Summary
Efficient delivery of toxic compounds to bacterial competitors is essential during interspecies microbial warfare. Rhamnolipids (RLPs) are glycolipids produced by Pseudomonas and Burkholderia species involved in solubilization and uptake of environmental aliphatic hydrocarbons and perform as biosurfactants for swarming motility. Here, we show that RLPs produced by Pseudomonas aeruginosa associate to form micelles. Using high-resolution microscopy, we found that RLP micelles serve as carriers for self-produced toxic compounds, which they deliver to Staphylococcus aureus cells, thereby enhancing and accelerating S. aureus killing. RLPs also potentiated the activity of lincosamide antibiotics, suggesting that RLP micelles may transport not only self-produced but also heterologous compounds to target competing bacterial species
Subject areas: Lipid, Microbiology
Graphical abstract

Highlights
-
•
Pseudomonas aeruginosa rhamnolipids form micelles
-
•
Rhamnolipid micelles delivery pyochelin into S. aureus cells
-
•
Rhamnolipid micelles potentiate activity of lincosamide antibiotics against S. aureus
Lipid; Microbiology
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen causing acute infections in immunocompromised hosts and chronic infections in patients with cystic fibrosis. Owing to intrinsic and acquired antibiotic resistance determinants, P. aeruginosa remains a challenge when choosing optimal antimicrobial therapies. P. aeruginosa possesses a variety of host-directed virulence factors, including cell-contact dependent (T3SS, T6SS) and independent mechanisms (secreted toxins and secondary metabolites). Furthermore, its metabolic versatility and the production of bacteriocins (pyocins), confer a competitive advantage during inter- and intra-species competition, explaining its success as an opportunistic pathogen and niche colonizer (Atanaskovic et al., 2020; Gonzalez and Mavridou, 2019; Wood et al., 2019).
We have recently defined an antimicrobial cocktail consisting of proteins and several metabolites secreted by P. aeruginosa, which showed broad-spectrum killing activity against both Gram-positive and Gram-negative bacterial species (Gdaniec et al., 2020). An essential component of this cocktail was rhamnolipids (RLPs), which belong to the chemically diverse group of glycolipids. RLPs are composed of a hydrophilic rhamnose sugar moiety, which is linked through a β-glycosidic bond to a hydrophobic fatty acid moiety (Hauser and Karnovsky, 1957). RLPs have multiple roles in metabolite uptake, community and host interactions, and microbial competition (Chrzanowski et al., 2012). The initially reported function of RLPs in P. aeruginosa was the solubilization and uptake of aliphatic hydrocarbons from the environment, that P. aeruginosa uses as a carbon and energy source (Noordman and Janssen, 2002). In the free form, their hydrophilic moiety can interact with the O-antigen component of lipopolysaccharides (LPS) of Gram-negative bacteria, thereby increasing the hydrophobicity of the outer membrane (Zhong et al., 2014).
One interesting feature of RLPs is their ability to form micelles above a critical micelle concentration (Haba et al., 2014). RLP micelles can remove LPS from the outer membrane of Gram-negative bacteria, which also leads to an increase in hydrophobicity due to the resulting lack of polar sugar residues on the cell surface (Al-Tahhan et al., 2000; Sotirova et al., 2009). However, the interactions between RLP micelles and Gram-positive bacteria are not well understood. Because P. aeruginosa is particularly efficient in killing S. aureus cells, we sought to get insight into the molecular interactions between P. aeruginosa RLPs and S. aureus using biochemical analysis and super-resolution microscopy. We found that RLP micelles are able to incorporate metabolites produced by P. aeruginosa and to deliver them to S. aureus cells. We further show that RLP micelles accelerate and potentiate the activity of lincosamide antibiotics directed against S. aureus.
Results
Rhamnolipids secreted by P.aeruginosa form micelles
We have previously shown that supernatants of a wspF mutant of P. aeruginosa contain elevated concentrations of several secondary metabolites, including RLPs. RLPs were an essential ingredient of a synthetic cocktail, including the siderophores pyoverdine and pyochelin, as well as the alkylquinoline 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO), able to cause a 5-log reduction of viable counts of S. aureus (Gdaniec et al., 2020) within 24 h. We wondered whether the RLPs concentration previously measured at 300–400 μg/mL in the wspF mutant supernatant (Gdaniec et al., 2020) would be sufficient to form micelles. We therefore analyzed by electron microscopy the supernatants of P. aeruginosa PA14ΔwspF and an isogenic ΔwspFΔrhlA mutant, deficient in RLP production. As a control, we analyzed purified P. aeruginosa RLPs, containing a mix of mono- and di-RLPs with varying fatty acid chain lengths at a concentration of 250 μg/mL. Electron microscopy imaging revealed the presence of round shaped, probably spherical structures in the supernatant of the ΔwspF mutant with an average surface area of 0.15 μm2 (Figure 1A, left panel), similar to those observed with the purified RLPs solution (Figure 1A, right panel), but absent in the supernatant of the ΔwspFΔrhlA mutant (Figure 1A, middle panel). We therefore considered the structures observed in the ΔwspF supernatant as RLP micelles. To further confirm their lipidic nature, we performed selective fluorescent staining with Nile Red (NR). While NR is slightly fluorescent in water, its fluorescence signal is enhanced upon integration into a lipophilic environment like phospholipid membranes or vesicles (Greenspan et al., 1985) (Figure 1B). We measured the fluorescence emission of purified RLPs, added either to M14 medium or to RLP-deficient ΔwspFΔrhlA culture supernatant. This allowed us to determine the RLPs critical micelle concentration (CMC), which was comprised between 50 and 100 μg/mL (Figure 1C). Next, we measured NR fluorescence in bacterial supernatants of the PA14 wild type, ΔwspF, and ΔwspFΔrhlA mutants, as well as in the M14 growth medium. While fluorescence remained low for the wild type and the ΔwspFΔrhlA supernatants, indicating lack of micelles formation, the ΔwspF supernatant showed a strong fluorescence signal indicating micelle formation and an RLP concentration above the CMC (Figure 1D). Altogether, these data suggest that RLPs overproduced in a ΔwspF mutant supernatant form micelles, which have similar shape and NR fluorescence compared with purified RLPs.
Figure 1.
P. aeruginosa ΔwspF supernatant contains rhamnolipid micelles
(A) Electron microscopy imaging of RLP micelles present in the ΔwspF supernatant (left panel) and formed by purified RLPs (right panel).
(B) Schematic representation of RLP micelles staining with Nile Red (NR).
(C) Critical micellar concentration (CMC) was determined by adding purified RLPs to either medium or to RLP-deficient ΔwspFΔrhlA supernatant.
(D) Detection of RLP micelles by NR staining and fluorescence measurement in supernatants of P. aeruginosa wild type and mutants. Student test (∗∗∗∗, p value <0.0001). The scale bar indicates 1 μm. Six-thirteen images per condition, showing similar heterogeneous size distributions of RLP micelles, were analyzed.
