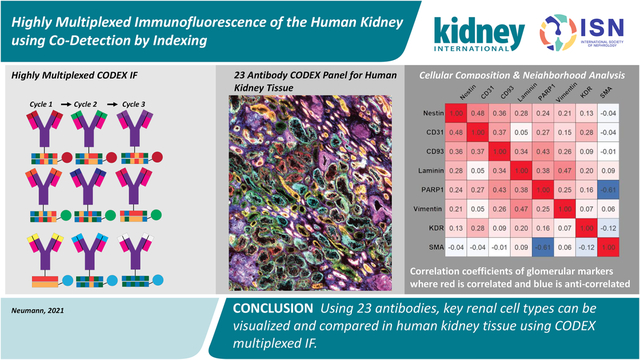
Abstract
The human kidney is composed of many cell types that vary in their abundance and distribution from normal to diseased organ. As these cell types perform unique and essential functions, it is important to confidently label each within a single tissue to accurately assess tissue architecture and microenvironments. Towards this goal, we demonstrate the use of co-detection by indexing (CODEX) multiplexed immunofluorescence for visualizing 23 antigens within the human kidney. Using CODEX, many of the major cell types and substructures, such as collecting ducts, glomeruli, and thick ascending limb, were visualized within a single tissue section. Of these antibodies, 19 were conjugated in-house, demonstrating the flexibility and utility of this approach for studying the human kidney using custom and commercially available antibodies. We performed a pilot study that compared both fresh frozen and formalin-fixed paraffin-embedded healthy non-neoplastic and diabetic nephropathy kidney tissues. The largest cellular differences between the two groups was observed in cells labeled with aquaporin 1, cytokeratin 7, and α-smooth muscle actin. Thus, our data show the power of CODEX multiplexed immunofluorescence for surveying the cellular diversity of the human kidney and the potential for applications within pathology, histology, and building anatomical atlases.
Keywords: CODEX, Multiplexed Imaging, Immunofluorescence, Microscopy, Cell-type, Cell neighborhoods
Graphical Abstract
INTRODUCTION
The kidney is composed of over 20 cell types that can be further subdivided into discrete subpopulations, each performing an essential function for human health.1,2 Often, these cell types are implicated in the transition to disease states, such as diabetic nephropathy.3–5 Immunohistochemical approaches are the standard for labeling cell types and structures within a tissue matrix because of their inherent specificity, application to a wide variety of protein targets, and insight into biochemical pathways.6,7 Fluorescently tagged antibodies are often used because they provide low background for high signal-to-noise imaging at spatial resolutions between 250 nm to 1 μm.8,9 Fluorescence experiments, however, are limited in plexity to ~4–7 antibodies because of spectral overlap between commonly used fluorophores.10–12 Traditional cyclic multiplexed immunofluorescence approaches consist of antibody application, imaging, and fluorescence inactivation/bleaching before starting a new cycle. These methods increase the number of imageable targets between 20–60 for large tissue areas but are ultimately limited by tissue distortion, antibody-antibody interactions, and longer incubation times.13
Recently, co-detection by indexing (CODEX) multiplexed immunofluorescence (IF) was developed to improve multiplexed imaging efforts to over 50 antigens.14,15 Briefly, oligonucleotide barcodes are conjugated to primary antibodies and all primary antibodies are incubated and fixed to either fresh frozen or paraffin embedded tissue. Complementary oligonucleotide barcodes attached to fluorophores are serially added, imaged, and removed to build highly multiplexed imaging datasets without significant tissue degradation. Here, we demonstrate the power of CODEX IF to image 23 proteins and their respective cell types within multiple human kidneys using a mixture of commercial and in-house conjugated antibodies. This multiplexed IF workflow has comparable or higher plexity than other multiplexed imaging approaches produced to date and can image larger tissue areas (on the order of mm2) than multiplexed technologies such as imaging mass cytometry and multiplexed ion beam imaging,16,17 demonstrating the potential of this technology for exploring cellular heterogeneity within the human kidney.
