Summary
The type 1 ryanodine receptor (RyR1)/calcium release channel on the sarcoplasmic reticulum (SR) is required for skeletal muscle excitation-contraction coupling and is the largest known ion channel, comprised of four 565 kDa protomers. Cryogenic electron microscopy (cryoEM) studies of the RyR have primarily used detergent to solubilize the channel; in the present study, we have used cryoEM to solve high-resolution structures of the channel in liposomes using a gel-filtration approach with on-column detergent removal to form liposomes and incorporate the channel simultaneously. This allowed us to resolve the structure of the channel in the primed and open states at 3.4 and 4.0 Å, respectively, with a single dataset. This method offers validation for detergent-based structures of the RyR and offers a starting point for utilizing a chemical gradient mimicking the SR, where Ca2+ concentrations are millimolar in the lumen and nanomolar in the cytosol.
Graphical Abstract
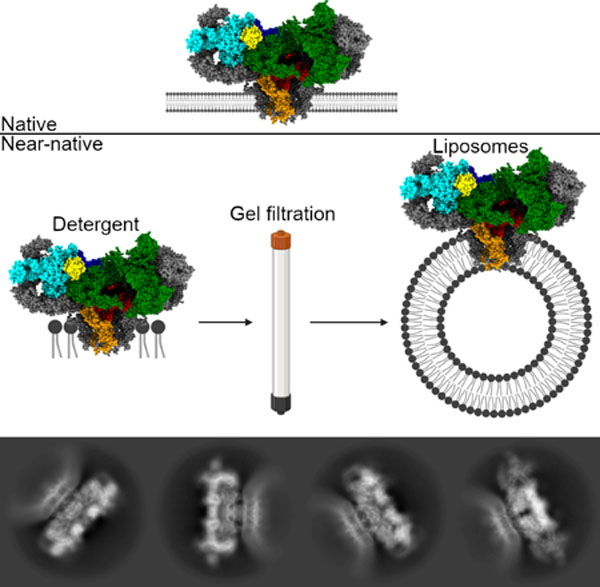
Blurb
Melville et al. show the cryo-electron microscopy structure of the ryanodine receptor calcium release channel in a lipid membrane. This is significant in the validation of previously published structures, resolved in detergent, and in providing an avenue for studying the structure and dynamics of this channel under a chemical gradient.
Introduction
Located on the sarco/endoplasmic reticulum (SR/ER) membrane, the two megadalton ryanodine receptor (RyR) is the largest known ion channel (Kushnir et al., 2018; Santulli et al., 2018). In skeletal muscle, RyR1 is required for SR Ca2+ release during excitation-contraction coupling (EC coupling). There are three RyR isoforms, RyR1 is the primary isoform in skeletal muscle while RyR2 is the cardiac isoform, though both are found in numerous tissues including neurons (Furuichi et al., 1994; Giannini et al., 1995; Marks et al., 1989). RyR3 is widely expressed but at significantly lower levels (Lanner et al., 2010). In skeletal muscle, RyR1 is thought to be activated by direct interaction with the dihydropyridine receptor (DHPR) (Kushnir and Marks, 2010; Marx and Marks, 2013), whereas RyR2 is activated by Ca2+ in a process termed calcium-incuded calcium release (CICR) (Fabiato, 1983; Fabiato and Fabiato, 1978). In short, Ca2+ influx via the DHPR triggers the release of Ca2+ via RyR2, which in turn creates a high local concentration of Ca2+ that activates neighboring RyR2 tetramers. RyR channels are tightly packed in checkerboard arrays on the SR and exhibit cooperative activation/deactivation through the process termed coupled gating (Marx et al., 2001; Marx et al., 1998; Porta et al., 2012; Zalk and Marks, 2017). Leaky RyR channels are associated with numerous diseases and disorders including muscular dystrophy (Bellinger et al., 2009), heart failure (Marx et al., 2000), cardiac arrhythmias (Dridi et al., 2020a), diabetes (Santulli et al., 2015), Huntington’s Disease (Chen et al., 2011; Dridi et al., 2020b; Suzuki et al., 2012), and Alzheimer’s (Bussiere et al., 2017; Kushnir et al., 2018; Lacampagne et al., 2017; Santulli et al., 2018).
The high resolution structures of individual, detergent solubilized RyR1 (Bai et al., 2016; des Georges et al., 2016; Wei et al., 2016; Zalk et al., 2015) and RyR2 (Chi et al., 2019) channels have been solved using cryogenic electron microscopy (cryoEM) (Zalk and Marks, 2017); however, solving the structure of proteins in native or near-native membranes remains challenging. Most cryoEM structures of membrane proteins use detergent micelles, amphipols, or nanodiscs (Bayburt and Sligar, 2010; Efremov et al., 2017; Efremov et al., 2014; Leitz et al., 2006; Nath et al., 2007). Liposomes, along with linear and circularized nanodiscs, allow for membrane protein incorporation while polymers such as DIBMA and SMA allow for native membrane extractions into so called native nanodiscs (Grethen et al., 2017; Oluwole et al., 2017a; Oluwole et al., 2017b; Swainsbury et al., 2017); however, critically, liposomes are the only method that allows for the creation of asymmetric environments, such as that of the SR. In the lumen of the SR, millimolar concentrations of Ca2+ are stored and maintained by the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) and calsequestrin, while the cytosol of the cell remains at approximately 150 nM Ca2+ at rest and low micromolar following Ca2+ release through the RyR. Liposomes form spontaneously as detergent is removed from solution during dialysis, or through the use of Bio-beads that adsorb detergent molecules. Lige Tonggu & Liguo Wang have shown that liposomes can also be formed via gel filtration (Tonggu and Wang, 2020). This method allows for significantly greater control over the size of the liposomes formed compared to dialysis1 and allowed for the successful incorporation of the large conductance calcium-activated potassium (BK) channel into liposomes (Wang and Sigworth, 2009).
Given the critical role of the RyR in normal and pathologic physiology, it is important to understand the structure and dynamics of the channel in the native context and to compare these structures to detergent solubilized channels. To this end, the structure and dynamics of proteoliposomal RyR1 was found to match that of detergent, offering validation for the detergent-resolved structures. As with detergent-resolved structures of the RyR, the dynamics of the cytosolic shell undergoes dramatic conformation changes in both the open and primed states of RyR1 channels. These movements are separate from the opening of the channel, marked by the dilation of the pore; however, these changes are difficult to understand in the context of an array of channels, each of which is connected to its neighbors at the four corners of the cytosolic domain (Franzini-Armstrong and Jorgensen, 1994; Franzini-Armstrong et al., 1998; Sun et al., 1995). In a truly native context, these dynamics should be muted, but it seems a fully-native approach may be necessary to capture these coupled channels. While we were unable to observe any such coupled channels in liposomes, we have successfully resolved the structure of RyR1 in liposomes in both the primed and open states to 3.4 and 4.0 Å, respectively, through a simple adaption of the gel filtration approach of liposome formation and incorporation. In doing so, we have established a framework for studies more closely mimicking the native environment.
Results
Our approach, detailed in Methods, is based on that of Tonggu & Wang (Tonggu and Wang, 2020) and resulted in small, consistent liposomes compared to forming liposomes by dialysis, which resulted in a wide range of liposome sizes (Figure 1A). Notably, the smallest liposomes were found in the centers of grid holes, where the ice is thinnest, and these liposomes were far more likely to contain one or more channels compared to larger liposomes found along the edges. This method also offered a greater than 20-fold increase in the RyR1 incorporation rate compared to a dialysis-based approach with approximately 8 channels per micrograph compared to 1 particle per 3 micrographs. While many particles of unincorporated RyR1 were still present, these were successfully discarded during 2d classification by increasing the box size to ensure the membrane would be clear and present as well as increasing the number of classes to weed out unincorporated RyR1. Despite the lack of top and bottom views of the channel, 360° of side views were present in addition to oblique views (Figure S3), allowing for a full 3d reconstruction of the channel in a lipid membrane, showing the potential of this method to achieve high-resolution cryoEM structures of proteins in membranes. Finally, the inclusion of 30 µM Ca2+free, 2 mM ATP, and 5 mM caffeine allowed us to capture RyR1 in the primed state as well as the open state from a single dataset.
Figure 1.
