Abstract
Quantitative neuroimaging studies in twin samples can investigate genetic contributions to brain structure and microstructure. Diffusion tensor imaging (DTI) studies with twin samples have shown moderate to high heritability in white matter microstructure. This study investigates the genetic and environmental contributions of another widely used diffusion MRI model not yet applied to twin studies, neurite orientation dispersion and density imaging (NODDI). The NODDI model is a multicompartment model of the diffusion-weighted MRI signal, providing estimates of neurite density (ND) and the orientation dispersion index (ODI). A cohort of monozygotic (MZ) and same-sex dizygotic (DZ) twins (N=460 individuals) between 13–24 years of age were scanned with a multi-shell diffusion weighted imaging protocol. Select white matter (WM) regions of interest (ROI) were extracted. Biometric structural equation modeling estimated the relative contributions from additive genetic (A) and common (C) and unique environmental (E) factors. Genetic factors for the NODDI measures accounted for 91% and 65% of the variation of global ND and ODI, respectively, compared with 83% for FA. We observed higher heritability for ND than both FA and ODI in 25 of 30 discrete white matter regions that we examined, suggesting ND may be more sensitive to underlying genetic sources of variation. This study demonstrated that genetic factors play a key role in the development of white matter microstructure using both DTI and NODDI.
Keywords: Diffusion MRI, Heritability, Imaging Genetics, NODDI, Twin Study, White matter microstructure
1. Introduction
Much of the vast spectrum of differences in human brain anatomy may be captured using magnetic resonance imaging (MRI). Understanding the genetic and environmental sources of individual differences in human brain structure holds considerable promise to better comprehend both typical and atypical brain development (Bigos et al., 2016); however, the specific neuronal microstructural properties that contribute to such individual differences are unclear. From a population-based twin cohort, we conducted comprehensive quantitative genetic analysis of white matter (WM) microstructural properties using measures of neurite density and dispersion (Zhang et al., 2012) to quantify the degree of genetic influences.
Diffusion tensor imaging (DTI) has been widely used to probe microstructural properties of white matter pathways (Alexander et al., 2007). DTI studies of twins and family pedigrees show that additive genetic factors account for substantial portions of variability in WM microstructure (Brouwer et al., 2010; Chiang et al., 2009) represented by DTI measures. The Human Connectome Project (HCP) identified twins and their siblings from the Missouri Family and Twin Registry and scanned these individuals using high resolution diffusion weighted imaging (DWI), finding heritabilities of 0.53–0.90 for FA in various white matter regions (Kochunov et al., 2015). Recent research initiatives that combine multiple cohorts from around the world using meta- and mega-analytic approaches further corroborate the significance of genetic contributions, with heritability ranging from 0.40 to 0.80 for FA in a majority of regions investigated (Jahanshad et al., 2013). Although FA is the most commonly reported measure in the literature, recent studies have further demonstrated substantial heritability among other DTI diffusivity metrics: mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD) (Hatton et al., 2018; Vuoksimaa et al., 2017). Heritabilities of DTI measures have also been investigated across the life span, from infancy to early and late adulthood (Chiang et al., 2011; Jansen et al., 2015). For example, genetic influences were observed in infant twins as early as 40 days after birth (Geng et al., 2012; Lee et al., 2015), and comparable estimates were also observed in an elderly twin sample (Vuoksimaa et al., 2017).
Although DTI measures are sensitive to individual differences in microstructure, the assumed Gaussian diffusion distribution model is inadequate in regions of crossing white matter tracts and for diffusion-weighted imaging (DWI) studies with increased diffusion-weighting (b > 1,500 s/mm2). As a result, more complex DWI signal models with non-Gaussian distributions or multiple compartments have been proposed (Assaf and Basser, 2005; Jensen and Helpern, 2010; Zhang et al., 2012). One widely used DWI method is Neurite Orientation Dispersion and Density Imaging (NODDI) (Zhang et al., 2012), which models the diffusion-weighted signals from three distinct compartments: (1) collections of restricted diffusion sticks or so-called “neurites” (axons, dendrites), (2) anisotropic Gaussian diffusion attributed to the extracellular tissue matrix, and (3) a fixed diffusion free-water isotropic compartment, which is attributed to cerebrospinal fluid (CSF). The NODDI model provides measures of the relative signal fraction of the restricted diffusion sticks, ND (for neurite density), and the orientation dispersion index (ODI) of the modeled restricted diffusion sticks (Zhang et al., 2012). In white matter, intra-axonal diffusion is often assumed to comprise the neurite signal and can be characterized by ND and ODI. The NODDI technique has been widely used to investigate white matter microstructural changes with brain development and maturation (Lynch et al., 2020) and a range of neurological conditions including brain injuries, epilepsy and Alzheimer’s disease (Colgan et al., 2016; Grussu et al., 2017; Mastropietro et al., 2019). However, NODDI based measures have not been widely applied in imaging genetics studies. To our knowledge, the only published investigation of the heritability of DTI and NODDI measures examined an Amish family pedigree (Kochunov et al., 2016). Using the corpus collosum as the sole WM region of interest, researchers found higher heritability for ND (0.70) than for ODI (0.42). This finding invites a more systematic whole brain investigation of genetic basis of NODDI measures in WM.