RLP micelles are essential for the killing activity of a ΔwspF mutant supernatant
We next analyzed the role attributable to RLPs in the S. aureus killing activity of P. aeruginosa culture supernatants. For this, we investigated the S. aureus killing activity of supernatants produced by the PA14 wild type, the ΔwspF, and ΔwspFΔrhlA mutants. As expected, the ΔwspF supernatant showed a potent S. aureus killing activity, not found in supernatants produced by the other strains, confirming the role for RLPs promoting S. aureus killing (data not shown). To further characterize this killing activity, we separated the crude culture supernatants (crude-S) by ultracentrifugation into an ultracentrifuged pellet (ultra-P) and an ultracentrifuged supernatant (ultra-S). Determined by Orcinol assays, the ΔwspF ultra-P fraction contained 88% of the RLPs, while 12% remained in the ultra-S fraction (Figure 2A), suggesting enrichment of RLPs in the ultra-P fraction. We next compared the killing kinetics of the crude-S and ultra-S supernatants, with those of resuspended ultra-P. PA14 crude-S, ultra-S, and ultra-P resuspended in M14 medium to the initial supernatant volume (1x ultra-P) had no killing activity (Figure 2B). In contrast, ultra-P resuspended in M14 medium to 1/15 of the initial supernatant volume (15x ultra-P) presented moderate killing during the initial 1.5 h (1.5-log decrease in S. aureus CFU), which was followed by a further extended killing period (3.5-log reduction over the next 21 h). ΔwspF crude-S and ultra-S, as well as 15x ultra-P showed similar killing kinetics (4-log reduction during first 3 h and additional 2-log reduction at 24 h), while 1x resuspended ultra-P had no killing activity (Figure 2C). The ΔwspFΔrhlA crude-S and ultra-S, and resuspended ultra-P (both 1x and 15x) showed no killing during the first 3 h. However, at 24 h, the crude-S and ultra-S showed a 2-log, and the 15x ultra-P a 5-log reduction in S. aureus CFUs, respectively (Figure 2D). To quantitatively assess the effect of the individual mutations as well as RLP supplementation, we considered the log decrease per hour as the primary readout for killing activity. We restricted this analysis to the first 3 h as at longer timepoints many experiments reached the detection limit and could not be quantitatively interpreted (Figure S1A). We observe statistically highly significant enhancement of killing in the various supernatant fractions of the ΔwspF mutant (Figure S1B), which is again highly significantly abolished in the ΔwspFΔrhlA mutant (Figure S1C), and restored by addition of exogenous RLPs (Figure S1D). This confirms the importance of metabolites present in the ΔwspF supernatant and the specific requirement of RLPs for S. aureus killing.
Figure 2.
RLP micelles are essential for initial S. aureus killing activity of ΔwspF mutant supernatants
(A) Scheme of isolation for crude and ultracentrifuged supernatants, and ultracentrifuged pellets.
(B–G) Killing curves of S. aureus exposed to crude or ultracentrifuged supernatants, or resuspended ultracentrifuged pellets produced by PA14 wild type (B), ΔwspF mutant (C), and ΔwspFΔrhlA mutant (D). ΔwspFΔrhlA mutant crude or ultracentrifuged supernatants, or resuspended ultracentrifuged pellets complemented with commercial rhamnolipids (E), RLP C10C10 (F) and RLP C12C12 (G). Parts of the data (E–G) are imported from (C) for comparison. Additional data and statistical details are given in Figure S1.
In the ultra-P fraction, the situation is more subtle because both in the wild type and the ΔwspF mutant the 15x concentrated ultra-P shows significant killing activity, whereas the 1x ultra-P does not. Nevertheless, abrogation of RLP production prevents killing also in the 15x ultra-P (Figure S1C), with restoration by exogenous RLPs (Figure S1D).
To further substantiate the specific role of RLPs in S. aureus killing, we supplemented the ultra-S and 15x ultra-P of the ΔwspFΔrhlA mutant with commercial RLPs. Our previous metabolomic analysis of ΔwspF supernatant had shown a large diversity of RLPs, dominated by the C10-C10 di-RLP species (Gdaniec et al., 2020). We therefore used a mix of purified mono- and di-RLPs (Figure 2E), but also C10-C10 (Figure 2F) or C12-C12 mono-RLPs (Figure 2G) alone for the supplementation experiments. ΔwspFΔrhlA ultra-S and 15x ultra-P supplemented with RLPs showed strong S. aureus killing. The killing activities differed significantly between the RLP types (p<0.001), but not between the ultra-S and 15x ultra-P fractions (p = 0.155) (Figure S1D). The effect of purified RLPs was most pronounced with the C10-C10 mono-RLPs, suggesting that the shorter acyl chains provide a more efficient killing (Figure S1E). Altogether, these results suggest that RLPs are crucial for the initial killing of S. aureus and that a single species of RLPs is sufficient for this activity.
Rhamnolipids interact with S. aureus cell membranes and cross their peptidoglycan
To further explore the mechanisms underlying the killing activity and to identify the localization of RLPs in S. aureus cells, we covalently labeled a mix of purified commercial RLPs with the fluorescent dye Abberior STAR 555 (Schermelleh et al., 2019). We performed comparative confocal microscopy of unlabeled and Abberior STAR 555 labeled RLP micelles, which showed a similar micelle area for the most abundant micelle fractions, confirming that labeled RLPs still form micelles (Figure S2).
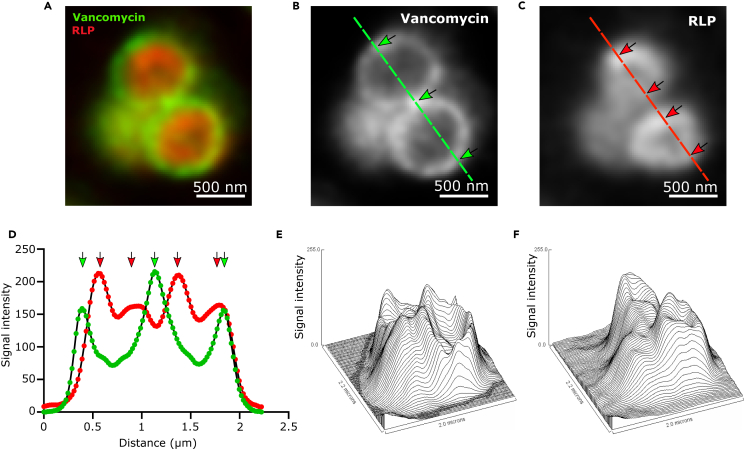
We then labeled the S. aureus peptidoglycan layer using vancomycin coupled with the Abberior STAR 488 NHS ester dye and visualized the cells using stimulated emission depletion (STED) nanoscopy to study the interaction between the labeled RLPs and S. aureus cells. As expected, we observed a fluorescence signal corresponding to the S. aureus cell wall, visualized by the incorporation of fluorescent vancomycin into the peptidoglycan layer (Watanakunakorn, 1984) (Figures 3A–3F). For up to 66.5% of the cells, the maximum RLPs signal formed a circular shape on the cytosolic side of the peptidoglycan, while the signal intensity decreased toward the cell center (Figures 3A–3F). This result suggests that the RLPs cross the peptidoglycan layer of S. aureus, accumulating in or next to the cell membrane.