METHODS
Normal portions of renal cancer nephrectomies from adult patients were studied in addition to kidneys from diabetic nephropathy patients. Tissue blocks were frozen over an isopentane dry ice slurry, and 10 μm sections were thaw-mounted onto glass cover slips. These sections were then fixed and incubated with a mixture containing all primary antibodies. Secondary oligonucleotide sequences were automatically added and removed serially for multiplexed visualization on a single section without spectral overlap or over saturation (Figure S1) using the commercial Akoya platform (Akoya Biosciences, Marlborough, MA). Resulting images were background subtracted, cycle aligned, processed using extended depth of field, and stitched using ZEN (Carl Zeiss AG, Oberkochen, Germany). Full images were interactively visualized in QuPath after conversion to the pyramidal OME.TIFF open standard format.18 Cell segmentation was performed using default QuPath. Fluorescence intensity greater than background was used as a positive label. Cell neighborhood analysis and Pearson’s coefficients were calculated using CytoMAP19 and MATLAB. Further and complete experimental details are in the Supplemental Information.
RESULTS
Multiplexed IF Imaging using CODEX
We have generated CODEX multiplexed IF images from human kidney tissue using 23 barcoded antibodies (Table 1). The major structures within the kidney were imaged, such as the endothelial layer within glomeruli, proximal tubules, and collecting ducts by targeting CD93, aquaporin 1, and calbindin, respectively. These structures are further visually subdivided by targeting antigens that localize to different tubules or tubular layers (e.g. endothelium [CD90], tubular epithelium [β-catenin], distal convoluted tubules [E-cadherin], and epithelium [calbindin]). Several disease markers were chosen to verify the health of the tissues, such as CD 7 which stains T cells, and their low abundance staining suggests healthy tissue. While single-plex fluorescence experiments can probe these structures, only multiplexed experiments, such as this, can probe these structures in the greater tissue context, such as tubular segments or diseased functional units, such as in diabetic nephropathy. Moreover, many of these selected antigens assist in defining cellular structures and have been previously studied by standard immunofluorescence studies, allowing comparison with previous results. When possible, we selected commercial, recombinant, and purified primary antibodies for conjugation. These criteria enable reproducible staining and constant supply, limiting the commonly cited weaknesses of antibody-based approaches (e.g. irreproducible staining and lot-to-lot variability).20,21 We also determined that indirect immunofluorescence of an unconjugated and a conjugated antibody appears the same (Figure S2, details in SI methods). The tissue remained intact through the eleven cycles performed in this study (two blank cycles and nine antibody-containing cycles, Figures S3–5), indicating that additional cycles can be incorporated with by increasing the number of antibodies. Because of cycle dependence and carry over concerns,22 cycles were kept consistent and vimentin was imaged separately because it was highly intense. Finally, there are imaging artifacts associated with corner bleaching that can be addressed in future experiments by installing a variable rectangular diaphragm on the microscopy system or exploring different image processing workflows.
Table 1:
Summary of the cell types visualized by CODEX multiplexed IF. Italicized rows indicate commercial CODEX markers. Other antibodies were conjugated to CODEX oligonucleotide barcodes by us.
| Antigen | Cell Type | Barcode | Channel | Cycle | Product Code | Lot |
|---|---|---|---|---|---|---|
| CD7 | Natural killer cells, T-cells | BX025 | FITC | 1 | Akoya Biosciences 4150022 | B311698 |