In the context of neuroimaging studies, data from monozygotic [MZ] and same-sex dizygotic [DZ] twins can allow for the statistical decomposition of the total variance of brain imaging measures into three sources: additive genetics (A), common environmental influences (C), and unique environmental influences (E) (Neale & Cardon, 1992). The correlation of MZ twins, with their 100% structural genetic similarity, sets an upper bound to heritability in this model. Doubling the difference between MZ and DZ correlations estimates heritability (Falconer & Mac Kay, 1998). The extent to which DZ twins, with their 50% overlap in segregating genes, are phenotypically more similar than their genetic factors alone can account for is a key factor in estimating common (i.e., shared by cotwins) environmental factors. Unique environmental factors are estimated, in part, by the extent to which the MZ correlation falls below 1.0. Here, as well as in practice (Neale and Cardon, 1992), these genetic and environmental variance components are estimated from structural equation models (SEM) in which a series of equations capturing the ideas just described are solved simultaneously and the fit of competing models evaluated.
Capitalizing on one of the largest single-site twin imaging cohorts with a multi-shell diffusion weighted imaging (Schmidt et al., 2019), we aimed to quantitatively model similarities of DTI and NODDI white matter in adolescent MZ and DZ twins to examine genetic and environmental influences on variation in whole-brain white matter microstructure. Specifically, the current work aimed to (1) replicate previous findings of DTI heritability in WM, and (2) further investigate the heritability of neurite properties of WM using NODDI-based measures.
Methods
Participants
Participants were recruited from a birth registry-based sample of MZ and DZ twin pairs from the Wisconsin Twin Project at the University of Wisconsin–Madison (Schmidt et al., 2019, 2013). Twins who were invited for the neuroimaging study had previously participated in extensive assessments that included behavioral batteries, cognitive testing, and structured psychiatric interviews. Exclusion criteria for the MRI assessment included the following conditions: claustrophobia, seizure disorder, metal orthodontic braces, dermal piercings, traumatic brain injury, and developmental disabilities.
After excluding opposite sex DZ twins, a total of 460 twin individuals were included in the analysis. The sample consisted of 238 females (52%) and 222 (48%) males, with a mean age of 17.9 years (SD=2.3) who ranged from age 13–24 years at the time of scan. Final twin modeling included 129 MZ and 90 DZ twin pairs, after excluding 22 unpaired twins whose cotwin’s DWI images were not obtained or usable. Based on self-report, the sample is 84% white and 16% other races/ethnicities. An Institutional Review Board (IRB) at the University of Wisconsin–Madison approved all study protocols, and both twins and their legal guardian provided informed assent (in the case of minors) and consent, respectively.
Image Acquisition
Twins were scanned in a 3.0 Tesla GE Discovery MR750 scanner with a 32-channel receive-only head coil (NOVA Medical, Wakefield, MA). Diffusion weighted imaging (DWI) used a spin-echo echo-planar imaging (EPI) sequence with three diffusion weighting shells. A total of 6 non-DWIs (b=0 s/mm2) and 63 DWIs with non-collinear, multi-b diffusion encoding directions were collected at b=500 s/mm2 (9 directions), b=800 s/mm2 (18 directions), and b=2000 s/mm2 (36 directions). 74 contiguous axial slices DWI (2 mm thick) were prescribed to cover the entire brain. Other protocol parameters were TR/TE = 8575/76.6 ms; parallel imaging (ASSET with acceleration factor = 2); isotropic 2 mm in-plane resolution (128×128 matrix with 256 mm field-of-view).
