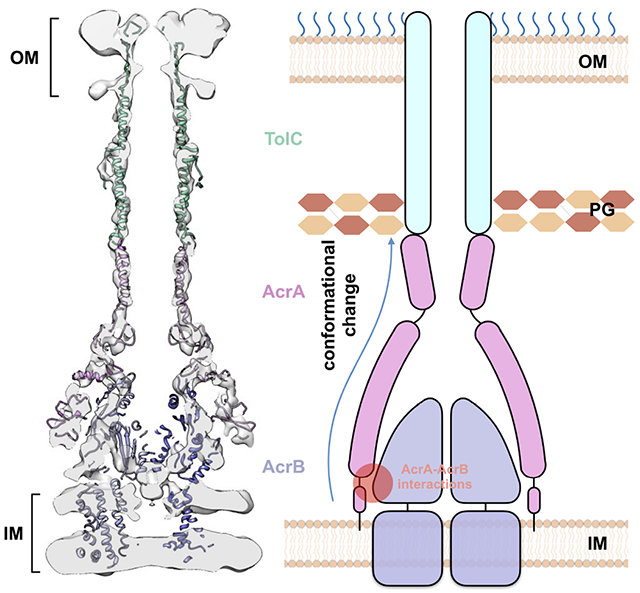
Summary
The tripartite AcrAB-TolC assembly, which spans both the inner and outer membranes in Gram-negative bacteria, is an efflux pump that contributes to multidrug resistance. Here, we present the in situ structure of full-length Escherichia coli AcrAB-TolC determined at 7 Å resolution by electron cryo-tomography. The TolC channel penetrates the outer membrane bilayer through to the outer leaflet and exhibits two different configurations that differ by a 60-degree rotation relative to the AcrB position in the pump assembly. AcrA protomers interact directly with the inner membrane and with AcrB via an interface located in proximity to the AcrB ligand-binding pocket. Our structural analysis suggests that these AcrA-bridged interactions underlie an allosteric mechanism for transmitting drug-evoked signals from AcrB to the TolC channel within the pump. Our study demonstrates the power of in situ electron cryo-tomography, which permits critical insights into the function of bacterial efflux pumps.
In Brief
The AcrAB-TolC assembly is a key efflux pump that contributes to multidrug resistance within Gram-negative bacteria. Spanning the cell envelope, it is a challenging target for in vitro structural studies. Chen et al. solve the subnanometer in situ structure of AcrAB-TolC, suggesting an allosteric mechanism during drug efflux.
Graphical Abstract
Introduction
Antibiotic resistance is an emerging crisis in healthcare. Bacterial efflux pumps, which actively expel a broad range of substances toxic to bacteria, play a major role in intrinsic and acquired drug resistance (Scheper et al., 1996). In Gram-negative bacteria, efflux pumps are multicomponent assemblies that span the inner and outer membranes (Du et al., 2015), making them challenging targets for in vitro structural studies. Isolation of the pumps requires stabilization of different transmembrane domains in detergents and disruption of the peptidoglycan layer that helps to constrain the components. It is a formidable challenge to form native complexes under in vitro conditions and expose them to the same conditions they experience in the cell, given that bacterial pumps span and function in the bacterial cell envelope comprising the inner and outer membranes with peptidoglycan wall between them and the lipopolysaccharide covering the outer membrane. In this study, we focus on the Escherichia coli tripartite efflux pump AcrAB-TolC, which is comprised of the inner membrane transporter AcrB, the outer membrane channel TolC, and the periplasmic membrane fusion protein AcrA. AcrAB-TolC is essential for intrinsic bacterial antibiotic resistance and when overexpressed facilitates acquired resistance as well as promoting other resistance mechanisms (Andersen et al., 2015; Fitzpatrick et al., 2017; Hocquet et al., 2003; Li et al., 2015; Masuda et al., 2000; Swick et al., 2011). Overexpression of AcrAB-TolC also occurs naturally and has been observed in numerous clinical isolates from patients with antibiotic-resistant bacterial infections (Li et al., 2015; Llanes et al., 2004; Narita et al., 2003; Poole, 2005; Swick et al., 2011; Webber and Piddock, 2002). Critically, the direct efflux of drugs and other substrates across the Gram-negative bacterial envelope via AcrAB-TolC requires its proper assembly in the bacteria to permit drug passage through periplasm to the extracellular medium (Eswaran et al., 2004; Morgan-Linnell et al., 2009; Nikaido, 2009).
Previously, we have determined in vitro structures of AcrAB-TolC complexes using cryo-EM single particle analysis (SPA) (Wang et al., 2017). These maps produced detailed molecular models of the entire pump assembly, providing important mechanistic insights. However, since the AcrAB-TolC assembly spans the entire cell envelope, the conditions used for in vitro structure determination of the isolated complexes do not capture the complexity of the in vivo environment. In our previous in vivo study, we observed that the AcrB inhibitor MBX3132 stabilizes the “open-TolC” conformation as observed in vitro, whereas the TolC unexpectedly has a closed state conformation in the presence of antibiotics (Shi et al., 2019; Sjuts et al., 2016) (Table S1). These results suggest that in situ interactions between the pump components and the surrounding membrane environment may play a critical role in the crosstalk between channel gating at TolC and substrate binding at AcrB through AcrA to activate the efflux process. Thus, high-resolution structures within the cell would greatly aid in understanding how the native bacterial environment impacts the pump components and how they allosterically communicate.
