Abstract
Rapid and reliable detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody can provide immunological evidence in addition to nucleic acid test for the early diagnosis and on-site screening of coronavirus disease 2019 (COVID-19). All-solid-state biosensor capable of rapid, quantitative SARS-CoV-2 antibody testing is still lacking. Herein, we propose an electronic labelling strategy of protein molecules and demonstrate SARS-CoV-2 protein biosensor employing colloidal quantum dots (CQDs)-modified electrode. The feature current peak corresponding to the specific binding reaction of SARS-CoV-2 antigen and antibody proteins was observed for the first time. The unique charging and discharging effect depending on the alternating voltage applied was ascribed to the quantum confinement, Coulomb blockade and quantum tunneling effects of quantum dots. CQDs-modified electrode could recognize the specific binding reaction between antigen and antibody and then transduce it into significant electrical current. In the case of serum specimens from COVID-19 patient samples, the all-solid-state protein biosensor provides quantitative analysis of SARS-CoV-2 antibody with correlation coefficient of 93.8% compared to enzyme-linked immunosorbent assay (ELISA) results. It discriminates patient and normal samples with accuracy of about 90%. The results could be read within 1 min by handheld testing system prototype. The sensitive and specific protein biosensor combines the advantages of rapidity, accuracy, and convenience, facilitating the implement of low-cost, high-throughput immunological diagnostic technique for clinical lab, point-of-care testing (POCT) as well as home-use test.
Keywords: Colloidal quantum dots, Ligand exchange, Protein biosensor, SARS-CoV-2, Electronic labelling
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a worldwide health threat (Wu et al., 2020). Globally, as of 4:14 p.m. CET, 29 December 2021, there have been 281,808,270 confirmed cases of COVID-19, including 5,411,759 deaths, according to World Health Organization (WHO Coronavirus (World Health Organization WHO, 2021). The strong infectiousness and mutability of SARS-CoV-2, combined with the presence of asymptomatic carriers have brought great difficulties to epidemic prevention and control (Hao et al., 2020; Zou et al., 2020). There is an urgent demand for rapid, accurate, and convenient testing of SARS-CoV-2 to promote early diagnosis and long-term surveillance of COVID-19 (Guan et al., 2020; Kevadiya et al., 2021; Yin et al., 2020; Sethuraman et al., 2020). In addition to nucleic acid test, testing of SARS-CoV-2 antigen and antibody can provide immunological evidence for the clinical diagnosis and on-site screening of COVID-19. When the virus is replicated, the detectable antigens are expressed and thus can be used to confirm acute or early infection. The presence of targeting immunoglobulins that are produced in response to SARS-CoV-2 during the onset of disease has been demonstrated (Chua et al., 2020; Huang et al., 2019). The level of IgM antibody increase during the first week after the infection, peaks after about 10 days and then usually falls back to near background level. IgG antibody could be detected after 1 week and is maintained at a relatively high level for a long period. Hence IgM would potentially be as a reference for early diagnosis. SARS-CoV-2 antibody test remains an effective tool for surveillance and can provide scientific evidence for the epidemiological investigation and vaccination evaluation (Wang et al., 2021; Bryan et al., 2020; Seo et al., 2020; Amanat et al., 2020).
Among the immunological techniques for SARS-CoV-2 antibody testing (Table S1), the enzyme-linked immunosorbent assay (ELISA) and chemiluminescence (CL) method generally take several hours or more because of multiple sample processing steps carried out in laboratory (Bampoe et al., 2020; Lequin, 2005; Li et al., 2020; Grossberg et al., 2021). For on-site use without requiring sophisticated sample preparation, the colloidal gold immunoassay (CGIA) technique takes advantage of the high electron density of gold particles to achieve immunolabeling and tracer for the electrostatically adsorbed proteins. CGIA can give qualitative results within ten to 15 min (min) via the visual observation of the test Kit (Margolin et al., 2020; Jiang et al., 2011).
Biosensors are featured by their molecule recognition and signal transduction capabilities which convert the biochemical information into optical or electrical signals (Elledge et al., 2021; Chen et al., 2020; Huang et al., 2021; Winkler et al., 1982). The biosensors based on spectroscopy techniques such as fluorescence, surface plasmon resonance (SPR), and surface enhanced Raman scattering (SERS) have been widely developed for the rapid and specific testing of SARS-CoV-2 (Yadav et al., 2021; Cheong et al., 2020; Zhang et al., 2021). The above techniques generally require multiple preparation and processing steps, and the support of sophisticated instruments or apparatus, limiting the on-site use of humoral testing. The demand for ubiquitous and reliable detection of SARS-CoV-2, one of the largest RNA viruses spurring the design of materials that exhibit engineered sensing properties and that can enable new fabrication methods for high-performance biosensors (Yin et al., 2020; Sadique et al., 2021a, Sadique et al., 2021b). All-solid-state SARS-CoV-2 electrochemical biosensors that uses solid electrodes (transducer) and solid sensing element (sensing film) typically based on chemically modified electrodes (CME) have been successfully developed for SARS-CoV-2 protein detection in Table S2 (Seo et al., 2020; Ali et al., 2021; Beduk et al., 2021; Hashemi et al., 2021; Yakoh et al., 2021; Torres et al., 2021; Yousefi et al., 2021; Zhou et al., 2021). Unlike glucose test, it is difficult to measure the long-range charge transfer and directional electron transport between proteins in which the corresponding reactive sites are surrounded by polypeptide of the biological macromolecules (Winkler et al., 1982). The electrochemical biosensors based on biomolecule-modified electrodes or molecularly imprinted polymers have been exploited for the detection of SARS-CoV-2 (Singhal et al., 2022; Yousefi et al., 2021). For instance, Yousefi et al. (2021) proposed an electrochemical sensor based on an analyte-recognizing antibody attached to negatively-charged DNA linker, in which the ferrocene redox probe was attached to the DNA linker through the cross-linker of NHS Ester. Zhou et al. (2021) developed an electrochemical resonance assay using the peptides with a terminal thiol group to tether on gold electrode surface. The electrochemical point-of-care (POC) immunosensors have potential to open broad prospect in biosensor applications for the detection of SARS-CoV-2 biomarkers (Ranjan et al., 2021). And they could be easily integrated with microfluidics, IoT system, and intelligent terminal, which could detect the multiple biomarkers simultaneously and can improve the portability (Sadique et al., 2021a, Sadique et al., 2021b).
In general, efficient enrichment and strong adhesion of proteins on the electrode favors the sensitive and specific protein detection. The construction of these electrochemical biosensors have relied on the organic cross-linkers, such as PBASE, EDC: NHS (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, N-Hydroxy-Succinimide) or glutaraldehyde. The modified electrodes need to be activated by the cross-linkers for a certain period (1–24 h) to allow for subsequent binding with biomolecules (Table S2). The organic cross-linkers as the medium-assisted labelling may set a barrier for the charge transfer across the solid-fluid interface. The use of cross-linkers also increases the complexity and the treatment time of the manufacturing process (Seo et al., 2020; Beduk et al., 2021; Hashemi et al., 2021; Yakoh et al., 2021; Torres et al., 2021). For traditional fluorescence labelling, the foreign crosslinkers have to be used as the medium for the anchoring of biomolecules, and it generally take hours or even days because of multi-step crosslinking processes in liquid phase atmosphere.
For practical application, the signal-to-noise ratio (SNR) must be high enough to achieve sensitive and specific detection with serum specimens. Owing to the large specific surface area and high carrier mobility, graphene has been used in all-solid-sate biosensors to enhance the enrichment and immobilization of SARS-CoV-2 proteins (Beduk et al., 2021; Hashemi et al., 2021; Yakoh et al., 2021). For instance, Ali et al. (2021) proposed the biosensing platform created by 3D nanoprinting of three-dimensional electrodes coated with reduced-graphene-oxide (rGO) nanoflakes for SARS-CoV-2 antibody testing. The coupling chemistry of EDC: NHS facilitated the formation of C–N covalent bonding between rGO and antigens via an amidation reaction. The device was able to detect SARS-CoV-2 antibodies to spike S1 antigen and RBD antigen in recombinant protein samples with limit-of-detection (LOD) of 2.8 × 10−15 and 16.9 × 10−15 M, respectively. Seo et al. (2020) reported a field-effect transistor (FET)-based biosensor in which a specific antibody against SARS-CoV-2 spike protein was conjugated to a graphene sheet through the cross-linker of 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE), as the sensing area. The sensor was able to detect SARS-CoV-2 virus in clinical samples rapidly without any preparation or preprocessing.