Image Processing
Processing of DWIs was previously described in detail (Adluru et al. 2017). Briefly, FSL software package tools were used to correct for eddy current-related distortions and head motion (Andersson & Sotiropoulos, 2016). To account for main magnetic field inhomogeneities, a fieldmap was created using a three echo, spoiled gradient echo acquisition and the iterative decomposition of water and fat with echo asymmetry and the least-squares estimation (IDEAL) technique (Reeder et al., 2005). The FSL tool ‘fugue’ was subsequently used to undistort the DWI dataset using fieldmaps (Cusack et al., 2003; Jezzard & Balaban, 1995).
DTI and NODDI Fitting
Estimation of the diffusion tensors at each voxel used non-linear tensor estimation in the CAMINO software package (Jones & Basser, 2004). From the diffusion tensors, the eigenvalues were estimated, and maps of FA, AD, RD, and MD were computed. Next, DWI data were fit to the three-compartment NODDI tissue model to provide estimates of ND and ODI (Zhang et al., 2012). To perform the fitting, we used the Accelerated Microstructure Imaging via Convex Optimization (AMICO) framework (Daducci et al., 2015), which reformulates the NODDI model into a system of linear equations that can be rapidly solved and has been used in similar large population based studies (Fukutomi et al., 2018). Such a strategy greatly speeds up the mapping of NODDI parameters while maintaining a high correlation with the original implementation (Daducci et al., 2015).
Study-specific template creation and regions of interest extraction
A study-specific template was created from all processed twins’ DTI images with DTI-TK using affine and diffeomorphic diffusion tensor registration (Zhang et al., 2006). The same transformation is subsequently applied to native-space ND and ODI images. Within the template space, an intraclass correlation (ICC) is calculated at each voxel among MZ and DZ twin pairs as initial indices of twin similarity.
The JHU/ICBM (Mori et al., 2008) WM atlas was co-registered to the study-specific template space using diffeomorphic registration algorithms from the Advanced Normalization Tools (ANTs) package (Avants et al., 2008). White matter regions of interest (ROIs) from the JHU/ICBM WM atlas were then warped into each subject’s native space by applying the inverse of the spatial transformations estimated from the registration process. Mean values of native-space DTI and NODDI parameters of each ROI were subsequently calculated. Each subject’s ROI alignment in individual space was visually inspected.
Global WM measures
We created a composite mask of all ROIs investigated in the current study, which was used to calculate each twin’s global WM measure for each of the DTI and NODDI indices. These global measurements are included in the subsequent twin modeling with JHU ROIs.
Quantitative Genetic Analyses
Genetic analyses with structural equation modeling were conducted with the publicly available R package umx (Bates et al., 2019), which employs maximum-likelihood variance decomposition methods to estimate the additive genetic and environmental influences on WM microstructure. The umx package offered concise syntax and matrix-based twin modeling that are appropriate for the proposed analysis.
Using univariate structural equation modeling, the phenotypic variances in DTI or NODDI measures were decomposed into additive genetic (A), common environmental (C), and unique environmental (E) contributions by contrasting the covariance among MZ and DZ twin pairs (Neale and Cardon, 1992). The path diagram for twin modeling is shown in Figure 1. The “A” components were assumed to correlate at 1.0 for MZ twins and 0.5 for DZ twins. Both MZ and DZ twins’ “C” components were assumed to correlate at 1.0, as dictated by the equal environment assumption.
