Abstract
Fecal excretion of Salmonella enterica serovar Typhimurium organisms was observed in patients and in people not showing symptoms who were involved in an outbreak of food-borne infection with this organism. Excretion of organisms was prolonged in the patients who were given antimicrobial drugs compared with those who were not. The isolates were indistinguishable by their pulsed-field gel electrophoresis patterns and biotyping from the strain recovered from the roast pork that had been consumed by all of the people. This indicates that these isolates obtained from the infected people had originated in the contaminated pork.
Prolonged fecal excretion of the organisms is well known as a consequence of intestinal Salmonella infection. Buchwald and Blaser (2) reviewed 32 reports of persistent excretion of Salmonella organisms following nontyphoid salmonellosis before 1984 and showed that such excretion was more prolonged in patients younger than 5 years and in persons with symptomatic infections. Although minor deviations in pulsed-field gel electrophoresis (PFGE) patterns were reported for enterohemorrhagic Escherichia coli O157:H7 isolates obtained from patients during the shedding period (5, 6), the possible genetic changes in Salmonella isolates recovered from individuals who showed prolonged excretion have not been investigated. A large food-borne outbreak of Salmonella enterica serovar Typhimurium infection with more than 100 cases, also involving people showing no symptoms, occurred in 1993 in Kanagawa, Japan. We describe here the duration of fecal excretion of organisms following infection with Salmonella serovar Typhimurium and the characteristics of the isolates obtained.
In October 1993, an outbreak of food-borne Salmonella serovar Typhimurium infections occurred in a home for mentally handicapped students in Kanagawa Prefecture, Japan. Among the foods consumed by 107 students (7 to 33 years old; 33 females and 74 males) and 33 staff members on 28 October was roast pork prepared by a caterer in Kanagawa. A total of 89 of the students (27 females and 62 males) and 16 of the staff (10 females and 6 males) exhibited symptoms of diarrhea, fever, and vomiting within 10.5 to 121 h after eating the pork. Sixty-two of the 89 students were hospitalized and given fosfomycin and norfloxacin. Eight staff members were also hospitalized, although details of their treatment are uncertain. Twenty-seven of the students with symptoms were treated only with antidiarrheal drugs and were not hospitalized. Eighteen students and 17 staff members showed no symptoms and were not given antimicrobial drugs. Stool samples from 51 individuals with symptoms and 6 without symptoms, vomit specimens from 12 patients, and 32 environmental specimens, including meals stored in refrigerators that were obtained between 29 October and 31 October, were subjected to bacteriological analysis. Cultures of stool specimens from 47 symptomatic and 5 asymptomatic persons, 4 vomit specimens, and a sample of roast pork were all positive for Salmonella serovar Typhimurium. Most-probable-number methods were performed using three tubes for each dilution. The most probable number in the roast pork was estimated as 2.6 × 105/g. Also, Salmonella serovar Typhimurium isolates were recovered from a sample of roast pork stored at the caterer's facility (4.3 × 104/g) and a fecal sample from a family member of the caterer who had eaten the pork. Based on these data, the roast pork was identified as the cause of the outbreak.
Stool specimens were continuously collected from the students and staff, at intervals ranging from 5 days to 2 months, until two consecutive specimens from each person were negative for Salmonella serovar Typhimurium. For isolation, specimens were enriched in a Hajna tetrathionate medium at 42°C for 20 h. The enrichment cultures were streaked onto deoxycholate hydrogen sulfide lactose agar and brilliant green agar and incubated at 37°C for 20 h. Suspect colonies were identified as Salmonella by standard procedures, and all isolates were serologically typed with antisera to O and H antigens (Denka-Seiken Co. Ltd., Tokyo, Japan). Salmonella serovar Typhimurium clinical isolates and those from roast pork were subtyped by using PFGE with XbaI- and BlnI-digested chromosomal DNA (8) and by biotyping (4). Antimicrobial susceptibilities were determined by disk diffusion tests.