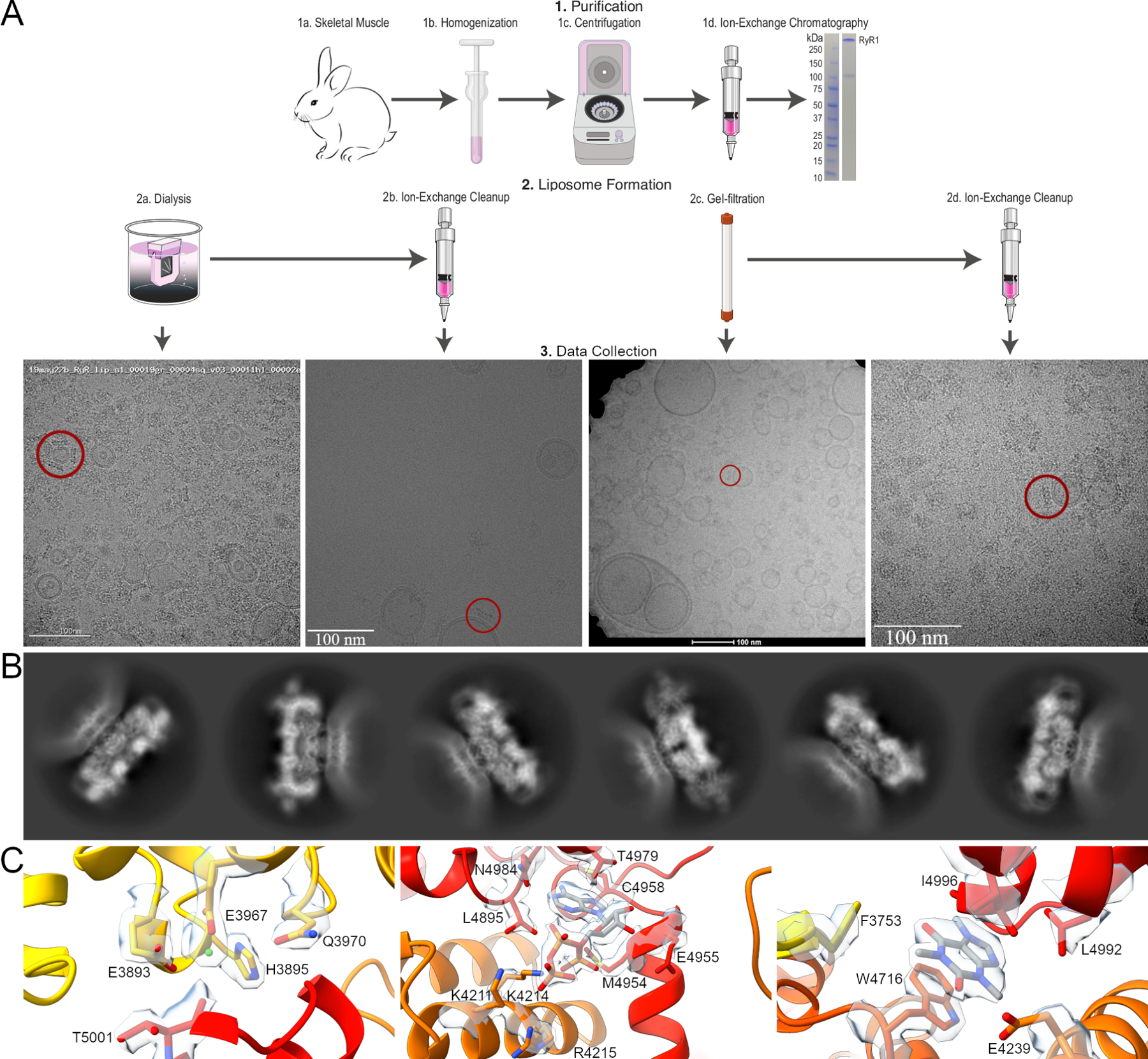
Workflow diagram for the preparation of proteoliposomal RyR1. A. RyR1 was purified from rabbit skeletal muscle using ion-exchange chromatography prior to liposome formation and incorporation by dialysis or gel filtration. For both methods, a second round of ion-exchange chromatography was employed to eliminate unincorporated RyR1 and empty liposomes. An embedded channel is highlighted in representative micrographs. B. 2D classes of proteoliposomal RyR1 showing the flattening of the lipid membrane around the TM domain of the channel. The first three show the primed state of the channel with a flat lipid membrane while the second three show the open state with the bending of the surrounding membrane. C. Representative density for the Ca2+, ATP, and caffeine binding sites of RyR1 in the primed state of proteoliposomal RyR1, resolved to 3.4 Å.
The juxtamembrane helices at the top of the TM interface in each protomer make several contacts with the lipid membrane, however the lipid membrane is too dynamic to clearly identify these interactions. These helices are likely responsible for the flattening of the lipid membrane around the TM domain of the RyR present in 2D classes (Figure 1B). Likewise, the dynamics of the cytosolic shell of the liposome embedded channel conform to what has previously been observed. The detergent solubilized RyR1 structure undergoes dramatic conformation changes that are separate from channel opening, in both the primed and open states (Movies 1&2) (des Georges et al., 2016; Zalk et al., 2015). The cytosolic shell of RyR1 rises and falls analogous to flapping wings. This movement is matched in the liposome embedded channels (Movies 3&4), though these movements are likely to be exaggerated in the absence of an array of channels. Analysis of the calstabin binding site, along with the binding sites for calcium, ATP, and caffeine showed no significant changes between detergent solubilized and liposome embedded RyR1 channels for both the primed and open states, however the improved resolution in the primed state allows for unambiguous assignment of the orientation of caffeine within the binding site (Figure 1C).
The overall structure of proteoliposomal RyR1 matches well with detergent-resolved structures with RMSD values <1 Å when aligning at the pore (residues 4866–5037) (Figure 2); however, the 7th transmembrane helix is not visible. Local refinement allowed for significant resolution improvements in many regions (Figure 3, Movies 5-8). Additional assignments were made in many unstructured loops and in the SPRY domain (residues 1420–1572, Figure 3C & Movie 9). Although the conformation of the bridging-solenoid is conserved, a dramatic shift was found as a result of the improved resolution. The bridging-solenoid was resolved at 3.1 Å in the primed state (3.45 Å in the open state), making it possible to assign residues 3064–3113 and revealing a shift of up to fifty residues compared to previously published structures (Figure 4, Movie 10) (des Georges et al., 2016; Iyer et al., 2020). The shifted region (residues 3021–3550) was unassigned in des Georges et al but was assigned in Iyer et al with the exception of residues 3064–3113.
Figure 2.
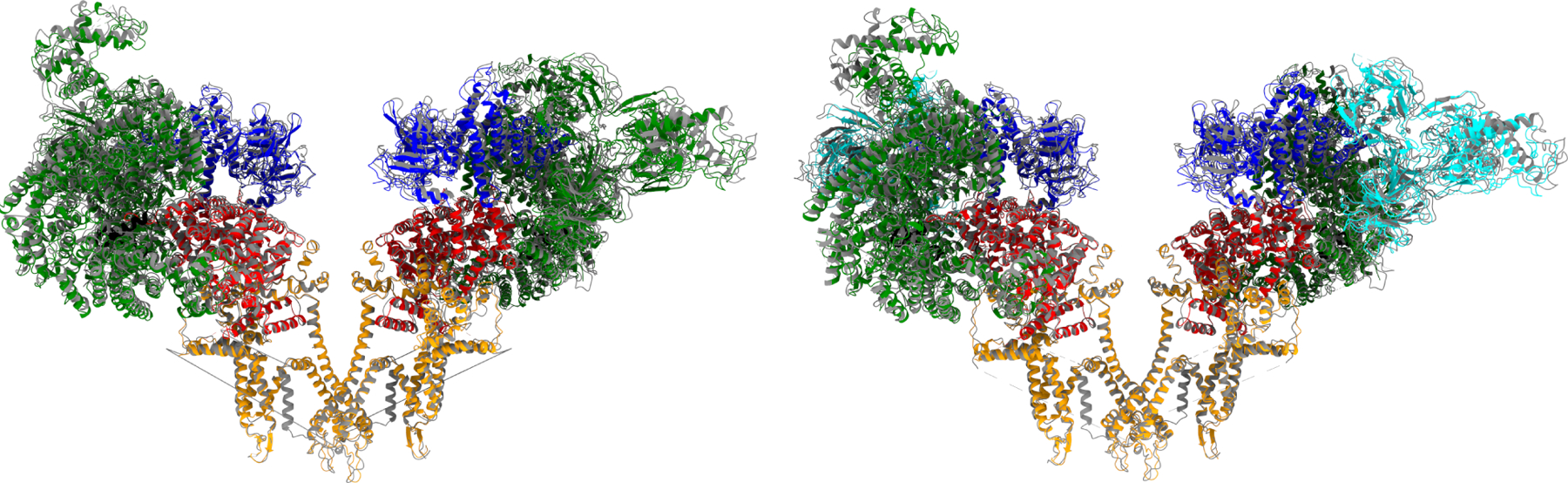
Side views of opposing RyR1 subunits. Ribbon diagrams are shown in the primed (left) and open (right) states overlayed at the pore (residues 4866–5037) with detergent. Detergent structures are 5TAQ and 5TAL, respectively, and colored in grey. Proteoliposomal RyR1 is colored by subunits (Zalk and Marks, 2017). RMSD values are 0.90 and 0.93 Å for aligning at the pore.