Fig. 1:
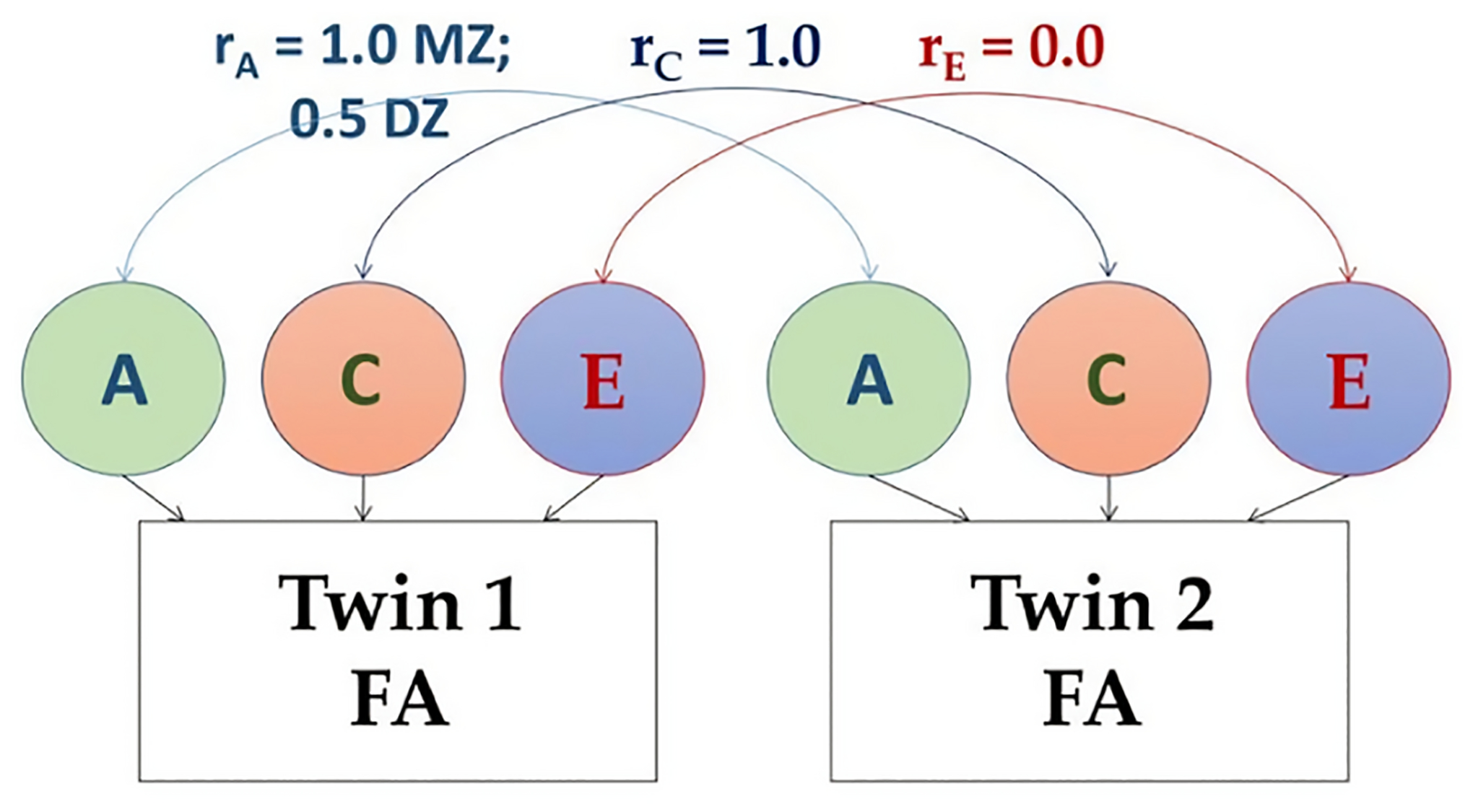
Path diagram of the structural equation modeling (SEM), showing additive genetic (A), common (“shared”) environment (C), and unique environment (E) variance components
Significance tests of genetic and environmental effects compared the full model against partial models obtained by dropping A or E estimates. The change in model fit was calculated using the likelihood-ratio test as the twice the log likelihood difference between the full and partial models, and the 95% confidence intervals (CIs) for the A, C, and E estimates were generated.
Results
Group level DTI and NODDI measures
Figure 2 shows the templates of FA, ND, and ODI maps generated from 460 twin individuals. Our brain microstructural measures did not show any sex or age dependencies in the ROIs investigated. No significant sex differences between males and females emerged for any of the tracts investigated (p>0.05). Also, no significant linear relationships occurred between age and microstructural measures (p>0.05).
