1-score, and the area under curve (AUC) are reported for comparison. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Method | Optimizer | AF | LR Scheduling | Images size | Pre-processing step | Dataset | Technique | DSC | F1-Score | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| DL [135] | Adam | Sigmoid | lr = 0.001 that is decreased by factor of 10 when validation loss is not improved | 224 224 | Image normalization | CXR-14 | DenseNet-121, transfer learning | – | 43.5 | – |
| DL [77] | SGD | – | lr = 0.00105 | 512 512 | Random scaling, shift in coordinate space, brightness and contrast adjustment, blurring with Gaussian blur | RSNA | ResNet50, ResNet101, mask-RCNN, data augmentation | – | – | – |
| DL [133] | Gradient Descent | ReLu, Softmax | lr = 0.0003 | 224 224, 227 227 | Image resize and augmentation | Kaggle [112] | AlexNet, ResNet-18, DenseNet-201, SqueezeNet, transfer learning, data augmentation, cross-validation | – | 93.5 | 95.0 |
| DL [43] | Adam | – | lr = 0.00001 with learning rate decrease factor of 0.2 | 512 512 | Image resizing and data augmentation techniques such as scaling, shear and rotation | CXR-14 | single-shot detector RetinaNet with Se-ResNext101, cross-validation | – | – | – |
| DL [37] | – | ReLu, Softmax | lr = 0.00001 | 227 227 | Image resizing | CXR-14 | VGG-19, CWT, DWT, GLCM, transfer learning, SVM-linear, SVM-RBF, KNN classifier, RF, DT | – | 92.15 | – |
| DL [158] | Adam | ReLu, Sigmoid | lr = 0.001 with = 0.9 and = 0.999 | 224 224 | Image normalization, resizing, cropping and data augmentation | CheXpert | DenseNet-122, transfer learning | – | – | 70.8 |
| DL [176] | Adam | ReLu, Softmax | lr = 0.0005 with = 0.9, = 0.999 | 768 768, 1024 1024 | Image normalization and data augmentation with random Gamma correction, random brightness and contrast change, CLAHE, motion blur, median blur, horizontal flip, random shift, random scale, and random rotation | SIIM-ACR, MC | UNet, SE-Resnext-101, EfficientNet-B3, transfer learning | 88.0 | – | – |
| DL [1] | Adam | ReLu, Sigmoid | lr = 0.001 which is relatively dropped per epoch using the cosine annealing learning rate technique | 256 256, 512 512 | Image resizing, normalization and data augmentation using horizontal flip, one of random contrast, random gamma, and random brightness, one of elastic transform, grid distortion, and optical distortion | SIIM-ACR | UNet, ResNet-34, transfer learning, stochastic weight averaging | 83.56 | – | – |
Table 7.
Quantitative comparison of COVID-19 detection methods. Accuracy (Acc.), F1-score, and the area under curve (AUC) are reported for comparison. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Method | Optimizer | AF | LR Scheduling | Images size | Pre-processing step | Dataset | Technique | Acc. | F1-Score | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| DL [125] | Adam | ReLu | lr = 0.0001 | 224 224 | Image resizing and data augmentation with rotation | COVID-19 | VGG-16, transfer learning | 97.97 | – | – |
| DL [107] | Adam | ReLu, Softmax | lr = 0.0001 and it reduces when there is no improvement for continuous three epochs | 224 224 | Image resizing, scaling, and data augmentation | COVID-19 | EfficientNet-B4, transfer learning, cross-validation | 96.70 | 97.11 | 96.66 |
| DL [76] | Adam | LeakyReLU, Softmax | lr = 0.001 | 128 128 | Image resizing, and data augmentation such as rotation and zoom | COVID-19, Kaggle [112] | InceptionNet-V3, XceptionNet, ResNext, transfer learning | 97.0 | 95.0 | – |
| DL [58] | Adam | ReLu | lr = 0.00001 with rate decay of 0.1 | 224 224 | Image resizing and data augmentation | COVID-19, Kermany [83] | SE-ResNext-50, transfer learning | 97.55 | – | – |
| DL [190] | Adam | ReLu | lr = 0.0001 | 224 224 | Image resizing, normalization, and data augmentation technique such as random rotation, width shift, height shift, horizontal flip | COVID-19, Kaggle[112] | VGG16, ResNet50, EfficientNetB0, synthetic image generation, cross-validation | 96.8 | – | – |
| DL [127] | Adam | – | lr = 0.0001 | 512 512 | Image normalization and resizing | CheXpert, Private | DenseNet-121, PCAM, Vision Transformer | 86.4 | – | 94.1 |
| DL [171] | Adam | ReLu, Softmax | lr = 0.000001 for 20 epochs and then 0.0000001 | 224 224 | Image resizing, normalization and data augmentation | COVID-19, RSNA, CheXpert, MC | ResNeXt-50, Inception-v3, DenseNet-161, transfer learning | 98.1 | – | – |
Pneumonia
Pneumonia can be life-threatening if it is not diagnosed at the time. It nearly killed 800,000 children under the age of five-year worldwide in 2018. The main reason for pneumonia is viral/bacterial infections or fungi that result in inflammation of the lungs. It is normally found in small children or old age people (above 65). Chest X-rays can help the radiologist to identify white spots (infection) or fluid surrounding the lungs. In the following, we have discussed research studies that have used deep learning models for pneumonia detection.
Rajpurkar et al. [135] deployed DenseNet-121 for pneumonia detection which exceed the radiologist interpretation. They compared the test set with the annotation of four radiologists. The proposed method performed (0.435) better than the radiologist (0.387) on the F1 metric. Jaiswal et al. [77] experimented with a deep neural network based on mask-region CNN (RCNN) that does pixel classification for pneumonia detection. It incorporates global and local features for pixel-wise segmentation. ResNet-101 and Resnet-50 are used as backbone detectors in masked-RCNN. They also experimented with YOLO3 [137] and UNet [141], but both models failed to give better predictions results on the test set. Rahman et al. [133] used the different pre-trained deep learning models for the classification of normal, bacterial, and viral pneumonia. They trained the AlexNet, ResNet18, DenseNet201, and SqueezeNet [67] on the augmented dataset to deal with overfitting. The DenseNet-201 achieved the highest accuracy for pneumonia detection.
Gabruseva et al. [43] proposed the model for automatic pneumonia detection using the deep learning. The proposed model uses a single-shot detector RetinaNet with SE-ResNext101 [60] encoder that had been pre-trained on the ImageNet dataset. On the ImageNet dataset, the SE-ResNext architectures performed best, with a good trade-off between accuracy and complexity [14]. The images are resized to 512 512 as the proposed model on 256 256 resolution yielded the poor performance and 1024 1024 increased the computational complexity. Dey et al. [37] proposed the deep learning system for early pneumonia detection using chest X-rays. They used the ensemble feature scheme (EFS) for the feature extraction. They combined the features extracted with the assistance of Complex Wavelet Transform (CWT), Discrete Wavelet Transform (DWT), and Gray-Level Co-occurrence Matrix (GLCM) and deep learning models. They used the pre-trained VGG-19 for the deep feature extraction. Further, they tested the different classifiers such as SVM-linear, SVM-RBF, kNN classifier, Random-Forest (RF), and Decision-Tree (DT) to obtain better accuracy. At last, VGG-19 with a random-forest classifier achieved the highest pneumonia detection accuracy. They also compared their proposed model with ResNet50, VGG-16, and AlexNet.
COVID-19
COVID-19 is a kind of pneumonia caused by coronavirus. This disease has killed more than a million people, with 58 million confirmed cases worldwide. RT-PCR and antigen tests are recommended for the diagnosis but are expensive. The increasing load of patients can crumble down the medical infrastructure. Chest X-rays can help in the early detection and isolation of COVID-19 patients. Deep learning can assist the medical community with automatic detection. Many research studies are proposed for COVID19 detection in chest X-rays, and in the following, we have discussed a few studies.
Panwar et al. [125] proposed the deep learning-based nCOVnet for detecting the COVID-19. The model contains 23 layers in which 18 are of pre-trained VGG-16 model and the rest are fine-tuned on COVID-19 and other datasets. The total number of trainable parameters in the proposed models is 14,846,520. Another work reported on COVID-19 is Marques et al. [107] that have used the pre-trained EfficientNetB4 for detection. The proposed model is validated through 10-fold stratified cross-validation. The total number of trainable parameters in the proposed models is 17,913,755. Jain et al. [76] evaluated the three different models namely, InceptionNet-V3 [160], XceptionNet [29] and ResNext [179] model for COVID-19 diagnosis. The images are downsampled to 128 128 for faster training. Further, data augmentation is used to prevent the models from overfitting. Among all three models, XceptionNet outperformed the other two models for accurately detecting the COVID-19.
Hira et al. [58] evaluated the nine pre-trained models for the COVID-19 detection. Among all the models, SE-ResNeXt-50 achieved the highest accuracy for binary and multi-class classification. The total number of parameters in the SE-ResNext-50 is 27.56 million. Vantaggiato et al. [171] proposed the ensemble-CNN for the three-class and five-class COVID-19 classification. They ensemble pre-trained ResNeXt-50 [179], Inception-v3 [160], and DenseNet-161 [61] networks for the classification. The proposed ensemble network achieves the higher performance for COVID-19 detection in comparison with the other CNN architectures. Again in Zebin et al. [190] three pre-trained CNN models (VGG16 [152], ResNet50 [56], EfficientNetB0 [162]) are used for COVID-19 detection. In this paper, synthetic images are generated with CycleGAN to augment the minority COVID-19 class. Among all these three CNN models, EfficientNetB0 achieves the best accuracy for COVID-19 detection. The number of parameters in EfficientNetB0 architecture is 5.3M, while in ResNet50 is 26M.