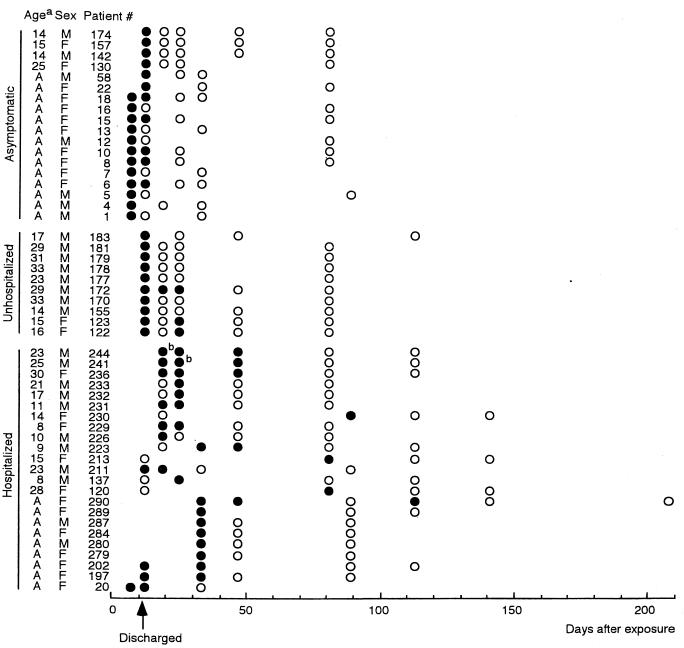
Salmonella serovar Typhimurium isolates were recovered from a total of 33 symptomatic patients and 18 asymptomatic carriers (Fig. 1). Fecal specimens from 14 students and 8 staff members were positive for Salmonella serovar Typhimurium, even after the patients were discharged from hospitals. Salmonella serovar Typhimurium was not recovered 12 days postexposure from people with no symptoms. Positive samples were obtained for 25 days from three of the students who were not hospitalized and were not given antimicrobial drugs but who had exhibited symptoms of diarrhea or fever at the onset of infection. In the specimens from patients 241 and 233 at respective days 19 and 25 postexposure, H2S-negative strains of Salmonella serovar Typhimurium, identified on a sulfide-indole motility medium (Eiken Chemical Co. Ltd., Tokyo, Japan), were obtained, as were H2S-positive strains. All strains from symptomatic and asymptomatic patients, including H2S-negative strains, were classified as biotype 7. They were susceptible to chloramphenicol, kanamycin, amikacin, gentamicin, ampicillin, trimethoprim-sulfamethoxazole, fosfomycin, cefazolin, and cephaloridine and resistant to tetracycline, streptomycin, and nalidixic acid. The H2S-positive strains showed a PFGE pattern identical to that of the strain obtained from the pork (Fig. 2). The H2S-negative strains gave a BlnI-digested PFGE pattern that varied by two bands from with the pattern of the H2S-positive strains. The XbaI-digested patterns of the H2S-negative strains were distinguished from those of the H2S-positive strains by one or four bands.
FIG. 1.
Isolation of Salmonella serovar Typhimurium from patients and asymptomatic persons. ●, feces positive for Salmonella serovar Typhimurium; ○, negative feces; a, all the staff members were adult (A); b, H2S-negative strains as well as H2S-positive strains were isolated (see the text).
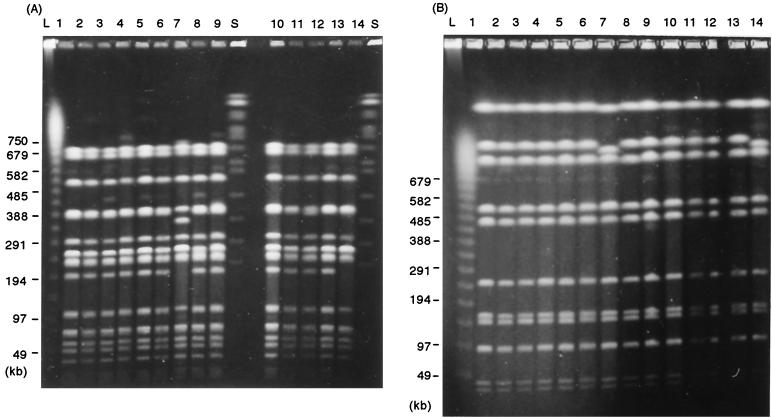
FIG. 2.
XbaI-digested (A) and BlnI-digested (B) PFGE patterns of Salmonella serovar Typhimurium isolates. Lanes: 1, the strain from roast pork; 2 to 5, strains from patient 4, 5, 6, and 7, respectively, at day 7 postexposure; 6, 8, and 9, strains from patient 241 at days 19, 25, and 47, respectively; 7, H2S-negative strain from patient 241 at day 19; 10 to 12, strains from patient 244 at days 19, 25, and 47, respectively; 13 and 14, H2S-positive strain and H2S-negative strain, respectively, from patient 233 at day 25; L, lambda 48.5-kb ladder; S, Saccharomyces cerevisiae size standards. The sizes of the markers in kilobase pairs are indicated to the left of each panel.