Fig. 2:
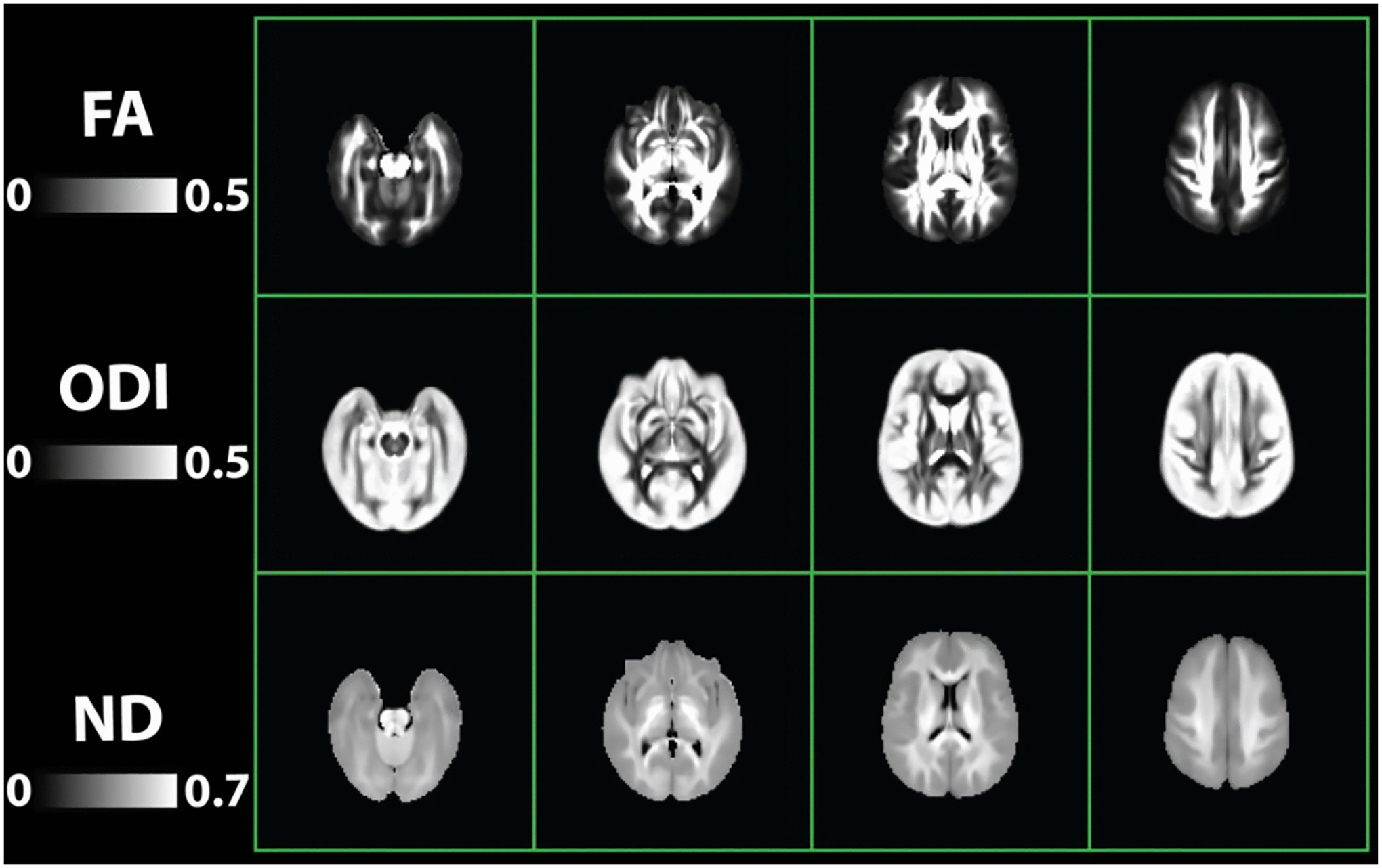
Template images obtained from 460 twin individuals. Top row: Diffusion tensor imaging (DTI) measure of Fractional anisotropy (FA); Middle row: Neurite orientation dispersion and density imaging (NODDI) measures of Orientation dispersion index (ODI); Bottom row: NODDI Neurite density (ND)
Table 1 shows the mean and standard deviation of DTI and NODDI indices of corresponding regions; these indices are consistent with known brain anatomy. For example, the corpus collosum, where commissural fibers are most coherently orientated between the left and right hemispheres with few crossing fibers, showed the lowest ODI values. Cerebral peduncles, which contain densely packed ascending and descending cortical WM projections, and middle cerebellar peduncle showed the highest ND.