The transformer is a deep neural network based on a self-attention mechanism that produces very wide receptive fields. It was first introduced in the field of natural language processing (NLP) [172]. Because it allows modeling long-range dependency inside images, it has spurred the vision community to examine its applications in computer vision after attaining spectacular results in NLP. Park et al. [127] proposed the Vision Transformer model for diagnosing the COVID-19 in chest X-rays. In the proposed model, the transformer uses the low-level chest X-ray feature corpus obtained from the backbone network trained to extract aberrant chest X-ray features. The DenseNet-121 is used as backbone network along with probabilistic class activation map poling is used to extract low-level features from an image. The proposed model outperformed the state-of-the-art models for the COVID-19 detection.
Pneumothorax
Pneumothorax can be life-threatening if not diagnosed at a time. The airspace between the lung and the chest wall causes pneumothorax. It can be diagnosed by X-rays, but the interpretation requires an expert radiologist. This can be time-consuming, so Sze-To et al. [158] proposed a tChexNet for the automatic detection. The proposed model is 122 layers deep and uses a training strategy to transfer knowledge learned in CheXNet [135] to detect the pneumothorax. The AUC obtained by the tCheXnet on the test set is 10% better than CheXNet.
In the literature, most papers perform the image classification for pneumothorax detection. But, Wang et al. [176] employed the two-stage model for the pneumothorax segmentation using deep learning models. In the first stage, image classification is done using the pre-trained encoder backbone of UNet [141]. If the image is predicted as pneumothorax positive, then in the second stage pneumothorax segmentation is performed to identify the affected regions. They have also performed different ablation studies to show the design benefits of a two-stage network. In this study, the proposed model obtained the 0.88 DC value.
Similarly, Abedalla et al. [1] also performed the pneumothorax segmentation using the two-stage using the modified UNet architecture. Wang et al. [176] use the SE-ResNext50, SE-Resnext101 [60], EfficientNet-B3, and EfficientNet-B5 [162], while Abedalla et al. [1] employed the ResNet-34 [56] as CNN backbone for UNet architecture. It trained the five different modified UNet architecture with two different image size and three binarization thresholds. Obtained results show that two-stage training improves the results and DC value of 0.8356 is obtained.
Multi-class classification
Chest X-rays are commonly used low-cost clinical examinations for pulmonary disease detection. Interpretation of X-rays needs a radiologist with years of experience. Therefore, CAD is needed to automatically detect different pulmonary diseases. CAD can improve efficiency and reduce the error in interpretation. Deep learning methods need a large dataset for training, otherwise, there is the possibility of model overfitting. With the availability of the CXR-14 [175] dataset, many models have been proposed for multi-classification using chest X-rays. Table 8 enlists the multi-classification studies for comparison. In the following, we have discussed some multi-classification studies performed on chest X-rays using the deep learning models.
Table 8.
Quantitative comparison of multi-class disease classification. Area under curve (AUC) is reported for comparison. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Method | LR Scheduling | Optimizer | AF | Image size | Pre-processing step | Dataset | Technique | AUC |
|---|---|---|---|---|---|---|---|---|
| DL [99] | lr = 0.0001 and a decay of 0.00001 over each update | Adam | Sigmoid | 224 224 | Images are resized and horizontally flipped | CXR-14 | DenseNet-121, CAM, UNet architecture | 81.5 |
| DL [175] | – | SGD | – | 1024 1024 | Image are resized and intensity ranges are rescaled | CXR-14 | ResNet-50, AlexNet, GoogLeNet, VGG-16, transfer learning | 69.62 |
| DL [16] | lr = 0.1 | – | Sigmoid | 512 512, 1024 1024 | Image are downsampled using bilinear interpolation | CXR-14 | ResNet, AM, CAM, knowledge preservation | – |
| DL [52] | lr = 0.001 and is reduced by factor of 10 when validation loss not improved | Adam | ReLu, Sigmoid | 256 256, 512 512 | Images are resized using the bilinear interpolation | CXR-14, PLCO | DenseNet, spatial knowledge, lung segmentation | 88.3 |
| DL [51] | lr = 0.01 and is divided by 10 after 20 epochs | SGD | Sigmoid | 224 224, 256 256 | Images are resized, cropped, and horizontally flipped. ImageNet mean is also subtracted from the image | CXR-14 | ResNet-50, DenseNet-121, AG-CNN. AG-Mask Interface | 87.1 |
| DL [136] | lr = 0.0001 | Adam | Softmax | 224 224 | Images are resized using the bilinear interpolation and are also horizontally flipped | CXR-14 | ResNet-18, Transfer learning, CAM, Data augmentation | 84.94 |
| DL [126] | lr = 0.00005 | SGD | Sigmoid | 1024 1024 | Images are resized using the bilinear interpolation and additional techniques such as rotation, zoom and shifting are performed for data augmentation | AMC, SNUBH | ResNet-50, fine-tuning, CAM, curriculum learning | 98.3 |
| DL [178] | lr = 0.0001 | Adam | ReLu, Sigmoid | 128 128 | Images are normalized to enhance the contrast. Further, rotation, scaling and flipping is performed for reducing the overfitting | CXR-14 | Inception-ResNet-v2, dilated ResNet, transfer Learning, cross-validation | 90.0 |
| DL [44] | lr = 0.0001 and it is decreased by 10 times after every 3 epochs | Adam | Softmax, Sigmoid | 224 224 | Images normalization is performed | CXR-14 | ResNet-18, DenseNet-121, MSML | 84.32 |
| DL [140] | – | – | – | 236 236 | Images are resized, cropped, flipped and rotated | MURA, CheXpert, CXR-14 | ResNet-50, DenseNet-121, one cycle training, transfer learning | 80.0 |
| DL [53] | lr = 0.001 and is reduced by factor of 10 when validation loss not improved | Adam | Sigmoid | 1024 1024 | Images in PLCO dataset are resized | CXR-14, PLCO | DenseNet-121, transfer learning | 84.1 |
| DL [185] | lr = 0.0001 and is reduced by factor of 10 when validation loss not improved | Adam | ReLu, Sigmoid | 512 512 | Image are resized, cropped, and horizontally flipped for augmentation | CXR-14 | DenseNet-121, SE, multi-map layer, max–min pooling | 83.0 |
| DL [154] | – | Adam | ReLu, Sigmoid | 224 224 | Images are resized and data augmentation is performed | CXR-14 | MobileNet-V2, transfer learning | 80.99 |
| DL [13] | lr = 0.01, 0.001 and is reduced by a factor of 2 when validation loss not improve | Adam | Sigmoid | 256 256, 480 480 | Image resizing and data augmentation techniques such as rotation and flipping | CXR-14 | ResNet-38, ResNet-50, ResNet-101, MLP, transfer learning | 82.2 |
Wang et al. [175] released the largest chest X-ray dataset in 2017 and as well performed the multi-classification. They used pre-trained DCNN for identifying the diseases with an average AUC of 69.62%. In the proposed method fully connected layers are replaced with transition, global pooling, prediction, and a loss layer. Yan et al. [185] proposed a weakly supervised deep learning method for the classification. They have used the DenseNet-121 with three modifications. First, using the squeeze and excitation (SE) block in between the convolution-pooling operator for feature recalibration. Second, using the multi-map layer by replacing the fully connected layers. At last, using the max–min pooling operator to multi-maps spatial activations for the final prediction. The results obtained show that these modifications have made an impact on the results. The proposed model outperformed the [175] on AUC mean by 10%.
The random data split can influence the performance of the models as the same patient may appear in both training and test set. So, Guendel et al. [53] have demonstrated the effect on the result by using the patient-wise split and random split for disease classification on the CXR-14 dataset. Further, they combined the CXR-14 [175] and PLCO [166] to have the dataset of 297,541 medical images with 86,876 patients. They created the location-aware Dense Network (DNetLoc) by using the location information available for pathologies. DNetLoc outperformed the Wang et al. [175] on the CXR-14 test set on mean AUC by 5%.
In all the papers discussed above, none have performed the lung segmentation before the disease classification. They have performed the image resizing, normalization and data augmentation to prevent the model from overfitting, but Guendel et al. [52] have demonstrated that additional spatial knowledge (localization), normalization, and segmentation can increase the classification performance. They do the patient-wise splitting to train and test the proposed model. When the additional features are used, the AUC increased by 2.4%. Further, they also studied the relation between the accuracy and the number of patients in training data.
Another research study that has done lung segmentation before classification is Liu et al. [99]. It has proposed a segmentation-based classification model. Using the JSRT dataset, the lung segmentation model (UNet) is trained for extracting the lung regions. After lung segmentation, two fine-tuned DenseNet-121 are used for the feature extraction. First DenseNet is fine-tuned on whole X-rays, while the second is on the segmented lungs. At last, both are concatenated for the final prediction. Park et al. [126] proposed the method consisting of two steps. First, regional patterns of abnormality are identified by learning with patch images, and second by fine-tuning the model to detect the disease pattern on the entire X-ray image. Unlike other studies, they evaluated the model on Asan Medical Center (AMC) and Seoul National University Bundang Hospital (SNUBH) datasets.