| Aquaporin 1* | Proximal tubule cells, some endothelial cells, descending limb of the loop of Henle | BX002 | DsRed | 1 | Abcam b9566 | GR3330284–1 |
| Aquaporin 2* | Principal cells of the collecting duct | BX015 | Cy5 | 1 | Abcam ab230170 | GR3309659–1 |
| CD90 | Proximal tubules, fibroblasts, activated endothelial cells | BX022 | FITC | 2 | Akoya Biosciences 4150021 | B311697 |
| CD38* | T cells, B cells, NK cells, erythrocytes | BX007 | FITC | 3 | Akoya Biosciences 4150007 | B275925 |
| Ki67 | Proliferating cells | BX026 | DsRed | 3 | Akoya Biosciences 4250019 | B305585 |
| Nestin* | Some podocytes, endothelial cells, basement membrane | BX024 | Cy5 | 3 | Abcam ab221660 | GR3234351 |
| CD45 | All hematopoietic cells, except erythrocytes and platelets | BX001 | FITC | 4 | Akoya Biosciences 4150003 | B304055 |
| Uromodulin* | Epithelial cells of the thick ascending limb of Henle’s loop | BX047 | DsRed | 4 | Abcam ab207170 | GR3291001 |
| ß-Catenin* | Tubular epithelium | BX003 | Cy5 | 4 | Abcam ab32572 | GR3269711 |
| Vimentin* | Mesenchymal cells, tubular cells, glomerular epithelial cells | BX043 | Cy5 | 5 | Abcam ab92548 | GR3238979 |
| PARP1* | Glomerular endothelium | BX013 | FITC | 6 | Abcam ab191217 | GR3281797–1 |
| CD31 | Endothelial cells | BX032 | DsRed | 6 | Akoya Biosciences 4250009 | B298655 |
| CD93* | Peritubular capillary and glomerular endothelium | BX006 | Cy5 | 6 | Abcam ab134079 | GR33666664–1 |
| E-Cadherin | Distal convoluted tubules, collecting duct, loop of Henle | BX016 | FITC | 7 | Cell Signaling 3195S | Cyg0419101 |
| Laminin | Basement membrane | BX023 | DsRed | 7 | DSHB 2E8 | 3/26/20–536 |
| KDR/Vegf * | Peritubular capillary and glomerular endothelium | BX030 | Cy5 | 7 | Abcam ab237634 | GR3256123–1 |
| Calbindin* | Epithelium in distal convoluted tubules, connecting segment, and cortical collecting duct | BX004 | FITC | 8 | Abcam ab108404 | GR3264008 |
| Cytokeratin 7* | Epithelial cells | BX005 | DsRed | 8 | Abcam ab68459 | GR3249684–1 |
| α-Smooth Muscle Actin* | Vascular smooth muscle, myofibroblasts | BX021 | Cy5 | 8 | Abcam ab7817 | GR3273998–2 |
| Renin* | Juxtaglomerular cell | BX010 | FITC | 9 | Abcam ab212197 | GR3366010–1 |
| Tryptase* | Mast cells | BX014 | DsRed | 9 | Abcam ab151757 | GR3366012–1 |
| MARCKS* | Peritubular capillary | BX027 | Cy5 | 9 | Abcam ab184546 | GR3309791–1 |
| Synaptopodin | Glomeruli | BX026 | DsRed | 3 | GeneTex GTX39067 | 822001354 |
| Collagen IV | Basement Membrane | BX033 | Cy5 | n/a | Abcam ab256353- did not work | GR3283654 |
CODEX imaging was performed on three normal non-neoplastic portions of nephrectomy samples, two men and one woman, 47, 66, and 77 years old, respectively (Figures 1, S6, and S7). Images from large sections were acquired (Figure 1A, comparison histological stain Figure S8, enlarged single channels Figures S9–31), including areas of cortex (Figure 1B) and medulla (Figure 1C), highlighting seven of the markers. The distribution of interstitial cells was also visualized, such as mast cells (Figure 1F). Special markers of the juxtaglomerular apparatus, key for nephron hemodynamic responses, were also assessed (renin, Figure 2A, Figure S28). Further, different cell types and regions within a single glomerulus were also resolved (Figure S32), specifically, the glomerular endothelium (PARP1), podocytes (calbindin, nestin), and basement membrane (laminin, nestin). The data demonstrate the unique distribution of these cells within the glomeruli. Each of these markers were located in different cycles and, while they could be condensed into a single plex experiment, this would require removing all other markers that are expressed in the glomeruli that provide spatial context but are not as variable (e.g. CD31, CD93, MARCKS, and vimentin; Figure 2A, Figures S15, S19, S25, S31, respectively). Initial experiments were performed on fresh frozen tissue but was adapted to work on formalin fixed paraffin embedded tissues as well (Figure S33).