Figure 3.
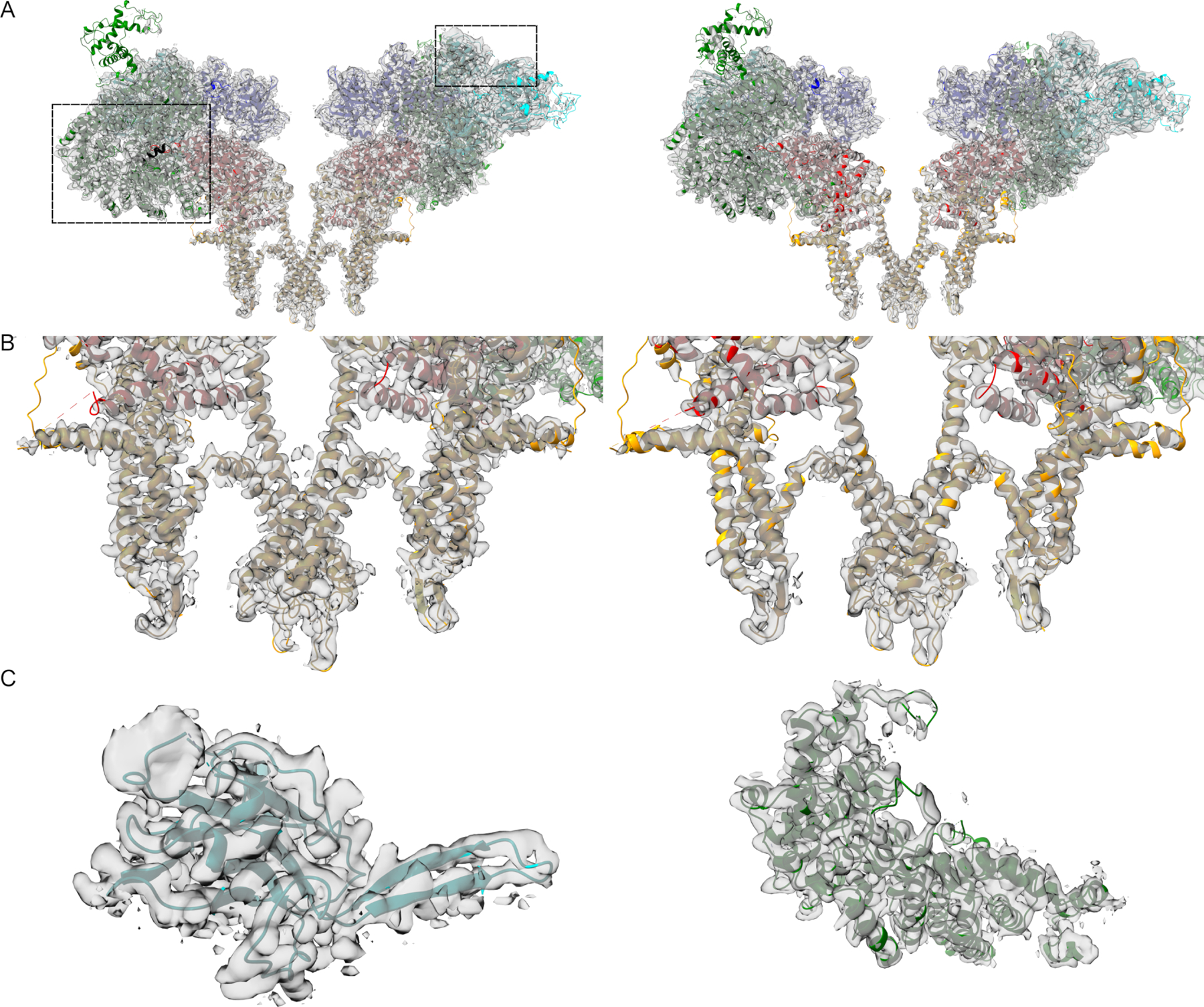
Coulombic density and fitted models. Proteoliposomal RyR1 is shown in the primed state (left) and open state (right) as side views and close-up views of the pore. Only two protomers are shown for clarity. C. Additional assignments in the SPRY (left) and Br-sol (right) domains. Residues 1420–1565 are shown in the SPRY domain and residues 3021–3550 in the Br-sol.
Figure 4.
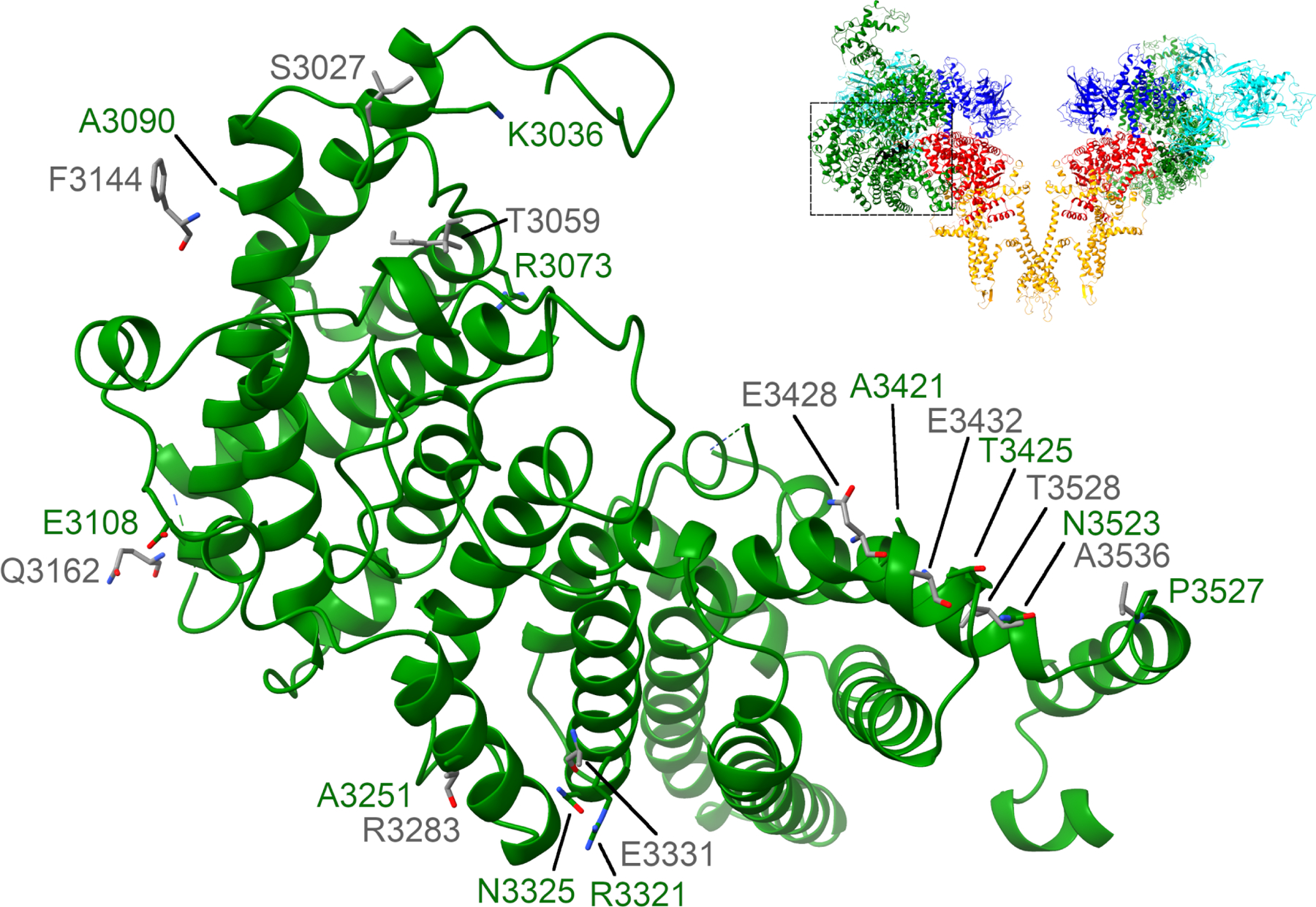
Bridging-solenoid shift. Residues 3021–3550 are shown with proteoliposomal RyR1 in the primed state in green and the corresponding sidechains of 6WOT in grey (Iyer et al., 2020). The detergent structures of RyR1 (6WOT and 5TAQ) were found to be in the same conformation but with a shift of up to fifty residues (des Georges et al., 2016).