Herein, we hypothesized that colloidal quantum dots (CQDs), which are semiconductor nanocrystals with a typical diameter of 2- to 20- nanometers, may offer the receptor-and-transducer functional properties to define an electronic labelling of protein molecules. CQDs possess abundant active sites owing to their extremely large surface area with dangling bonds, making them intrinsically ideal receptors for foreign ligands of proteins without the treatment of activation, which may enable one-step connect between CQDs modification layer and biomolecules without assisted medium (Kagan et al., 2016; Liu et al., 2014, 2015; Choi et al., 2016). The surface ligands surrounding CQDs are exchangeable, which potentially enables their sufficient adsorption, direct binding, and stable presence on protein surface (Boles et al., 2016). Attractively, CQDs possess unique quantum effect such as quantum confinement, Coulomb blockade effect and quantum tunneling, which may modulate the charge transfer and carrier transport across the CQD-protein interfaces under external electric fields (Arquer et al., 2021; Liu et al., 2021). Moreover, CQDs can be solution-processed at room temperature onto a wide variety of substrates, so that their materials properties are well preserved in real devices. QDs fluorescence biosensing is mainly based on the principle that the change of optical properties of QDs such as fluorescence intensity, fluorescence spectral position and shape, excited state lifetime or fluorescence polarization due to the interaction between the fluorescent material and the analyte. The antigen-antibody specific binding reaction could be converted into optical signal by the fluorescence quenching process of donor-acceptor electron transfer or energy transfer.
In the work reported here, we have demonstrated an all-solid-state SARS-CoV-2 protein biosensor employing lead sulfide (PbS) CQDs-modified electrode. We utilize the surface ligand exchangeability of CQDs to construct the electronic labelling of SARS-CoV-2 proteins on Au work electrode, through which the specific binding of SARS-CoV-2 antigen and antibody is transduced into significant current signals. The all-solid-state SARS-CoV-2 protein biosensor discriminates patient and normal samples with an accuracy of about 90%. The results could be read within 1 min by a handheld testing system prototype. We studied the mechanism that enable these remarkable protein biosensors. We found that the direct binding of CQD to protein surface realized by ligand exchange treatment, and the unique charging and discharging effect of semiconductor nanocrystals in quantum regime depending on the alternating voltage applied were the key parameters that accounts for the rapid, sensitive, and portable SARS-CoV-2 specific protein testing.
2. Experimental
All the experimental details including ethics statement, materials, the synthesis process of PbS CQDs, the calculations of the size diameter of PbS CQDs, characterizations, electrochemical characterization, the extraction of response signal, the fitting of limit-of-detection, the optimization of blocking time, and the reproducibility of CQDs-modified electrode, the preparation and testing of serum samples, the operating process of handheld testing system prototype are in Supporting Information.
3. Results and discussion
3.1. Fabrication and characterization of CQDs-modified electrode
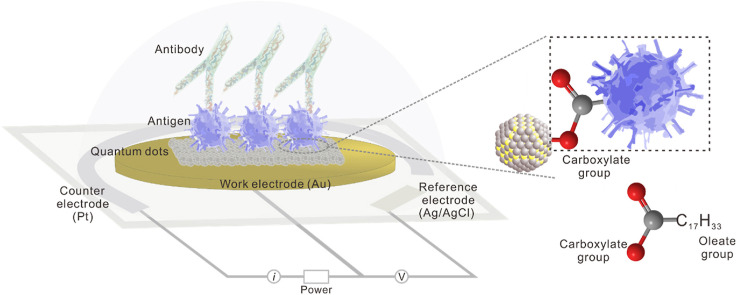
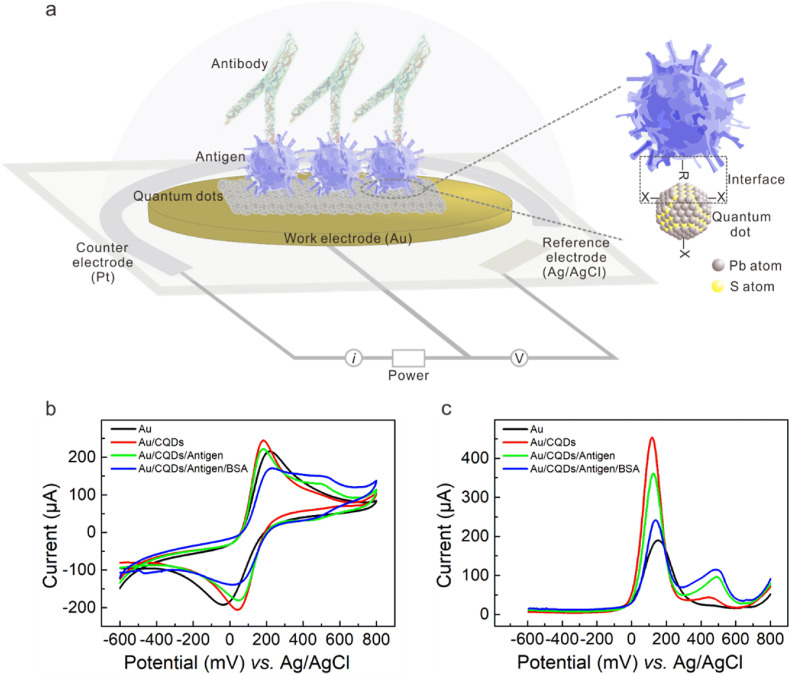
The all-solid-state SARS-CoV-2 protein biosensor was based on the CQDs-modified electrode schematically shown in Fig. 1 a. The dimension of the whole electrochemical planar three-electrode is 35 × 12.5 × 0.5 mm3 (L × W × D), in which the diameter size of Au work electrode (WE) is 6.5 mm (Fig. S1). Pt and Ag/AgCl are respectively used as the counter electrode (CE) and reference electrode (RE). PbS CQDs synthesized using organo-hot injection method were spin-coated onto the WE surface, forming a thin film of CQD solids as the chemically modification layer via Au–S bond (Tang et al., 2010). For the testing of SARS-CoV-2 antibody, the sensitivity and specificity of antigen protein are the premise. By comparing the affinity of various commercial SARS-CoV-2 antigen products via ELISA method (Fig. S2), we selected SARS-CoV-2 S-RBD protein from China AtaGenix Laboratories Co., Ltd., which exhibited optimal affinity for antibody binding. SARS-CoV-2 S-RBD protein is located on the spike protein. Its main function is to recognize receptors on the surface of host cell and mediate fusion with host cells. For the construction of electronic labelling of proteins, the sufficient adsorption and stable presence of CQDs with antigen protein is essential. PbS CQDs, antigen, BSA were successively coated onto the surface of WE to fabricate CQDs-modified electrode (details in Supporting Information).
Fig. 1.