Table 1:
Mean and (standard deviation) of DTI and NODDI indices (N=460)
| FA | AD (m2ms−1) | RD (m2ms−1) | MD (m2ms−1) | ND | ODI | |
|---|---|---|---|---|---|---|
| Projection fibers: | ||||||
| Anterior corona radiata-L | 0.46 (0.03) | 1.05 (0.04) | 0.49 (0.03) | 0.68 (0.03) | 0.55 (0.03) | 0.21 (0.02) |
| Anterior corona radiata-R | 0.47 (0.03) | 1.06 (0.04) | 0.48 (0.03) | 0.68 (0.03) | 0.54 (0.03) | 0.20 (0.02) |
| Superior corona radiata-L | 0.50 (0.02) | 1.02 (0.04) | 0.45 (0.02) | 0.64 (0.02) | 0.62 (0.03) | 0.21 (0.02) |
| Superior corona radiata-R | 0.49 (0.03) | 1.00 (0.04) | 0.44 (0.02) | 0.62 (0.02) | 0.62 (0.03) | 0.22 (0.02) |
| Posterior corona radiata-L | 0.48 (0.03) | 1.11 (0.05) | 0.49 (0.03) | 0.70 (0.03) | 0.56 (0.04) | 0.19 (0.02) |
| Posterior corona radiata-R | 0.50 (0.03) | 1.13 (0.05) | 0.48 (0.03) | 0.70 (0.03) | 0.55 (0.03) | 0.17 (0.02) |
| Anterior limb of internal capsule-L | 0.57 (0.02) | 1.13 (0.03) | 0.40 (0.02) | 0.64 (0.01) | 0.61 (0.02) | 0.08 (0.02) |
| Anterior limb of internal capsule-R | 0.58 (0.02) | 1.14 (0.03) | 0.39 (0.02) | 0.64 (0.01) | 0.59 (0.02) | 0.09 (0.02) |
| Posterior limb of internal capsule-L | 0.66 (0.02) | 1.20 (0.03) | 0.35 (0.02) | 0.63 (0.01) | 0.68 (0.03) | 0.11 (0.01) |
| Posterior limb of internal capsule-R | 0.67 (0.02) | 1.23 (0.03) | 0.35 (0.02) | 0.64 (0.01) | 0.68 (0.03) | 0.10 (0.01) |
| Posterior thalamic radiation-L | 0.63 (0.02) | 1.39 (0.06) | 0.42 (0.03) | 0.74 (0.03) | 0.55 (0.03) | 0.06 (0.01) |
| Posterior thalamic radiation-R | 0.63 (0.03) | 1.37 (0.06) | 0.41 (0.03) | 0.73 (0.03) | 0.54 (0.03) | 0.06 (0.01) |
| Association fibers: | ||||||
| Superior longitudinal fasciculus-L | 0.51 (0.02) | 1.03 (0.03) | 0.44 (0.02) | 0.64 (0.02) | 0.62 (0.03) | 0.19 (0.02) |
| Superior longitudinal fasciculus-R | 0.51 (0.02) | 1.03 (0.03) | 0.44 (0.02) | 0.64 (0.02) | 0.62 (0.03) | 0.19 (0.02) |
| Superior fronto-occipital fasciculus-L | 0.52 (0.04) | 1.02 (0.05) | 0.42 (0.03) | 0.62 (0.02) | 0.64 (0.04) | 0.18 (0.03) |
| Superior fronto-occipital fasciculus-R | 0.52 (0.04) | 1.04 (0.04) | 0.42 (0.03) | 0.63 (0.02) | 0.61 (0.03) | 0.18 (0.03) |
| Cingulum-L | 0.56 (0.03) | 1.18 (0.04) | 0.43 (0.03) | 0.68 (0.02) | 0.56 (0.03) | 0.08 (0.03) |
| Cingulum-R | 0.51 (0.03) | 1.11 (0.04) | 0.46 (0.03) | 0.67 (0.02) | 0.54 (0.02) | 0.13 (0.04) |
| Uncinate fasciculus-L | 0.47 (0.04) | 1.13 (0.05) | 0.51 (0.03) | 0.72 (0.03) | 0.48 (0.03) | 0.17 (0.03) |
| Uncinate fasciculus-R | 0.49 (0.03) | 1.22 (0.06) | 0.53 (0.03) | 0.76 (0.03) | 0.46 (0.03) | 0.13 (0.03) |
| Sagittal Stratum-L | 0.61 (0.02) | 1.34 (0.05) | 0.44 (0.03) | 0.74 (0.02) | 0.53 (0.03) | 0.08 (0.01) |