Figure 3.
Interaction between RLPs and S. aureus membrane
(A–C) Merged STED nanoscopy image of S. aureus cells. Peptidoglycans were visualised using Abberior dye STAR 488 (artificially in green) labeled vancomycin (also shown in B). RLPs were labeled with the dye Abberior STAR 555 (artificially in red) (also shown in C).
(D) Plot profile (FIJI software) of the corresponding green and red lines shown in (B) and (C). Green arrows show the presence of the cell wall, while red arrows show the localization of RLPs.
(E and F) Surface profiles (FIJI software) of respectively vancomycin (E) and RLPs (F).
Rhamnolipid micelles serve as a carrier for P.aeruginosa metabolites
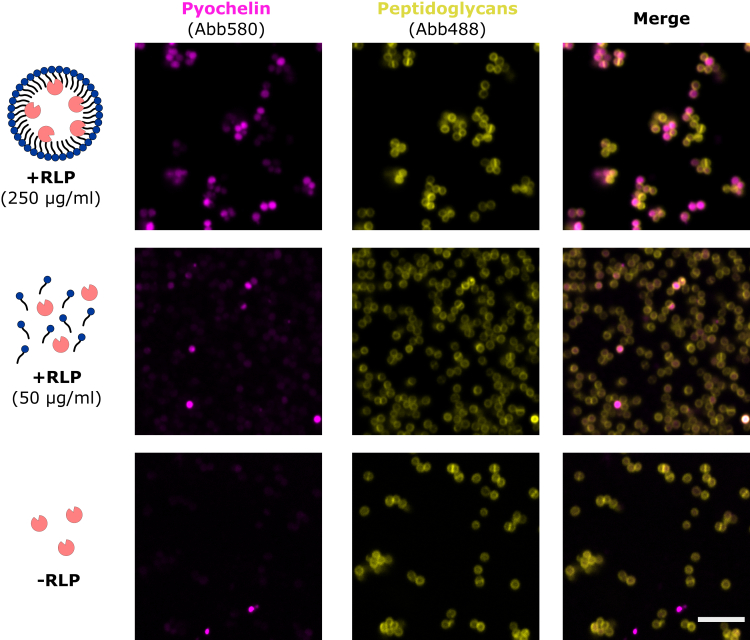
To decipher the role of RLP micelles as potential carriers for P. aeruginosa metabolites into S. aureus cells, we labeled the P. aeruginosa siderophore pyochelin with the fluorescent dye Abberior 580, and performed confocal microscopy of S. aureus cells with peptidoglycan visualized with Abberior 488 labeled vancomycin, in the absence or presence of commercial RLPs (Figure 4). To distinguish delivery of labeled pyochelin between free RLP molecules and micelles, we choose two RLPs concentrations: 50 μg/mL (below CMC) and 250 μg/mL (above CMC). In the presence of RLPs below CMC or without RLPs (Figure 4, second and third rows, respectively), uptake of labeled pyochelin reached 25% and 4%, respectively. When RLPs were added at a concentration above the CMC, labeled pyochelin was visible in the cytosol of 66% of S. aureus cells (Figure 4, first row).
Figure 4.
Rhamnolipid micelles serve as a vehicle for pyochelin
Confocal microscopy was performed on S. aureus Newman cells incubated in the presence of pyochelin labeled with Aberrior dye 580 (artificially colored in magenta) in the presence of RLP concentrations below (50 μg/mL) or above (250 μg/mL) the CMC. S. aureus peptidoglycan layer was visualized using Aberrior 488 labeled vancomycin (artificially colored in yellow). Cells were observed under a Zeiss confocal microscope LSM800. The scale bar corresponds to 5 μm and applies to all images.
Chemically inactivated Abberior 580 dye itself did not enter the cells even at the highest RLPs concentration (Figure S3). These data suggest that RLP micelles serve as a carrier for pyochelin in vitro. They also highlight the specificity of translocation of pyochelin in S. aureus, as opposed to non-specific increase in cellular permeability.
Rhamnolipids potentiate the activity of lincosamide antimicrobials
As RLPs are essential for killing of S. aureus in combination with other metabolites present in the P. aeruginosa ΔwspF mutant supernatants (Gdaniec et al., 2020), we investigated whether RLP micelles might potentiate the effect of antibiotics. We tested the effect of azithromycin, novobiocin, rifampicin, nalidixic acid, ciprofloxacin, ampicillin, and sulfamethoxazole on S. aureus in the presence/absence of RLPs, as representative members of the main antibiotic classes. The antimicrobial activity of these compounds on S. aureus Newman (MSSA) and COL (MRSA) strains were not affected by the presence of RLPs at concentrations above the CMC (250 μg/mL) (data not shown). In contrast, the minimal inhibitory concentrations (MICs) of lincomycin and clindamycin, two members of the lincosamide antibiotic family, decreased by 4–8 times in the presence of RLPs at concentrations above the CMC, whereas the MICs were not affected by RLPs at concentrations below the CMC (25 μg/mL) (Table 1).
Table 1.
Lincosamide MICs for S. aureus strains Newman and COL in the presence of RLPs
| Antibiotics/compound | MIC (μg/mL) |
|
|---|---|---|
| S. aureus Newman | S. aureus COL | |
| Rhamnolipids (RLPs) | 1000 | 1000 |
| Clindamycin | 0.25 | 0.25 |
| Clindamycin + RLPs 25 μg/mL | 0.125 | 0.125 |
| Clindamycin + RLPs 250 μg/mL | 0.032 | 0.032 |
| Lincomycin | 4 | 4 |
| Lincomycin + RLPs 25 μg/mL | 2 | 2 |
| Lincomycin + RLPs 250 μg/mL | 0.5 | 0.5 |
To evaluate the effect of RLPs on antibiotic activity, we performed checkerboard MIC assays and calculated the fractional inhibitory concentration (FIC) (Table S1). For the S. aureus Newman strain, the combination of RLPs with lincomycin revealed an additive effect (FIC 0.5–1.0), while combination of RLPs with clindamycin showed a synergistic effect (FIC ≤0.5). For the S. aureus COL strain, the combination of RLPs with clindamycin or lincomycin also resulted in a synergistic effect (FIC ≤0.5).