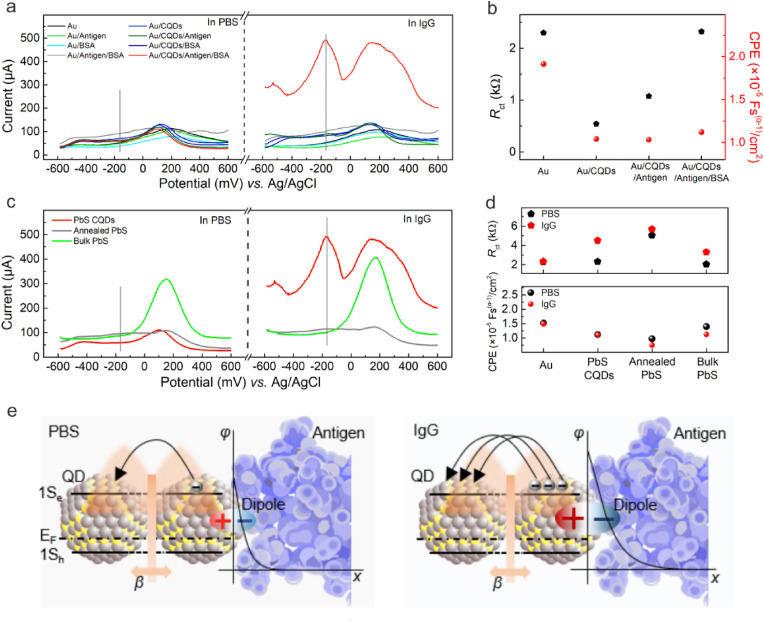
Fabrication and electrochemical characterization of the colloidal quantum dots-modified electrode as the all-solid-state SARS-CoV-2 protein biosensor. (a) Schematic diagram of CQDs-modified electrode as SARS-CoV-2 protein biosensor. The planar electrochemical three-electrode system consists of BT (Bismaleimide Triazine) resin substrate, Au work electrode, Pt counter electrode, and Ag/AgCl reference electrode. Lead sulfide colloidal quantum dots (PbS CQDs) were utilized as modified materials. (b) CV and (c) DPV characterizations of bare gold electrode (Au), Au/CQDs, Au/CQDs/Antigen, and Au/CQDs/Antigen/BSA in the presence of 1 × PBS containing 5 mM [Fe(CN)6]3−/4− and 0.5 M KCl as REDOX mediator. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We conducted cyclic voltammetry (CV) measurement to examine the basic electrochemical characteristics of the modified electrodes. For comparison, unmodified electrode (Au), pristine CQDs-modified electrode (Au/CQDs), antigen-coated CQDs-modified electrode before (Au/CQDs/Antigen) and after BSA-blocking treatment (Au/CQDs/Antigen/BSA) were prepared and measured in the presence of 1 × PBS (phosphate buffer) containing 5 mM [Fe(CN)6]3−/4− and 0.5 M KCl as REDOX mediator. PBS is one of the most widely used buffer in biochemistry, in which phosphate ions could adjust salt balance and construct suitable environment for antigen-antibody reaction. As shown in Fig. 1b, all these electrodes (Au, Au/CQDs, Au/CQDs/Antigen, Au/CQDs/Antigen/BSA) exhibited obvious REDOX peaks, indicating the electrochemical planar three-electrode could convert the charge transfer at the solid-liquid interface into current output. We also observed that the modification layer had influence on the current intensity of these electrodes, suggestive of the difference in the charge transfer capability of the modification layer. Compared to the peak current (215.3 μA) of the unmodified Au electrode, the Au/CQDs sample has higher peak current (244.5 μA), suggestive of the satisfactory charge transfer capability of the CQDs modification layer. The peak current decreased to 221.8 μA and 170.4 μA in the case of Au/CQDs/Antigen and Au/CQDs/Antigen/BSA sample, respectively. The electrochemical response of [Fe(CN)6]3−/4− is still very well, suggestive of the practicable function of biomolecules recognition and signal transduction of the CQDs-modified electrode. The result suggested that the network structure of protein molecules could not completely block the transport and reaction of Fe2+/3+ ions in the electrolyte (Cui et al., 2020). Although the successive coating with Antigen and BSA affected the conduction capability, the electrode is still capable of transducing the charge transfer across the solid-liquid across into appreciable current. With the successive coating of SARS-CoV-2 S-RBD protein and BSA protein, the conduction capabilities of the electrode became worse. The results suggested the stable adhesion of CQDs to proteins since the conduction capability of biomacromolecules is weaker compared to the inorganic thin film based on CQD solids.
For electrochemical measurement, the voltage conditions applied have important influence on the output current signal, which could be adjusted by designing measure parameters including scan rate, pulse width and pulse period. Differential pulse voltammetry (DPV) mode is usually used for the testing of targets with lower concentration, and provide improved selectivity for observing different REDOX processes compared with CV. We continued to compare the above samples under DPV mode (Fig. 1c) and found that the trend in peak current was in good agreement with CV measurement result. The CQD modification layer helps to increase the charge transfer capability of the Au electrode, the peak current increased from 189.6 μA to 453.5 μA; the peak current decreased down to 361.3 μA and 239.3 μA in the case of Au/CQDs/Antigen and Au/CQDs/Antigen/BSA, respectively. The results indicated that CQDs favor the charge transfer across the liquid-solid interface. After the modification with antigen and BSA, the transduction capability of the Au/CQDs/Antigen/BSA electrode is better than that of Au/Antigen/BSA. According to previously reported works, the equilibrium potential in a range of 400–500 mV vs. Ag/AgCl corresponded to the standard potential of the Fe2+/3+ couple. Here the appearance of new redox potential peak at ∼500 mV could be ascribed to the oxidation reactions of Fe2+/3+ ([Fe(CN)6]3−/4− REDOX mediator) (Lowson, 1982; Córdoba et al., 2008). We can thereby expect an improvement in the sensitivity, specificity and signal-to-noise ratio when used as the antibody biosensor.
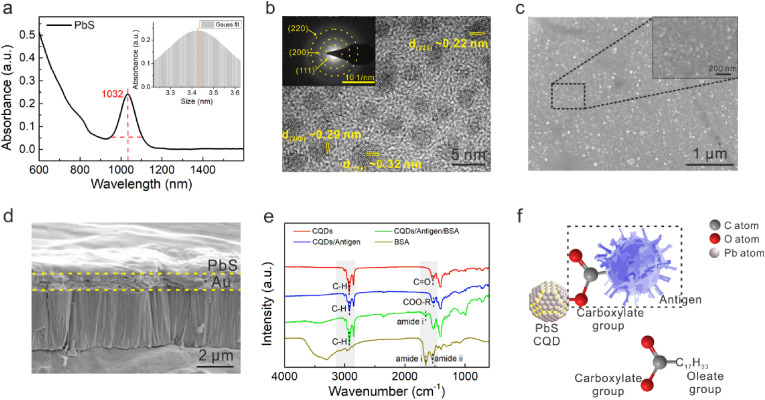
We further investigated the microstructure and morphology of the CQDs-modified electrode. PbS CQDs exhibited a sharp exciton absorption peak at the wavelength of 1032 nm in UV–vis–NIR (ultraviolet–visible–near infrared) absorption spectrum (Fig. 2 a); the diameter was estimated to be 3.4 ± 0.3 nm according to the Moreels method (details in Supporting Information), which is well below the exciton Bohr radius of PbS ∼18 nm. Corresponding size histogram plotted in the inset indicated a narrow size distribution (Moreels et al., 2009). High resolution transmission electron microscopy (HRTEM) analysis further revealed that PbS CQDs were near spherical nanocrystals with diameter of 3–4 nm (Fig. 2b). The selected area electron diffraction (SAED) image exhibited three clear diffraction rings corresponded to the (111), (200) and (220) facets of PbS, suggestive of the good crystallinity. The small size of CQDs is beneficial to the receptor function of the biosensor in improving the surface-to-volume ratio for enhanced enrichment of protein molecules.
Fig. 2.
Characterization of CQDs-modification layer and CQD-protein interface. (a) UV–Vis–NIR absorption spectrum of PbS CQDs (The inset shows the average diameter size of 3.4 nm). (b) HRTEM images of PbS CQDs (The inset depicts selected area electron diffraction, showing the lattice planes of (111), (200), and (220)). (c) SEM images of antigen-bonding CQDs film with 2h blocking, revealing uniform morphology. (d) The cross-sectional SEM images of antigen-bonding CQDs film on the WE. (e) The FTIR spectra of BSA, Au/CQDs, Au/CQDs/Antigen, Au/CQDs/Antigen/BSA. (f) Carboxylate group between PbS CQD and antigen.
We provide more details of the morphology of CQDs-modification layer using scanning electron microscope (SEM) and atomic force microscope (AFM) characterizations. The CQDs solid film had a flat surface with regular gaps and holes (Figs. S3a and b). After coating with proteins, the quantity of gaps and holes on the film surface was significantly reduced (Fig. 2c, Fig. S3c). Energy dispersive spectrometer (EDS) spectra indicated that the lead and sulfur elements of CQDs were uniformly distributed (Figs. S3d and e). The cross-sectional image suggested the uniform thickness of the CQDs-modification layer (Fig. 2d). The elements distribution (Figs. S3f–h) analysis at the cross section also indicated that the Au-CQDs and CQDs-protein interfaces were continuous and compact. The AFM images further confirmed that the BSA treatment reduced the roughness of CQDs film, in which the surface roughness decreased from ∼145 nm to ∼100 nm in the region of 2 μm2 (Fig. S3i and Figs. S4a and b); in a larger view, the film roughness decreased from ∼2.4 μm to ∼1.0 μm (Figs. S4c–f). Owing to the excellent solution-processability of CQDs, the CQDs-modification layer was successfully deposited on Au electrode via the spin-coating method. Their flat surface, uniform thickness and homogenous composition distribution are desirable for improving the consistency of the biosensor in mass production and practical use.