The hydrophobic-gate residue, I4937, also matches previous structures (Figure 5). In liposomes and detergent, the pore helix rotates such that I4937 is no longer within the pore in the open state of the channel. In the open state in liposomes, the radius of the pore residue could not be measured at I4937 (Smart et al., 1993), though by estimation it remains at approximately 4 Å, which is consistent with the 4 Å radius or 8 Å diameter observed in detergent. This matches the conserved I4866 in RyR2 as well (Iyer et al., 2020). The lumen of RyR, at the base of the channel, varies slightly between liposomes and detergent but this region consists of unstructured loops so dynamic behavior is to be expected.
Figure 5.
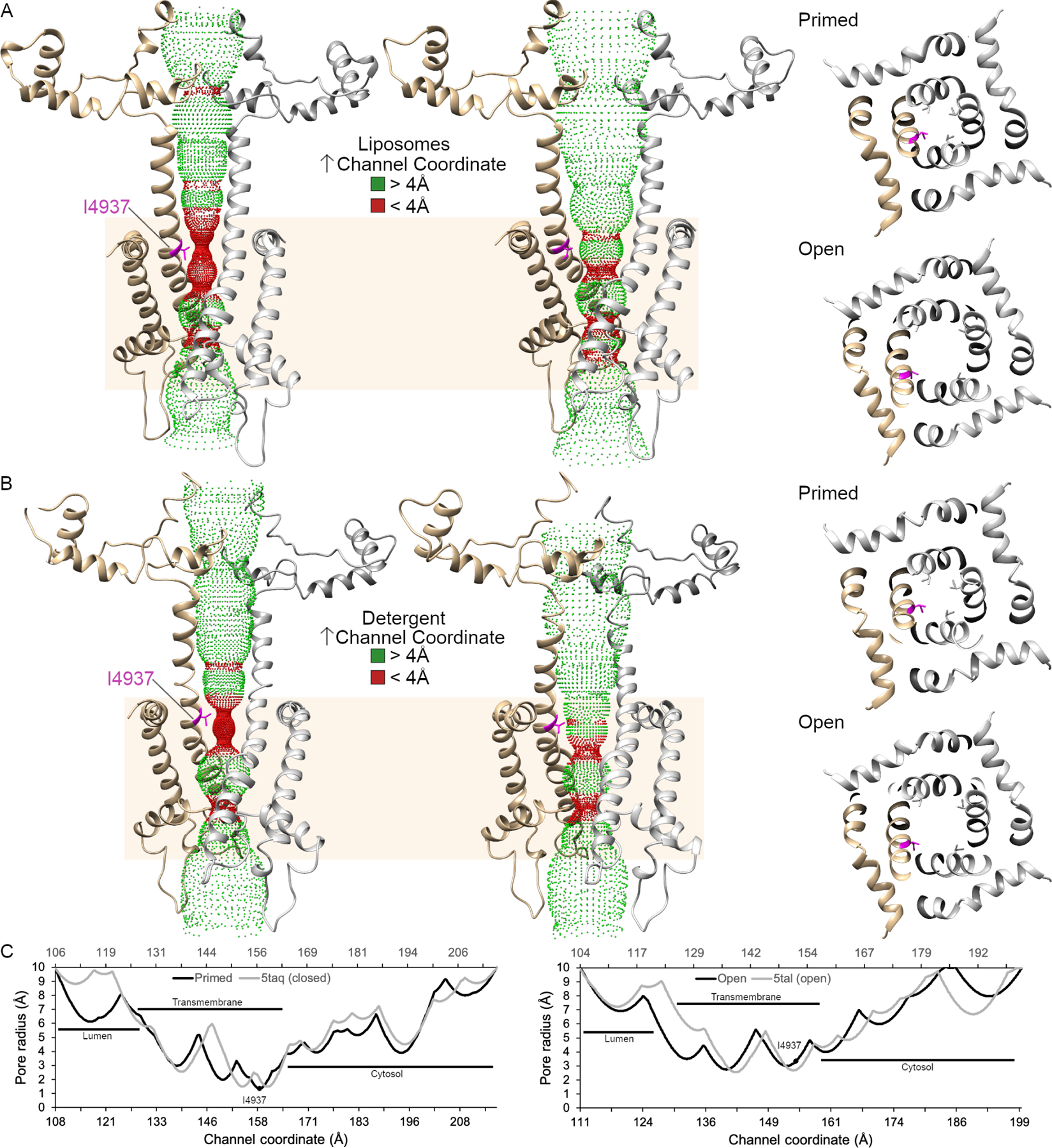
Accessible inner surface of the RyR1 pore. The transmembrane pore (residues 4820–5037) is depicted as a ribbon diagram of two protomers with the hydrophobic gate residue, I4937, in magenta. The dotted representation of the accessible inner surface of the channel was calculated using HOLE and is colored green where the radius exceeds 4 Å and red when the radius is less than 4 Å. The transmembrane domain of RyR1 is highlighted in tan. A. Side and cytoplasmic views of the inner surface of the pore of proteoliposomal RyR1 in the primed and open states. B. Side and cytoplasmic views for the RyR1 in detergent in the primed (5taq) and open (5tal) state. C. Graphical representation of the pore radius of the channel in liposomes (black) and detergent (gray) in the primed and open states. The channel coordinates follow from the lumen to the cytosolic side of the RyR1 with the liposomal structure on the primary x-axis and the detergent structures on the secondary x-axis.
Discussion
The present study seeks to provide an alternative methodology that can be applied to other membrane proteins including receptors and channels. Ion-exchange clearing provided a significant improvement to liposomes formed via dialysis but this effect was diminished in liposomes formed by gel-filtration (Figure 1A), thus additional cleanup steps may provide a larger improvement, including dialysis prior to ion-exchange, to ensure the removal of residual detergent. The possibility of remaining detergent falls in line with ongoing efforts to minimize the detergent concentration during purification, which has seen stable, detergent-solubilized RyR1 in even as low as 0.1% CHAPS. These efforts have proven to be unsuccessful in preserving the 7th transmembrane helix and, likewise, in both the primed and open states in proteoliposomes, the extra transmembrane helix is entirely absent (Figures 2 & 3), suggesting that the loss during detergent-based purification may be irreversible.
Though dynamic, the liposome membrane compares well with the work of Chen et al (Chen and Kudryashev, 2020), which suggested that activation of the RyR was associated with changes in the curvature of the SR membrane. Specifically, while the liposome membrane is flattened around the transmembrane domain of the RyR in the primed and open states, the surrounding membrane appears flat in the primed state and curved to approximately 38° in the open state (Figure 6). 37% of the channels were found to be in the open state, similar to the 50% observed in detergent-solubilized RyR1 channels examined under the same conditions (Ca2+, ATP, and caffeine) (des Georges et al., 2016). Planar lipid bilayer studies of RyR1 show a roughly 90% open probability under the same conditions (Bezprozvanny et al., 1991; Brillantes et al., 1994a); however, this disparity may simply be the result of different methodology, with samples on grids showing a preference for the closed state of the channel. Likewise, the final Ca2+ concentration may change as a result of of wicking during blotting require to prepare the grids for flash freezing.
Figure 6.