In chest X-rays, the region of interest (ROI) is very less, while the entire X-ray may contain noisy areas. Further, irregular lung boundaries can affect performance. So, Guan et al. [51] proposed the model integrating the global and local cues to classify the thorax disorders using the attention-guided convolutional neural network (AG-CNN). The proposed model is divided into three parts: global, local, and fusion branches. The global branch is fine-tuned using the entire image; then, the local branch uses the lesion area for classification. The fusion branch combines the output of the global and local branches. Both local and global branch uses the variant of ResNet-50 architecture. They have also evaluated their method with DenseNet-121 and compared it with the other state-of-the-art models. They have also compared the localization accuracy for the different values of T(IoU), but the achieved results are not good as obtained by Li et al.[96]. The training time taken by the local and global branches is approximately 6 hours. Cai et al. [16] proposed an attention mining (AM) model related to adversarial erasing scheme [177]. The difference is that AM drops the corresponding pixels in the activation map to maintain the original radiographs. AM helps the model to localize abnormalities, but the model may overfit because of learning from the specific region, not from the actual pattern. This is prevented using the knowledge preservation method where a part of the dataset is used for the AM and while the remaining for knowledge preservation.
The major issue with deep learning is that the model needs a large dataset for training. Few large medical datasets are available, but all are not publicly available. Using the small datasets can lead to the overfitting [188]. Transfer learning can be used to prevent overfitting. It uses the learned knowledge from one task and then applies it to other but related tasks. All the pre-trained models are generally trained on the ImageNet dataset. Romero et al. [140] have reviewed, evaluated, and compared the state-of-the-art training techniques on the small datasets. Deep CNN model performance has been checked on small datasets for emphysema detection, pneumonia detection, hernia detection, and CXR-14 classification with or without transfer learning. They have used two training strategies to train the models: regular training, and one-cycle training.
Ge et al. [44] proposed an error function based on the Softmax concept named multi-label softmax loss (MSML) and correlation loss to deal with the multi-disease and imbalanced data. Further, they have used the bilinear pooling scheme to increase model discrimination. MSML captures the characteristics of multi-label learning, while bilinear pooling allows the end-to-end training with image labels. They have evaluated the proposed function on various pre-trained models such as ResNet, DenseNet, and ensemble variants. Rakshit et al. [136] used the pre-trained ResNet-18 model for label classification. The pre-trained model first extracts the feature from the X-rays. These features are used by the coming dense layers for classification. Dense layers take the feature vector as input from the pre-trained layers and produce the vector of size 14 (equal to the number of labels in the dataset). The proposed model has 11,183,694 trainable parameters.
Wong et al.[178] proposed a deep neural network for finding the normal/healthy radiographs. They had used the pre-trained Inception-ResNet-v2 model for finding the features which are further used for training the model on X-rays labeled by the radiologist. A total of 3000 AP images are used of which 1300 are normal/healthy and the rest have one or more abnormalities labeled. The result obtained by the model is quite promising. It achieved the ROC-AUC value of 0.92 and the precision–recall curve AUC value of 0.91. The main issue that remains with deep learning is domain shift. Domain shift is an artificial intelligence problem that means the difference between training and testing data distribution. Chest radiography data also have the same problem because different machines with various parameters generate the data, and it results in high heterogeneity distribution. In Pooch et al.[130] they have shown the same by experimenting on three publicly available chest X-ray datasets. Experiments show that cross-data generalization (e.g., train with MIMIC-CXR and test with CheXpert) affects the model performance.
Souid et al. [154] proposed the automated method for abnormalities identification from chest x-rays. They used the modified pre-trained MobileNet-V2 for feature extraction and classification. The proposed model obtained good results on AUC metrics in comparison with other methods. However, reported the lower sensitivity due to dataset bias (the difference in the class distribution). The proposed model has 2.11 M parameters. Baltruschat et al. [13] used the pre-trained ResNet with and without fine-tuning for chest x-ray classification. They have also evaluated the model with training from scratch strategy. Further, they trained a multi-layer perceptron (MLP) classifier with non-images features to improve the classification results. For this, they concatenated the image feature vector with the new non-image feature vector. When integrated with the non-image features, the proposed ResNet-38 achieved the best results.
Discussion
Training on large datasets brings the computation problem. A large dataset requires high computation power for the training. Transfer learning plays important role in deep learning projects where it prevents computational cost and overfitting. Pre-trained and fine-tuned networks are two different transfer learning used in deep learning models. Kim et al.[84] and Antony et al.[7] has given conflicting results about the performance of both transfer learning approaches. Recently, Oakden-Rayner[122] investigated that the CXR-14 dataset does not accurately reflect the correct labeling. It has 10-30% less positive predictive values than presented in the published document [175]. So, data-generating procedures and labeling rules should be included in the documentation of the dataset. In this section, we have discussed the various deep learning models for nodule, TB, pneumonia, COVID-19, pneumothorax detection. Further, multi-class classification studies have also been discussed. The nodule detection with deep learning needs exploration as very few studies has been reported. In COVID-19 detection, most of the research studies has used the same dataset source. More diversified COVID-19 dataset is needed to truly explore the COVID-19 using DL. We have also discussed the models that do not employ deep learning, so the readers can have a deeper insight into the field.
Generative Adversarial network
Goodfellow et al.[48] proposed a generative model via an adversarial setting called Generative Adversarial Network (GAN). It is based on two neural network models that compete to analyze, capture and copy the variations within a dataset. It has received huge attention from the research community, and there are various variants proposed for the generation of natural images [132]. Few GAN applications are the image to image translation [72, 118] and image inpainting [186].
Many works have been proposed on medical image analysis using the GAN, such as lung and heart segmentation [34], brain tumor segmentation [184], and for the image to image translation [118]. Nie et al. [118] trained a GAN network for the translation between brain CT and MRI images. One of the major problems in deep learning is to train the network with a large dataset. A study done by Madani et al. [105] shows that semi-supervised GAN can be trained with a less amount of data, and gives better results than conventional supervised CNN networks. One of the advantages of the GAN is its resistance to overfitting, but the problem is its instability during the training process because of the Nash equilibrium between generator and discriminator. DCGAN [132] and WGAN [8] architecture have reported the stable training phase. Figure 7 shows the GAN architecture and Table 9 enlists the mentioned segmentation and classification research studies.
Fig. 7.
GAN Architecture
Table 9.
Quantitative comparison of segmentation and classification methods using GAN models. Accuracy, area under curve (AUC), and dice similarity coefficient (DSC) are reported for comparison. SGD is the abbreviation for stochastic gradient descent, ReLu for rectified linear unit, lr for learning rate, and AF for activation function
| Method | Optimizer | AF | LR Scheduling | Images size | Dataset | Pre-processing step | Technique | Accuracy | AUC | DSC |
|---|---|---|---|---|---|---|---|---|---|---|
| DL [106] | Adam | ReLu, Leaky ReLU | lr = 0.001 | 1204 1024 | CXR-14 | Data augmentation | CycleGAN, VGG-16, ResNet, JRS | – | – | 88.9 |
| DL [21] | Adam | – | lr = 0.002 which is reduced after every 100 epochs | 512 512 | MC, JSRT | Image resizing and rescaling | Semantic aware GAN, ResNet-101, dilated convolutions | – | – | 94.5 |
| DL [34] | – | ReLu | lr = 0.01 | 400 400 | MC, JSRT | Image resizing | Residual FCN, Critic FCN | – | – | 97.3 |
| DL [117] | Adam, Nesterov | – | lr = 0.0004 with = 0.5, = 0.999, 0.00001 with a momentum of 0.99 | 128 128 | JSRT | Image resizing | DCGAN, UNet architecture | – | – | 94.6 |
| DL [115] | Adam | ReLu, Leaky ReLu | lr = 0.0002 and decay rate of 0.5 | 256 256, 512 512 | MC, Shenzhen, JSRT | Image resizing | Skip connections, conditional GAN, pixel GAN, patch GAN | – | – | 97.4 |
| DL [42] | SGD | ReLu | – | 512 512 | MC, Shenzhen, JSRT | Image resizing, histogram equalization | Attention UNet, Critic FCN, Focal Tversky Loss | – | – | 97.5 |
| DL [68] | Adam | ReLu | lr = 0.00001 | 128 128 | MC, Shenzhen, JSRT | Image resizing and normalization | Adversarial Pyramid Progressive Attention UNet, KL divergence with Tversky loss | 75.8 | – | 97.6 |
| DL [105] | – | Leaky ReLu | – | 128 128 | PLCO, Indiana | Image resizing | GAN, CNN | 93.7 | – | – |
| DL [163] | Adam | Softmax | lr = 0.0002 for first 100 epochs | 512 512 | RSNA, CXR dataset [83] | Image resizing | ResNet-18, ResNet-50, CycleGAN, semantic modeling | 93.1 | 96.3 | – |
| DL [165] | Adam | Leaky ReLu | lr = 0.0005 | 64 64 | CXR-14 | Image resizing and no augmentation is performed | GAN, U-Net autoencoder, CNN discriminator, One class learning | – | 84.1 | – |
| DL [174] | Adam | ReLu, Leaky Relu | lr = 0.0002 with = 0.5 | 112 112 | COVID-19 | Image resizing and normalization | VGG-16, Auxiliary Classifier Generative Adversarial Network, PCA | 95.0 | – | – |
Segmentation
There is a scarcity of datasets for segmentation in medical image analysis. The ground-truth preparation needs expertise and is a laborious task. So, Neff et al. [117] modified the DCGAN architecture that generates new synthetic images and the segmentation mask from random noise. The discriminator has to decide whether the image and segmentation pair are real or synthetic. This helps both the generator and the discriminator to learn about the ground-truth structure. The use of real images makes the discriminator improve the image quality [144], and the authors claim that it is the first of its work that has generated synthetic images as well ground-truth masks. The proposed method is evaluated by threefold cross-validation, and each fold took approximately 24 hours for training. At last, separate modified UNet architecture is used for the segmentation results.