We have now resolved a 7 Å resolution structure of the AcrAB-TolC complex within E. coli cells in the presence of the MBX3132 inhibitor by cellular electron cryotomography (cryo-ET). With a combination of improved sample preparation, data collection parameters, a new subtomogram averaging software (Chen et al., 2019), and collection of a much larger data set, it was possible to achieve subnanometer resolution and provide a detailed view of the pump in its native environment. This structure has sufficient resolution to accurately fit the high-resolution models of AcrAB-TolC (Wang et al., 2017), permitting visualization of the interactions of the pump with both the inner and outer membranes and identification of inter-protomer interactions. These findings suggest how allosteric communications within the pump underlie the mechanism of drug efflux across the bacterial envelope.
Results
In situ structure of the AcrAB-TolC pump
From the reconstructed tomograms, densely packed arrays of AcrAB-TolC pumps are clearly visualized and show particles in the top and side views that appear as small circles and long thin rods spanning the cell envelope, respectively (Figure 1). From 102 tomograms, we manually selected 27,932 particles located in the envelope of intact cells. Following de novo initial model generation, the structure of the fully assembled pump was determined to 7 Å resolution with C3 symmetry using the subtilt refinement pipeline in EMAN2 (Chen et al., 2019) (Figure S1, Methods). At this resolution, we were able to identify sufficient alpha helices in both AcrA and TolC (Figures 2A and 2B) to accurately anchor available high-resolution structure (PDB: 5NG5) within cryo-ET density maps. Subtomogram classification analysis does not show a subpopulation of AcrAB subcomplexes without TolC which were observed in a large proportion in our previous in situ study in the presence of antibiotics (Shi et al., 2019) (Table S1). This implies that the inhibitor MBX3132 promotes and stabilizes the full-pump assembly.
Figure 1. Cryo-ET of E. coli expressing the AcrAB-TolC multidrug efflux pump.
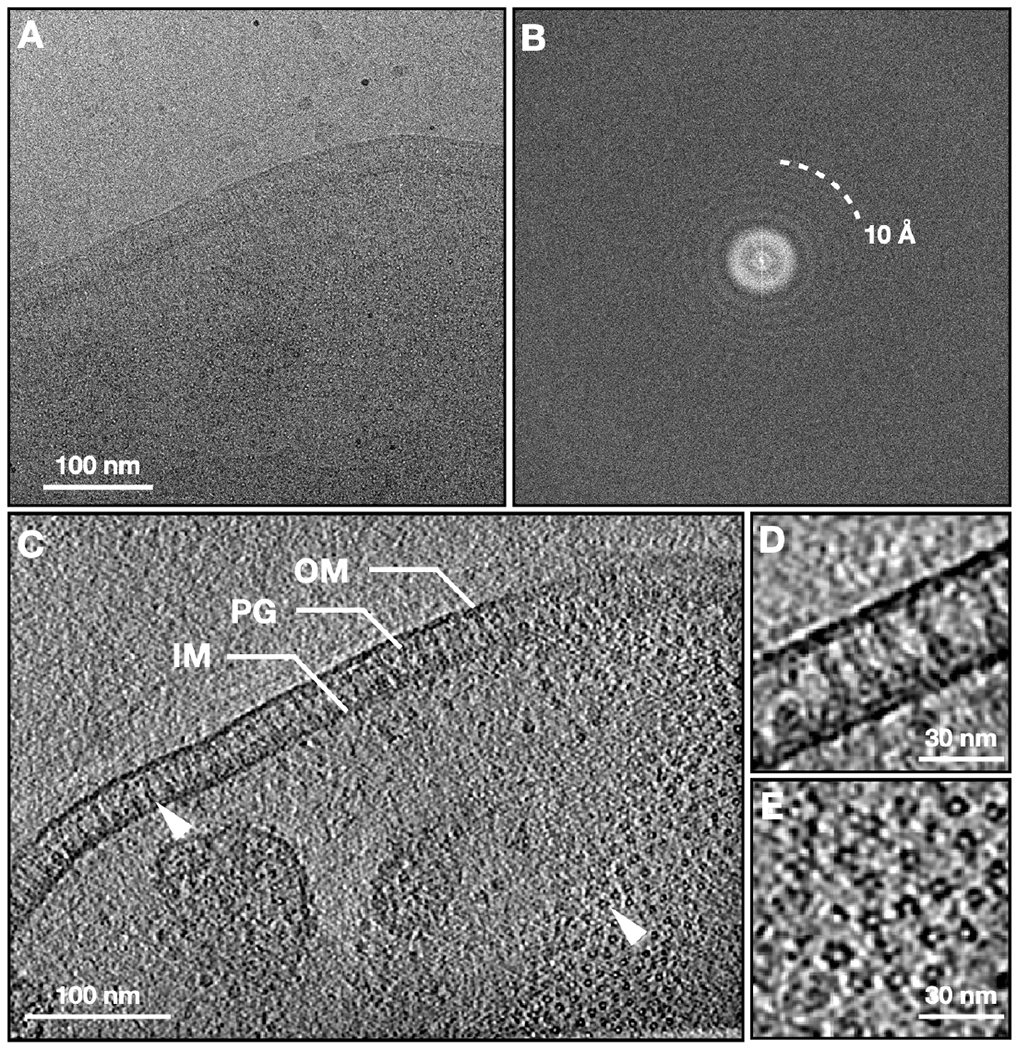
(A) Zero-degree tilt image of a representative tilt series. (B) Fourier transform of (A). (C) Slice view of the reconstructed tomogram, with arrowheads showing the side and top view particles. (D, E) Zoomed in view of the side and top view particles. OM, outer membrane; IM, inner membrane; PG, peptidoglycan.