| Sagittal Stratum-R | 0.61 (0.02) | 1.33 (0.05) | 0.44 (0.03) | 0.74 (0.02) | 0.53 (0.03) | 0.08 (0.01) |
| Commissural WM: | ||||||
| Genu of corpus callosum | 0.65 (0.02) | 1.41 (0.04) | 0.37 (0.03) | 0.72 (0.03) | 0.58 (0.03) | 0.06 (0.01) |
| Body of corpus callosum | 0.65 (0.02) | 1.42 (0.04) | 0.36 (0.03) | 0.71 (0.02) | 0.63 (0.03) | 0.05 (0.01) |
| Splenium of corpus callosum | 0.69 (0.02) | 1.50 (0.06) | 0.32 (0.03) | 0.72 (0.03) | 0.66 (0.03) | 0.05 (0.01) |
| Tracts in the brainstem: | ||||||
| Corticospinal tracts-L | 0.52 (0.03) | 1.11 (0.05) | 0.44 (0.03) | 0.67 (0.03) | 0.66 (0.03) | 0.13 (0.04) |
| Corticospinal tracts-R | 0.54 (0.03) | 1.09 (0.05) | 0.43 (0.03) | 0.65 (0.03) | 0.68 (0.03) | 0.12 (0.05) |
| Cerebral peduncle-L | 0.66 (0.02) | 1.42 (0.05) | 0.35 (0.03) | 0.71 (0.02) | 0.70 (0.03) | 0.08 (0.01) |
| Cerebral peduncle-R | 0.67 (0.02) | 1.51 (0.06) | 0.35 (0.03) | 0.74 (0.03) | 0.71 (0.03) | 0.08 (0.01) |
| Middle cerebellar peduncle | 0.52 (0.02) | 1.07 (0.03) | 0.41 (0.03) | 0.63 (0.02) | 0.72 (0.03) | 0.19 (0.01) |
| Global Measures: | ||||||
| All fiber tracts | 0.57 (0.02) | 1.27 (0.02) | 0.47 (0.02) | 0.74 (0.02) | 0.62 (0.02) | 0.16 (0.01) |
Preliminary phenotypic analyses: Individual level phenotypic correlation between DTI and NODDI in WM
Table 2 illustrates the direction and degree of correlation between global DTI and NODDI measures. All correlations are significant at p<0.01 level with the exception of AD with FA and AD with ODI. For specific ROI correlations, the heat map in Supplemental Figure 2 depicts correlations between each of the 30 regions of interest for DTI and NODDI measures. Results show FA is negatively correlated with ODI and positively correlated with ND, whereas ND and ODI are only weakly correlated (Table 2, Figure S2).
Table 2.
Correlations between global DTI and NODDI measures.
| FA | ND | ODI | AD | RD | MD | |
|---|---|---|---|---|---|---|
| FA | 1 | |||||
| ND | 0.62 | 1 | ||||
| ODI | −0.55 | 0.22 | 1 | |||
| AD | −0.07n.s | −0.21 | −0.08n.s | 1 | ||
| RD | −0.78 | −0.52 | 0.46 | 0.65 | 1 | |
| MD | −0.54 | −0.44 | 0.26 | 0.87 | 0.94 | 1 |
n.s. = not significant, p>0.05
Initial twin similarity analyses and preliminary Falconer’s heritability maps
We computed ROI-based intraclass correlations (ICCs) among MZ and DZ twin pairs using NODDI and DTI metrics (see Figure 3 for the voxel-wise metrics; see Supplement Tables S1–S6 for ROI-specific ICCs). Among all the ROIs investigated, MZ twin pairs had higher ICCs, suggesting the presence of genetic contributions (Supplement Tables S1–S6).