We then investigated the killing kinetics of clindamycin and lincomycin in the presence of RLPs mix. RLPs accelerated and increased killing by both lincosamides as shown by a 2–3 log decrease in S. aureus CFUs (Figure 5). Of note, the effect of RLPs was more pronounced on the MSSA strain Newman than on the MRSA strain COL.
Figure 5.
Rhamnolipids accelerate the killing of S. aureus by lincosamides
(A) S. aureus Newman killing kinetics.
(B) S. aureus COL killing kinetics. RLPs and lincosamides (CLINDA, clindamycin; LINCO, lincomycin) were added at t = 0 and CFU counts were determined at indicated time points. Values are the average of triplicate determinations. Error bars are standard deviations.
Discussion
Antibiotic resistance is a worldwide threat to human health that requires major efforts to develop new antimicrobial strategies and/or increase the efficacy of already existing antibiotics (ref: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance). Consequently, drug delivery systems enhancing the efficacy of antibiotics, while reducing off-target toxicity, have attracted attention. While synthetic liposomes (Chen et al., 2018) or siderophore-drug conjugates have been studied to increase antibiotic delivery to their targets (Schalk, 2018), little is known about natural bacterial delivery tools. Here, we demonstrate that P. aeruginosa uses naturally produced rhamnolipid micelles to enhance the killing of S. aureus. Our results reveal new properties of rhamnolipids acting as carriers for toxic metabolites, increasing their delivery to their targets in competing microorganisms. Therefore, our study provides evidence for natural antimicrobial delivery systems used by pathogenic bacteria in interspecies competition.
The properties of biosurfactants were intensively investigated over the last 50 years since the first biosurfactant “surfactin” was purified and characterized from Bacillus subtilis (Arima et al., 1968). Among the various categories of biosurfactants, rhamnolipids belong to the glycolipids group and are studied not only as a virulence factor of P. aeruginosa but also as an interesting compound for industrial processes (Sekhon Randhawa and Rahman, 2014). P. aeruginosa synthesizes a variety of RLPs composed of mono- and di-RLP species, which carry either one or two acyl chains of variable chain length (C8 to C14), respectively. The dominant species in P. aeruginosa are the di-RLPs C10-C10 and C12-C12 (Abdel-Mawgoud et al., 2010; Déziel et al., 1999). RLPs play an essential role as biosurfactant for swarming motility (Köhler et al., 2000), and help maintain the biofilm architecture (Davey et al., 2003; Tremblay et al., 2007). RLPs also play a role in protecting P. aeruginosa from the host defense by inhibiting phagocytosis by macrophages (Alhede et al., 2009; McClure and Schiller, 1996; Van Gennip et al., 2009b) and by lysing polymorphonuclear neutrophils (Jensen et al., 2007; Van Gennip et al., 2009a). At high concentrations, RLPs possess modest intrinsic antimicrobial activity against both Gram-positive and Gram-negative bacteria (Haba et al., 2003; Nitschke et al., 2010; Samadi et al., 2012), as well as ameba (Cosson et al., 2002) and fungi (Goswami et al., 2015). Moreover, it has been shown that RLPs increase the fluidity of fungal membranes (Monnier et al., 2019).
While low concentrations of rhamnolipids (below critical micellar concentration) were shown to enhance the action of aminoglycoside antibiotics (Radlinski et al., 2019), the mode of action of RLPs above the CMC has not been investigated. Importantly, the molecular details of how RLP micelles interact with the bacterial membrane are poorly understood. The amphiphilic nature of free rhamnolipids points to the membrane as their hypothetical site of action. Ortiz et al. showed that di-RLPs intercalate into artificial phosphatidylcholine bilayers and produce structural perturbations, which might affect the function of the membrane (Ortiz et al., 2006). Our data support the notion that the affinity of RLPs micelles for bacterial membranes allows them to interact with the S. aureus cell surface and to release toxic compounds inside the cells. We show that modified fluorescent RLPs cross the peptidoglycan layer and intercalate into the membrane. Notably, RLPs interacted with various Gram-positive pathogens, whose membranes contain a range of diverse phospholipid compounds and molecules (Sohlenkamp and Geiger, 2015), suggesting a “broad-spectrum” activity. Fluorescent pyochelin labeling revealed the RLPs' ability to deliver pyochelin to S. aureus cells, where it generates reactive oxygen species and participates in cell killing (Adler et al., 2012). However, the physical nature of S. aureus membrane and RLP micelle interactions will require further investigations. We further demonstrate the synergistic effect of RLPs with lincosamide antimicrobials, accelerating the killing activity of clindamycin and lincomycin against S. aureus by enhancing their delivery to their intracellular targets. The origin of the apparent selectiveness of RLP micelles for some cargo compounds remains to be elucidated.
The RLPs cargo function described in our study yields novel insights on the pathogenesis of P. aeruginosa infections and the remarkable capacities of this pathogen during interspecies competition. We previously described the selection of ΔwspF mutants of P. aeruginosa during a co-evolution experiment with S. aureus (Tognon et al., 2017). Notably, supernatants of ΔwspF mutants had the ability to kill not only S. aureus but also other Gram-positive as well as Gram-negative bacterial species. The killing activity requires the combined action of four different molecules, namely alkyl quinoline N-oxides (AQNOs), pyoverdine, pyochelin, and rhamnolipids (Gdaniec et al., 2020). Previous studies reported that outer membrane vesicles (OMVs) of P. aeruginosa could fuse to the membranes of other Gram-negative bacteria, as well as to S. aureus cells (Kadurugamuwa and Beveridge, 1995). OMVs incorporate hydrophobic molecules, like AQNOs and the QS signal molecule 3-oxo-C12-HSL (Calfee et al., 2005; Mashburn-Warren et al., 2008a, 2008b). While we confirmed high levels of AQNO molecules in ΔwspF mutant supernatants, the concentrations of 3-oxo-C12-HSL were lower when compared with PA14 wild type supernatants (Gdaniec et al., 2020). Notably, synthesis of AQNOs and 3-oxo-C12-HSL were strongly reduced in a ΔwspFΔrhlA mutant. Thus, in the absence of rhamnolipids, AQNOs and 3-oxo-C12-HSL may remain attached to the P. aeruginosa membrane, leading to reduced release into culture supernatants. However, whether RLP micelles can also deliver AQNOs, phenazines, or QS molecules to target bacteria remains to be determined. Our data suggest that RLPs, provided as a mixture or as single species (C10-C10, C12-C12), exert their cargo effect at concentrations above the CMC, suggesting the importance of micelles formation for the carrier function.
Altogether, our findings support a novel role for RLPs as a remarkable strategy of P. aeruginosa to transport self-produced or environmentally available metabolites to target competitor microorganisms. Our data highlight the potential for enhancing antimicrobial activity via well-designed drug delivery strategies.