Figure 1:
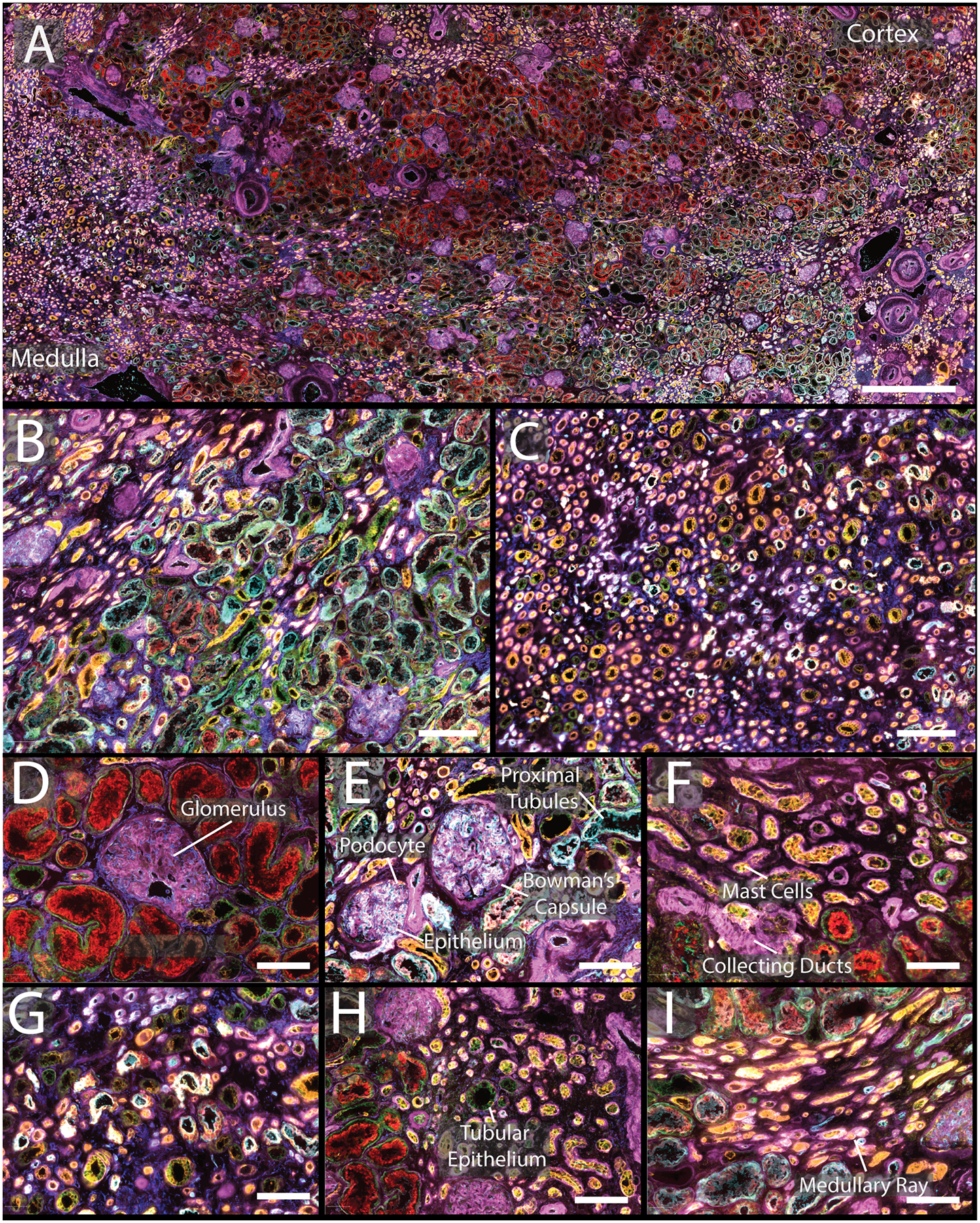
A) CODEX multiplexed IF staining of a human kidney from a 66-year-old male. B) Enlargement of a region of the cortex with several glomeruli present as well as proximal tubules. C) Medullary region of the kidney with many tubules present. D&E) Enlargement of glomeruli surrounded by different tubular segments with high expression of α-smooth muscle actin and aquaporin 1, respectively. F) Tubules within the cortex with high expression of cytokeratin 7 and tryptase. G) Representative view of medulla. H) Portion of cortex with increased numbers of mast cells. I) Enlargement of medullary rays. The color legend for all panels is as follows: cytokeratin 7 (pink), tryptase (orange), nestin (yellow), ß-catenin (green), aquaporin 1 (teal), vimentin (dark blue). Scale bars are 1 mm for panel A, 200 μm for panels B–C, and 50 μm for panels D–I.