Figure 2. In situ structure of the AcrAB-TolC multidrug efflux pump.
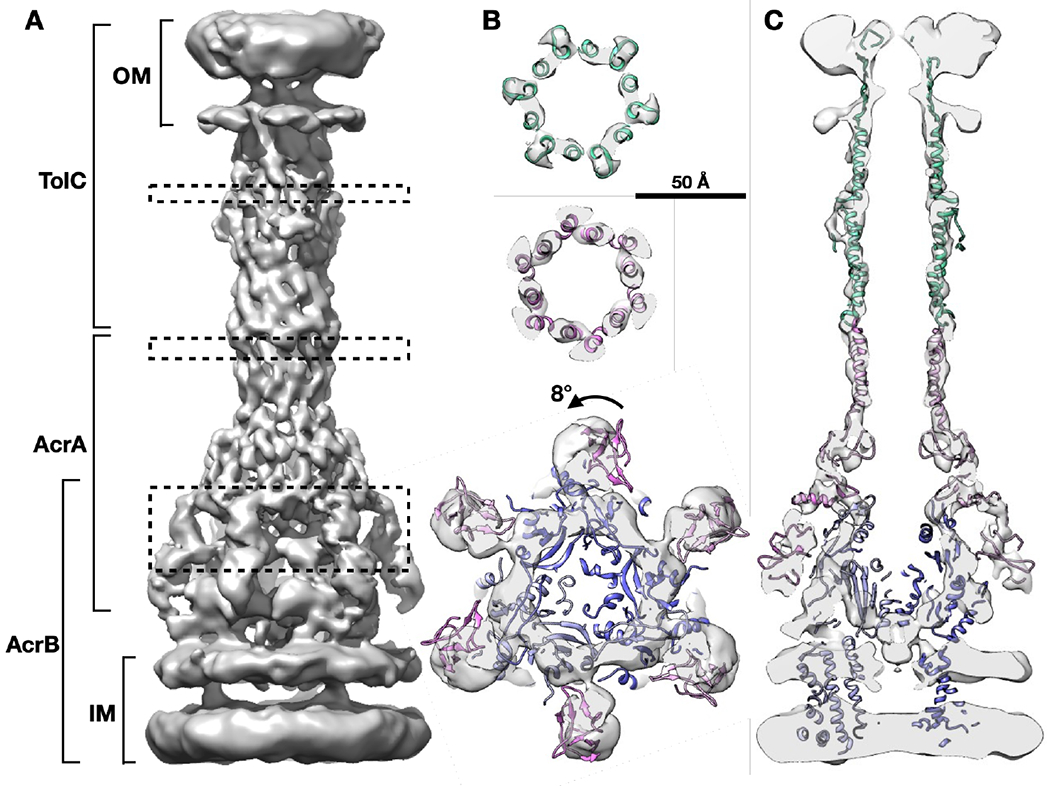
(A) Side view of the efflux pump density map. (B) Cross section views of the pump at the dashed box positions in (A), with the model of in vitro pump (PDB: 5NG5) fitted in density. (C) Cross section view through the center of the pump with the fitted model. OM, outer membrane; IM, inner membrane.
The pump forms an open conduit from the funnel domain of AcrB toward the tip of TolC (Figures 2A and 2C), which validates our previous prediction that MBX3132 will lock the assembled pumps in transporting state in vivo due to the saturation of the deep binding pocket (distal binding pocket) on AcrB (Shi et al., 2019; Sjuts et al., 2016). The bacterial peptidoglycan layer shows a weak density at the interface between AcrA and TolC that is consistent with our previous in situ studies (Shi et al., 2019). The alpha-helices identified in the TolC and AcrA region interacting with the TolC match well with the SPA structure of the AcrAB-TolC treated with inhibitor (PDB: 5NG5) (Wang et al., 2017). However, the membrane proximal (MP) domain of AcrA in the cryo-ET structure is rotated by 8 degrees (Figure 2B) compared with the aforementioned in vitro structure (PDB: 5NG5).
Focused refinement of the AcrAB-TolC pump reveals membrane-protein interactions
The density map is best resolved near the AcrA-TolC interface, but less well resolved in AcrB and the upper portion of TolC (Figure S1). Although the structure of the complex was determined with C3 symmetry, the TolC equatorial domains together with AcrA hexamer show a pseudo C6 symmetry. Thus, we performed focused refinements of the TolC region and the AcrB region separately, first with a C6 symmetry for a precise alignment then relaxed from C6 to C3 symmetry. This resulted in two density maps of the upper and lower parts of the pump.
In the focused refinement of TolC, the outer membrane beta-barrel, and equatorial helices are better resolved (Figure 3A). The resulting density map suggests that the outer leaflet of the outer membrane is thicker than the inner leaflet, indicating the presence of the polysaccharide layer above the outer membrane. The loop at the top of the TolC beta-barrel is buried inside the polysaccharide layer (Figure 3A), suggesting an interaction of the loop with the polysaccharide. It has been suggested by several studies that the interiors of outer membrane proteins are often partially or completely occluded (Deisenhofer et al., 1999; Ferguson et al., 1998; Locher et al., 1998; Pautsch and Schulz, 1998) and those exposed loops have conformational mobility and may close over as a partial external ‘lid’ (Koronakis et al., 2000). According to our structure, the outer mouth is open with a diameter >10 Å, enabling the unimpeded exit of substrates (Figure 3A).
Figure 3. The conformation of TolC and interactions of AcrA protomers with AcrB.