Limitations of the study
We focused here on the interaction of RLP micelles with S. aureus and did not investigate other Gram-positive bacteria. Our work was not designed to identify all endogenous molecules, which could be transported by RLP micelles from P. aeruginosa. However, this could be achieved by mass-spectroscopy analyses of purified RLP micelles.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Pseudomonas aeruginosa PA14 | (He et al., 2004) | N/A |
| Pseudomonas aeruginosa PA14 ΔwspF | (Tognon et al., 2017) | N/A |
| Pseudomonas aeruginosa PA14 ΔwspFΔrhlA | (Gdaniec et al., 2020) | N/A |
| Staphylococcus aureusNewman | (Baba et al., 2008) | N/A |
| Staphylococcus aureus COL (MRSA) | A. Renzoni, University of Geneva | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Rhamnolipids 90% | AGAE Technologies | Corvallis, OR 97333, USA |
| Rhamnolipids C10-C10 | GlycoSurf | Salt Lake City, UT 84103, USA |
| Rhamnolipids C12-C12 | GlycoSurf | Salt Lake City, UT 84103, USA |
| Pyochelin | G. Mislin, I. Schalk; University of Strasbourg, France | N/A |
| Lincomycin | Merck-Sigma-Aldrich | Cat#62143 |
| Clindamycin | Chemie Brunschwig | Cat#ACR44351-0100 |
| Vancomycin | Merck-Sigma-Aldrich | Cat#PHR1732 |
| Abberior STAR 580 (RED-NHS) | Merck-Sigma-Aldrich | Cat#38377 |
| Abberior STAR 488 | Merck-Sigma-Aldrich | Cat#61408 |
| Nile Red | Merck-Sigma-Aldrich | Cat#72485 |
| Orcinol | Merck-Sigma-Aldrich | Cat#447420 |
| Adipic acid dihydrazide | Merck-Sigma-Aldrich | Cat#A0638 |
| N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimid-hydrochlorid | Merck-Sigma-Aldrich | Cat#BCBZ8350 |
| 2-(N-morpholino)ethanesulfonic acid | Merck-Sigma-Aldrich | Cat#M3671 |
| Deposited data | ||
| Raw and analyzed data | This paper | Zenodo; https://doi.org/10.5281/zenodo.5159447 |
| Software and algorithms | ||
| Fiji open-source software | https://imagej.net/orgs/loci | https://fiji.sc/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Thilo Köhler (thilo.kohler@unige.ch).
Materials availability
This study did not generate any new biological material.
Experimental model and subject details
Bacterial strains, growth conditions and supernatant preparation
Bacterial strains, plasmids and primers used in this study are listed in the key resources Table. M14 medium was adapted from the literature (Rudin et al., 1974) and is based on M9 salts (Na2HPO4 6 g/L; KH2PO4 3 g/L; NaCl 0.5 g/L; NH4Cl 1 g/L) supplemented with casamino acids (BD™, USA) 10 g/L, magnesium sulfate (MgSO4·7H2O) 1 mM, thiamine (vitamin B1) 2 mg/L, niacin (vitamin B3) 2 mg/L, calcium pantothenate (vitamin B5) 2 mg/L, biotin (vitamin B9) 0.1 mg/L and glucose 2 g/L. Casamino acids, vitamins and glucose solutions were sterilized by filtration and stored separately at 4°C. M9 salts and magnesium sulfate were sterilized by autoclaving at 121°C for 15 min (Tognon et al., 2017). Conditioned medium of bacterial cultures were recovered after 24 h of static growth at 37°C in microtiter plates (TPP, Switzerland). The cultures were pooled and centrifuged at 8′000 rpm for 5 min. Supernatants were sterilized by filtration (0.22 μm filters, Millipore, Switzerland) and stored at −20°C. Medium conditioned in this way is also referred to as crude supernatant. To determine the critical micellar concentration (CMC) of commercial RLPs or in crude supernatants, sterilized supernatants of ΔwspFΔrhlA mutant or M14 medium were supplemented with RLPs (90% purity, AGAE Technologies, USA) to the given concentrations and stained with Nile Red (Sigma Aldrich) at a final concentration of 1 mg/mL for 30 min. Crude supernatants were directly stained with Nile Red (Sigma Aldrich) at a final concentration of 1 mg/mL for 30 min. Next, 200μL of solution was deposited in a 96 well plate in triplicates. Fluorescence of Nile Red (ex 559 nm/em 635nm) was measured in a plate reader (Synergy 1, Bio Tek®, USA). To concentrate the micelles and obtain the RLP pellet, 50 mL of supernatant was stained with Nile Red at a final concentration of 1 mg/mL for 30 min. Samples were centrifuged at 150,000 x g for 4 h at 4°C (Optima XPN, Beckman Coulter ROTOR TYPE 45 Ti). After ultracentrifugation supernatants were collected and the pellets resuspended in 1 mL of M14 medium yielding the 50x concentrated pellet sample, while 15x and 1x concentrated samples were prepared by dilution of 50x sample in M14 medium.
Method details
Orcinol assay for rhamnolipids quantification
Rhamnolipid quantification was performed as described (Wittgens et al., 2011) with slight modifications. Samples containing 50 mL of filtered supernatant before and after ultracentrifugation were treated with equal volumes of ethyl acetate. A standard curve was established by dissolving a mix of mono and di-rhamnolipids (R90-10G, Sigma Aldrich, Switzerland) in 2 mL ddH2O and subsequent extraction with equal volumes of ethyl acetate. After vortexing for 1 min, samples were centrifuged for 1 min at 5000 rpm to separate the phases. The organic solvent was evaporated in a SpeedVac (Thermo Scientific) and residues containing rhamnolipids were resuspended in 100 μL of ddH2O and mixed with an equal volume of 1.6% orcinol (Sigma Aldrich, Switzerland) in ddH2O. Subsequently, 800 μL of 60% sulfuric acid (vol/vol) were added, and samples were incubated for 45 min at 80°C. A 100 μL aliquot of the reaction mix was transferred to a microtitre plate, and OD420 was measured in a Synergy H1 Multi-Mode plate reader (BioTek®).
Transmission electron microscopy
Supernatant samples and RLP suspensions were loaded onto 2 mm single slot copper grids (Electron Microscopy Sciences) coated with 1% Pioloform plastic support film. Grids were dried for 30 min and then stained with a 1% aqueous uranyl acetate solution for 3 min and examined using a Tecnai 20 TEM (FEI) electron microscope operating at an acceleration voltage of 80 kV and equipped with a side-mounted MegaView III CCD camera (Olympus Soft-Imaging Systems) controlled by iTEM acquisition software (Olympus Soft-Imaging Systems). Grids for EM were prepared twice independently and multiple pictures were acquired.