Both registration and segmentation methods are supportive of each other, and using both methods side by side can improve the results. Mahapatra et al. [106] proposed a deep learning GAN network that performs registration and segmentation on chest X-rays. The proposed method does not need to train a separate network for segmentation. GAN is used to register the floating and referenced image by combining the segmentation and conventional feature maps. The proposed model took 36 hours for training on the augmented dataset. Dai et al. [34] have proposed the lung and heart semantic segmentation using the GAN discriminative model. The model consists of two networks: segmentation and critic network. The segmentation network is the fully convolutional network (FCN), and it works as the generator. The critic network is the mirror of the segmentation network that does the discrimination between the ground truth and segmentation network masks. It learns the high order structure for needful discrimination and produces accurate segmentation without using any pre-trained models. The model is validated through cross-data generalization. The prediction time for the proposed model is 0.84 second for each X-ray. The segmentation architecture has 271,000 parameters and the critic architecture has 258,000 parameters.
Chen et al. [21] proposed a segmentation with unsupervised domain adaption for medical images. The proposed unsupervised domain shift approach uses the semantic aware generative adversarial network. The proposed method separates the segmentation neural network from adapting the domain and does not need any label from the test dataset. It does the image-to-image translation to generate the source dataset images. Model is tested on two public datasets: the MC dataset and the JSRT dataset are used as source and target, respectively. Modified ResNet-101 is used for the segmentation where convolution layers are replaced by dilated convolutions in high-level residual blocks. CycleGAN is improved to preserve the information with a novel semantic-aware loss.
Munawar et al. [115] trained and evaluated the GAN-based lung segmentation method using four different discriminators. The generator used in the proposed model is similar to UNet architecture. They have trained the discriminators with different patch sizes to determine the best match. Discriminator (D3) outperformed all others with 70 70 patch-GAN. Gaal et al. [42] used the attention-based UNet architecture to generate the segmentation mask with a resolution similar to input images. The use of attention gates results in the reduction of false positives, and better segmentation accuracy. Further, to improve evaluation they used adversarial techniques. They created the adversarial network similar to [34], where the FCN is replaced by the proposed architecture. Moreover, they have not introduced any modifications to the critic network. The proposed model outperformed the other state-of-the-art models on different datasets.
Classification
Labeling or annotating the image dataset is the first and most important task for classification. Manual annotation is most accurate, but it is time-consuming and expensive. The automatic annotation takes less time but is error-prone. In this regard, Tang et al. [163] proposed a model to transfer the information gained from labeled data to unlabeled data. It will avoid the labeling of the new dataset. The proposed model can maintain the micro-level details along with high-level semantic information. During the image to image translation process, it also maintains the mid-level feature representation to accurately recognize the target disease, and enhance the function of generalizing the unlabeled target domain. The proposed TUNA-Net model is tested on two datasets which consist of a subset of adult X-rays from CXR-14 (source domain) and pediatric X-rays obtained from Guangzhou Women and Children Medical Center (target domain). The TUNA-Net performance has been compared with other states of the art models including the supervised models. The proposed TUNA-Net in cross-domain X-rays for the pneumonia classification outperformed the other models.
GAN models are gaining popularity as these can learn from real data distributions. Imran et al. [68] proposed a model for combined segmentation and classification from a limited labeled dataset. The proposed method consists of a segmentor and discriminator. Segmentor is made of a pyramid encoder and modified UNet decoder. The encoder helps the model to learn the locally aware features. The decoder generates the side output at different levels and combines it for the final segmentation. They applied the proposed model in a semi-supervised setting for the classification. The discriminator takes the image labels for classification. Madani et al. [105] proposed the model to show that semi-supervised GAN requires less data than conventional deep CNN. The proposed semi-supervised GAN takes advantage of both labeled and unlabeled data as it is capable of learning from both types of data. They have used the PLCO [123] dataset and the Indiana dataset [35] for cardiac classification.
Tang et al. [165] proposed the generative adversarial one-class classifier that takes input only the healthy radiographs, unlike the other approaches. The three deep neural networks (UNet autoencoder, CNN discriminator, and encoder network) competes with each other to produce the structure of healthy X-rays. The approach is that since the network has seen only the normal radiographs it will not reconstruct the abnormal x-ray correctly. Thus, it will help in the identification of abnormal X-rays. Waheed et al. [174] proposed CovidGAN to improve the COVID-19 detection. They developed the Auxiliary Classifier Generative Adversarial Network (ACGAN) for synthetic image generation to overcome the scarcity of small datasets. They used the fine-tuned VGG-16 for the COVID-19 detection. The COVID-19 detection accuracy increased when the synthetic images were used in comparison to the real images. The proposed CovidGAN has 24 million parameters, and it took nearly 5 hours for the training.
Discussion
Collecting and annotating a large image dataset is a tedious task. It also increases the computation when used on deep learning models. Not much work had been done in the label transferring from labeled data to unlabeled data. GAN models are effectively used for some time to create synthetic images. The same has also been used for creating chest X-rays. One problem associated with the GAN is its instability during training, while they also avoid overfitting. Many published studies have used the synthetic chest dataset and using them for segmentation and classification purpose.
Conclusion
In traditional CV algorithms, feature extraction is done via handcraft methods, while the deep learning methods extract features automatically which is extremely effective compared to the models that manually extract features. Recent advancements in large datasets and computational power have made possible the training of deep learning models. As a result, the focus of most research is turning to deep learning. A standard dataset is a salient requirement for the evaluation of various models designed for the segmentation and disease classification in chest X-rays. Therefore, we have given a description of the various publicly available datasets which has been used by various research studies.
In this paper, we have conducted a study for lung segmentation and disease detection using chest X-ray images in three phases: segmentation, classification, and GAN models. In the first phase, a detailed review of the lung segmentation studies is performed. The majority of papers have trained, validated, and tested their models on the same dataset. It is generally seen that the model trained on one dataset fails on a different test dataset. Therefore, the model should be evaluated using the cross-dataset for generalization. In the second phase, the nodule, TB, pneumonia, COVID-19, pneumothorax, and multi-class classification research studies have been reviewed. Most of the classification studies have done image-level classification except nodule detection. There are very few studies that have identified the disease-affected regions in the lungs. In the third phase, lung segmentation, and disease classification studies involving the GAN models are reviewed. These models are attracting a lot of attention. The requirement of large labeled data is the biggest challenge for the deep learning community. GAN models can generate synthetic data to overcome scarcity. Some studies have also shown that GAN requires less data for learning than conventional CNN. There is a lot of scope for improvement in segmentation and classification using GAN. Our aim for this study is to provide an integrated, synthesized overview of the current state of knowledge of lung segmentation and disease classification in chest X-rays.
Biographies
Tarun Agrawal
is currently pursing the Ph.D. degree at Department of Computer Science & Engineering with National Institute of Technology Hamirpur, HP, India. He did his B.Tech and M.Tech from the Zakir Hussain College of Engineering & Technology, Aligarh Muslim University, in 2014 and 2016, respectively. He served as Assistant Professor for 3 years before enrolling to Ph.D. His research interests are Computer Vision, Medical Imaging, Machine and Deep Learning.
Prakash Choudhary
is an Assistant Professor at the Department of Computer Science & Engineering, at National Institute of Technology Hamirpur, India, since December 21, 2018. He was an Assistant Professor from December 2013 to December 2018 at the Department of Computer Science & Engineering at National Institute of Technology, Manipur. He received Ph.D. degree from Malaviya National Institute of Technology, Jaipur, in 2016, the M.Tech degree from Visvesvaraya National Institute of Technology, Nagpur, in 2012, and B.E. in Information Technology from Govt. Engineering College Ajmer (Rajasthan University) in 2009. His areas of Research include Computer Vision and Image Processing: Content-Based Image Retrieval, Automatic Image Annotation, Medical Imaging, Biometrics, Multilingual Text Recognition in Indian Scripts, Machine Learning and Pattern Recognition.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tarun Agrawal, Email: tarunagrawal@nith.ac.in.
Prakash Choudhary, Email: pc@nith.ac.in.