In our all-solid-sate biosensor, the CQD-protein interface is the key parameter that determines the success in realizing the electronic labelling of SARS-CoV-2 antigen for future antibody molecule recognition and signal transduction. In the oleic acid (OA)-capped PbS CQDs, the long-chain (18 carbons) OA ligands serve as metal coordinating groups (Pb–O) as well as solvophilic groups which enable the stabilization of quantum dots in the form of colloids (Liu et al., 2015; Boles et al., 2016; Arquer et al., 2021). Traditionally, these long-chain ligands are replaced by shorter ligands via ligand exchange process to achieve application-specific electronic and optoelectronic properties for the large-area, solution-based assembly of semiconductor devices (Liu et al., 2021).
Here we demonstrated that CQD surfaces directly functionalized with biological macromolecule like the SARS-CoV-2 S-RBD protein could be achieved via facile one-step ligand exchange. The Fourier transform infrared spectroscopy (FTIR) spectra of different samples were show in Fig. 2e. Compared to the pristine PbS CQDs, the C–H stretching vibration at 2812–2987 cm−1 of OA ligands were slightly attenuated in CQDs/Antigen sample, and the carboxylate group (COO-R) appeared at 1546 cm−1 instead of the carbonyl group (C O) at 1531 cm−1 of OA ligands. The results suggested that OA ligands were stripped off and replaced by protein ligands which may have stronger affinity to CQD surface (Liu et al., 2015). After successive coating with BSA, while the characteristic peak of amide ii in the CQDs/Antigen/BSA sample intensified because of the superposition of amide ii peak and the carboxyl group peak, the characteristic peak at 1647 cm−1 corresponding to the amide i of BSA was clearly observed, confirming the presence of BSA on the all-solid-state sensor (Bandekar, 1992; Wang et al., 2011).
Therefore, we successfully labelled CQDs to the target proteins and sealed them with BSA. As shown in Fig. 2f, the SARS-CoV-2 S-RBD protein kicks out the original OA ligands and binds to Pb2+ through O in the carboxylate group of the protein molecule itself. CQDs synthesized in a solution contain surface ligands attached to their surface atoms. Unlike the development of CQDs fluorescence labelling or medium-assisted labelling for biomolecules where the foreign crosslinkers have to be used and generally take hours or even days because of multiple crosslinking or activation processes in liquid phase atmosphere, a solid-state CQD-protein interface which is direct connected via Pb–O coordinate bond was constructed simply through one-step ligand exchange treatment on the CQD solids film (Medintz et al., 2005). This labelling method offers more advantages in high-efficiency and low-cost scale production of biosensors.
3.2. Electrochemical measurement of SARS-CoV-2 antibody
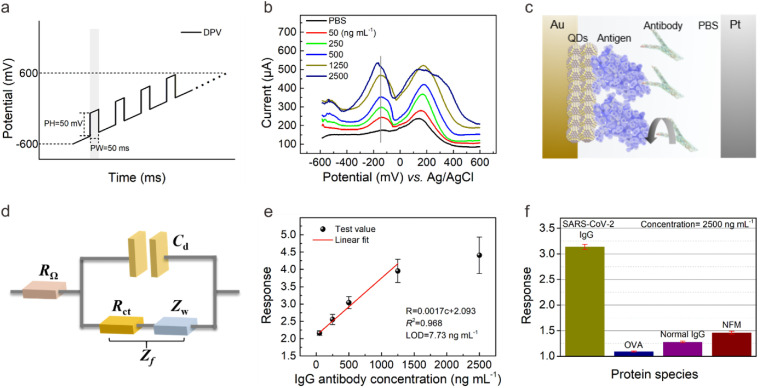
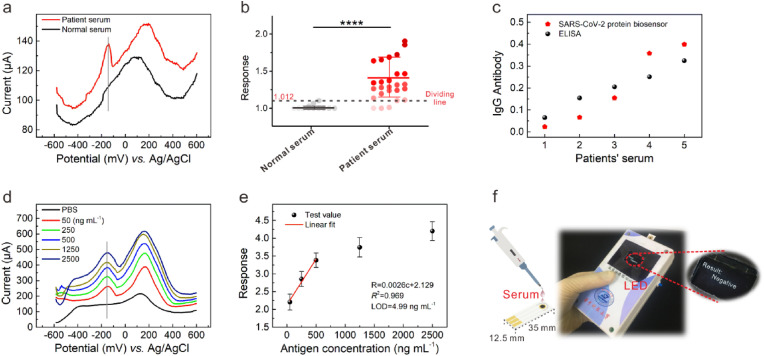
Now, we study the sensing performance of the CQDs-modified electrode as the SARS-CoV-2 antibody biosensor. The devices from different batches were tested in 1 × PBS (details in Supporting Information) to explore the batch-to-batch variation, slight variation in the current with a small relative standard deviation (5.66%) indicated the desirable reproducibility. These results suggested that high-quality CQD solids film could be acquired through spin-coating technique, which is very attractive for achieving mass production and wide application. We began by preparing standard SARS-CoV-2 IgG antibody solutions in 1 × PBS with concentrations ranging from 50 to 2500 ng mL−1. In the DPV measurement mode, the voltage applied consists of short pulses superponated to a linearly increasing direct current (DC) ramp (the superposition of a linearly increasing DC ramp with a rectangular pulse), through which an enhanced discrimination of Faradaic currents (electron transfer to and from an electrode) can be obtained (Scott, 2016). Here, the pulse height (PH) and pulse width (PW) were set as 50 mV and 50 ms, respectively (Fig. 3 a). The current-voltage curves measured at different IgG antibody concentrations (0–2500 ng mL−1) were shown in Fig. 3b. As expected, the curve measured in PBS without IgG antibody consists of only one current peak at ∼145 mV (background peak), which could be ascribed to the redox potential of the Ag/AgCl reference electrode in PBS solution, which have been observed in existing literatures (Suzuki et al., 1999; Shanmugam et al., 2014; Fang et al., 2005). Attractively, in addition to background peak, new current peak appeared at ∼ -145 mV as the antibody was added. With the increased antibody concentration, the charged substances in the PBS buffer atmosphere increased, enhancing the electrochemical activity of the CQDs-modified electrode (Chikkaveeraiah et al., 2012; Madrakian et al., 2016). At DPV mode, the current corresponding to each potential increased, and the current corresponding to the redox potential of the Ag/AgCl reference electrode increased more, causing the increase of peak current at ∼145 mV. Compared with the peak current to the redox potential of the reference electrode used as the signal in some literatures, we speculated that this peak current might be susceptible to the cross interference or nonspecific reactions (Hashemi et al., 2021; Yakoh et al., 2021). Attractively, we observed for the first time the feature current peak at ∼ -145 mV corresponding to the specific binding of SARS-CoV-2 antigen and antibody, through which high sensitivity and specificity detection of antibody could be achieved. According to Supporting Note 1, we thereby concluded that the current peak at ∼ -145 mV (response peak) was induced by the specific binding reaction of SARS-CoV-2 antigen with antibody, as shown in Fig. 3c (Bard and Fualkneret, 1980). When the antibody and antigen bind specifically at the electrode surface (solid-fluid interface), the induced charge transfer through the CQD-protein interface may be transduced into the current signal at DPV mode.
Fig. 3.
The sensing performance of the CQDs-modified electrode for SARS-CoV-2 antibody testing. (a) The voltage applied in DPV measurement. (b) The current-voltage curves with the increased IgG antibody (recombinant antibody) concentrations (0–2500 ng mL−1). (c) The working principle of SARS-CoV-2 protein biosensor employing colloidal quantum dots-modified electrode. (d) The equivalent circuit model of chemically modified electrode. (e) Recombinant antibody concentration-dependent response curve using linear fit. (f) The selectivity against OVA, normal IgG and NFM. The concentration was 2500 ng mL−1 in 1 × PBS.
According to the equivalent circuit model of electrochemical three-electrode (Fig. 3d), the current intensity is influenced by R Ω, C d, and R ct, which represent the internal resistance of the system including electrolyte solution and electrode, the capacitance of the electrical double layer located at electrode-electrolyte interface, and the resistance for the charge transfer across the fluid-solid interface to overcome. The Fermi level of Au (E Fm = 5.1 eV) is slightly lower than that of PbS CQDs (E Fs = 4.61 eV), enabling the appropriate energy band matching and the ohmic contact between Au electrode and CQDs (Kim et al., 2013). It helps to reduce the internal resistance R Ω. Z f, Faradaic impedance, represents the additional impedance caused by electrochemical polarization and concentration polarization when the current passes through the electrolyte and the fluid-solid (electrolyte-work electrode) interface. Z w represents Warburg impedance, which is an impedance due to the process of diffusion. The influence of Z w representing the impedance caused by the transfer process of inherent reactants in electrolyte solution is negligible in DPV mode of this study. Under the influence of the pulse in the voltage applied at DPV mode, the charging and discharging effect of C d contribute to the Faraday current, the current arising from the charge transfer of REDOX reaction at fluid-solid interface. The CQDs-modification layer may influence the Faraday current as well as the sensor response by changing the value of R ct and C d of the electrode. This explains the observation that the output current of the sensor increased with increasing SARS-CoV-2 IgG antibody concentration in this study. The key factors that determine the specificity of our biosensor includes the specific reaction of paired antigen and antibody at the micro level, and the feature response peak at the macro level. The detailed signal transduction mechanism of the CQDs-modified electrode and the role of CQDs will be discussed later.