Killing assays and kinetics
Killing assays were performed on S. aureus cells grown for 6 h in M14 medium under static growth conditions in microtiter plates as described (Tognon et al., 2017). After the 6 h incubation, 100 μL of S. aureus culture was removed and replaced with either 100 μL M14 medium or crude or fractionated P. aeruginosa supernatant. Growth (OD600) was monitored in a plate reader (BioTek®, USA) for 24 h. At the end of the killing assay, viable plate counts were performed to determine S. aureus survival. For killing kinetics, samples were taken at different time intervals during growth and surviving S. aureus cells determined by viable plate counts. Other bacterial strains were grown on LB-plates or specific growth media. Cells were scraped from the plate and a suspension was prepared and adjusted to obtain 108 CFU /mL. After 24 h incubation in the presence of culture supernatants, surviving cells were determined by plate counts.
Minimal inhibitory concentration determinations
To determine antibiotic susceptibility, minimal inhibitory concentrations were performed in triplicates by two-fold serial dilutions in Müller-Hinton broth (Becton Dickinson, Franklin Lakes, NJ, USA) according to Clinical and Laboratory Standard Institute (CLSI) guidelines.
Fractional inhibitory concentration (FIC)
For all of the wells of the 96wells plates that corresponded to an MIC, the sum of the FICs (ΣFIC) was calculated for each well with the equation ΣFIC = FICA + FICB = (CA/MICA) + (CB/MICB), where MICA and MICB are the MICs of drugs A and B alone, respectively, and CA and CB are the concentrations of the drugs in combination, respectively, in all of the wells corresponding to an MIC (isoeffective combinations) (Hall et al., 1983).
Covalent labeling of rhamnolipids, pyochelin and vancomycin with Abberior STARNHS ester dye
A stock solution of rhamnolipids (RLP) (90% purity, AGAE Technologies, USA) was prepared at 100 mg/mL in DMSO. When necessary, sonication was performed until complete dissolution of the RLP aggregates. When required, the stock solution was further diluted in DMSO. Then sequentially 800 μL of a solution of adipic acid dihydrazide (AAD, Sigma Aldrich, A0638, Switzerland) at 31.25 mg/mL in ddH2O and 50 μL of Abberior STAR 488 or 580 NHS-ester (depending on the experiment, respectively Sigma Aldrich, 61,048 and 38,377, Switzerland) at a concentration of 2 mg/mL in DMSO were added. After 1 h incubation at room temperature under constant agitation (vortex on the lowest speed), 200 μL of a solution of N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, Sigma Aldrich, E1769, Switzerland) at 6 mg/mL (final concentration of EDC at 0.8 mg/mL) in MES buffer 0.5 M (pH 5.5) was added. Reaction was incubated for 20 min at room temperature under constant agitation.
Finally, RLP extraction was performed with ethyl acetate as described previously with slight modifications (Loiseau et al., 2018). Briefly, 2 mL of ethyl acetate (Sigma Aldrich, 270,989, Switzerland) was added to the solution and vortexed for 30 s. After complete separation, the upper phase containing the RLP was collected and 20 μL of DMSO was added before drying for at least 2 h in a SpeedVac Vacuum (Thermofisher, Germany). RLP were suspended in the 100 μL of DI and sonication was performed until complete dissolution. Pyochelin covalent modification with Abberior STAR RED-NHS ester dye was performed as follow: 16 μg of pyochelin were incubated with 5 mg of AAD and 40 μg of Abberior STAR red-NHS ester in 200 μL of ddH2O for 1 h at room temperature. Then, 300 μg of EDC solubilized in MES buffer (0.5 M, pH 5.5) were added to the reactive mix and incubated for 20 min. Pyochelin was then extracted as previously described (Hoegy et al., 2014). Briefly, adjustment to pH 3 was performed by adding 5 mg of citric acid monohydrate (Sigma Aldrich, G1909, Switzerland) before pyochelin extraction using 1 mL of dichloromethane. After vortexing and complete phase separation, the lower phase was collected. A second extraction was carried out when necessary. Extracted pyochelin was dried for at least 2 h under nitrogen flow. Labeling of vancomycin was performed by resuspending 0.2 mg of vancomycin in 500 μL of ddH2O, addition of 0.1 mg of Abberior STAR 488 NHS-ester and incubation at room temperature for 2 h.
Confocal imaging and pyochelin RLP-assisted delivery
Inactivation of the Abberior STAR RED-NHS dye was performed to prevent any reaction with cell surface compounds. For this, Abberior STAR RED-NHS ester dye was incubated with NaOH 0.1 M for 10 min prior incubation with or without RLP. S. aureus cells were incubated in absence or presence of RLPs at different concentration 25 μg/mL (below the CMC) or 250 μg/mL (above the CMC) either with labeled pyochelin (12 ng/condition) or with inactivated dye (20 ng/conditions) in 200 μL of PBS, for 10 min at room temperature. Then cells were centrifuged and washed with 200 μL of PBS. After centrifugation, cells were incubated with 0.01 μg of labeled vancomycin in 200μL of PBS for 5 min. Cells were centrifuged and washed again in 200 μL of PBS. 1 μL of the cell suspension was spotted onto a microscopy glass slide with 1 μL of mounting buffer. Coverslips were sealed using nail polish. A Zeiss confocal microscope LSM800 was used for all observations.
STED nanoscopy
2D-STED dual color imaging was performed with a Leica TCS SP8 STED 3X microscope in a thermostatic chamber at 21°C and equipped with a STED motorized glycerol immersion objective (HC PL Apo 93×/N.A. 1.30 motCOR). Fluorescently-labelled samples were mounted in Prolong Antifade Gold (Thermofisher Scientific) between a coverslip (0.170 ± 0.01 mm thick, Hecht-Assistent) sealed on a microscope slide with nail polish. Excitation was performed with a White Light Laser (WLL), depletion with either a continuous 592 nm laser (STED 592) or a 775 nm pulsed laser (STED 775). Excitation and depletion lasers were calibrated with the STED Expert Alignment Mode and Abberior gold nanoparticles (80 nm in diameter) before starting each imaging session, or with the STED Auto Beam Alignment tool during imaging sessions (Leica LAS X software, Leica Microsystems CMS GmbH). 2D-STED dual color was made using an excitation at 559 nm (WLL) and a STED 775 depletion laser line for Nile Red, followed by an excitation at 488 nm (WLL) and a STED 592 depletion laser line for Abberior STAR 488. Detection signals were collected from 650 nm to 740 nm for Nile Red and from 500 nm to 564 nm for Abberior STAR 488 using highly sensitive Leica Hybrid Detectors (HyDs) with a fixed gain and offset (100 mV and 0, respectively). Time-gated detection was used for Nile Red (0.50–5.92 ns). Acquisitions were performed sequentially with a line average of 4, a speed of 400 Hz and an optimized pixel size (20 nm). Images were deconvolved using the Leica Lightning Mode (LAS X software) and analyzed with Fiji open-source software (https://fiji.sc/).