References
- 1.Abedalla, A., Abdullah, M., Al-Ayyoub, M., Benkhelifa, E.: 2st-unet: 2-stage training model using u-net for pneumothorax segmentation in chest x-rays. In: 2020 International Joint Conference on Neural Networks (IJCNN), pp. 1–6. IEEE (2020)
- 2.Agrawal A, Mittal N. Using cnn for facial expression recognition: a study of the effects of kernel size and number of filters on accuracy. Vis. Comput. 2020;36(2):405–412. doi: 10.1007/s00371-019-01630-9. [DOI] [Google Scholar]
- 3.Ahmed MN, Yamany SM, Mohamed N, Farag AA, Moriarty T. A modified fuzzy c-means algorithm for bias field estimation and segmentation of mri data. IEEE Trans. Med. Imaging. 2002;21(3):193–199. doi: 10.1109/42.996338. [DOI] [PubMed] [Google Scholar]
- 4.Al Aseri Z. Accuracy of chest radiograph interpretation by emergency physicians. Emerg. Radiol. 2009;16:111–114. doi: 10.1007/s10140-008-0763-9. [DOI] [PubMed] [Google Scholar]
- 5.Andermatt, S., Pezold, S., Cattin, P.: Multi-dimensional gated recurrent units for the segmentation of biomedical 3d-data. In: Deep Learning and Data Labeling for Medical Applications, pp. 142–151. Springer (2016)
- 6.Annangi, P., Thiruvenkadam, S., Raja, A., Xu, H., Sun, X., Mao, L.: A region based active contour method for x-ray lung segmentation using prior shape and low level features. In: 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 892–895. IEEE (2010)
- 7.Antony, J., McGuinness, K., O’Connor, N.E., Moran, K.: Quantifying radiographic knee osteoarthritis severity using deep convolutional neural networks. In: 2016 23rd International Conference on Pattern Recognition (ICPR), pp. 1195–1200. IEEE (2016)
- 8.Arjovsky, M., Chintala, S., Bottou, L.: Wasserstein generative adversarial networks. In: International Conference on Machine Learning, pp. 214–223. PMLR (2017)
- 9.Armato SG, III, Giger ML, MacMahon H. Automated lung segmentation in digitized posteroanterior chest radiographs. Acad. Radiol. 1998;5(4):245–255. doi: 10.1016/S1076-6332(98)80223-7. [DOI] [PubMed] [Google Scholar]
- 10.Austin J, Romney B, Goldsmith L. Missed bronchogenic carcinoma: radiographic findings in 27 patients with a potentially resectable lesion evident in retrospect. Radiology. 1992;182(1):115–122. doi: 10.1148/radiology.182.1.1727272. [DOI] [PubMed] [Google Scholar]
- 11.Ayaz M, Shaukat F, Raja G. Ensemble learning based automatic detection of tuberculosis in chest x-ray images using hybrid feature descriptors. Phys. Eng. Sci. Med. 2021;44(1):183–194. doi: 10.1007/s13246-020-00966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 13.Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A. Comparison of deep learning approaches for multi-label chest x-ray classification. Sci. Rep. 2019;9(1):1–10. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianco S, Cadene R, Celona L, Napoletano P. Benchmark analysis of representative deep neural network architectures. IEEE Access. 2018;6:64270–64277. doi: 10.1109/ACCESS.2018.2877890. [DOI] [Google Scholar]
- 15.Brown MS, Wilson LS, Doust BD, Gill RW, Sun C. Knowledge-based method for segmentation and analysis of lung boundaries in chest x-ray images. Comput. Med. Imaging Graph. 1998;22(6):463–477. doi: 10.1016/S0895-6111(98)00051-2. [DOI] [PubMed] [Google Scholar]
- 16.Cai, J., Lu, L., Harrison, A.P., Shi, X., Chen, P., Yang, L.: Iterative attention mining for weakly supervised thoracic disease pattern localization in chest x-rays. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 589–598. Springer (2018)
- 17.Candemir, S., Jaeger, S., Palaniappan, K., Antani, S., Thoma, G.: Graph-cut based automatic lung boundary detection in chest radiographs. In: IEEE Healthcare Technology Conference: Translational Engineering in Health & Medicine, pp. 31–34 (2012)
- 18.Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Xue Z, Karargyris A, Antani S, Thoma G, McDonald CJ. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging. 2013;33(2):577–590. doi: 10.1109/TMI.2013.2290491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Candemir, S., Palaniappan, K., Akgul, Y.S.: Multi-class regularization parameter learning for graph cut image segmentation. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, pp. 1473–1476. IEEE (2013)
- 20.Caselles V, Catté F, Coll T, Dibos F. A geometric model for active contours in image processing. Numer. Math. 1993;66(1):1–31. doi: 10.1007/BF01385685. [DOI] [Google Scholar]
- 21.Chen, C., Dou, Q., Chen, H., Heng, P.A.: Semantic-aware generative adversarial nets for unsupervised domain adaptation in chest x-ray segmentation. In: International Workshop on Machine Learning in Medical Imaging, pp. 143–151. Springer (2018)
- 22.Chen, J., Yang, L., Zhang, Y., Alber, M., Chen, D.Z.: Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation. In: Advances in Neural Information Processing Systems, pp. 3036–3044 (2016)
- 23.Chen S, Han Y, Lin J, Zhao X, Kong P. Pulmonary nodule detection on chest radiographs using balanced convolutional neural network and classic candidate detection. Artif. Intell. Med. 2020;107:101881. doi: 10.1016/j.artmed.2020.101881. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Suzuki K. Computerized detection of lung nodules by means of virtual dual-energy radiography. IEEE Trans. Biomed. Eng. 2012;60(2):369–378. doi: 10.1109/TBME.2012.2226583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Suzuki K, MacMahon H. Development and evaluation of a computer-aided diagnostic scheme for lung nodule detection in chest radiographs by means of two-stage nodule enhancement with support vector classification. Med. Phys. 2011;38(4):1844–1858. doi: 10.1118/1.3561504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Zhang D. Robust image segmentation using fcm with spatial constraints based on new kernel-induced distance measure. IEEE Trans. Syst. Man Cybernet. Part B (Cybernet.) 2004;34(4):1907–1916. doi: 10.1109/TSMCB.2004.831165. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Giger ML, Bick U. A fuzzy c-means (fcm)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced mr images1. Acad. Radiol. 2006;13(1):63–72. doi: 10.1016/j.acra.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 28.Cheng, D., Goldberg, M.: An algorithm for segmenting chest radiographs. In: Visual Communications and Image Processing’88: Third in a Series, vol. 1001, pp. 261–268. International Society for Optics and Photonics (1988)
- 29.Chollet, F.: Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1251–1258 (2017)
- 30.Clevert, D.A., Unterthiner, T., Hochreiter, S.: Fast and accurate deep network learning by exponential linear units (elus). arXiv preprint arXiv:1511.07289 (2015)
- 31.Cohen, J.P., Morrison, P., Dao, L., Roth, K., Duong, T.Q., Ghassemi, M.: Covid-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006.11988 (2020)
- 32.Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models-their training and application. Comput. Vis. Image Underst. 1995;61(1):38–59. doi: 10.1006/cviu.1995.1004. [DOI] [Google Scholar]
- 33.Coppini G, Diciotti S, Falchini M, Villari N, Valli G. Neural networks for computer-aided diagnosis: detection of lung nodules in chest radiograms. IEEE Trans. Inf Technol. Biomed. 2003;7(4):344–357. doi: 10.1109/TITB.2003.821313. [DOI] [PubMed] [Google Scholar]
- 34.Dai, W., Dong, N., Wang, Z., Liang, X., Zhang, H., Xing, E.P.: Scan: Structure correcting adversarial network for organ segmentation in chest x-rays. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 263–273. Springer (2018)
- 35.Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, McDonald CJ. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 2016;23(2):304–310. doi: 10.1093/jamia/ocv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: Imagenet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. Ieee (2009)
- 37.Dey N, Zhang YD, Rajinikanth V, Pugalenthi R, Raja NSM. Customized vgg19 architecture for pneumonia detection in chest x-rays. Pattern Recogn. Lett. 2021;143:67–74. doi: 10.1016/j.patrec.2020.12.010. [DOI] [Google Scholar]
- 38.Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
- 39.Duong LT, Le NH, Tran TB, Ngo VM, Nguyen PT. Detection of tuberculosis from chest x-ray images: boosting the performance with vision transformer and transfer learning. Expert Syst. Appl. 2021;184:115519. doi: 10.1016/j.eswa.2021.115519. [DOI] [Google Scholar]
- 40.Duryea J, Boone JM. A fully automated algorithm for the segmentation of lung fields on digital chest radiographic images. Med. Phys. 1995;22(2):183–191. doi: 10.1118/1.597539. [DOI] [PubMed] [Google Scholar]
- 41.Fukushima, K., Miyake, S.: Neocognitron: A self-organizing neural network model for a mechanism of visual pattern recognition. In: Competition and cooperation in neural nets, pp. 267–285. Springer (1982)
- 42.Gaál, G., Maga, B., Lukács, A.: Attention u-net based adversarial architectures for chest x-ray lung segmentation. arXiv preprint arXiv:2003.10304 (2020)
- 43.Gabruseva, T., Poplavskiy, D., Kalinin, A.: Deep learning for automatic pneumonia detection. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 350–351 (2020)
- 44.Ge, Z., Mahapatra, D., Sedai, S., Garnavi, R., Chakravorty, R.: Chest x-rays classification: A multi-label and fine-grained problem. arXiv preprint arXiv:1807.07247 (2018)
- 45.Giger ML, Chan HP, Boone J. Anniversary paper: history and status of cad and quantitative image analysis: the role of medical physics and aapm. Med. Phys. 2008;35(12):5799–5820. doi: 10.1118/1.3013555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gleason, S., Paulus, M., Johnson, D., Sari-Sarraf, H., Abidi, M.: Statistical-based deformable models with simultaneous optimization of object gray-level and shape characteristics. In: 4th IEEE Southwest Symposium on Image Analysis and Interpretation, pp. 93–95. IEEE (2000)