We proceeded to extract the sensor response according to the current at response peak. Here, we defined the sensor response as the value of I PT/I NT, where I PT and I NT represented the current at response peak measured in recombinant antibody solution and PBS, respectively (details in Supporting Information). As shown in Fig. 3e, the sensor response depended approximately linearly on concentration of recombinant antibody in the range 50–1250 ng mL−1. Through the least-squares method of fitting, we obtained the response slop in the linear regime to be 0.001 (mL ng−1) with a fitting quality of 0.968. The limit-of-detection (LOD) of the biosensor for SARS-CoV-2 IgG antibody testing was estimated to be 7.73 ng mL−1 (details in Supporting Information).
The selectivity of the biosensor is critical to its accuracy, as the sample composition is often very complex. We examined the response of the sensor toward non-specific proteins in this study. As shown in Fig. 3f, the response toward ovalbumin (OVA), human normal immunoglobulin G (normal IgG), nonfat-dried milk (NFM) were rather low compared to SARS-CoV-2 IgG protein. The results suggested the success of CQDs as the electronic labelling of SARS-CoV-2 S-RBD protein in contributing the immobilization efficacy and following sensitive and specific signal. In our all-solid-state biosensor, the presence of response peak which is characteristic of the specific bonding of antigen and antibody protein offer better discrimination against nonspecific disturbance and may improve the detection specificity with serum samples.
3.3. The signal transduction mechanism of CQDs-modified electrode
We now explore the sensing mechanism of the protein biosensor employing CQDs-modified electrode. First, to clarify the receptor-and-transducer function of CQDs as the electronic labelling of proteins, we compared the sensing performance of Au, Au/Antigen, Au/BSA, Au/Antigen/BSA electrode samples with and without CQDs modification (Fig. 4 a). In the case of Au, Au/Antigen, Au/BSA and Au/Antigen/BSA electrode samples no matter whether there were CQDs or not, the variation of the current-voltage curves compared to those measured in PBS were indistinguishable when the SARS-CoV-2 IgG antibody was added. The CQDs modification layer plays a key role in the biomolecule recognition and signal transduction. The use of CQDs enable the stable immobilization and enrichment of specific antigen on CQD solids film via chemical bonds through ligand exchange, which is prerequisite for the following transducer function and achieving sensitive and specific sensor response. Au/CQDs/Antigen electrode showed a weak response current. In addition to prevent the non-specific protein reaction, the blocking treatment of BSA also encapsulated the CQDs-modified electrode, further improving the immobilizing stability of CQDs with antigen protein, which helped to improve the robustness of the electrode interfaces. Only the Au/CQDs/Antigen/BSA electrode was sensitive to the SARS-CoV-2 IgG antibody; when the antibody was added, the current intensity increased with the arise of the response peak. The results indicated that the CQDs-modification layer in this study is indispensable for the protein biosensor. Our work experimentally demonstrated the excellent receptor-and-transducer functional properties of CQDs to define an electronic labelling of protein molecules. As inherently receptors for foreign ligands, CQDs enable the stable immobilization and enrichment of specific antigen on CQD solids film via chemical bonds through ligand exchange, which is prerequisite for achieving sensitive and specific sensor response toward target antibody.
Fig. 4.
The electrical model and signal transduction of CQDs-modified electrode. (a) The DPV curves of various electrodes measured in PBS and target antibody in 1 × PBS: Au, Au/Antigen, Au/BSA, Au/Antigen/BSA, Au/CQDs, Au/CQDs/Antigen, Au/CQDs/BSA, Au/CQDs/Antigen/BSA. (b) Rct (charge transfer resistance, black dot) and CPE (constant phase angle element, proportional to Cd, red dot) of Au, Au/CQDs, Au/CQDs/Antigen, and Au/CQDs/Antigen/BSA in the presence of 1 × PBS containing 5 mM [Fe(CN)6]3−/4− and 0.5 M KCl as REDOX mediator. (c) The DPV characterizations of PbS CQDs-modified electrode, annealed PbS-modified electrode, and bulk PbS-modified electrode measured in PBS and target antibody in 1 × PBS. (d) Rct and RΩ of PbS CQDs-modified electrode, annealed PbS-modified electrode, and bulk PbS-modified electrode measured in 1 × PBS and target antibody in 1 × PBS. (e) The mechanism of signal transduction, displaying the formation of dipole at the CQD-antigen interface and the process of hopping or tunneling. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The transduction mechanism of CQDs-modified electrode was discussed according to a equivalent circuit model. At DPV mode, smaller R ct and higher C d values are favorable to improve the sensor signal (Supporting Note 2 and Note 3). We utilized electrochemical impedance spectroscopy (EIS) to extract the electrical parameters of the samples during electrode preparation (Fig. S5). As shown in Fig. 4b, the Au/CQDs sample had lowest R ct value (black dot), suggesting that CQDs favor the charge transfer across the fluid-solid interface, which reduced the resistance to overcome. We observed a decrease trend in the internal resistance with successively coating Au with CQDs, Antigen and BSA (Fig. S6). These results suggested that the modification of Au electrode with CQD solids are helpful for the enrichment of [Fe(CN)6]3−/4−. The internal resistance slightly decreased after proteins were further added, which indicated that the proteins around the surface of CQDs do not affect the transport of [Fe(CN)6]3−/4−, possibly due to the network of protein macromolecules. The result also suggested the key role of CQD solids that determines the internal resistance of the electrode. As expected, the value of R ct increased with the successive coating of antigen and BSA due to the worse conduction capabilities of proteins. It is interesting to find that for the final Au/CQDs/Antigen/BSA electrode, it had highest R ct and a relatively lower CPE (constant phase angle element, proportional to C d, red dot) values among those electrodes but exhibited obvious sensor response. This seems abnormal when we expect lowest R ct and highest C d values for the most sensitive electrode. Instead, the unmodified Au electrode which had almost the same R ct but with higher C d values had no sensor response at all. The results suggested a unique charging and discharging effect of the CQDs, which enables the successful transduction of the specific binding reaction of antigen and antibody into current output signal.
Second, to elucidate the mechanism underlying the unique role of CQDs in the signal transduction, we prepared two bulk PbS materials (without quantum effects)-modified electrode samples for comparison. The ‘Annealed-PbS’ sample stands for the sample obtained by annealing the control electrode (here renamed as PbS-CQDs) at 120 celsius degree for 2 h; the ‘Bulk PbS’ sample was based on the commercial lead sulfide material with other preparation conditions being kept the same. As shown in Fig. 4c, when IgG antibody was added into PBS, unlike the PbS-CQDs sample where the output current increased accompanied with the arise of response peak at ∼ -145 mV, neither of the annealed-PbS and bulk PbS-modified electrode samples exhibited obvious current change. The very low and broad current peaks at the potential of ∼ -145 mV for the ‘Bulk PbS’ sample, observed for both PBS and IgG solutions, was almost negligible (Fig. S7).
Fig. 4d summarized the electrical parameters extracted from EIS (Fig. S8) measurement of these three samples conducted in PBS and with IgG antibody added, respectively. Compared to Annealed-PbS and Bulk-PbS samples, the obvious increase of Rct for the control sample modified with CQDs, while not favorable for charge transfer, indicated the effective antigen-antibody binding. The results suggested the ability of CQDs to immobilize and enrich the antigen protein for subsequent antibody recognition. Again, the unique charging and discharging effect of the CQDs was demonstrated since the value of CPE for the control sample was relatively low among these samples, yet capable of output a sharp response peak with high signal-to-noise ratio for IgG antibody testing. In addition, the change of CPE value was neglectable for the control sample when measured in PBS with and without IgG, suggesting that the capacitance value itself is not sensitive to IgG antibody. The unique charging and discharging effect could be well maintained under the antibody testing environment.