Quantification and statistical analysis
All statistical analysis was performed with GraphPad Prism version 8.0 and R package. To compare killing activities of crude-S, ultra-S and ultra-P fractions of PA14 wild type and mutants, we extracted the slope in a log-diagram up to 3 h by linear regression. Saturation due to detection limits did not allow evaluation after 3 h. The data consisted of three replica per condition (Figure S1A). All triplets showed similar variance (Bartlett test failed to reject the hypothesis of identical variance in all groups of three killing curves, p = 0.17), therefore, unpaired t-tests assuming equal sample variance were used for individual comparisons (Figure S1). To compare the rescue efficiency of different rhamnolipid supplements (Figure S1E), rescue efficiency was calculated as the enhancement of killing activity over the corresponding unsupplemented controls. Given the presence of both supernatant and pellet fractions, ANOVA was then performed, followed by Tukey's test for honest significant differences to analyze the effect of different RLP supplementations. Experiments were performed with three biological replicates from at least two independent experiments when possible. Statistical significance is reported in Figure Legends, and data are presented as mean +/- SD as indicated.
Acknowledgments
We are grateful to I. Schalk and G. Mislin (CNRS, University of Strasbourg, France) for providing pyochelin. We thank A. Renzoni (University of Geneva) for providing S. aureus strains and for helpful discussions. We are grateful to B. Maco (University of Geneva) for help with electron microscopy studies. Authors are grateful for help with image acquisition provided by the Bioimaging Core Facility, Faculty of Medicine, University of Geneva. This study was funded by grantNo. 32473B-179289 from the Swiss National Science Foundation to C.v.D. and supported by a grant to B.G.G. from the Bertarelli Foundation (Geneva, Switzerland), as well as SNSF grant PP00P2_163684 and PP00P2_194813 to T.B.
Authorcontributions
Conceptualization and methodology, B.G.G., F.B., T.K., T.B., C.v.D., and F.P.; Investigation, B.G.G., F.B., and F.P.; Writing, B.G.G., F.B., T.K., C.v.D., and T.B.; Visualization, B.G.G. and F.B.; Funding Acquisition, B.G.G, C.v.D., and T.B.; Supervision, T.K., T.B., and C.v.D.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103669.
Contributor Information
Thomas Braschler, Email: thomas.braschler@unige.ch.
Thilo Köhler, Email: thilo.kohler@unige.ch.
Supplemental information
Data and code availability
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
All original code and raw data supporting the conclusions of this study have been deposited at Zenodo and are publicly available from the lead contact upon request (https://doi.org/10.5281/zenodo.5159447).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- Abdel-Mawgoud A.M., Lepine F., Deziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol.Biotechnol. 2010;86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler C., Corbalan N.S., Seyedsayamdost M.R., Pomares M.F., de Cristobal R.E., Clardy J., Kolter R., Vincent P.A. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS One. 2012;7:e46754. doi: 10.1371/journal.pone.0046754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tahhan R.A., Sandrin T.R., Bodour A.A., Maier R.M. Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl. Environ. Microbiol. 2000;66:3262–3268. doi: 10.1128/aem.66.8.3262-3268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhede M., Bjarnsholt T., Jensen P.O., Phipps R.K., Moser C., Christophersen L., Christensen L.D., van Gennip M., Parsek M., Hoiby N., et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- Arima K., Kakinuma A., Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem.Biophys. Res. Commun. 1968;31:488–494. doi: 10.1016/0006-291x(68)90503-2. [DOI] [PubMed] [Google Scholar]
- Atanaskovic I., Mosbahi K., Sharp C., Housden N.G., Kaminska R., Walker D., Kleanthous C. Targeted killing of Pseudomonas aeruginosa by pyocin G occurs via the hemin transporter hur. J. Mol. Biol. 2020;432:3869–3880. doi: 10.1016/j.jmb.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Bae T., Schneewind O., Takeuchi F., Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee M.W., Shelton J.G., McCubrey J.A., Pesci E.C. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 2005;73:878–882. doi: 10.1128/IAI.73.2.878-882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Xie S., Wei J., Song X., Ding Z., Li X. Antibacterial micelles with vancomycin-mediated targeting and pH/Lipase-Triggered release of antibiotics. ACS Appl. Mater. Inter. 2018;10:36814–36823. doi: 10.1021/acsami.8b16092. [DOI] [PubMed] [Google Scholar]
- Chrzanowski L., Lawniczak L., Czaczyk K. Why do microorganisms produce rhamnolipids? World J. Microbiol.Biotechnol. 2012;28:401–419. doi: 10.1007/s11274-011-0854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P., Zulianello L., Join-Lambert O., Faurisson F., Gebbie L., Benghezal M., Van Delden C., Curty L.K., Köhler T. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 2002;184:3027–3033. doi: 10.1128/jb.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.E., Caiazza N.C., O'Toole G.A. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003;185:1027–1036. doi: 10.1128/jb.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E., Lépine F., Dennie D., Boismenu D., Mamer O.A., Villemur R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta. 1999;1440:244–252. doi: 10.1016/s1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- Gdaniec B.G., Allard P.M., Queiroz E.F., Wolfender J.L., van Delden C., Köhler T. Surface sensing triggers a broad-spectrum antimicrobial response in Pseudomonas aeruginosa. Environ.Microbiol. 2020;22:3572–3587. doi: 10.1111/1462-2920.15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D., Mavridou D.A.I. Making the best of aggression: the many dimensions of bacterial toxin regulation. Trends Microbiol. 2019;27:897–905. doi: 10.1016/j.tim.2019.05.009. [DOI] [PubMed] [Google Scholar]
- Goswami D., Borah S.N., Lahkar J., Handique P.J., Deka S. Antifungal properties of rhamnolipid produced by Pseudomonas aeruginosa DS9 against Colletotrichum falcatum. J. Basic Microbiol. 2015;55:1265–1274. doi: 10.1002/jobm.201500220. [DOI] [PubMed] [Google Scholar]
- Greenspan P., Mayer E.P., Fowler S.D. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haba E., Pinazo A., Jauregui O., Espuny M.J., Infante M.R., Manresa A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol.Bioeng. 2003;81:316–322. doi: 10.1002/bit.10474. [DOI] [PubMed] [Google Scholar]
- Haba E., Pinazo A., Pons R., Perez L., Manresa A. Complex rhamnolipid mixture characterization and its influence on DPPC bilayer organization. Biochim.Biophys. Acta. 2014;1838:776–783. doi: 10.1016/j.bbamem.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Hall M.J., Middleton R.F., Westmacott D. The fractional inhibitory concentration (FIC) index as a measure of synergy. J. Antimicrob. Chemother. 1983;11:427–433. doi: 10.1093/jac/11.5.427. [DOI] [PubMed] [Google Scholar]