Because these isolates showed identical PFGE patterns, it was proven that Salmonella serovar Typhimurium isolates obtained from symptomatic and asymptomatic shedders of the organism originated with the strain that contaminated the roast pork, that is, the outbreak strain. According to the established criteria for bacterial strain typing by PFGE (9), one of the H2S-negative strains (Fig. 2, lane 14) was considered to be closely related to the outbreak strain (H2S-positive strain). The other (Fig. 2, lane 7) was interpreted as possibly being part of the outbreak. A similar observation has previously been reported for fecal samples from patients involved in an outbreak of food poisoning due to Salmonella enterica serovar Enteritidis (7).
Our observation suggested that administration of antimicrobial drugs prolongs fecal excretion of Salmonella serovar Typhimurium organisms. It has been known that prolonged excretion of salmonellae is caused by antimicrobial treatment of acute salmonellosis (1), possibly because in their intracellular site, Salmonella organisms are protected from the action of antibiotics (3). There was no relation between the duration of excretion and the age of infected persons. Because children older than 7 years and adults were involved in the outbreak, our results are consistent with the previous observation (2) that the duration of excretion of organisms did not differ between children of 5 to 14 years and adults. In food-borne infections among mentally handicapped persons, secondary infections due to their behavior are considered a possible cause of prolonged excretion. Our results show that the duration of excretion of organisms for the students was almost the same as that for the staff. No recurrence of excretion was observed for the asymptomatic students. Therefore, it is unlikely that the prolonged occurrence of positive fecal samples was caused by secondary infections. However, frequent checking of fecal specimens after the acute phase of Salmonella infection is considered necessary to prevent further infections.
Acknowledgments
We are grateful to Jane K. A. Cook of Intervet U.K.
REFERENCES
- 1.Aserkoff B, Bennett J V. Effect of antibiotic therapy in acute salmonellosis on the fecal excretion of salmonellae. N Engl J Med. 1969;281:636–640. doi: 10.1056/NEJM196909182811202. [DOI] [PubMed] [Google Scholar]
- 2.Buchwald D S, Blaser M J. A review of human salmonellosis. II. Duration of excretion following infection with nontyphi Salmonella. Rev Infect Dis. 1984;6:345–356. doi: 10.1093/clinids/6.3.345. [DOI] [PubMed] [Google Scholar]
- 3.Chiu C-H, Lin T-Y, Ou J T. In vitro evaluation of intracellular activity of antibiotics against non-typhoid Salmonella. Int J Antimicrob Agents. 1999;12:47–52. doi: 10.1016/s0924-8579(99)00038-2. [DOI] [PubMed] [Google Scholar]
- 4.Duguid J R, Anderson E S, Alfredsson G A, Baker R, Olg D C. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J Med Microbiol. 1975;8:149–166. doi: 10.1099/00222615-8-1-149. [DOI] [PubMed] [Google Scholar]
- 5.Gouveia S, Proctor M E, Lee M-S, Luchansky J B, Kaspar C W. Genomic comparisons and Shiga toxin production among Escherichia coli O157:H7 isolates from a day care center outbreak and sporadic cases in southeastern Wisconsin. J Clin Microbiol. 1998;36:727–733. doi: 10.1128/jcm.36.3.727-733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karch H, Russman H, Schmidt H, Schwarzkopf A, Heesemann J. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J Clin Microbiol. 1995;33:1602–1605. doi: 10.1128/jcm.33.6.1602-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murase T, Nakamura A, Matsushima A, Yamai S. An epidemiological study of Salmonella enteritidis by pulsed-field gel electrophoresis (PFGE): several PFGE patterns observed in isolates from a food poisoning outbreak. Microbiol Immunol. 1996;40:873–875. doi: 10.1111/j.1348-0421.1996.tb01153.x. [DOI] [PubMed] [Google Scholar]
- 8.Murase T, Okitsu T, Suzuki R, Morozumi H, Matsushima A, Nakamura A, Yamai S. Evaluation of DNA fingerprinting by PFGE as an epidemiologic tool for Salmonella infections. Microbiol Immunol. 1995;39:673–676. doi: 10.1111/j.1348-0421.1995.tb03255.x. [DOI] [PubMed] [Google Scholar]
- 9.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]