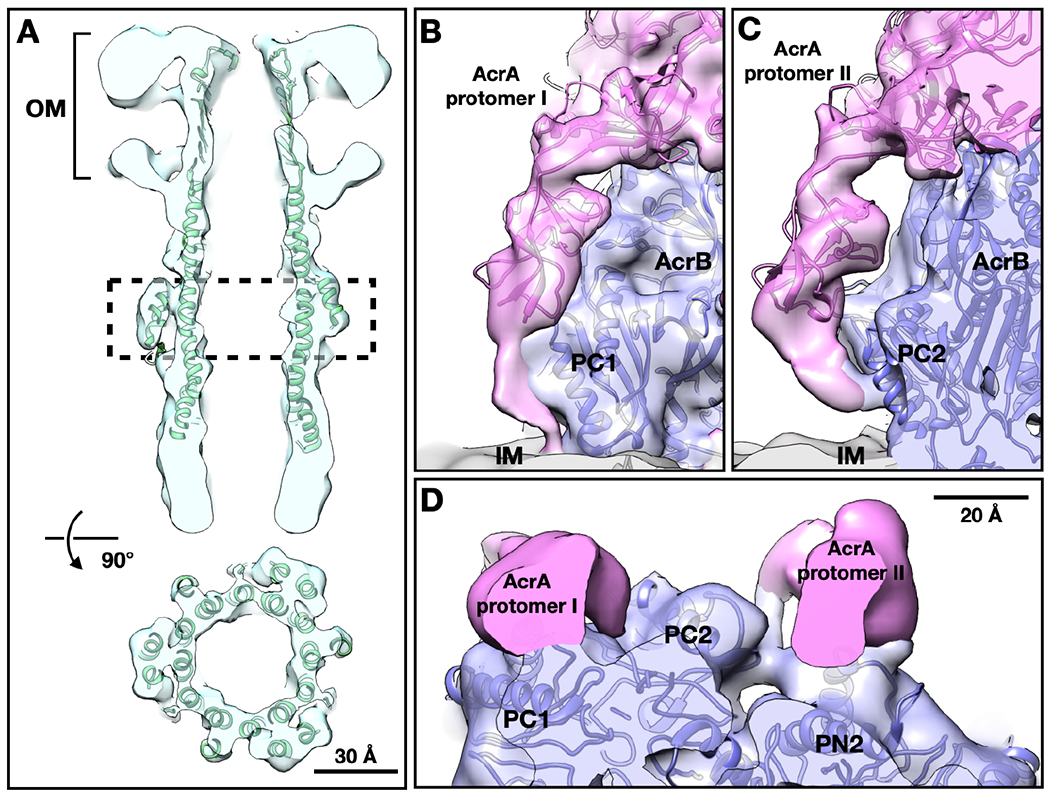
(A) Cross section views of TolC focused refinement result, with the model of in vitro pump (PDB: 5NG5) fitted in density. (B, C) Side view of the termini of the two AcrA protomers (pink) from the focused refinement result of the lower part of the pump. (D) Top view of the termini of AcrA protomer I on the left and II on the right. OM, outer membrane; IM, inner membrane.
The TolC, AcrA, and AcrB form a complex in a 3:6:3 ratio (Wang et al., 2017), which immediately leads to two possible relative orientations of TolC with respect to AcrB, with a 60-degree rotation. Both possible complexes are observed in our data with roughly equal occurrence (Figure S2), indicating that the AcrAB subcomplex does not have a preference in recruiting the TolC in either orientation. These two different conformations of the overall complex also likely result in subtly different interactions between all three elements. Two relative orientations between the channel component have also been observed in the homologous MexAB-OprM analyzed by single particle analyses (Glavier et al., 2020; Tsutsumi et al., 2019).
Compared to the in vitro structures of the entire pump, the focused refinement of the lower part of the pump suggests a clear C3, not C6, symmetry. Densities of the transmembrane helix bundles are visible in the map. Notably, the density of the AcrA terminus, which was not visible in all previous AcrAB-TolC structures, appears and strongly breaks the 6-fold symmetry (Figures 3B-3D). The AcrA termini have been previously shown to be critical in pump assembly and efflux activity (Ge et al., 2009; Zgurskaya et al., 2009). We found that the cryo-ET density formed by the N- and C-termini of six AcrA protomers exhibits two distinct conformations interacting either only with AcrB or with AcrB and the membrane bilayer (Figures 3B-3D). The AcrA membrane anchor has not been previously observed in the AcrAB-TolC pump. A membrane fusion protein (MFP)-membrane anchor, as well as different conformations of the MFP protomers were modeled in the MexAB-OprM tripartite complex reconstituted in a nanodisc (Glavier et al., 2020). Models, generated by Rosetta, for the AcrA terminal region fit well into these densities, where hydrophobic residues near the N-terminus can reasonably interact with the membrane (Zgurskaya et al., 2009) (Figure S3). These models do not consider the surrounding membrane and protein densities, but provide a conceptual description of our interpretation of the structure. Based on these models, it is plausible that the AcrA protomer I termini closely interacts with the groove of the porter domain of AcrB (G639-S656) and is anchored into the inner membrane (Figure 3B and 3D). The AcrA protomer II terminus sits between two AcrB subunits, contacting one AcrB subunit at the loop near S319, and interacts with the PC2 subregion in the porter domain of the neighboring AcrB (S834-K850) (Figure 3C and 3D).