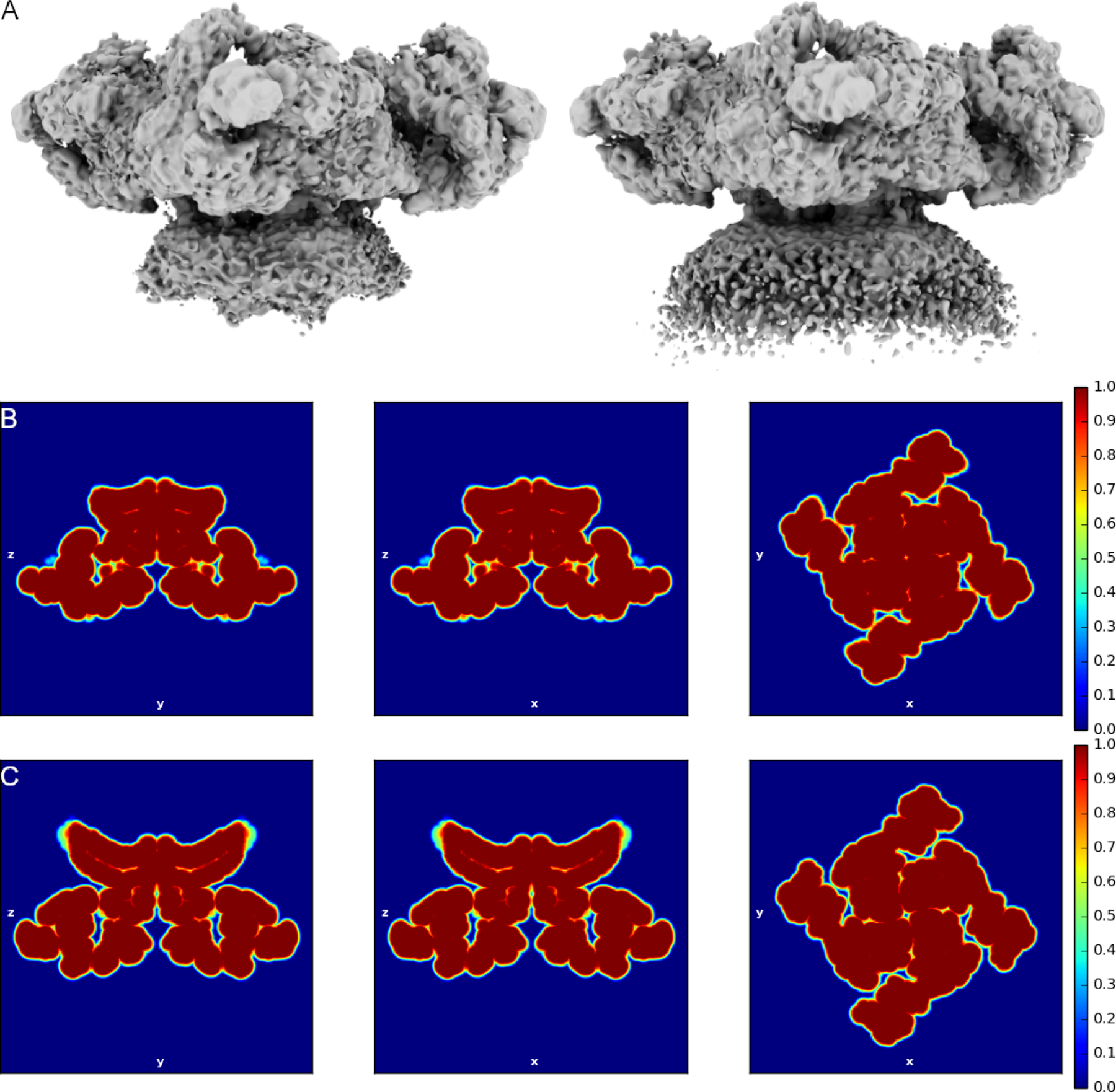
Proteoliposomal RyR1 maps highlighting the lipid membrane. A. Composite maps of RyR1 in the primed (left) and open (right) state. Both maps are low pass filtered to 6 Å. The membrane is highly dynamic but appears flat in the primed state and curved at approximately 38° in the open state. B-C. Real space mask slices low pass filtered to 5 Å for the primed and open states, respectively.
As functional studies of RyR1 are often performed using planar lipid bilayers (Santulli et al., 2017), solving the structure in a similar membrane establishes that these channels are likely in physiologically relevant conformations. The ability to study the structure of the channel in proteoliposomes now provides the opportunity to resolve structures of membrane proteins under markedly more physiological conditions. In the case of RyR1, the Ca2+ concentration in the lumen of the SR is millimolar, but the cytosolic shell is exposed to nanomolar Ca2+ at rest. Ca2+ within the SR is not simply free Ca2+, but is significantly bound to Ca2+ binding proteins including calsequestrin and going forward it may be possible to incorporate calsequestrin into the lumen of proteoliposomes for structural studies.
One limitation of the present study is that the structure of liposome embedded RyR1 channels remains limited to single RyR1 channels, while native RyR1 exists in an array of channels in a checkerboard pattern on the terminal cisternae of the SR membrane (Franzini-Armstrong and Jorgensen, 1994; Franzini-Armstrong et al., 1998; Sun et al., 1995). Although as many as three molecules of RyR1 were sometimes present in a single liposome, no intermolecular interactions were observed (Flucher et al., 1993; Inui et al., 1987; Wang et al., 2004). Curiously, multiple channels were only observed in small liposomes (<100 nm diameter); however, no more than a single channel was observed in liposomes >100 nm. Instead, larger liposomes were more likely to be empty. A structure of RyR1 in native SR membranes will be necessary to observe RyR1 arrays and the native intermolecular dynamics. In addition to validation of existing structures of detergent solubilized RyR1, the present study provides a road map for studies of RyR1 structures in native membranes at high resolution using cryoEM (Chen and Kudryashev, 2020), including studies mimicking the chemical gradient between the cytoplasm of the cell and the lumen of the SR.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrew R. Marks (arm42@cumc.columbia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
cryoEM data have been deposited at PDB, EMDB, and EMPIAR and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| New Zealand white rabbit | BioIVT | Cat#RAB00MSCUZN |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphatidylcholine | Avanti | Cat#441601G |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine | Avanti | Cat#850375C |
| Phosphatidylethanolamine | Avanti | Cat#850725P |
| Critical Commercial Assays | ||
| HiTrap Q HP | Cytiva | Cat#17115401 |
| Deposited Data | ||
| Proteoliposomal RyR1 primed model | This paper | PDB ID: 7M6A |
| Proteoliposomal RyR1 primed map | This paper | EMDB ID: 23692 |
| Proteoliposomal RyR1 open model | This paper | PDB ID: 7M6L |
| Proteoliposomal RyR1 popen map | This paper | EMDB ID: 23699 |
| Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | Iyer et al., 2020 | PDB ID: 6WOT |
| Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3&4) | des Georges et al., 2016 | PDB ID: 5TAQ |
| Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1&2) | des Georges et al., 2016 | PDB ID: 5TAL |
| Software and Algorithms | ||
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| Relion | Scheres, 2012 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| Leginon | Suloway et al., 2005 | https://sbgrid.org/software/titles/leginon |
| Phenix | Adams et al., 2004 | https://www.phenix-online.org/ |
| Coot | Emsley and Cowtan, 2004 | https://www.ccpem.ac.uk/ |
| HOLE | Smart et al., 1993 | http://www.holeprogram.org/ |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| ChimeraX | Pettersen et al., 2021 | https://www.cgl.ucsf.edu/chimerax/ |
| Other | ||
| UltrAuFoil Holey gold grids | Quantifoil | R 0.6/1.0, Au 300 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rabbit tissue used for the purification of RyR1 was New Zealand white rabbit, age and gender unspecified, purchased from BioIVT.
METHOD DETAILS
The workflow for solving the high-resolution structure of RyR1 embedded in liposomes is outlined in Figure S1.
RyR1 purification:
All purification steps were performed on ice unless otherwise stated. RyR1 was purified from rabbit skeletal muscle as described previously with modifications (Brillantes et al., 1994b), primarily the use of an ion-exchange column, which allowed for faster purification without the need for overnight to binding to a GST-calstabin column and improved the final yield by up to six-fold, allowing significantly less rabbit tissue to be used for each purification. This method also makes it possible to omit the final gel-filtration column in detergent-purifications; however, this results in leftover lipid in cryoEM samples. In brief, rabbit back and thigh skeletal muscle, purchased from BioIVT, was snap frozen in liquid nitrogen immediately following euthanasia prior to storage at −80°C. 20 g of rabbit skeletal muscle was homogenized in a Waring blender and lysed in buffer A [10 mM tris maleate pH 6.8, 1 mM EGTA, 1 mM benzamidine hydrochloride, 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF)]. The resulting suspension was pelleted by centrifugation for ten minutes at 11,000 X g.
The supernatant was filtered through cheesecloth to remove debris and the membranes were then pelleted by centrifugation for thirty minutes at 36,000 X g. The membranes were then solubilized in buffer B [10 mM HEPES pH 7.4, 0.8 M NaCl, 1% CHAPS, 0.1% phosphatidylcholine (Avanti, Cat#441601G), 1 mM EGTA, 2 mM DTT, 0.5 mM AEBSF, 1 mM benzamidine hydrochloride, 1 protease inhibitor tablet (Pierce)] prior to homogenization using a glass tissue grinder (Kontes). Homogenization was repeated following the addition of buffer C (buffer B with no NaCl) at a 1:1 mixture with buffer B. The resulting homogenate was subjected to centrifugation for thirty minutes at 100k x g. The supernatant was then vacuum filtered and loaded at 1 mL/min onto a HiTrap Q HP column (5 mL, GE Healthcare Life Sciences) pre-equilibrated with buffer D [10 mM HEPES pH 7.4, 400 mM NaCl, 1.0% CHAPS, 1 mM EGTA, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), 0.5 mM AEBSF, 1 mM benzamidine hydrochloride, 0.01% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti, Cat#850375C)]. DOPC (25 mg/mL), dissolved in chloroform, was evaporated under nitrogen gas and resuspended in buffer D. Contaminating proteins were washed away with six column volumes (CV) of buffer D prior to elution of RyR1 with a linear gradient from 480 to 550 mM NaCl using buffers D and E (buffer D with 600 mM NaCl) over the course of 10 CV.