- 47.Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp. 249–256. JMLR Workshop and Conference Proceedings (2010)
- 48.Goodfellow, I.J., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville, A., Bengio, Y.: Generative adversarial nets. In: Proceedings of the 27th International Conference on Neural Information Processing Systems, vol. 2, pp. 2672–2680 (2014)
- 49.Gozes, O., Greenspan, H.: Deep feature learning from a hospital-scale chest x-ray dataset with application to tb detection on a small-scale dataset. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 4076–4079. IEEE (2019) [DOI] [PubMed]
- 50.Gu J, Wang Z, Kuen J, Ma L, Shahroudy A, Shuai B, Liu T, Wang X, Wang G, Cai J, et al. Recent advances in convolutional neural networks. Pattern Recogn. 2018;77:354–377. doi: 10.1016/j.patcog.2017.10.013. [DOI] [Google Scholar]
- 51.Guan Q, Huang Y, Zhong Z, Zheng Z, Zheng L, Yang Y. Thorax disease classification with attention guided convolutional neural network. Pattern Recogn. Lett. 2020;131:38–45. doi: 10.1016/j.patrec.2019.11.040. [DOI] [Google Scholar]
- 52.Guendel, S., Ghesu, F.C., Grbic, S., Gibson, E., Georgescu, B., Maier, A., Comaniciu, D.: Multi-task learning for chest x-ray abnormality classification on noisy labels. arXiv preprint arXiv:1905.06362 (2019)
- 53.Guendel, S., Grbic, S., Georgescu, B., Liu, S., Maier, A., Comaniciu, D.: Learning to recognize abnormalities in chest x-rays with location-aware dense networks. In: Iberoamerican Congress on Pattern Recognition, pp. 757–765. Springer (2018)
- 54.ter Haar Romeny, B.M., Titulaer, B., Kalitzin, S., Scheffer, G., Broekmans, F., Staal, J., te Velde, E.: Computer assisted human follicle analysis for fertility prospects with 3d ultrasound. In: Biennial International Conference on Information Processing in Medical Imaging, pp. 56–69. Springer (1999)
- 55.Hasegawa, A., Lo, S.C.B., Freedman, M.T., Mun, S.K.: Convolution neural-network-based detection of lung structures. In: Medical Imaging 1994: Image Processing, vol. 2167, pp. 654–662. International Society for Optics and Photonics (1994)
- 56.He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
- 57.Henschke CI, McCauley DI, Yankelevitz DF, Naidich DP, McGuinness G, Miettinen OS, Libby DM, Pasmantier MW, Koizumi J, Altorki NK, et al. Early lung cancer action project: overall design and findings from baseline screening. Lancet. 1999;354(9173):99–105. doi: 10.1016/S0140-6736(99)06093-6. [DOI] [PubMed] [Google Scholar]
- 58.Hira S, Bai A, Hira S. An automatic approach based on cnn architecture to detect covid-19 disease from chest x-ray images. Appl. Intell. 2021;51(5):2864–2889. doi: 10.1007/s10489-020-02010-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hooda, R., Sofat, S., Kaur, S., Mittal, A., Meriaudeau, F.: Deep-learning: A potential method for tuberculosis detection using chest radiography. In: 2017 IEEE International Conference on Signal and Image Processing Applications (ICSIPA), pp. 497–502. IEEE (2017)
- 60.Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern recognition, pp. 7132–7141 (2018)
- 61.Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
- 62.Huang, X., Liu, M.Y., Belongie, S., Kautz, J.: Multimodal unsupervised image-to-image translation. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 172–189 (2018)
- 63.Huang Y, Cheng Y, Bapna A, Firat O, Chen D, Chen M, Lee H, Ngiam J, Le QV, Wu Y, et al. Gpipe: efficient training of giant neural networks using pipeline parallelism. Adv. Neural. Inf. Process. Syst. 2019;32:103–112. [Google Scholar]
- 64.Huang, Z., Wang, X., Huang, L., Huang, C., Wei, Y., Liu, W.: Ccnet: Criss-cross attention for semantic segmentation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 603–612 (2019)
- 65.Hwang, S., Kim, H.E., Jeong, J., Kim, H.J.: A novel approach for tuberculosis screening based on deep convolutional neural networks. In: Medical imaging 2016: computer-aided diagnosis, vol. 9785, p. 97852W. Int Soc Optics Photonics (2016)
- 66.Hwang, S., Park, S.: Accurate lung segmentation via network-wise training of convolutional networks. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 92–99. Springer (2017)
- 67.Iandola, F.N., Han, S., Moskewicz, M.W., Ashraf, K., Dally, W.J., Keutzer, K.: Squeezenet: Alexnet-level accuracy with 50x fewer parameters and< 0.5 mb model size. arXiv preprint arXiv:1602.07360 (2016)
- 68.Imran, A.A.Z., Terzopoulos, D.: Semi-supervised multi-task learning with chest x-ray images. In: International Workshop on Machine Learning in Medical Imaging, pp. 151–159. Springer (2019)
- 69.Inunganbi S, Choudhary P, Manglem K. Meitei mayek handwritten dataset: compilation, segmentation, and character recognition. Vis. Comput. 2021;37(2):291–305. doi: 10.1007/s00371-020-01799-4. [DOI] [Google Scholar]
- 70.Ioffe, S., Szegedy, C.: Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning, pp. 448–456. PMLR (2015)
- 71.Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S., Chute, C., Marklund, H., Haghgoo, B., Ball, R., Shpanskaya, K., et al.: Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 590–597 (2019)
- 72.Isola, P., Zhu, J.Y., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1125–1134 (2017)
- 73.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014;4(6):475. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaeger, S., Karargyris, A., Antani, S., Thoma, G.: Detecting tuberculosis in radiographs using combined lung masks. In: 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 4978–4981. IEEE (2012) [DOI] [PMC free article] [PubMed]
- 75.Jaeger S, Karargyris A, Candemir S, Folio L, Siegelman J, Callaghan F, Xue Z, Palaniappan K, Singh RK, Antani S, et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging. 2013;33(2):233–245. doi: 10.1109/TMI.2013.2284099. [DOI] [PubMed] [Google Scholar]
- 76.Jain R, Gupta M, Taneja S, Hemanth DJ. Deep learning based detection and analysis of covid-19 on chest x-ray images. Appl. Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jaiswal AK, Tiwari P, Kumar S, Gupta D, Khanna A, Rodrigues JJ. Identifying pneumonia in chest x-rays: a deep learning approach. Measurement. 2019;145:511–518. doi: 10.1016/j.measurement.2019.05.076. [DOI] [Google Scholar]
- 78.Jangam, E., Rao, A.: Segmentation of lungs from chest x rays using firefly optimized fuzzy c-means and level set algorithm. In: International Conference on Recent Trends in Image Processing and Pattern Recognition, pp. 303–311. Springer (2018)
- 79.Johnson, A.E., Pollard, T.J., Berkowitz, S.J., Greenbaum, N.R., Lungren, M.P., Deng, C.y., Mark, R.G., Horng, S.: Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Scientific data 6(1), 1–8 (2019) [DOI] [PMC free article] [PubMed]
- 80.Joseph A, Geetha P. Facial emotion detection using modified eyemap-mouthmap algorithm on an enhanced image and classification with tensorflow. Vis. Comput. 2020;36(3):529–539. doi: 10.1007/s00371-019-01628-3. [DOI] [Google Scholar]
- 81.Kalinovsky, A., Kovalev, V.: Lung image ssgmentation using deep learning methods and convolutional neural networks (2016)
- 82.Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int. J. Comput. Vision. 1988;1(4):321–331. doi: 10.1007/BF00133570. [DOI] [Google Scholar]
- 83.Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 84.Kim, E., Corte-Real, M., Baloch, Z.: A deep semantic mobile application for thyroid cytopathology. In: Medical Imaging 2016: PACS and Imaging Informatics: Next Generation and Innovations, vol. 9789, p. 97890A. Int Soc Optics Photonics (2016)
- 85.Kim M, Lee BD. Automatic lung segmentation on chest x-rays using self-attention deep neural network. Sensors. 2021;21(2):369. doi: 10.3390/s21020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kimmel R, Amir A, Bruckstein AM. Finding shortest paths on surfaces using level sets propagation. IEEE Trans. Pattern Anal. Mach. Intell. 1995;17(6):635–640. doi: 10.1109/34.387512. [DOI] [Google Scholar]
- 87.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process. Syst. 2012;25:1097–1105. [Google Scholar]
- 88.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 89.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 90.Le Cun, Y., Boser, B., Denker, J.S., Henderson, D., Howard, R.E., Hubbard, W., Jackel, L.D.: Handwritten digit recognition with a back-propagation network. In: Proceedings of the 2nd International Conference on Neural Information Processing Systems, pp. 396–404 (1989)
- 91.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 92.Leung CC. Reexamining the role of radiography in tuberculosis case finding. Int. J. Tuberc. Lung Dis. 2011;15(10):1279–1279. doi: 10.5588/ijtld.11.0425. [DOI] [PubMed] [Google Scholar]
- 93.Li C, Zhu G, Wu X, Wang Y. False-positive reduction on lung nodules detection in chest radiographs by ensemble of convolutional neural networks. IEEE Access. 2018;6:16060–16067. doi: 10.1109/ACCESS.2018.2817023. [DOI] [Google Scholar]
- 94.Li L, Zheng Y, Kallergi M, Clark RA. Improved method for automatic identification of lung regions on chest radiographs. Acad. Radiol. 2001;8(7):629–638. doi: 10.1016/S1076-6332(03)80688-8. [DOI] [PubMed] [Google Scholar]
- 95.Li X, Shen L, Xie X, Huang S, Xie Z, Hong X, Yu J. Multi-resolution convolutional networks for chest x-ray radiograph based lung nodule detection. Artif. Intell. Med. 2020;103:101744. doi: 10.1016/j.artmed.2019.101744. [DOI] [PubMed] [Google Scholar]
- 96.Li, Z., Wang, C., Han, M., Xue, Y., Wei, W., Li, L.J., Fei-Fei, L.: Thoracic disease identification and localization with limited supervision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8290–8299 (2018)