Discussion
Mechanisms of the AcrAB-TolC pump active efflux process
The elongated AcrA molecule is anchored in the inner membrane and interacts with AcrB, while also reaching TolC and contacting the peptidoglycan layer. This architecture allows AcrA to bridge and communicate conformational changes in AcrB in the inner membrane to TolC in the outer membrane. In the in situ structure from this study, AcrA is observed to interact extensively with AcrB. Specifically, the AcrA protomer II terminal region contacts AcrB’s PC2 subregion (Figure 3C), which undergoes substantial conformational changes during transition from the binding to the extrusion state, and from the extrusion to the access state (Wang et al., 2017). This implies an allosteric mechanism for the transmission of drug-triggered conformational changes from AcrB to AcrA, which is supported by numerous functional and structural studies from different groups (Abdali et al., 2016; Du et al., 2014; Ge et al., 2009; Murakami et al., 2006; Seeger et al., 2006; Wang et al., 2017).
To compare structure conformational features, we made a superimposition focusing on the MFP and secondary transporter interacting region of the cryo-EM structure of nanodisc-reconstituted MexAB-OprM, the cryo-EM structure of purified AcrAB-TolC, and the in situ structure of AcrAB-TolC (Figure S4). We found that MexA and in vitro AcrA exhibit similar overall conformations within the pump assemblies (Figure S4A and S4C), however the N-terminal region was not resolved in in vitro AcrA. The in situ AcrA have deviations from the MexA conformations (Figure S4B and S4D), with the AcrA termini forming more secondary structures, which could favor it forming interactions with AcrB and the inner membrane and accomplish its functions.
In our cryo-ET density map, the in situ AcrA shows an 8-degree rotation in the AcrA membrane-proximal (MP) domain (Figure 2B) and interacts with the membrane bilayer, compared with the in vitro structure of AcrAB-TolC (PDB: 5NG5). This rotation is likely caused by the AcrA termini being in its native environment, anchored into the inner membrane, rather than bound to free surfactant as in the in vitro system. This result implies that lipid-protein interactions play an important role in maintaining the overall pump architecture as well as in providing a structural flexibility to support conformational changes as the pump goes between different states.
In isolation, TolC assumes a closed state (Koronakis et al., 2000; Wang et al., 2017) (Table S1) that could prevent leakage occurring from the periplasm. Antibiotic efflux requires TolC transitions to an open state to form a continuous conduit through the periplasmic space. Based on all currently available structural and functional data (Abdali et al., 2016; Du et al., 2014; Ge et al., 2009; Murakami et al., 2006; Seeger et al., 2006; Wang et al., 2017), we propose a model wherein drug binding in AcrB triggers a conformational change in the AcrB porter domain which is propagated to TolC through the TolC-AcrA interaction interface; this movement in the AcrB porter domain drives conformational changes in the AcrA assembly leading to a twisting of AcrA protomers that results in opening of the TolC channel (Figure 4). Based on our earlier studies, the TolC opening is coordinated via the tightly meshed hairpin regions of AcrA and TolC (Wang et al., 2017).
Figure 4. Model for the transmission of ligand-induced conformational signals from AcrB to TolC through AcrA during substrate transport.
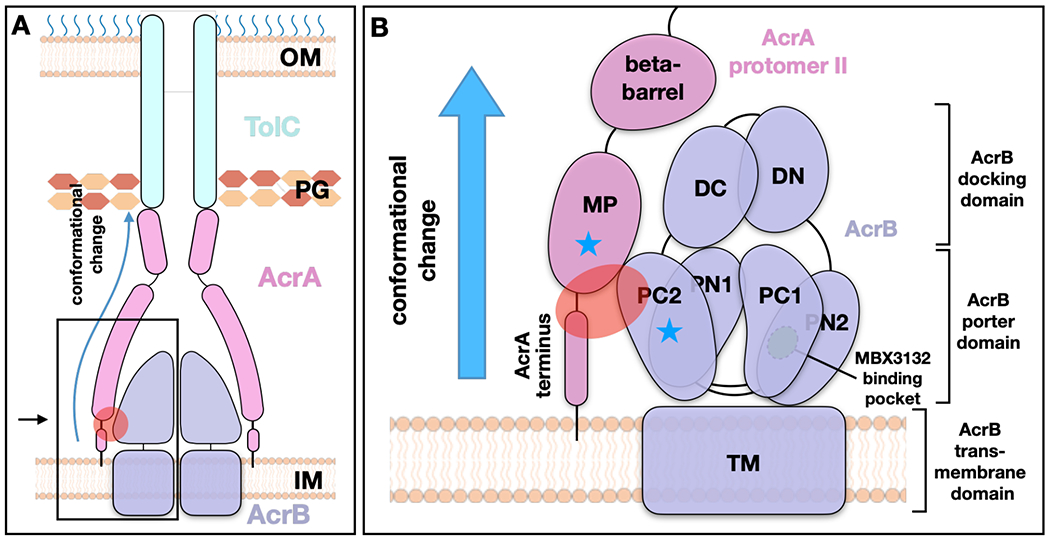
(A) Schematic view of the AcrAB-TolC efflux pump. (B) Zoomed in view of the boxed area in (A). The red oval indicates the interaction zone between AcrA protomer II and AcrB, and the blue stars mark the subregions that undergoes conformational changes. The MP domain of AcrA protomer II, anchored into the inner membrane, connects to the AcrB PC2 domain which has substantial conformational changes during transition from the binding to the extrusion state, and from the extrusion to the access state. The conformational change can be communicated through AcrA to TolC to affect the channel gating. OM, outer membrane; IM, inner membrane; PG, peptidoglycan.