- Hauser G., Karnovsky M.L. Rhamnose and rhamnolipide biosynthesis by Pseudomonas aeruginosa. J. Biol. Chem. 1957;224:91–105. [PubMed] [Google Scholar]
- He J., Baldini R.L., Deziel E., Saucier M., Zhang Q., Liberati N.T., Lee D., Urbach J., Goodman H.M., Rahme L.G. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegy F., Mislin G.L., Schalk I.J. Pyoverdine and pyochelin measurements. Methods Mol. Biol. 2014;1149:293–301. doi: 10.1007/978-1-4939-0473-0_24. [DOI] [PubMed] [Google Scholar]
- Jensen P.O., Bjarnshold T., Phipps R., Rasmussen T.B., Calum H., Christoffersen L., Moser C., Williams P., Pressler T., Givskov M., Høiby N. Rapid necrotic killing of polymorphunuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153:1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa J.L., Beveridge T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T., Curty L.K., Barja F., van Delden C., Pechère J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau C., Portier E., Corre M.-H., Schlusselhuber M., Depayras S., Berjeaud J.-M., Verdon J. Highlighting the potency of biosurfactants produced by Pseudomonas strains as anti-Legionella agents. Biomed.Res. Int. 2018;2018:8194368. doi: 10.1155/2018/8194368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L., Howe J., Garidel P., Richter W., Steiniger F., Roessle M., Brandenburg K., Whiteley M. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol.Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L., McLean R.J., Whiteley M. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology. 2008;6:214–219. doi: 10.1111/j.1472-4669.2008.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure C.D., Schiller N.L. Inhibition of macrophage phagocytosis by Pseudomonas aeruginosa rhamnolipids in vitro and in vivo. Curr.Microbiol. 1996;33:109–117. doi: 10.1007/s002849900084. [DOI] [PubMed] [Google Scholar]
- Monnier N., Furlan A.L., Buchoux S., Deleu M., Dauchez M., Rippa S., Sarazin C. Exploring the dual interaction of natural rhamnolipids with plant and fungal biomimetic plasma membranes through biophysical studies. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke M., Costa S.G., Contiero J. Structure and applications of a rhamnolipid surfactant produced in soybean oil waste. Appl. Biochem. Biotechnol. 2010;160:2066–2074. doi: 10.1007/s12010-009-8707-8. [DOI] [PubMed] [Google Scholar]
- Noordman W.H., Janssen D.B. Rhamnolipid stimulates uptake of hydrophobic compounds by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2002;68:4502–4508. doi: 10.1128/aem.68.9.4502-4508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A., Teruel J.A., Espuny M.J., Marqués A., Manresa Á., Aranda F.J. Effects of dirhamnolipid on the structural properties of phosphatidylcholine membranes. Int. J. Pharm. 2006;325:99–107. doi: 10.1016/j.ijpharm.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Radlinski L.C., Rowe S.E., Brzozowski R., Wilkinson A.D., Huang R., Eswara P., Conlon B.P. Chemical induction of aminoglycoside uptake overcomes antibiotic Tolerance and resistance in Staphylococcus aureus. Cell Chem. Biol. 2019;26:1355–1364 e1354. doi: 10.1016/j.chembiol.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin L., Sjostrom J.E., Lindberg M., Philipson L. Factors affecting competence for transformation in Staphylococcus aureus. J. Bacteriol. 1974;118:155–164. doi: 10.1128/jb.118.1.155-164.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadi N., Abadian N., Ahmadkhaniha R., Amini F., Dalili D., Rastkari N., Safaripour E., Mohseni F.A. Structural characterization and surface activities of biogenic rhamnolipid surfactants from Pseudomonas aeruginosa isolate MN1 and synergistic effects against methicillin-resistant Staphylococcus aureus. Folia Microbiol. 2012;57:501–508. doi: 10.1007/s12223-012-0164-z. [DOI] [PubMed] [Google Scholar]
- Schalk I.J. Siderophore–antibiotic conjugates: exploiting iron uptake to deliver drugs into bacteria. Clin.Microbiol. Infect. 2018;24:801–802. doi: 10.1016/j.cmi.2018.03.037. [DOI] [PubMed] [Google Scholar]
- Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O., Drummen G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019;21:72–84. doi: 10.1038/s41556-018-0251-8. [DOI] [PubMed] [Google Scholar]
- Sekhon Randhawa K.K., Rahman P.K.S.M. Rhamnolipid biosurfactants—past, present, and future scenario of global market. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C., Geiger O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 2015;40:133–159. doi: 10.1093/femsre/fuv008. [DOI] [PubMed] [Google Scholar]
- Sotirova A., Kronevska M., Vasileva-Tonkova E., Galabova D. Effects of rhamnolipid-biosurfactant on cell surface of Pseudomonas aeruginosa. Microbiol. Res. 2009;164:297–303. doi: 10.1016/j.micres.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Tognon M., Köhler T., Gdaniec B.G., Hao Y., Lam J.S., Beaume M., Luscher A., Buckling A., van Delden C. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 2017;11:2233–2243. doi: 10.1038/ismej.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J., Richardson A.-P., Lépine F., Déziel E. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 2007;9:2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- Van Gennip M., Christensen L.D., Alhede M., Phipps R., Jensen P.O., Christophersen L., Pamp S.J., Moser C., Mikkelsen P.J., Koh A.Y., et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117:537–546. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gennip M., Christensen L.D., Alhede M., Phipps R., Jensen P.O., Christophersen L., Pamp S.J., Moser C., Mikkelsen P.J., Koh A.Y., et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117:537–546. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanakunakorn C. Mode of action and in-vitro activity of vancomycin. J. Antimicrob.Chemother. 1984;14:7–18. doi: 10.1093/jac/14.suppl_d.7. [DOI] [PubMed] [Google Scholar]
- Wittgens A., Tiso T., Arndt T.T., Wenk P., Hemmerich J., Muller C., Wichmann R., Kupper B., Zwick M., Wilhelm S., et al. Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb.Cell Fact. 2011;10:80. doi: 10.1186/1475-2859-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.E., Howard S.A., Förster A., Nolan L.M., Manoli E., Bullen N.P., Yau H.C.L., Hachani A., Hayward R.D., Whitney J.C., et al. The Pseudomonas aeruginosa T6SS delivers a periplasmic toxin that disrupts bacterial cell morphology. Cell Rep. 2019;29:187–201.e187. doi: 10.1016/j.celrep.2019.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Liu Y., Liu Z., Jiang Y., Tan F., Zeng G., Yuan X., Yan M., Niu Q., Liang Y. Degradation of pseudo-solubilized and mass hexadecane by a Pseudomonas aeruginosa with treatment of rhamnolipid biosurfactant. Int. Biodeterior. Biodegradation. 2014;94:152–159. doi: 10.1016/j.ibiod.2014.07.012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
All original code and raw data supporting the conclusions of this study have been deposited at Zenodo and are publicly available from the lead contact upon request (https://doi.org/10.5281/zenodo.5159447).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request