Taking together the above experimental observations, we are able to propose a more detailed model for the transduction mechanism of CQDs. The diameter (3.4 ± 0.3 nm) of PbS CQDs was estimated above, which is below the exciton Bohr radius (∼18 nm) of PbS. Decreasing the size of particles to less than their Bohr radius results in confinement of electron and hole wavefunctions and a significant increase in bandgap. As depicted in Fig. 4e, individual PbS CQD, subjected to quantum confinement, was equivalent to a dipole function as an insulative capacitor. At DPV mode, the charge transfer from solution to electrode charge CQD dipoles along the CQD-protein interface; due to Coulomb Blockade effect, the CQD dipole will discharge under appropriate voltage. On the other hand, the quantum mechanical coupling between two adjacent CQDs can be expounded in terms of coupling energy β (Tang et al., 2010, Tang and Sargent, 2010). Here, the lowest CQD states (1S h for holes and 1S e for electrons) were depicted. The charge transfer of PbS CQD solids film may occur through possible hopping (like electronic sharing of bulk materials) because of the leakage of wave function (Fig. S9a) or quantum tunneling. Therefore, the CQDs possess a unique charging and discharging effect depending on the alternating voltage applied, through which the charge transfer across CQD-protein interface may be converted into Faraday current. It should not be ignored that this response peak has a higher specificity compared with the peak at ∼145 mV. When IgG antibody and antigen bind specifically at the electrode surface, the increased charge amount from IgG antibody combined with the charge redistribution at the antigen-antibody interface will induce charge increase at the CQD-protein interface, enhancing the charge intensity of CQD dipoles (Fig. S9b) which thereby leads to the response peak with higher output current as observed in CQDs-modified electrode, which is not observed in bulk PbS electrode samples.
According to Marcus theory, most biological reactions belong to outer sphere electron transfer (Marcus, 1997). For antigen and antibody binding reaction, the volume of the two biomolecules is usually larger and the REDOX active center is buried deep within the protein, which could not establish the ligand bridge (Winkler et al., 1995). Long-range electron transfer between antigen and antibody is generally difficult and often requires external assistance like “springboard”. The size of PbS CQD is much smaller than its Debye length, so their ability to transmit electric field maybe strong enough to ‘sensing’ distant charge redistribution or electric quantity change, which may contribute to the transduction function from the antigen-antibody specific binding reaction to electrical signal. Our model may explain why the electrodes without CQDs-modification had poor response due to insufficient receptor-and-transducer functions. The C d of the Au electrode, which was fixed in the inherent system, causes the non-faradaic current that is ineffective signal for protein testing at DPV mode.
From a macroscopic angle, the unique charging and discharging effect of CQDs as the electronic labelling, ascribed to their quantum confinement and Coulomb blockade effect, is analog to a ‘Faraday current dam’, which combines an antenna function collecting signal from biological reactions as well as a switch function to allow for enhanced Faraday current, which is characteristic of a REDOX on the CQDs-modified electrode surface, e.g. the specific binding reaction of SARS-CoV-2 antigen and antibody proteins in this study. The charging and discharging effect of CQDs, combined with efficient signal conditioning circuit design, could contribute to the specificity and high signal to noise ratio of output signal.
3.4. Serological studies and functional extension
We further investigated the performance of the sensor using serum samples from COVID-19 patients and normal people. The preparation and testing of serum samples were detailed in Supporting Information. The SARS-CoV-2 IgG antibody levels have been measured by ELISA method (Table S3) for comparison. As shown in Fig. 5 a, we clearly observed the response peak for a typical patient sample, with much higher current compared to the normal sample where no response peak could be identified. Fig. 5b showed the significances test with 26 positive serum samples randomly selected from 11 patients and 12 normal serum samples (negative) (Figs. S10 and S11). The input current signal of the sensor is relatively low because the serum specimens contain lots of impurities. The response with the value of 1.102 was regarded as the threshold between positive and negative samples. The sensor can detect the SARS-CoV-2 antibody of the serum sample from those who have been clinically diagnosed as COVID-19 patient. When the highest response value in the case of the negative serum was used as the dividing line, 23 of the 26 positive samples could be identified using the all-solid-state biosensor, indicating an accuracy of about 90% in discrimination of positive/negative samples with a statistical significance (P value: less than 0.0001) using unpaired t-test. We randomly selected 5 patient serum samples and compared the quantitative level of IgG antibody obtained with our sensor and ELISA method, respectively. As shown in Fig. 5c, the sensor provided a quantitative analysis of SARS-CoV-2 antibody with a correlation coefficient of 93.8% compared to ELISA value through Pearson correlation. Through the repeatability DPV testing on the above 5 patient serum samples, our device possessed reliable reproducibility (Fig. S12). Taken together, the CQDs-modified electrode has the potentials to be used for the SARS-CoV-2 antibody biosensor.
Fig. 5.
The verification of serum specimens, SARS-CoV-2 RBD protein, and the handheld testing system prototype. (a) Comparison of DPV curves between patient and normal serum specimens. (b) The significances test of normal serum (negative) and patient serum (positive). (c) The IgG testing comparison of protein biosensor and ELISA. (d) The current-voltage curves with the increased SARS-CoV-2 S-RBD antigen (recombinant antigen) concentrations (0–2500 ng mL−1). (e) Recombinant antigen concentration-dependent response curve using linear fit. (f) The testing process of the handheld testing system prototype.
According to the basic principle of immunoassay, we proposed further that the CQDs-modified electrode could be also utilized for SARS-CoV-2 antigen detection. Direct testing the viral antigens in clinical specimen using nasopharyngeal and oropharyngeal swabs without multiple sample preparation is of great significance for rapid and accurate SARS-CoV-2 screening. Here, the antibody was dip-coated onto the CQD solids film and then incubated to prepare antibody-bonding films. With BSA treatment, antigen protein biosensor employing Au/CQDs/Antibody/BSA electrode was prepared for the recombinant antigen testing. As expected, response peaks at ∼ -145 mV were observed only in samples with standard SARS-CoV-2 recombinant antigen solutions added (Fig. 5d); the output current of the sensor increased with increasing recombinant antigen concentration ranging from 50 to 2500 ng mL−1. The linearity and LOD of concentrations-response curve were calculated to be 0.969 and 4.99 ng mL−1, respectively (Fig. 5e). The results suggested the universality of the CQDs-modified electrode as a versatile platform for the development of sensitive and specific protein biosensor. Interestingly, in contrast with the DPV I–V curves in the case of antibody testing, here we found that the current magnitude of response peak was lower to that of the background peak, which is possibly due to the difference in spatial conformation and distribution of active sites between antigen and antibody proteins. The differences of spatial conformation between antigen and antibody lead to different labeling efficiency of CQDs/protein in the existing preparation method. The results suggested that the design, adjustment, and construction of a more effective interface based on the differences between antigen and antibody molecules could be utilized to improve sensitivity (Cathou and Haber, 1967; Ricci et al., 2012). The influence of the protein structure on the electronic labelling efficiency with CQDs is beyond the range of this study.
Based on the rapidity and reliable accuracy of antibody testing in above lab study, we explored the feasibility of field-testing application through the design of a handheld testing system prototype (Fig. 5f and Fig. S13). The data could be transmitted to the upper computer through Wi-Fi module, and the LED display module was set to display the test results in real-time (Fig. S14). We integrated the circuit system into the handheld testing system prototype and reserved the interface for CQDs-modified electrode (Fig. S15). The dimensions of the whole system are 12 × 7 × 4 cm3 (L × W × D), closer to the size of household glucometer. The operating process was detailed in Supporting Information. For comparison, the testing procedure of positive serum specimen using the handheld testing system prototype and commercial CGIA Kit was illustrated in the Supporting Video 1. Traditional immunological techniques such as ELISA and CGIA need pairs of matching antibodies, which increases the complexity of preparation and takes more time. Our prototype exhibits testing time (about 30 s) which is about 10 times faster than CGIA Kit (about 5 min).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.bios.2022.113974
The following is/are the supplementary data related to this article:
Multimedia component 2
Rapid, accurate, and portable testing of SARS-CoV-2 antibody can provide immunological evidence for the whole viral stages including infection (symptomatic and asymptomatic cases), prognosis and after vaccination, which may find wide applications in disease diagnosis, epidemiological investigation, and physiological management. The electronic labelling of proteins with CQDs enables sensitive and specific protein biosensors, which combines the advantages of rapidity, accuracy, and convenience. Successful construction of more efficient Au/CQDs as well as CQDs/protein interfaces may further improve the activity, stability and reproducibility of the CQDs-modified electrode, which are directly affected by the design of the modification materials, the rationality of the modification method and the quality of the modification layer. For instance, the receptor-and-transducer functional properties could be controlled by adjusting film microstructure and depth. Attractively, the physical and chemical properties of CQDs are highly programmable by tailoring its size, shape, and composition, which could be adapted to various biomolecules such as proteins, polypeptides, and DNA. Meanwhile, we may expect a direct aqueous assembly of inorganic/biological complex would enhance the charge transfer across the solid-liquid interface of electrochemical sensor. In addition, the optimization of driving voltage, the adaptive design of signal acquisition and amplifier circuit module can also improve the detection performance. Our work may pave a way to colloidal quantum dot immunoassay (CQDIA) method, which have promising potentials in the development of low-cost, high-throughput diagnostic instrument in clinical lab as well as user-friendly system for POCT and home-use test.