Figure 2:
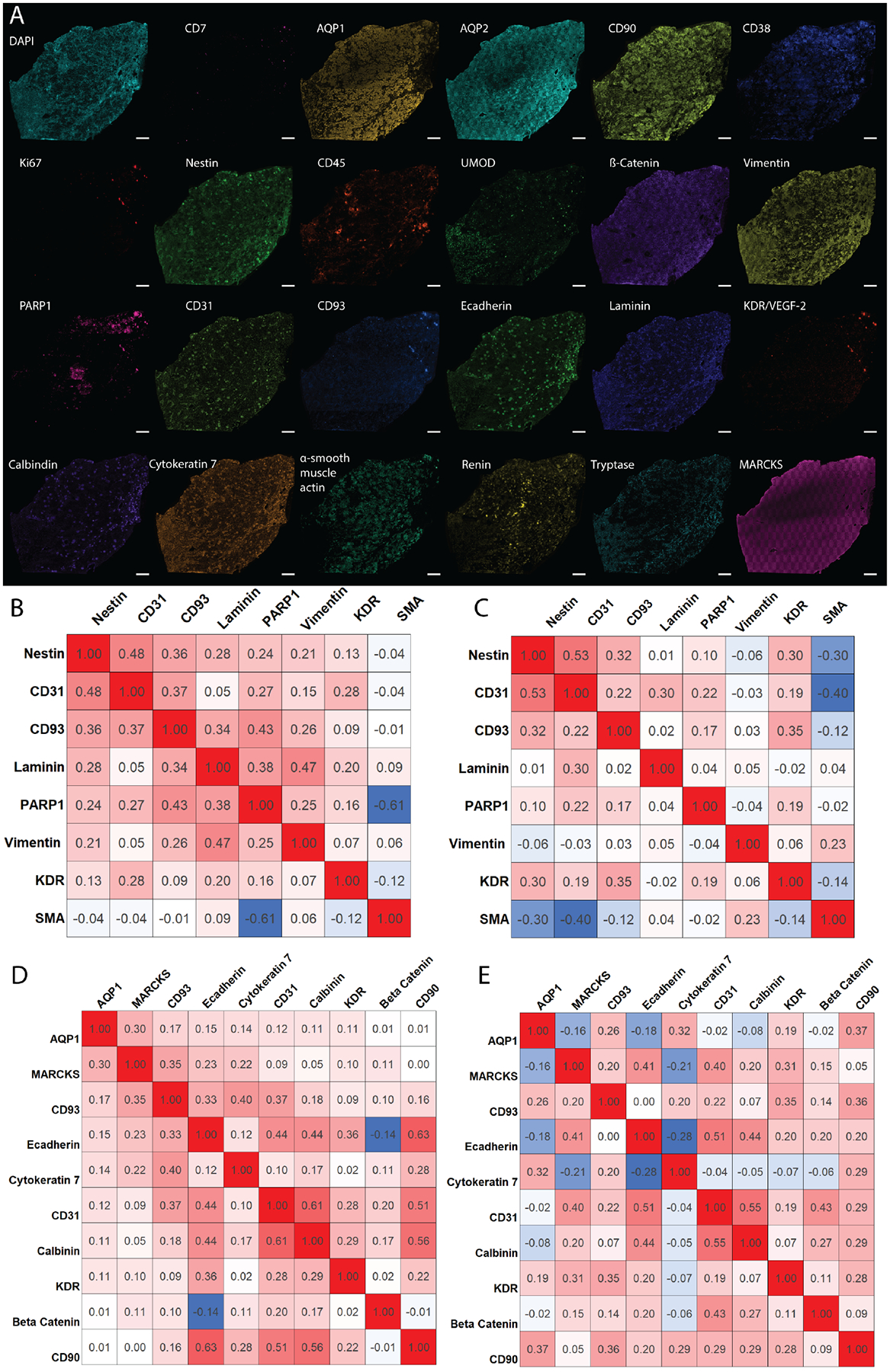
A) Individual images of assayed antigens showing the diversity of structures within the kidney. B) Pearson’s correlation coefficients from three healthy kidney samples for glomerular markers, indicating cell types that are highly correlated (red), noncorrelated (white), and anti-correlated (blue). C) Correlation plot derived from three diabetic nephropathy kidneys, showing cellular redistribution resulting from disease. D) Similar correlation plot for tubular markers from healthy and E) diabetic nephropathy patients. Correlations were established between 0 and 40 μm from the center of the cell for the entire tissue and consist of ~1 million cells.