Liposome formation using dialysis and initial screening:
Purified RyR1 (1 mg/mL) was incubated at a molar ratio of 1:2,000 with 5:3 phosphatidylethanolamine and phosphatidylcholine (Avanti Cat#850725P and 441601G, respectively) and dialyzed into buffer F [10 mM HEPES pH 7.4, 400 mM NaCl, 1 mM EGTA, 0.5 mM TCEP] for 48 hours replacing the buffer with fresh buffer twice per day. Initial screening was performed on a Tecnai F20 (FEI) microscope at Columbia University. Liposomes formed by dialysis were found to have widely variable sizes and even with passage through an extruder, many liposomes were not unilammelar.
Liposome formation using gel filtration:
Purified RyR1 was concentrated to <0.5 mL and incubated with a molar ratio of 1:1,000 5:3 phosphatidylethanolamine and phosphatidylcholine in 10% CHAPS for 10 minutes prior to loading onto a 24 mL hand-packed G50 column pre-equilibrated with buffer F. Liposome formation and RyR1 incorporation occurred on the column and proteoliposomal RyR1 was eluted at 0.20–0.25 mL/min in buffer F.
Cleanup:
Proteoliposomal RyR1 was cleared via passage over a second HiTrap Q column, washed at 425 mM NaCl then eluted at 600 mM NaCl. Proteoliposomal RyR1 was concentrated to 2.5 mg/mL, determined by spectroscopy using a NanoDrop 1000 (ThermoFisher, 1 abs @ 280 nm = 1 mg/mL).
Grid preparation:
UltrAuFoil holey gold grids (Quantifoil R 0.6/1.0, Au 300) were plasma cleaned for thirty seconds with H2 and O2 (Gatan). Prior to setting grids, proteoliposomal RyR1 was incubated for 10 min with 2 mM NaATP, 5 mM caffeine, and 30 µM Ca2+free (MaxChelator) (Bers et al., 1994). These conditions were chosen to ensure both the primed and open states would be visible. 3.0 µL of 2.5 mg/mL proteoliposomal RyR1 was applied to each grid. Grids were then blotted with ashless blotting paper (Whatman) for 7.5 sec at blot force 3, with a wait time of thirty seconds prior to vitrification by plunge freezing into liquid ethane chilled with liquid nitrogen (Jacques Dubochet, 1988; Wagenknecht et al., 1988) with a Vitrobot Mark IV (ThermoFisher) operated at 4°C with 100% relative humidity.
Data Collection:
Grids prepared with liposomes formed by gel filtration were screened at City University of New York (CUNY) using a 120-kV G2 Spirit Twin microscope (FEI Tecnai). Microscope operations and data collection were carried out using the SerialEM software (Mastronarde, 2003). High resolution data collection was performed at Columbia University on a Titan Krios 300-kV (ThermoFisher) microscope equipped with an energy filter (slit width 20 eV) and a K3 direct electron detector (Gatan). Data were collected using Leginon (Suloway et al., 2005) and at a nominal magnification of 105,000X in electron counting mode, corresponding to a pixel size of 0.826 Å. The electron dose rate was set to 16 e−/pixel/sec with 2.5 second exposures, for a total dose of 58.34 e/Å2. These grids showed small, consistent liposomes, similar to those reported by by Tonggu & Wang (Tonggu and Wang, 2020) and significantly greater incorporation rate of RyR1; however, empty liposomes and unincorporated and aggregated RyR1 also remained.
Data Processing:
CryoEM data processing was performed using cryoSPARC (Punjani et al., 2017; Rohou and Grigorieff, 2015; Rubinstein and Brubaker, 2015; Stagg et al., 2014; Tan et al., 2017) with the exception of 3D classes, which were performed in Relion 3.1 (Scheres, 2012) to ensure accurate comparisons of the dynamics between liposome embedded channels and the previously published detergent-resolved channels (des Georges et al., 2016). Image stacks were aligned using Patch motion correction and defocus value estimation by Patch CTF estimation. Initial particle picking was performed manually for >100 particles to create templates prior to template-based picking. This involved increasing the box size to ensure a portion of the lipid membrane would be included. Two million particles were initially picked from 11,187 micrographs and these were subjected to several iterations of 2D classification in cryoSPARC with 200 classes each to separate proteoliposomal RyR1 from empty liposomes and free RyR1. Classes comprised of 175,000 particles of unincorporated RyR1 were eliminated during iterative rounds of 2D classification, leaving 85,000 liposome-embedded particles. This represents a greater than 20-fold increase in RyR1 incorporation rate over the dialysis-based method which resulted in 2,600 particles from over 7,000 micrographs. The particles from the highest-resolution classes were pooled for ab initio 3D reconstruction with a single class followed by homogenous refinement with per-particle defocus refinement and C4 symmetry imposed. 3D variability analysis revealed the presence of the open state and heterogenous refinement was performed in order to separate the two states, with 63% of particles (53,882) in the primed state and 37% in the open state (31,599).
Symmetry expansion and local refinement, were performed using cryoSPARC to improve local resolution. Local refinement was performed using three separate masks. The first mask was composed of the N-terminal domain, the SPRY domains, the RY1&2 domain, and calstabin. The second mask surrounded the bridging solenoid, and the third mask surrounded the RyR1 pore without symmetry expansion. The resulting maps were combined in Chimera (Pettersen et al., 2004) to generate a composite map prior to calibration of the pixel size using correlation coefficients with a map generated from the crystal structure of the N-terminal domain (2XOA) (Tung et al., 2010). The pixel size was altered by 0.001 Å per step, up to 10 steps in each direction with an initial and final pixel size of 0.826 and 0.831 Å, respectively. Model building was performed in Coot (Emsley and Cowtan, 2004) with refinement in Phenix (Adams et al., 2010; Liebschner et al., 2019) and figures were generated using Chimera and ChimeraX (Goddard et al., 2018). The graphical abstract was made in BioRender. The pore aperture of RyR1 was calculated using HOLE (Smart et al., 1993). Movies of the dynamics of the channel were made in ChimeraX using maps generated by 3D classes for liposomes and the corresponding maps from 3D Classes in detergent (5TAN, 5TAM, 5TA3, 5T9V). Each map was filtered to 5 Å and aligned at the pore. CryoEM statistics are summarized in Figure S2 and Table 1.
Table 1.
cryoEM statistics.
| Data collection | Primed | Open |
|---|---|---|
| Microscope | FEI Titan Krios | |
| Detector | Gatan K3 | |
| Voltage (kV) | 300 | |
| Magnification | 105,000 | |
| Exposure (e−/Å2) | 58.34 | |
| Defocus range (µm) | −1–2 | |
| Pixel size (Å) | 0.831 | |
| Processing | ||
| Software | cryoSPARC | |
| Symmetry | C4 | |
| Initial particles (no.) | 85,481 | |
| Final particles (no.) | 53,882 | 31,599 |
| Map resolution (Å) | 3.4 | 4.0 |
| Map resolution range (Å) | 3.07–3.66‡ | 3.45–4.15‡ |
| Model Composition | ||
| Peptide chains | 12 | |
| Nonhydrogen | 140,676 | 140,432 |
| Protein residues | 17,652 | 17,624 |
| Ligands | 16 | |
| Mean B factors (Å2) | ||
| Protein | 83.94 | 97.75 |
| Ligands | 149.86 | 127.35 |
| R.m.s. deviations | ||
| Bond length (Å) | 0.009 | 0.007 |
| Bond angles (°) | 0.970 | 0.899 |
| Ramachandran | ||
| Favored (%) | 95.76 | 92.50 |
| Allowed (%) | 4.19 | 7.41 |
| Disallowed (%) | 0.05 | 0.09 |
| Validation | ||
| MolProbity score | 2.05 | 2.26 |
| Clashscore | 18.21 | 19.72 |
| Rotamer outliers (%) | 0.72 | 0.94 |
| PDB ID | 7M6A | 7M6L |
| EMDB ID | 23692 | 23699 |
Map resolution ranges represent the range determined by local refinement in cryoSPARC using the local masks described in Methods.
QUANTIFICATION AND STATISTICAL ANALYSIS
Cryo-EM data were processed using cryoSPARC with 3d classification performed in Relion. Quantification of the pore radius was calculated using HOLE and visualized in Excel (Figure 5). Real space mask slices were analyzed using cryoSPARC (Figure 6). Cryo-EM structural statistics were analyzed using Phenix (Table 1). Reported map resolutions are based on the 0.143 Fourier Shell Correlation criterion. The reported map resolutions and viewing angle distribution were calculated using cryoSPARC (Figure S2, Table 1).