The special advantages of the all-solid-state biosensor employing CQDs-modified electrode are summarized in the following three points:
-
1)
The facile electronic labelling method through one-step ligand exchange treatment without organic cross-linkers we proposed is helpful to enhance the receptor function and the consistency of immobilizing process, which presents universality for other biomolecules.
-
2)
Owing to the remarkable surface effect, size effect and unique quantum effect of CQDs, which can improve the enrichment of biomolecules and the signal transduction of SARS-CoV-2 antigen-antibody binding reactions. We observed for the first time the feature current peak corresponding to the antigen-antibody specific binding of, through which we obtained high sensitivity and specificity in the case of serum specimens.
-
3)
The high-quality CQD solids film could be acquired through spin-coating technique, which is very attractive for achieving mass production and wide application, suggestive of the development potentials in silicon-based biosensors and integrated biochips.
4. Conclusions
We have proposed the electronic labelling strategy of protein with quantum dots, through which the CQD-protein interface is designed for achieving specific protein recognition and efficient electronic transduction. The PbS CQDs-modified electrodes have been demonstrated as sensitive and specific SARS-CoV-2 protein biosensors by converting the antigen-antibody specific binding reaction into significant current output signals. The electronic transduction mechanism is attributed to the quantum confinement and Coulomb blockade effect of quantum dots, which induce the unique charging and discharging effect depending on the alternating voltage applied. In the case of serum specimens from COVID-19 patient samples, the all-solid-state SARS-CoV-2 protein biosensor provides a quantitative analysis of SARS-CoV-2 antibody with a correlation coefficient of 93.8% compared to ELISA value. It discriminates patient and normal samples with an accuracy of about 90%. The results could be read within 1 min by a handheld testing system prototype.
CRediT authorship contribution statement
Yunong Zhao: Conceptualization, Investigation, Writing – original draft, , Visualization, Formal analysis, Data curation, Validation, fabricated the devices and conducted the characterizations. conducted the electrochemical experiments. conducted serum test, wrote the manuscript. Jianjun Chen: Methodology, Formal analysis, Writing – original draft, analyzed all the data and guided the writing of the manuscript. wrote the manuscript. Zhixiang Hu: Investigation, Formal analysis, conducted the synthesis of quantum dots and the analysis of FTIR spectra. Yan Chen: Investigation, Validation, Formal analysis, performed ELISA tests and analysis, and supported serum test. Yanbing Tao: Investigation, conducted serum test. Le Wang: Investigation, designed and built the handheld detector prototype. Long Li: Investigation, designed and built the handheld detector prototype. Ping Wang: Validation, Resources, performed ELISA tests and analysis, and supported serum test. Hua-Yao Li: Validation, Conceptualization, Supervision, guided the design of handheld detector prototype. Jianbing Zhang: Supervision, contributed to the anaysis of characterizations. Jiang Tang: Methodology, Supervision, contributed to the discussions of sensing mechanism. Huan Liu: Conceptualization, Investigation, Formal analysis, Writing – original draft, , Writing – review & editing, Project administration, Funding acquisition, conceived the concept and designed the research. analyzed all the data and guided the writing of the manuscript. wrote the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by National Natural Science Foundation of China (No. 61922032 and 61861136004) and “the Fundamental Research Funds for the Central Universities” (COVID-19 Emergency Special Fund of Huazhong University of Science and Technology, No. 2020kfyXGYJ076). We thank Program for HUST Academic Frontier Youth Team (2018QYTD06) and Innovation Fund of WNLO for equipment support. We thank Analytical and Testing Center of HUST for the characterization support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.113974.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Experimental Details; Supporting Note 1 to Note 3; Tables S1–S25; Figs. S1–S26; Supporting Video 1.
References
- Ali M., Hu C., Jahan S., Yuan B., Saleh M., Ju E., Gao S., Panat R. Adv. Mater. 2021;33(7):2006647. doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T., Chromikova V., McMahon M., Jiang K., Arunkumar G., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D., Lugo L., Kojic E., Liu J., Cunningham-Rundles C., Felgner P., Moran T., García-Sastre A., Caplivski D., Cheng A., Kedzierska K., Vapalahti O., Hepojoki J., Simon V., Krammer F. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquer F., Talapin D., Klimov V., Arakawa Y., Bayer M., Sargent E. Science. 2021;373(6555):eaaz8541. doi: 10.1126/science.aaz8541. [DOI] [PubMed] [Google Scholar]
- Bampoe S., Lucas D., Neall G., Sceales P., Aggarwal R., Caulfield K., Siassakos D., Odor P. 2020. Medrxiv. [DOI] [PubMed]
- Bandekar J. BBA-Protein Struct. M. 1992;1120(2):123–143. doi: 10.1016/0167-4838(92)90261-b. [DOI] [PubMed] [Google Scholar]
- Bard A., Fualkneret L. 1980. Wiley. [DOI]
- Beduk T., Beduk D., Filho J., Zihnioglu F., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Salama K., Timur S. Anal. Chem. 2021;93(24):8585–8594. doi: 10.1021/acs.analchem.1c01444. [DOI] [PubMed] [Google Scholar]
- Boles M., Ling D., Hyeon T., Talapin D. Nat. Mater. 2016;15(2):141–153. doi: 10.1038/nmat4526. [DOI] [PubMed] [Google Scholar]
- Bryan A., Pepper G., Wener M., Fink S., Morishima C., Chaudhary A., Jerome K., Mathias P., Greninger A. J. Clin. Microbiol. 2020;58(8):e00941–20. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathou R., Haber E. Biochemistry. 1967;6(2):513. doi: 10.1021/bi00854a020. [DOI] [PubMed] [Google Scholar]
- Chen X., Wang Y., Zhou J., Yan W., Li X., Zhu J. Anal. Chem. 2020;80(6):2133–2140. doi: 10.1021/ac7021376. [DOI] [PubMed] [Google Scholar]
- Cheong J., Yu H., Lee C., Lee J., Choi H., Lee J., Lee H., Cheon J. Nat. Biomed. Eng. 2020;4(12):1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikkaveeraiah B., Bhirde A., Morgan N., Eden H., Chen X. ACS Nano. 2012;6(8):6546–6561. doi: 10.1021/nn3023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Wang H., Oh S., Paik T., Sung P., Sung J., Ye X., Zhao T., Diroll B., Murray C., Kagan C. Science. 2016;352(6282):205–208. doi: 10.1126/science.aad0371. [DOI] [PubMed] [Google Scholar]
- Chua K., Vogrin S., Bittar I., Horvath J., Wimaleswaran H., Trubiano J., Holmes N., Lam Q. Pathology. 2020;52(7):778–782. doi: 10.1016/j.pathol.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdoba E., Muñoz J., Blázquez M., Gonzálezb F., Ballester A. Hydrometallurgy. 2008;93(3–4):88–96. [Google Scholar]
- Cui N., Guan M., Xu M., Fang W., Zhang Y., Zhao C., Zeng Y. Opt Express. 2020;28(11):16834–16844. doi: 10.1364/OE.393397. [DOI] [PubMed] [Google Scholar]
- Elledge S., Zhou X., Byrnes J., Martinko A., Lui I., Pance K., Lim S., Glasgow J., Glasgow A., Turcios K., Iyer N., Torres L., Peluso M., Henrich T., Wang T., Tato C., Leung K., Greenhouse B., Wells J. Nat. Biotechnol. 2021;39(8):928–935. doi: 10.1038/s41587-021-00878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B., Wang G., Zhang W., Li M., Kan X. Electroanalysis. 2005;17(9):744–748. [Google Scholar]