Cell Type and Neighborhood Analysis
The potential of highly multiplexed molecular analysis is demonstrated by the stark differences seen within each individual channel in the 23-plex experiment (Figure 2A). Further, we calculated the number of each cell type per tissue area. On average, the tissue contained 420± 19 cells/mm2. This analysis can be extended to include specific cell subtypes. Finally, we also assessed how cells are spatially organized within the tissue (Figure 2B–E) and how cellular neighborhoods are reorganized as a result of diabetes by analyzing diabetic nephropathy tissue donors, two men and one woman, who were 52, 65, and 75 years old, respectively. By subdividing the analysis into glomerular (Figure 2B&C) and tubular localizing markers (Figure 2D&E), CODEX multiplexed IF was used to determine the correlation (Pearson’s correlation coefficients >0.1) and anticorrelation (<−0.1) of different protein markers detected within a 40 μm neighborhood around each segmented cell. Within the glomerular endothelium, CD93 was found to correlate with other endothelium markers, such as PARP1. These glomerular endothelium markers are also correlated with the expression of nestin, which labels podocytes within the glomerulus. CD93 and nestin are less correlated within diabetic kidneys. Similar trends can be extrapolated for protein markers that localize within tubules. Moreover, diabetic samples show higher numbers of uncorrelated cell types compared to healthy samples (e.g. AQP1 becomes uncorrelated with MARCKS and e-cadherin enriched cells) suggesting that cellular neighborhoods are reorganized during disease states, such as diabetic nephropathy. While only some examples are discussed here, similar comparisons can be made for all the antigens stained using CODEX. These preliminary analyses importantly demonstrate the ability to survey cell populations with high sensitivity and selectivity using multiplexed molecular imaging and the potential of this technique to illuminate renal structure and function.
DISCUSSION
Using 23 antibodies, key renal cell types can be visualized and compared in human kidney tissue. This study incorporates a combination of commercial CODEX labels as well as conjugated purified antibodies from three different vendors, demonstrating the flexibility of the approach to accommodate a variety of antibody sources. Our studies support that well-validated, primary antibodies free from common preservations (e.g. bovine serum albumin, glycerol, and sodium azide) are compatible with this approach. Additionally, selecting recombinant antibodies reduces variability and is essential for creating large renal atlases, such as those pioneered by the Kidney Precision Medicine Project23 and Human BioMolecular Atlas Program,24 which require examination of renal sections from many patients over the course of years. Most of the antibodies used here are recombinant (Table 1, *) and can serve as an initial panel for applying CODEX IF to fresh frozen human kidney tissues or banked FFPE samples (Figure S33) for these critical atlas efforts. Moreover, we anticipate that study of kidney diseases will also benefit from the development of similar multiplexed panels that may aid in assessment of complex cell interplay and pathophysiology of a spectrum of injury as we also initially demonstrate here with diabetic nephropathy (Figures S34–S36). While it is known cellular neighborhoods are affected by disease, using highly multiplexed analyses, we can parse which cell types are particularly affected within their complex microenvironments. Diseases with similar light microscopic appearances may have varying phenotypes and alterations of specific cells, which may shed light on mechanisms and potential targets for intervention. Because we demonstrate CODEX IF on both healthy and diseased fresh frozen and FFPE tissue, we anticipate its adoption by both research and clinal laboratories. Such highly multiplexed analysis could pave the way for detailed phenotyping, classification and understanding of diseases, enhancing research and patient care.
Supplementary Material
TRANSLATIONAL STATEMENT:
In this manuscript, we describe the application of CODEX multiplexed immunofluorescence to fresh frozen and formalin fixed and paraffin embedded (FFPE) samples of both normal and diseased human kidney. While we have shown images for 23 antigen markers, this technology can be extended to include both custom and commercial antibodies for most protein targets. The ability to probe dozens of protein targets within a single tissue enables visualization and segmentation of fundamental cell types within the healthy and disease kidney on a larger scale than previously possible. Because the kidney has so many functional units and cell types, CODEX IF allows for a more complete picture of the cellular architecture and tissue microenvironments that are fundamental to normal renal function, transition to disease, impact of disease, and potential therapeutics.
ACKNOWLEDGEMENTS
Support was provided by the NIH Common Fund and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (U54DK120058 awarded to J.M.S. and R.M.C.) and NIH National Institute of Allergy and Infectious Disease (NIAID) (R01AI138581 awarded to J.M.S.). E.K.N. is supported by a National Institute of Environmental Health Sciences training grant (T32ES007028). Human tissues were acquired through the Cooperative Human Tissue Network at Vanderbilt University Medical Center which is supported by the NIH National Cancer Institute (5 UM1 CA183727-08).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
All authors declare no competing interests.