Supplementary Material
Movie 1. Detergent-resolved RyR1 primed state dynamics from 3d classes. This video was created using ChimeraX with the maps corresponding to 5TAN and 5TAM. Related to Figure 2.
Movie 2. Detergent-resolved RyR1 open state dynamics from 3d classes. This video was created using ChimeraX with the maps corresponding to 5TA3 and 5T9V. Related to Figure 2.
Movie 3. Proteoliposomal RyR1 primed state dynamics from 3d classes. Related to Figure 2.
Movie 4. Proteoliposomal RyR1 open state dynamics from 3d classes. Related to Figure 2.
Movie 5. Coulombic density and fitted model of the primed state. Only two protomers are shown for clarity. Related to Figure 3.
Movie 6. Coulombic density and fitted model of the open state. Only two protomers are shown for clarity. Related to Figure 3.
Movie 7. Coulombic density and fitted model of the pore in the primed state. Only two protomers are shown for clarity. Related to Figures 3 & 5.
Movie 8. Coulombic density and fitted model of the pore in the open state. Only two protomers are shown for clarity. Related to Figures 3 & 5.
Movie 9. Coulombic density and fitted model for additional assignments in the SPRY domain. Residues 1420–1572 are shown. Related to Figure 3.
Movie 10. Coulombic density and fitted model for additional assignments in the Br-sol. Residues 3064–3471 are shown. Related to Figures 3 & 4.
Highlights.
High-resolution structures of RyR1 resolved in liposomes
The cryo-EM structure of RyR1 in liposomes closely matches detergent structures
The channel remains dynamic in liposomes
Acknowledgements
These studies were supported by NIH grants R01HL145473, R01DK118240, R01HL142903, R01HL140934, R01AR070194 and T32 HL120826 (to A.R.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
A.R.M. is a consultant for ARMGO Pharma and A.R.M. and Columbia University own shares in ARMGO Pharma, a biotechnology company developing ryanodine receptor-targeted drugs.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Yan Z, Wu J, Li Z, and Yan N. (2016). The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening. Cell Res 26, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt TH, and Sligar SG (2010). Membrane protein assembly into Nanodiscs. FEBS Lett 584, 1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger AM, Reiken S, Carlson C, Mongillo M, Liu X, Rothman L, Matecki S, Lacampagne A, and Marks AR (2009). Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med 15, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers DM, Patton CW, and Nuccitelli R. (1994). A practical guide to the preparation of Ca2+ buffers. Methods Cell Biol 40, 3–29. [DOI] [PubMed] [Google Scholar]
- Bezprozvanny l., Watras J, and Ehrlich BE (1991). Bell-shaped calcium-response curves of lns(l,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature 351, 751–754. [DOI] [PubMed] [Google Scholar]
- Brillantes A-MB, Ondrias K, Scott A, Kobrinsky E, Ondriašová E, Moschella MC, Jayaraman T, Landers M, Ehrlich BE, and Marks AR (1994a). Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell 77, 513–523. [DOI] [PubMed] [Google Scholar]
- Brillantes AB, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Moschella MC, Jayaraman T, Landers M, Ehrlich BE, and Marks AR (1994b). Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell 77, 513–523. [DOI] [PubMed] [Google Scholar]
- Bussiere R, Lacampagne A, Reiken S, Liu X, Scheuerman V, Zalk R, Martin C, Checler F, Marks AR, and Chami M. (2017). Amyloid beta production is regulated by beta2-adrenergic signaling-mediated post-translational modifications of the ryanodine receptor. J Biol Chem 292, 10153–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, and Kudryashev M. (2020). Structure of RyR1 in native membranes. EMBO Rep 21, e49891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wu J, Lvovskaya S, Herndon E, Supnet C, and Bezprozvanny I. (2011). Dantrolene is neuroprotective in Huntington’s disease transgenic mouse model. Mol Neurodegener 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Gong D, Ren K, Zhou G, Huang G, Lei J, Zhou Q, and Yan N. (2019). Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators. Proc Natl Acad Sci U S A 116, 25575–25582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, Hendrickson WA, Marks AR, and Frank J. (2016). Structural Basis for Gating and Activation of RyR1. Cell 167, 145–157.e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi H, Kushnir A, Zalk R, Yuan Q, Melville Z, and Marks AR (2020a). Intracellular calcium leak in heart failure and atrial fibrillation: a unifying mechanism and therapeutic target. Nat Rev Cardiol. [DOI] [PMC free article] [PubMed]
- Dridi H, Liu X, Yuan Q, Reiken S, Yehia M, Sittenfeld L, Apostolou P, Buron J, Sicard P, Matecki S, et al. (2020b). Role of defective calcium regulation in cardiorespiratory dysfunction in Huntington’s disease. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Gatsogiannis C, and Raunser S. (2017). Chapter One - Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM. In Methods in Enzymology, Ziegler C, ed. (Academic Press; ), pp. 1–30. [DOI] [PubMed] [Google Scholar]
- Efremov RG, Leitner A, Aebersold R, and Raunser S. (2014). Architecture and conformational switch mechanism of the ryanodine receptor. Nature 517, 39. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallographica Section D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fabiato A. (1983). Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am J Physiol 245, C1–14. [DOI] [PubMed] [Google Scholar]
- Fabiato A, and Fabiato F. (1978). Calcium-induced release of calcium from the sarcoplasmic reticulum of skinned cells from adult human, dog, cat, rabbit, rat, and frog hearts and from fetal and new-born rat ventricles. Ann N Y Acad Sci 307, 491–522. [DOI] [PubMed] [Google Scholar]
- Flucher BE, Andrews SB, Fleischer S, Marks AR, Caswell A, and Powell JA (1993). Triad formation: organization and function of the sarcoplasmic reticulum calcium release channel and triadin in normal and dysgenic muscle in vitro. The Journal of cell biology 123, 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong C, and Jorgensen AO (1994). Structure and development of E-C coupling units in skeletal muscle. Annu Rev Physiol 56, 509–534. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Protasi F, and Ramesh V. (1998). Comparative ultrastructure of Ca2+ release units in skeletal and cardiac muscle. Ann N Y Acad Sci 853, 20–30. [DOI] [PubMed] [Google Scholar]
- Furuichi T, Furutama D, Hakamata Y, Nakai J, Takeshima H, and Mikoshiba K. (1994). Multiple types of ryanodine receptor/Ca2+ release channels are differentially expressed in rabbit brain. J Neurosci 14, 4794–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini G, Conti A, Mammarella S, Scrobogna M, and Sorrentino V. (1995). The ryanodine receptor/calcium channel genes are widely and differentially expressed in murine brain and peripheral tissues. J Cell Biol 128, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grethen A, Oluwole AO, Danielczak B, Vargas C, and Keller S. (2017). Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Scientific reports 7, 11517–11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Saito A, and Fleischer S. (1987). Purification of the ryanodine receptor and identity with feet structures of junctional terminal cisternae of sarcoplasmic reticulum from fast skeletal muscle. Journal of Biological Chemistry 262, 1740–1747. [PubMed] [Google Scholar]
- Iyer KA, Hu Y, Nayak AR, Kurebayashi N, Murayama T, and Samsó M. (2020). Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM. Sci Adv 6, eabb2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques Dubochet, M.A., Chang Jiin-ju, Homo Jean-Claude, Lepault Jean, McDowall Alasdair W., Patrick Shultz (1988). Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21, 129–228. [DOI] [PubMed] [Google Scholar]
- Kushnir A, and Marks AR (2010). The Ryanodine Receptor in Cardiac Physiology and Disease. Advances in pharmacology (San Diego, Calif) 59, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir A, Wajsberg B, and Marks AR (2018). Ryanodine receptor dysfunction in human disorders. Biochim Biophys Acta Mol Cell Res 1865, 1687–1697. [DOI] [PubMed] [Google Scholar]
- Lacampagne A, Liu X, Reiken S, Bussiere R, Meli AC, Lauritzen I, Teich AF, Zalk R, Saint N, Arancio O, et al. (2017). Post-translational remodeling of ryanodine receptor induces calcium leak leading to Alzheimer’s disease-likepathologies and cognitive deficits. Acta Neuropathologica 134, 749–767. [DOI] [PubMed] [Google Scholar]
- Lanner JT, Georgiou DK, Joshi AD, and Hamilton SL (2010). Ryanodine Receptors: Structure, Expression, Molecular Details, and Function in Calcium Release. Cold Spring Harbor Perspectives in Biology 2, a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, and Sligar SG (2006). Functional reconstitution of β2-adrenergic receptors utilizing self-assembling Nanodisc technology. BioTechniques 40, 601–612. [DOI] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung L-W, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallographica Section D 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks AR, Tempst P, Hwang KS, Taubman MB, Inui M, Chadwick C, Fleischer S, and Nadal-Ginard B. (1989). Molecular cloning and characterization of the ryanodine receptor/junctional channel complex cDNA from skeletal muscle sarcoplasmic reticulum. Proc Natl Acad Sci U S A 86, 8683–8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx S, Gaburjakova O, Gaburjakova J, Henrikson M, Ondrias C, K., and Marks Andrew R. (2001). Coupled Gating Between Cardiac Calcium Release Channels (Ryanodine Receptors). Circulation Research 88, 1151–1158. [DOI] [PubMed] [Google Scholar]