In summary, our subnanometer in situ structures clearly demonstrates the importance of cellular environment in maintaining the proper structural interface and demonstrates the ability of cellular cryo-ET to retrieve critical structural data for understanding efflux activity. The improvement of resolution in the current study was achieved through a combination of cryo-EM imaging with a new generation transmission electron microscope, TFS Titan Krios G3, which is proven to be a high-resolution and high-throughput instrument with a stable specimen stage. In our current study the cryo-ET data were acquired with better sampling (smaller pixel size), closer to focus, and image processing was performed with a new alignment algorithm that uses the large-scale geometry information of cells to guide the particle alignment. In situ structures fill in a critical knowledge gap of the tripartite pump assembly, and provide new insights into the mechanism of antibiotic resistance and drug efflux. The AcrA-AcrB in situ interaction sites revealed by the in situ structure may also lead to a new direction in the future for drug development.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Zhao Wang (zhaow@bcm.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Density maps are available at EMDB with accession codes EMD-24609. EMAN2.9 is a free and open-source software available from http://eman2.org with source code on GitHub (https://github.com/cryoem/eman2). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
E. coli BL21 (DE3) strain was used for protein overexpression. Cells were grown in 2xYT medium at 37 °C and protein expression was induced at an OD600 of 0.8, by adding 0.1 mM IPTG for 16-18 hours at 20 °C.
METHOD DETAILS
Plasmid construction and protein expression
To overexpress the AcrAB-TolC multidrug efflux pump, E. coli BL21 (DE3) cells (Invitrogen) were co-transformed with plasmids pAcBH and pRSF-tolC, which carry the acrAB and tolC genes respectively (Fujihira et al., 2002; Shi et al., 2019). Cells were cultured in 2xYT medium with antibiotics (100 μg/ml ampicillin and 50 μg/ml kanamycin) at 37 °C until an OD600 of 0.8 was reached and then induced by addition of 0.1 mM isopropyl β- d-1-thiogalactopyranoside at 20 °C overnight.
Sample preparation
E. coli cultures were harvested and washed by PBS buffer, then resuspended to an OD600 of 1.0. Cells were mixed with MBX3132 (1.4 μg/ml) and incubated at 37 °C for 0.5 h. Subsequently, cells were mixed with 6 nm BSA fiducial gold (Aurion) and then 3μl mixture was deposited onto freshly glow-discharged, continuous floating carbon film covered grids (Quantifoil Au R3.5/1, 200 mesh). The grids were blotted with filter paper for 4 s and plunge frozen in liquid ethane by using a Vitrobot Mark IV (FEI). Grids were stored in liquid nitrogen until data collection.
Cryo-ET data collection
Similar to our previous experiments (Shi et al., 2019), the cryo-ET data acquisition uses a bidirectional tilt scheme. The frozen-hydrated samples were imaged on a 300 kV Titan Krios transmission electron microscope (FEI) using a K2 Summit direct electron detector camera (Gatan), with a magnification of 81,000x. The pixel size is calibrated to be 1.67 Å. The tilt-series images were acquired at −1 to −2 μm defocus range with an average cumulative dose of ~90 e−/Å2 distributed over 34 images and covering an angular range of −51 to +51 degrees, with an angular increment of 3degree. 192 tilt series were collected in a 4-day imaging session.
Cryo-ET Data processing
First, movie frames of each individual tilt image are aligned using MotionCorr2. The tilt series alignment and tomogram reconstruction are performed using the automated workflow in EMAN2. Using the same workflow, defocus is determined for each tilt image, and CTF correction is performed at a per-particle-per-tilt level during particle extraction.
For the refinement of the full pump, 27,932 particles of the efflux pump are selected manually from 102 tomograms. The particles are then extracted from the tilt series with a box size of 256. 15 iterations of subtomogram refinement, followed by 4 iterations of sub-tilt refinement are performed using those particles and an initial model directly generated from the particles. The refinement process follows the “gold-standard” procedure, where particles are split into two independent subsets and aligned using different reference models that are phase randomized to 20 Å. At each iteration of the subtomogram refinement step, the worst 30% of the particles are excluded based on their similarity to the averaged structure. In the sub-tilt refinement step, images beyond 45 degrees are excluded, so are the worst 20% of the sub-tilt images.
During the subtomogram refinement, the large-scale geometry information of the cells is used to guide the alignment and prevent the particles from being aligned up-side-down. Because the pump particles are tightly packed on the cell envelope, we simply draw a vector from the center of all particles in a tomogram to each particle and rotate a particle by 180 degrees around the x axis if the particle is facing the opposite direction of the vector.
After subtomogram refinement of the full pump, particles of the upper and lower parts of the pump are re-extracted from the tilt series with a box size of 128, using the alignment information of the full pump. 10 iterations of subtomogram refinement are performed for each type of particle, using the corresponding part from the full pump structure as the initial model. The orientation search during the refinement is limited to 30 degrees from the Euler angle assigned in the full pump refinement. A symmetry breaking from C6 to C3 is also performed during the refinement. That is, for each particle after each round of the refinement, the similarity scores at the current orientation and at the C2 symmetrical orientation are compared, and the particle is assigned to its best orientation.
Since the entire refinement follows the “gold-standard” procedure (Scheres and Chen, 2012), the reported resolution is measured by the spatial frequency that the Fourier Shell Correlation (FSC) curve between the masked structures determined from the two subsets of particles dropping below 0.143. Local resolution is measured by tiling the 3D structure with small overlapping cubes and calculating the FSC using the same criteria.