Supplementary Material
Supplementary figures, tables, and methods listed below are located within the Supporting Information.
REFERENCES
- 1.Al-Awqati Q, Oliver JA. Stem cells in the kidney. Kidney International. 2002;61:387–395. [DOI] [PubMed] [Google Scholar]
- 2.Puelles VG, Hoy WE, Hughson MD, et al. Glomerular number and size variability and risk for kidney disease. Current Opinion in Nephrology and Hypertension. 2011;20. [DOI] [PubMed] [Google Scholar]
- 3.Gambara V, Mecca G, Remuzzi G, Bertani T. Heterogeneous nature of renal lesions in type II diabetes. Journal of the American Society of Nephrology. 1993;3:1458–1466. [DOI] [PubMed] [Google Scholar]
- 4.Toyota E, Ogasawara Y, Fujimoto K, et al. Global heterogeneity of glomerular volume distribution in early diabetic nephropathy. Kidney International. 2004;66:855–861. [DOI] [PubMed] [Google Scholar]
- 5.Najafian B, Alpers CE, Fogo AB. Pathology of Human Diabetic Nephropathy. Contrib Nephrol. 2011;170:36–47. [DOI] [PubMed] [Google Scholar]
- 6.Buchwalow IB, Böcker W. Immunohistochemistry. Basics and Methods. 2010;1:1–149. [Google Scholar]
- 7.Duraiyan J, Govindarajan R, Kaliyappan K, Palanisamy M. Applications of immunohistochemistry. Journal of pharmacy & bioallied sciences. 2012;4:S307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hell SW, Dyba M, Jakobs S. Concepts for nanoscale resolution in fluorescence microscopy. Current Opinion in Neurobiology. 2004;14:599–609. [DOI] [PubMed] [Google Scholar]
- 9.Bastiaens PIH, Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends in Cell Biology. 1999;9:48–52. [DOI] [PubMed] [Google Scholar]
- 10.Schubert W, Bonnekoh B, Pommer AJ, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nature Biotechnology. 2006;24:1270–1278. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes MJ, Sevinsky CJ, Sood A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proceedings of the National Academy of Sciences. 2013;110:11982–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsurui H, Nishimura H, Hattori S, et al. Seven-color Fluorescence Imaging of Tissue Samples Based on Fourier Spectroscopy and Singular Value Decomposition. Journal of Histochemistry & Cytochemistry. 2000;48:653–662. [DOI] [PubMed] [Google Scholar]
- 13.Lin J-R, Izar B, Wang S, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell. 2018;174:968–981.e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J, Liu C, Kim J, Susztak K. Understanding the kidney one cell at a time. Kidney International. 2019;96:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fribourg M, Anderson L, Fischman C, et al. T-cell exhaustion correlates with improved outcomes in kidney transplant recipients. Kidney International. 2019;96:436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh N, Avigan ZM, Kliegel JA, et al. Development of a 2-dimensional atlas of the human kidney with imaging mass cytometry. JCI Insight. 2019;4:e129477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Scientific Reports. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoltzfus CR, Filipek J, Gern BH, et al. CytoMAP: A Spatial Analysis Toolbox Reveals Features of Myeloid Cell Organization in Lymphoid Tissues. Cell Reports. 2020;31:107523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis MF, Williams M. Irreproducibility in Preclinical Biomedical Research: Perceptions, Uncertainties, and Knowledge Gaps. Trends in Pharmacological Sciences. 2016;37:290–302. [DOI] [PubMed] [Google Scholar]
- 21.Gong B, Murray KD, Trimmer JS. Developing high-quality mouse monoclonal antibodies for neuroscience research – approaches, perspectives and opportunities. New Biotechnology. 2016;33:551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taube JM, Akturk G, Angelo M, et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. Journal for ImmunoTherapy of Cancer. 2020;8(1):e000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Boer IH, Alpers CE, Azeloglu EU, et al. Rationale and design of the Kidney Precision Medicine Project. Kidney International. 2021;99:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder MP, Lin S, Posgai A, et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019;574:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