- 97.Lin, M., Chen, Q., Yan, S.: Network in network. arXiv preprint arXiv:1312.4400 (2013)
- 98.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 99.Liu H, Wang L, Nan Y, Jin F, Wang Q, Pu J. Sdfn: segmentation-based deep fusion network for thoracic disease classification in chest x-ray images. Comput. Med. Imaging Graph. 2019;75:66–73. doi: 10.1016/j.compmedimag.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lo SC, Lou SL, Lin JS, Freedman MT, Chien MV, Mun SK. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans. Med. Imaging. 1995;14(4):711–718. doi: 10.1109/42.476112. [DOI] [PubMed] [Google Scholar]
- 101.Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015) [DOI] [PubMed]
- 102.Luo, H., Gaborski, R., Acharya, R.: Automatic segmentation of lung regions in chest radiographs: a model guided approach. In: Proceedings 2000 International Conference on Image Processing (Cat. No. 00CH37101), vol. 2, pp. 483–486. IEEE (2000)
- 103.Luo, H., Lu, Q., Acharya, R., Gaborski, R.: Robust snake model. In: Proceedings IEEE Conference on Computer Vision and Pattern Recognition. CVPR 2000 (Cat. No. PR00662), vol. 1, pp. 452–457. IEEE (2000)
- 104.Ma J, Song Y, Tian X, Hua Y, Zhang R, Wu J. Survey on deep learning for pulmonary medical imaging. Front. Med. 2020;14(4):450–469. doi: 10.1007/s11684-019-0726-4. [DOI] [PubMed] [Google Scholar]
- 105.Madani, A., Moradi, M., Karargyris, A., Syeda-Mahmood, T.: Semi-supervised learning with generative adversarial networks for chest x-ray classification with ability of data domain adaptation. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 1038–1042. IEEE (2018)
- 106.Mahapatra, D., Ge, Z., Sedai, S., Chakravorty, R.: Joint registration and segmentation of xray images using generative adversarial networks. In: International Workshop on Machine Learning in Medical Imaging, pp. 73–80. Springer (2018)
- 107.Marques G, Agarwal D, de la Torre Díez I. Automated medical diagnosis of covid-19 through efficientnet convolutional neural network. Appl. Soft Comput. 2020;96:106691. doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McInerney T, Terzopoulos D. Deformable models in medical image analysis: a survey. Med. Image Anal. 1996;1(2):91–108. doi: 10.1016/S1361-8415(96)80007-7. [DOI] [PubMed] [Google Scholar]
- 109.McNitt-Gray, M.F., Sayre, J.W., Huang, H., Razavi, M.: Pattern classification approach to segmentation of chest radiographs. In: Medical Imaging 1993: Image Processing, vol. 1898, pp. 160–170. International Society for Optics and Photonics (1993)
- 110.Milletari, F., Navab, N., Ahmadi, S.A.: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
- 111.Mittal A, Hooda R, Sofat S. Lf-segnet: a fully convolutional encoder-decoder network for segmenting lung fields from chest radiographs. Wireless Pers. Commun. 2018;101(1):511–529. doi: 10.1007/s11277-018-5702-9. [DOI] [Google Scholar]
- 112.Mooney, P.: Chest x-ray images (pneumonia). kaggle. com (2018)
- 113.Mould RF. A century of X-rays and radioactivity in medicine: with emphasis on photographic records of the early years. Florida: CRC Press; 1993. [Google Scholar]
- 114.Munadi K, Muchtar K, Maulina N, Pradhan B. Image enhancement for tuberculosis detection using deep learning. IEEE Access. 2020;8:217897–217907. doi: 10.1109/ACCESS.2020.3041867. [DOI] [Google Scholar]
- 115.Munawar F, Azmat S, Iqbal T, Grönlund C, Ali H. Segmentation of lungs in chest x-ray image using generative adversarial networks. IEEE Access. 2020;8:153535–153545. doi: 10.1109/ACCESS.2020.3017915. [DOI] [Google Scholar]
- 116.Nair, V., Hinton, G.E.: Rectified linear units improve restricted boltzmann machines. In: Icml (2010)
- 117.Neff, T., Payer, C., Stern, D., Urschler, M.: Generative adversarial network based synthesis for supervised medical image segmentation. In: Proc. OAGM and ARW Joint Workshop, vol. 3, p. 4 (2017)
- 118.Nie, D., Trullo, R., Lian, J., Petitjean, C., Ruan, S., Wang, Q., Shen, D.: Medical image synthesis with context-aware generative adversarial networks. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 417–425. Springer (2017) [DOI] [PMC free article] [PubMed]
- 119.Nijiati, M., Zhang, Z., Abulizi, A., Miao, H., Tuluhong, A., Quan, S., Guo, L., Xu, T., Zou, X.: Deep learning assistance for tuberculosis diagnosis with chest radiography in low-resource settings. J. X-Ray Sci. Technol. (Preprint), 1–12 [DOI] [PubMed]
- 120.Noh, H., Hong, S., Han, B.: Learning deconvolution network for semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1520–1528 (2015)
- 121.Novikov AA, Lenis D, Major D, Hladvka J, Wimmer M, Bühler K. Fully convolutional architectures for multiclass segmentation in chest radiographs. IEEE Trans. Med. Imaging. 2018;37(8):1865–1876. doi: 10.1109/TMI.2018.2806086. [DOI] [PubMed] [Google Scholar]
- 122.Oakden-Rayner L. Exploring large-scale public medical image datasets. Acad. Radiol. 2020;27(1):106–112. doi: 10.1016/j.acra.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 123.Oken MM, Hocking WG, Kvale PA, Andriole GL, Buys SS, Church TR, Crawford ED, Fouad MN, Isaacs C, Reding DJ, et al. Screening by chest radiograph and lung cancer mortality: the prostate, lung, colorectal, and ovarian (plco) randomized trial. JAMA. 2011;306(17):1865–1873. doi: 10.1001/jama.2011.1591. [DOI] [PubMed] [Google Scholar]
- 124.Organization, W.H.: Global tuberculosis report 2013. World Health Organization (2013)
- 125.Panwar H, Gupta P, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of covid-19 in x-rays using ncovnet. Chaos, Solitons Fract. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park B, Cho Y, Lee G, Lee SM, Cho YH, Lee ES, Lee KH, Seo JB, Kim N. A curriculum learning strategy to enhance the accuracy of classification of various lesions in chest-pa x-ray screening for pulmonary abnormalities. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-51832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Park, S., Kim, G., Oh, Y., Seo, J.B., Lee, S.M., Kim, J.H., Moon, S., Lim, J.K., Ye, J.C.: Vision transformer for covid-19 cxr diagnosis using chest x-ray feature corpus. arXiv preprint arXiv:2103.07055 (2021) [DOI] [PMC free article] [PubMed]
- 128.Pasa F, Golkov V, Pfeiffer F, Cremers D, Pfeiffer D. Efficient deep network architectures for fast chest x-ray tuberculosis screening and visualization. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-42557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petinaux B, Bhat R, Boniface K, Aristizabal J. Accuracy of radiographic readings in the emergency department. Am. J. Emerg. Med. 2011;29(1):18–25. doi: 10.1016/j.ajem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 130.Pooch, E.H., Ballester, P., Barros, R.C.: Can we trust deep learning based diagnosis? the impact of domain shift in chest radiograph classification. In: International Workshop on Thoracic Image Analysis, pp. 74–83. Springer (2020)
- 131.Powell GF, Doi K, Katsuragawa S. Localization of inter-rib spaces for lung texture analysis and computer-aided diagnosis in digital chest images. Med. Phys. 1988;15(4):581–587. doi: 10.1118/1.596209. [DOI] [PubMed] [Google Scholar]
- 132.Radford, A., Metz, L., Chintala, S.: Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434 (2015)
- 133.Rahman T, Chowdhury ME, Khandakar A, Islam KR, Islam KF, Mahbub ZB, Kadir MA, Kashem S. Transfer learning with deep convolutional neural network (cnn) for pneumonia detection using chest x-ray. Appl. Sci. 2020;10(9):3233. doi: 10.3390/app10093233. [DOI] [Google Scholar]
- 134.Rahman T, Khandakar A, Kadir MA, Islam KR, Islam KF, Mazhar R, Hamid T, Islam MT, Kashem S, Mahbub ZB, et al. Reliable tuberculosis detection using chest x-ray with deep learning, segmentation and visualization. IEEE Access. 2020;8:191586–191601. doi: 10.1109/ACCESS.2020.3031384. [DOI] [Google Scholar]
- 135.Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan, T., Ding, D., Bagul, A., Langlotz, C., Shpanskaya, K., et al.: Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225 (2017)
- 136.Rakshit, S., Saha, I., Wlasnowolski, M., Maulik, U., Plewczynski, D.: Deep learning for detection and localization of thoracic diseases using chest x-ray imagery. In: International Conference on Artificial Intelligence and Soft Computing, pp. 271–282. Springer (2019)
- 137.Redmon, J., Farhadi, A.: Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767 (2018)
- 138.Rehman B, Ong WH, Tan ACH, Ngo TD. Face detection and tracking using hybrid margin-based roi techniques. Vis. Comput. 2020;36(3):633–647. doi: 10.1007/s00371-019-01649-y. [DOI] [Google Scholar]
- 139.Rochester N, Holland J, Haibt L, Duda W. Tests on a cell assembly theory of the action of the brain, using a large digital computer. IRE Trans. Inf. Theor. 1956;2(3):80–93. doi: 10.1109/TIT.1956.1056810. [DOI] [Google Scholar]
- 140.Romero M, Interian Y, Solberg T, Valdes G. Targeted transfer learning to improve performance in small medical physics datasets. Med. Phys. 2020;47(12):6246–6256. doi: 10.1002/mp.14507. [DOI] [PubMed] [Google Scholar]
- 141.Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241. Springer (2015)
- 142.Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958;65(6):386. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- 143.Saidy, L., Lee, C.C.: Chest x-ray image segmentation using encoder-decoder convolutional network. In: 2018 IEEE International Conference on Consumer Electronics-Taiwan (ICCE-TW), pp. 1–2. IEEE (2018)
- 144.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved techniques for training gans. Adv. Neural. Inf. Process. Syst. 2016;29:2234–2242. [Google Scholar]