- Grossberg A., Koza L., Ledreux A., Prusmack C., Krishnamurthy H., Jayaraman V., Granholm A., Linseman D. Nat. Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-21040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D., Du B., Li L., Zeng G., Yuen K., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. N. Engl. J. Med. 2020;58(4):711–712. [Google Scholar]
- Hao X., Cheng S., Wu D., Wu T., Lin X., Wang C. Nature. 2020;584(7821):420–424. doi: 10.1038/s41586-020-2554-8. [DOI] [PubMed] [Google Scholar]
- Hashemi S., Bahrani S., Mousavi S., Omidifar N., Behbahan N., Arjmand M., Ramakrishna S., Lankarani K., Moghadami M., Shokripour M., Firoozsani M., Chiang W. J. Electroanal. Chem. 2021;894(3):115341. doi: 10.1016/j.jelechem.2021.115341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Lu Q., Zhao H., Zhang Y., Sui Z., Fang L., Liu D., Sun X., Peng K., Liu W., Guan W. Cell Discov. 2019;6(1):64. doi: 10.1038/s41421-020-00209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Ding L., Zhou J., Chen S., Chen F., Zhao C., Xu J., Hu W., Ji J., Xu H., Liu G. Biosens. Bioelectron. 2021;171:112685. doi: 10.1016/j.bios.2020.112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Wang Z., Zhang H., Zhang X., Liu X., Wang S. J. Agric. Food Chem. 2011;59(18):9763–9769. doi: 10.1021/jf2012437. [DOI] [PubMed] [Google Scholar]
- Kagan C., Lifshitz E., Sargent E., Talapin D. Science. 2016;353(6302):aac5523. doi: 10.1126/science.aac5523. [DOI] [PubMed] [Google Scholar]
- Kevadiya B., Machhi J., Herskovitz J., Oleynikov M., Blomberg W., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C., Reid S., Punyadeera C., Chang L., Gendelman H. Nat. Mater. 2021;20(5):593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kim D., Lee J., Grossman J. Phys. Rev. Lett. 2013;110(19):196802. doi: 10.1103/PhysRevLett.110.196802. [DOI] [PubMed] [Google Scholar]
- Lequin R. Clin. Chem. 2005;51(12):2415–2418. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- Li H., Liu Z., He Y., Qi Y., Chen J., Ma Y., Liu F., Lai K., Zhang Y., Jiang L., Wang X., Ge J. Clin. Transl. Med. 2020;10(15):e90. doi: 10.1002/ctm2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li M., Voznyy O., Hu L., Fu Q., Zhou D., Xia Z., Sargent E., Tang J. Adv. Mater. 2014;26(17):2718–2724. doi: 10.1002/adma.201304366. [DOI] [PubMed] [Google Scholar]
- Liu H., Li M., Shao G., Zhang W., Wang W., Song H., Cao H., Ma W., Tang J. Sensor. Actuator. B Chem. 2015;212:434–439. [Google Scholar]
- Liu M., Yazdani N., Yarema M., Jansen M., Wood V., Sargent E. Nat. Electro. 2021;4:548–558. [Google Scholar]
- Lowson R. Chem. Rev. 1982;82(5):461–497. [Google Scholar]
- Madrakian T., Maleki S., Heidari M., Afkhami A. Mater. Sci. Eng. C Mater. Biol. Appl. 2016;63:637–643. doi: 10.1016/j.msec.2016.03.041. [DOI] [PubMed] [Google Scholar]
- Marcus R. J. Electroanal. Chem. 1997;438(1–2):251–259. [Google Scholar]
- Margolin E., Burgers W., Sturrock E., et al. Prospects for SARS-CoV-2 diagnostics, therapeutics and vaccines in Africa. Nat. Rev. Microbiol. 2020;18:690–704. doi: 10.1038/s41579-020-00441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz I., Uyeda H., Goldman E., Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Moreels I., Lambert K., Smeets D., Muynck D., Nollet T., Martins J., Vanhaecke F., Vantomme A., Delerue C., Allan G., Hens Z. ACS Nano. 2009;3(10):3023–3030. doi: 10.1021/nn900863a. [DOI] [PubMed] [Google Scholar]
- Ranjan P., Singhal A., Yadav S., Kumar N., Murali S., Sanghi S., Khan R. Int. Rev. Immunol. 2021;40(1–2):120–142. doi: 10.1080/08830185.2021.1872566. [DOI] [PubMed] [Google Scholar]
- Ricci F., Adornetto G., Palleschi G. Electrochim. Acta. 2012;84(12):74–83. [Google Scholar]
- Sadique A., Yadav S., Ranjan P., Verma S., Salammal T., Khan A., Kaushik A., Khan R. J. Mater. Chem. B. 2021;9(23):4620–4642. doi: 10.1039/d1tb00472g. [DOI] [PubMed] [Google Scholar]
- Sadique A., Bhirde A., Yadav S., Ranjan P., Khan A., Kumar A., Khan R. Mater. Lett. 2021;305:130824. doi: 10.1016/j.matlet.2021.130824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. Electrochemical principles and characterization of bioelectrochemical systems. Microb. Electrochem. Fuel Cell. 2016:29–66. [Google Scholar]
- Seo G., Lee G., Kim M., Baek S., Choi M., Ku K., Lee C., Jun S., Park D., Kim H., Kim S., Lee J., Kim B., Park E., Kim S. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S., Ryo A. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shanmugam R., Barathi P., Kumar A. Colloid. Surface. 2014;452:129–137. [Google Scholar]
- Singhal A., Parihar A., Kumar N., Khan R. Mater. Lett. 2022;306:130898. doi: 10.1016/j.matlet.2021.130898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Hirakawa T., Sasaki S., Karube I. Anal. Chim. Acta. 1999;387(1):103–112. [Google Scholar]
- Tang J., Sargent E. Adv. Mater. 2010;23(1):12–29. doi: 10.1002/adma.201001491. [DOI] [PubMed] [Google Scholar]
- Tang J., Brzozowski L., Barkhouse D., Wang X., Debnath R., Wolowiec R., Palmiano E., Levina L., Pattantyus-Abraham A., Jamakosmanovic D., Sargent E. ACS Nano. 2010;4(2):869–878. doi: 10.1021/nn901564q. [DOI] [PubMed] [Google Scholar]
- Torres M., Araujo W., Lima L., Ferreira A., Fuente-Nunez C. Matter. 2021;4(7):2403–2416. doi: 10.1016/j.matt.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ye F., Fang T., Niu W., Liu P., Min X., Li X. J. Colloid Interface Sci. 2011;355(1):9–14. doi: 10.1016/j.jcis.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H., Barnes C., Cipolla M., Ramos V., Oliveira T., Cho A., Schmidt F., Silva J., Bednarski E., Aguado L., Yee J., Daga M., Turroja M., Millard K., Jankovic M., Gazumyan A., Zhao Z., Rice C., Bieniasz P., Caskey M., Hatziioannou T., Nussenzweig M. Nature. 2021;595(7867):426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J., Nocera D., Yocom K., Bordignon E., Gray H. J. Am. Chem. Soc. 1982;104(21):5798–5880. [Google Scholar]
- Winkler J., Malmstrm B., Gray H. Biophys. Chem. 1995;54(3):199–209. doi: 10.1016/0301-4622(94)00156-e. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) WHO; 2021. Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/ URL. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y., Wang W., Song Z., Hu Y., Tao Z., Tian J., Pei Y., Yuan M., Zhang Y., Dai F., Liu Y., Wang Q., Zheng J., Xu L., Holmes E., Zhang Y. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Sadique A., Ranjan P., Kumar N., Singhal A., Srivastava A., Khan R. ACS Appl. Bio Mater. 2021;4(4):2974–2995. doi: 10.1021/acsabm.1c00102. [DOI] [PubMed] [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Behbahan N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176(14):112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y., Tian G., Jiang H., Tao S., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H. Science. 2020;368(6498):abc1560. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., McGeer A., Christie N., Gray-Owen S., Cochrane A., Rini J., Sargent E., Kelley S. J. Am. Chem. Soc. 2021;143(4):1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- Zhang M., Li X., Pan J., Zhang Y., Zhang L., Wang C., Yan X., Liu X., Lu G. Biosens. Bioelectron. 2021;190:113421. doi: 10.1016/j.bios.2021.113421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Hao P., Li H., Zhang Z. ACS Sens. 2021;6(4):1613–1620. doi: 10.1021/acssensors.1c00022. [DOI] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H., Peiris M., Wu J. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 2