Model building
Modeling was performed by rigid body fitting of the high resolution cryo-EM structures (PDB: 5NG5) into the cryo-ET density map using the Fit in Map tool in UCSF Chimera (Pettersen et al., 2004). To visualize the domain rotation of the full pump, the fitting focused on the helical hairpin domain of AcrA. To show the interface between AcrA terminus and AcrB, the model of AcrA membrane proximal domain and AcrB were fit into the focused refinement result respectively, and densities corresponding to the AcrB model were masked out from the averaged structure.
To further analyze the conformational differences between the in situ and in vitro pump structures, we flexibly fit our previous single particle structure of AcrAB-TolC complex with MBX3132 (PDB: 5NG5) into the in situ density map. First, the multimeric AcrA, AcrB and TolC portions of the 5NG5 were extracted and fit directly to the in situ map using UCSF Chimera. Models for the N-terminal portion of AcrA were generated using Rosetta, aligned to the larger AcrA subunit and refined with Phenix. The individual subunit complexes were then serially flexibly fit to the density map using the simulated annealing option in Phenix’s real space refinement tool, as well as FlexEM (Topf et al., 2008). Fittings with Phenix and FlexEM gave similar results. The individual complexes were then combined back into a single model and iteratively refined to maximize both fit to density and stereochemistry using real space refinement from Phenix (Adams et al., 2010) and manual optimization with Coot (Emsley et al., 2010). As a note, the beta-sheet domain of TolC was not refined to the density due to the lack of features in this region. Rather, the final model contains the TolC beta-sheet extracted from 5NG5 concatenated to the refined portion of TolC. The final model was then assessed using MolProbity (Chen et al., 2009) and fit to density.
QUANTIFICATION AND STATISTICAL ANALYSIS
The cryo-ET data was processed using EMAN2 (Chen et al., 2019). Model building was performed with UCSF Chimera (Pettersen et al., 2004), FlexEM (Topf et al., 2008), Phenix (Adams et al., 2010), Coot (Emsley et al., 2010), and MolProbity (Chen et al., 2009).
Supplementary Material
Movie S1. Visualization of the AcrAB-TolC efflux pump in situ, from cellular tomograms to averaged structures. Related to Figure 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 (DE3) | New England Biolabs | Cat#C2527H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MBX3132 | Sjuts et al., 2016 | N/A |
| Deposited Data | ||
| Subnanometer in situ structure of AcrAB-TolC bound with inhibitor | This study | EMD-24609 |
| Cryo-EM structure of AcrAB-TolC bound with inhibitor | Wang et al., 2017 | PDB: 5NG5 |
| Cryo-EM structure of AcrAB-TolC bound with antibiotics | Wang et al., 2017 | PDB: 5O66 |
| MexAB-OprM efflux pump reconstituted in nanodiscs | Glavier et al., 2020 | PDB: 6TA6 |
| Recombinant DNA | ||
| pAcBH | Fujihira et al., 2002 | N/A |
| pRSF-TolC | Shi et al., 2019 | N/A |
| Software and Algorithms | ||
| EMAN2 | Chen et al., 2019 | http://eman2.org |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| FlexEM | Topf et al., 2008 | http://topf-group.ismb.lon.ac.uk/flex-em/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MolProbity | Chen et al., 2009 | http://molprobity.biochem.duke.edu/ |
Highlights.
The in situ structure of AcrAB-TolC efflux pump is achieved at 7 Å resolution
The TolC channel penetrates the outer membrane bilayer through to the outer leaflet
Domain interactions underlie an allosteric mechanism for pump opening and close
Cellular cryo-ET reveals critical structural details relevant to pump activity
Acknowledgements
We thank Xinzhe Yu, Tong Huo and Hongjiang Wu for suggestions on sample preparation; Tom Dendooven and Angela Kirykowicz for helpful comments; and Snekalatha Raveendran for data backup. We thank Tim Opperman and colleagues for the kind gift of the AcrB inhibitor. This work was supported by the Welch Foundation (Q-1967-20180324; AU-2014-2019330), BCM BMB department seed and junior seed funds, NIH R01GM080139, R01GM072804, R01GM143380 and P01GM121203. B.F.L. is supported by an ERC Advanced Award (742210).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Abdali N, Parks JM, Haynes KM, Chaney JL, Green AT, Wolloscheck D, Walker JK, Rybenkov VV, Baudry J, Smith JC, et al. (2016). Reviving Antibiotics: Efflux Pump Inhibitors That Interact with AcrA, a Membrane Fusion Protein of the AcrAB-TolC Multidrug Efflux Pump. Acs Infect Dis 5, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biological Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, He G-X, Kakarla P, KC R, Kumar S, Lakra WS, Mukherjee MM, Ranaweera I, Shrestha U, Tran T, et al. (2015). Multidrug Efflux Pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus Bacterial Food Pathogens. Int J Environ Res Pu 12, 1487–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Bell JM, Shi X, Sun SY, Wang Z, and Ludtke SJ (2019). A complete data processing workflow for cryo-ET and subtomogram averaging. Nat Methods 16, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2009). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biological Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J, Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, and Helm D. van der (1999). Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol 6, 56–63. [DOI] [PubMed] [Google Scholar]
- Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, and Luisi BF (2014). Structure of the AcrAB–TolC multidrug efflux pump. Nature 509, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Veen H.W. van, Murakami S, Pos KM, and Luisi BF (2015). Structure, mechanism and cooperation of bacterial multidrug transporters. Curr Opin Struc Biol 33, 76–91. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr Sect D Biological Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Koronakis E, Higgins MK, Hughes C, and Koronakis V (2004). Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struc Biol 14, 741–747. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Hofmann E, Coulton JW, Diederichs K, and Welte W (1998). Siderophore-Mediated Iron Transport: Crystal Structure of FhuA with Bound Lipopolysaccharide. Science 282, 2215–2220. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AWP, Llabrés S, Neuberger A, Blaza JN, Bai X-C, Okada U, Murakami S, Veen H.W. van, Zachariae U, Scheres SHW, et al. (2017). Structure of the MacAB–TolC ABC-type tripartite multidrug efflux pump. Nat Microbiol 2, 17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihira E, Tamura N, and Yamaguchi A (2002). Membrane Topology of a Multidrug Efflux Transporter, AcrB, in Escherichia coli. J Biochem 131, 145–151. [DOI] [PubMed] [Google Scholar]
- Ge Q, Yamada Y, and Zgurskaya H (2009). The C-Terminal Domain of AcrA Is Essential for the Assembly and Function of the Multidrug Efflux Pump AcrAB-TolC ▿. J Bacteriol 191, 4365–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavier M, Puvanendran D, Salvador D, Decossas M, Phan G, Garnier C, Frezza E, Cece Q, Schoehn G, Picard M, et al. (2020). Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex. Nat Commun 11, 4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D, Vogne C, Garch FE, Vejux A, Gotoh N, Lee A, Lomovskaya O, and Plésiat P (2003). MexXY-OprM Efflux Pump Is Necessary for Adaptive Resistance of Pseudomonas aeruginosa to Aminoglycosides. Antimicrob Agents Ch 47, 1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, and Hughes C (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919. [DOI] [PubMed] [Google Scholar]
- Li X-Z, Plésiat P, and Nikaido H (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28, 337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanes C, Hocquet D, Vogne C, Benali-Baitich D, Neuwirth C, and Plésiat P (2004). Clinical Strains of Pseudomonas aeruginosa Overproducing MexAB-OprM and MexXY Efflux Pumps Simultaneously. Antimicrob Agents Ch 48, 1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, and Moras D (1998). Transmembrane Signaling across the Ligand-Gated FhuA Receptor. Cell 95, 771–778. [DOI] [PubMed] [Google Scholar]
- Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, and Nishino T (2000). Substrate Specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM Efflux Pumps in Pseudomonas aeruginosa. Antimicrob Agents Ch 44, 3322–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Linnell SK, Boyd LB, Steffen D, and Zechiedrich L (2009). Mechanisms Accounting for Fluoroquinolone Resistance in Escherichia coli Clinical Isolates▿. Antimicrob Agents Ch 53, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Matsumoto T, and Yamaguchi A (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179. [DOI] [PubMed] [Google Scholar]
- Narita S, Eda S, Yoshihara E, and Nakae T (2003). Linkage of the efflux-pump expression level with substrate extrusion rate in the MexAB–OprM efflux pump of Pseudomonas aeruginosa. Biochem Bioph Res Co 308, 922–926. [DOI] [PubMed] [Google Scholar]
- Nikaido H (2009). Multidrug Resistance in Bacteria. Biochemistry-Us 78, 119–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautsch A, and Schulz GE (1998). Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol 5, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Poole K (2005). Efflux-mediated antimicrobial resistance. J Antimicrob Chemoth 56, 20–51. [DOI] [PubMed] [Google Scholar]
- Scheper RJ, Scheffer GL, Flens MJ, Valk P. van der, Broxterman HJ, and Izquierdo MA (1996). Transporter molecules in multi drug resistance. Cytotechnology 19, 187–190. [DOI] [PubMed] [Google Scholar]
- Scheres SHW, and Chen S (2012). Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, and Pos KM (2006). Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism. Science 313, 1295–1298. [DOI] [PubMed] [Google Scholar]
- Shi X, Chen M, Yu Z, Bell JM, Wang H, Forrester I, Villarreal H, Jakana J, Du D, Luisi BF, et al. (2019). In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat Commun 10, 2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjuts H, Vargiu AV, Kwasny SM, Nguyen ST, Kim H-S, Ding X, Ornik AR, Ruggerone P, Bowlin TL, Nikaido H, et al. (2016). Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc National Acad Sci 113, 3509–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swick MC, Morgan-Linnell SK, Carlson KM, and Zechiedrich L (2011). Expression of Multidrug Efflux Pump Genes acrAB-tolC, mdfA, and norE in Escherichia coli Clinical Isolates as a Function of Fluoroquinolone and Multidrug Resistance ‡. Antimicrob Agents Ch 55, 921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, and Sali A (2008). Protein structure fitting and refinement guided by cryo-EM density. Struct Lond Engl 1993 16, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi K, Yonehara R, Ishizaka-Ikeda E, Miyazaki N, Maeda S, Iwasaki K, Nakagawa A, and Yamashita E (2019). Structures of the wild-type MexAB–OprM tripartite pump reveal its complex formation and drug efflux mechanism. Nat Commun 10, 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fan G, Hryc CF, Blaza JN, Serysheva II, Schmid MF, Chiu W, Luisi BF, and Du D (2017). An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber MA, and Piddock LJV (2002). The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemoth 51, 9–11. [DOI] [PubMed] [Google Scholar]
- Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, and Krishnamoorthy G (2009). Structural and functional diversity of bacterial membrane fusion proteins. Biochimica Et Biophysica Acta Bba - Proteins Proteom 1794, 794–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Visualization of the AcrAB-TolC efflux pump in situ, from cellular tomograms to averaged structures. Related to Figure 1.
Data Availability Statement
Density maps are available at EMDB with accession codes EMD-24609. EMAN2.9 is a free and open-source software available from http://eman2.org with source code on GitHub (https://github.com/cryoem/eman2). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