- 145.Schilham AM, Van Ginneken B, Loog M. A computer-aided diagnosis system for detection of lung nodules in chest radiographs with an evaluation on a public database. Med. Image Anal. 2006;10(2):247–258. doi: 10.1016/j.media.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 146.Sethian JA. Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 147.Shah PK, Austin JH, White CS, Patel P, Haramati LB, Pearson GD, Shiau MC, Berkmen YM. Missed non-small cell lung cancer: radiographic findings of potentially resectable lesions evident only in retrospect. Radiology. 2003;226(1):235–241. doi: 10.1148/radiol.2261011924. [DOI] [PubMed] [Google Scholar]
- 148.Shao Y, Gao Y, Guo Y, Shi Y, Yang X, Shen D. Hierarchical lung field segmentation with joint shape and appearance sparse learning. IEEE Trans. Med. Imaging. 2014;33(9):1761–1780. doi: 10.1109/TMI.2014.2305691. [DOI] [PubMed] [Google Scholar]
- 149.Shi Y, Qi F, Xue Z, Chen L, Ito K, Matsuo H, Shen D. Segmenting lung fields in serial chest radiographs using both population-based and patient-specific shape statistics. IEEE Trans. Med. Imaging. 2008;27(4):481–494. doi: 10.1109/TMI.2007.908130. [DOI] [PubMed] [Google Scholar]
- 150.Shi, Z., Zhou, P., He, L., Nakamura, T., Yao, Q., Itoh, H.: Lung segmentation in chest radiographs by means of gaussian kernel-based fcm with spatial constraints. In: 2009 Sixth International Conference on Fuzzy Systems and Knowledge Discovery, vol. 3, pp. 428–432. IEEE (2009)
- 151.Shiraishi, J., Katsuragawa, S., Ikezoe, J., Matsumoto, T., Kobayashi, T., Komatsu, K.I., Matsui, M., Fujita, H., Kodera, Y., Doi, K.: Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology 174(1), 71–74 (2000) [DOI] [PubMed]
- 152.Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
- 153.Smith, R.A., Cokkinides, V., Eyre, H.J.: American cancer society guidelines for the early detection of cancer, 2004. CA: A Cancer Journal for Clinicians 54(1), 41–52 (2004) [DOI] [PubMed]
- 154.Souid A, Sakli N, Sakli H. Classification and predictions of lung diseases from chest x-rays using mobilenet v2. Appl. Sci. 2021;11(6):2751. doi: 10.3390/app11062751. [DOI] [Google Scholar]
- 155.Souza JC, Diniz JOB, Ferreira JL, da Silva GLF, Silva AC, de Paiva AC. An automatic method for lung segmentation and reconstruction in chest x-ray using deep neural networks. Comput. Methods Programs Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 156.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Machine Learn. Res. 2014;15(1):1929–1958. [Google Scholar]
- 157.Stollenga MF, Byeon W, Liwicki M, Schmidhuber J. Parallel multi-dimensional lstm, with application to fast biomedical volumetric image segmentation. Adv. Neural. Inf. Process. Syst. 2015;28:2998–3006. [Google Scholar]
- 158.Sze-To, A., Wang, Z.: tchexnet: Detecting pneumothorax on chest x-ray images using deep transfer learning. In: International Conference on Image Analysis and Recognition, pp. 325–332. Springer (2019)
- 159.Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., Rabinovich, A.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
- 160.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
- 161.Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med. Image Anal. 2020;63:101693. doi: 10.1016/j.media.2020.101693. [DOI] [PubMed] [Google Scholar]
- 162.Tan, M., Le, Q.: Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114. PMLR (2019)
- 163.Tang, Y., Tang, Y., Sandfort, V., Xiao, J., Summers, R.M.: Tuna-net: Task-oriented unsupervised adversarial network for disease recognition in cross-domain chest x-rays. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 431–440. Springer (2019)
- 164.Tang, Y.B., Tang, Y.X., Xiao, J., Summers, R.M.: Xlsor: A robust and accurate lung segmentor on chest x-rays using criss-cross attention and customized radiorealistic abnormalities generation. In: International Conference on Medical Imaging with Deep Learning, pp. 457–467. PMLR (2019)
- 165.Tang, Y.X., Tang, Y.B., Han, M., Xiao, J., Summers, R.M.: Abnormal chest x-ray identification with generative adversarial one-class classifier. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 1358–1361. IEEE (2019)
- 166.Team, P.P., Gohagan, J.K., Prorok, P.C., Hayes, R.B., Kramer, B.S.: The prostate, lung, colorectal and ovarian (plco) cancer screening trial of the national cancer institute: history, organization, and status. Controlled clinical trials 21(6), 251S–272S (2000) [DOI] [PubMed]
- 167.Van Ginneken B, Frangi AF, Staal JJ, ter Haar Romeny BM, Viergever MA. Active shape model segmentation with optimal features. IEEE Trans. Med. Imaging. 2002;21(8):924–933. doi: 10.1109/TMI.2002.803121. [DOI] [PubMed] [Google Scholar]
- 168.Van Ginneken B, Katsuragawa S, ter Haar Romeny BM, Doi K, Viergever MA. Automatic detection of abnormalities in chest radiographs using local texture analysis. IEEE Trans. Med. Imaging. 2002;21(2):139–149. doi: 10.1109/42.993132. [DOI] [PubMed] [Google Scholar]
- 169.Van Ginneken B, Romeny BTH, Viergever MA. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans. Med. Imaging. 2001;20(12):1228–1241. doi: 10.1109/42.974918. [DOI] [PubMed] [Google Scholar]
- 170.Van Ginneken B, Stegmann MB, Loog M. Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database. Med. Image Anal. 2006;10(1):19–40. doi: 10.1016/j.media.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 171.Vantaggiato E, Paladini E, Bougourzi F, Distante C, Hadid A, Taleb-Ahmed A. Covid-19 recognition using ensemble-cnns in two new chest x-ray databases. Sensors. 2021;21(5):1742. doi: 10.3390/s21051742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I.: Attention is all you need. In: Advances in Neural Information Processing Systems, pp. 5998–6008 (2017)
- 173.Vittitoe NF, Vargas-Voracek R, Floyd CE., Jr Identification of lung regions in chest radiographs using markov random field modeling. Med. Phys. 1998;25(6):976–985. doi: 10.1118/1.598405. [DOI] [PubMed] [Google Scholar]
- 174.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. Covidgan: data augmentation using auxiliary classifier gan for improved covid-19 detection. Ieee Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R.M.: Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2097–2106 (2017)
- 176.Wang, X., Yang, S., Lan, J., Fang, Y., He, J., Wang, M., Zhang, J., Han, X.: Automatic segmentation of pneumothorax in chest radiographs based on a two-stage deep learning method. IEEE Transactions on Cognitive and Developmental Systems (2020)
- 177.Wei, Y., Feng, J., Liang, X., Cheng, M.M., Zhao, Y., Yan, S.: Object region mining with adversarial erasing: A simple classification to semantic segmentation approach. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1568–1576 (2017)
- 178.Wong, K.C., Moradi, M., Wu, J., Syeda-Mahmood, T.: Identifying disease-free chest x-ray images with deep transfer learning. In: Medical Imaging 2019: Computer-Aided Diagnosis, vol. 10950, p. 109500P. International Society for Optics and Photonics (2019)
- 179.Xie, S., Girshick, R., Dollár, P., Tu, Z., He, K.: Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1492–1500 (2017)
- 180.Xie, Y., Zhang, Z., Sapkota, M., Yang, L.: Spatial clockwork recurrent neural network for muscle perimysium segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 185–193. Springer (2016) [DOI] [PMC free article] [PubMed]
- 181.Xu, T., Mandal, M., Long, R., Basu, A.: Gradient vector flow based active shape model for lung field segmentation in chest radiographs. In: 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 3561–3564. IEEE (2009) [DOI] [PubMed]
- 182.Xu XW, Doi K. Image feature analysis for computer-aided diagnosis: accurate determination of ribcage boundary in chest radiographs. Med. Phys. 1995;22(5):617–626. doi: 10.1118/1.597549. [DOI] [PubMed] [Google Scholar]
- 183.Xu XW, Doi K. Image feature analysis for computer-aided diagnosis: Detection of right and left hemidiaphragm edges and delineation of lung field in chest radiographs. Med. Phys. 1996;23(9):1613–1624. doi: 10.1118/1.597738. [DOI] [PubMed] [Google Scholar]
- 184.Xue Y, Xu T, Zhang H, Long LR, Huang X. Segan: adversarial network with multi-scale l 1 loss for medical image segmentation. Neuroinformatics. 2018;16(3):383–392. doi: 10.1007/s12021-018-9377-x. [DOI] [PubMed] [Google Scholar]
- 185.Yan, C., Yao, J., Li, R., Xu, Z., Huang, J.: Weakly supervised deep learning for thoracic disease classification and localization on chest x-rays. In: Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp. 103–110 (2018)
- 186.Yeh, R.A., Chen, C., Yian Lim, T., Schwing, A.G., Hasegawa-Johnson, M., Do, M.N.: Semantic image inpainting with deep generative models. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5485–5493 (2017)
- 187.Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med. Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 188.Yosinski, J., Clune, J., Bengio, Y., Lipson, H.: How transferable are features in deep neural networks? arXiv preprint arXiv:1411.1792 (2014)
- 189.Yuan, X., Giritharan, B., Oh, J.: Gradient vector flowdriven active shape for image segmentation. In: 2007 IEEE International Conference on Multimedia and Expo, pp. 2058–2061. IEEE (2007)
- 190.Zebin T, Rezvy S. Covid-19 detection and disease progression visualization: deep learning on chest x-rays for classification and coarse localization. Appl. Intell. 2021;51(2):1010–1021. doi: 10.1007/s10489-020-01867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang J, Xia Y, Cui H, Zhang Y. Pulmonary nodule detection in medical images: a survey. Biomed. Signal Process. Control. 2018;43:138–147. doi: 10.1016/j.bspc.2018.01.011. [DOI] [Google Scholar]
- 192.Zoph, B., Vasudevan, V., Shlens, J., Le, Q.V.: Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8697–8710 (2018)