- Marx SO, and Marks AR (2013). Dysfunctional ryanodine receptors in the heart: New insights into complex cardiovascular diseases. Journal of molecular and cellular cardiology 58, 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx SO, Ondrias K, and Marks AR (1998). Coupled Gating Between Individual Skeletal Muscle Ca2+ Release Channels (Ryanodine Receptors). Science 281, 818–821. [DOI] [PubMed] [Google Scholar]
- Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, and Marks AR (2000). PKA Phosphorylation Dissociates FKBP12.6 from the Calcium Release Channel (Ryanodine Receptor): Defective Regulation in Failing Hearts. Cell 101, 365–376. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2003). SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microscopy and Microanalysis 9, 1182–1183. [Google Scholar]
- Nath A, Atkins WM, and Sligar SG (2007). Applications of Phospholipid Bilayer Nanodiscs in the Study of Membranes and Membrane Proteins. Biochemistry 46, 2059–2069. [DOI] [PubMed] [Google Scholar]
- Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, and Keller S. (2017a). Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angewandte Chemie (International ed in English) 56, 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluwole AO, Klingler J, Danielczak B, Babalola JO, Vargas C, Pabst G, and Keller S. (2017b). Formation of Lipid-Bilayer Nanodiscs by Diisobutylene/Maleic Acid (DIBMA) Copolymer. Langmuir 33, 14378–14388. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Porta M, Diaz-Sylvester PL, Neumann JT, Escobar AL, Fleischer S, and Copello JA (2012). Coupled gating of skeletal muscle ryanodine receptors is modulated by Ca2+, Mg2+, and ATP. Am J Physiol Cell Physiol 303, C682–C697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods 14, 290. [DOI] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N. (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL, and Brubaker MA (2015). Alignment of cryo-EM movies of individual particles by optimization of image translations. Journal of Structural Biology 192, 188–195. [DOI] [PubMed] [Google Scholar]
- Santulli G, Lewis D, des Georges A, Marks AR, and Frank J. (2018). Ryanodine Receptor Structure and Function in Health and Disease. Subcell Biochem 87, 329–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Nakashima R, Yuan Q, and Marks AR (2017). Intracellular calcium release channels: an update. J Physiol 595, 3041–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Pagano G, Sardu C, Xie W, Reiken S, D’Ascia SL, Cannone M, Marziliano N, Trimarco B, Guise TA, et al. (2015). Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J Clin Invest 125, 4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW (2012). RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Goodfellow JM, and Wallace BA (1993). The pore dimensions of gramicidin A. Biophysical Journal 65, 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, Noble AJ, Spilman M, and Chapman MS (2014). ResLog plots as an empirical metric of the quality of cryo-EM reconstructions. Journal of Structural Biology 185, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B. (2005). Automated molecular microscopy: The new Leginon system. Journal of Structural Biology 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Sun XH, Protasi F, Takahashi M, Takeshima H, Ferguson DG, and Franzini-Armstrong C. (1995). Molecular architecture of membranes involved in excitation-contraction coupling of cardiac muscle. J Cell Biol 129, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Nagai Y, Wada K, and Koike T. (2012). Calcium leak through ryanodine receptor is involved in neuronal death induced by mutant huntingtin. Biochem Biophys Res Commun 429, 18–23. [DOI] [PubMed] [Google Scholar]
- Swainsbury DJK, Scheidelaar S, Foster N, van Grondelle R, Killian JA, and Jones MR (2017). The effectiveness of styrene-maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes. Biochimica et biophysica acta Biomembranes 1859, 2133–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, and Lyumkis D. (2017). Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature Methods 14, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonggu L, and Wang L. (2020). Cryo-EM sample preparation method for extremely low concentration liposomes. Ultramicroscopy 208, 112849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CC, Lobo PA, Kimlicka L, and Van Petegem F. (2010). The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature 468, 585–588. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, Grassucci R, and Frank J. (1988). Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. Journal of Molecular Biology 199, 137–147. [DOI] [PubMed] [Google Scholar]
- Wang L, and Sigworth FJ (2009). Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature 461, 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SQ, Stern MD, Ríos E, and Cheng H. (2004). The quantal nature of Ca2+ sparks and in situ operation of the ryanodine receptor array in cardiac cells. Proceedings of the National Academy of Sciences of the United States of America 101, 3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Wang X, Zhang Y, Mukherjee S, Zhang L, Chen Q, Huang X, Jing S, Liu C, Li S, et al. (2016). Structural insights into Ca(2+)-activated long-range allosteric channel gating of RyR1. Cell Res 26, 977–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, Clarke OB, des Georges A, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, and Marks AR (2015). Structure of a mammalian ryanodine receptor. Nature 517, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, and Marks AR (2017). Ca(2+) Release Channels Join the ‘Resolution Revolution’. Trends Biochem Sci 42, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Detergent-resolved RyR1 primed state dynamics from 3d classes. This video was created using ChimeraX with the maps corresponding to 5TAN and 5TAM. Related to Figure 2.
Movie 2. Detergent-resolved RyR1 open state dynamics from 3d classes. This video was created using ChimeraX with the maps corresponding to 5TA3 and 5T9V. Related to Figure 2.
Movie 3. Proteoliposomal RyR1 primed state dynamics from 3d classes. Related to Figure 2.
Movie 4. Proteoliposomal RyR1 open state dynamics from 3d classes. Related to Figure 2.
Movie 5. Coulombic density and fitted model of the primed state. Only two protomers are shown for clarity. Related to Figure 3.
Movie 6. Coulombic density and fitted model of the open state. Only two protomers are shown for clarity. Related to Figure 3.
Movie 7. Coulombic density and fitted model of the pore in the primed state. Only two protomers are shown for clarity. Related to Figures 3 & 5.
Movie 8. Coulombic density and fitted model of the pore in the open state. Only two protomers are shown for clarity. Related to Figures 3 & 5.
Movie 9. Coulombic density and fitted model for additional assignments in the SPRY domain. Residues 1420–1572 are shown. Related to Figure 3.
Movie 10. Coulombic density and fitted model for additional assignments in the Br-sol. Residues 3064–3471 are shown. Related to Figures 3 & 4.
Data Availability Statement
cryoEM data have been deposited at PDB, EMDB, and EMPIAR and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| New Zealand white rabbit | BioIVT | Cat#RAB00MSCUZN |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphatidylcholine | Avanti | Cat#441601G |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine | Avanti | Cat#850375C |
| Phosphatidylethanolamine | Avanti | Cat#850725P |
| Critical Commercial Assays | ||
| HiTrap Q HP | Cytiva | Cat#17115401 |
| Deposited Data | ||
| Proteoliposomal RyR1 primed model | This paper | PDB ID: 7M6A |
| Proteoliposomal RyR1 primed map | This paper | EMDB ID: 23692 |
| Proteoliposomal RyR1 open model | This paper | PDB ID: 7M6L |
| Proteoliposomal RyR1 popen map | This paper | EMDB ID: 23699 |
| Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | Iyer et al., 2020 | PDB ID: 6WOT |
| Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3&4) | des Georges et al., 2016 | PDB ID: 5TAQ |
| Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1&2) | des Georges et al., 2016 | PDB ID: 5TAL |
| Software and Algorithms | ||
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| Relion | Scheres, 2012 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| Leginon | Suloway et al., 2005 | https://sbgrid.org/software/titles/leginon |
| Phenix | Adams et al., 2004 | https://www.phenix-online.org/ |
| Coot | Emsley and Cowtan, 2004 | https://www.ccpem.ac.uk/ |
| HOLE | Smart et al., 1993 | http://www.holeprogram.org/ |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| ChimeraX | Pettersen et al., 2021 | https://www.cgl.ucsf.edu/chimerax/ |
| Other | ||
| UltrAuFoil Holey gold grids | Quantifoil | R 0.6/1.0, Au 300 |