Fig. 3:
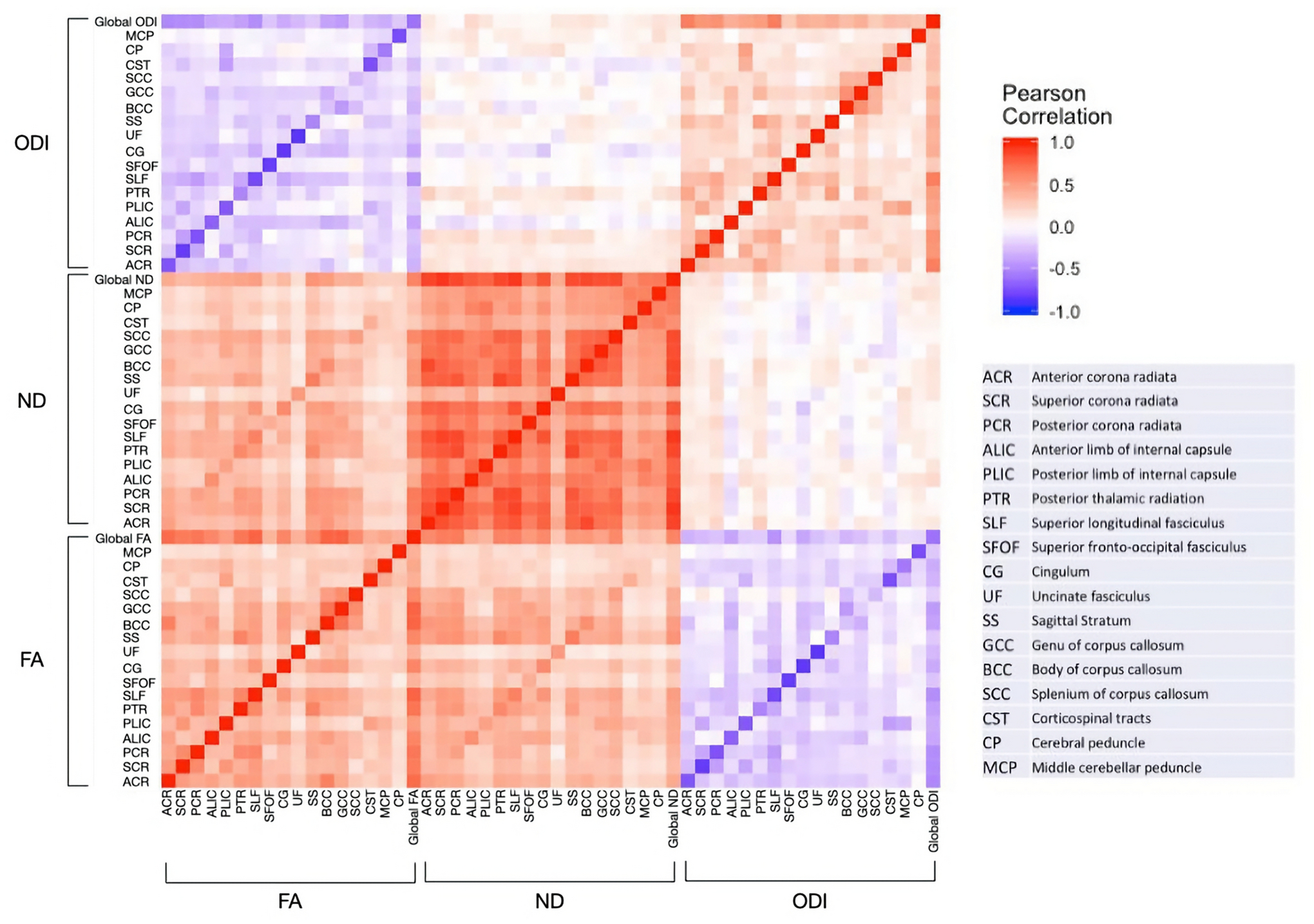
Heat map of Pearson Correlation between FA and NODDI metrics in WM
In the first and second rows of Figure 4, voxel-wise maps of ICCs for MZ and DZ twins using DTI and NODDI illustrate the gross resemblance of cotwins for WM microstructure. Qualitatively, MZ twins have widespread regions of WM with higher ICCs compared with DZ twins. Falconer’s formula (h2 = 2[ICCMZ − ICCDZ]) (Falconer & Mac Kay, 1998) was used to calculate a preliminary heritability estimate at the voxel level. Figure 4, row 3 illustrates voxel-wise heritability maps where the color spectrum indicates levels of heritability in ND, ODI and FA. For consistency, we did not set the upper bound of Falconer heritability at the MZ ICC. This decision led to the anomalous result that heritabilities were sometimes higher than MZ ICCs, which violates the underlying assumptions of the Falconer model. Still, Figure 4 depicts highly variable heritable effects across brain regions. We also note that ND ICCs are more homogeneous, especially at grey matter and WM boundaries, whereas ODI and FA ICCs drastically decrease near the edge of WM.
Fig. 4:
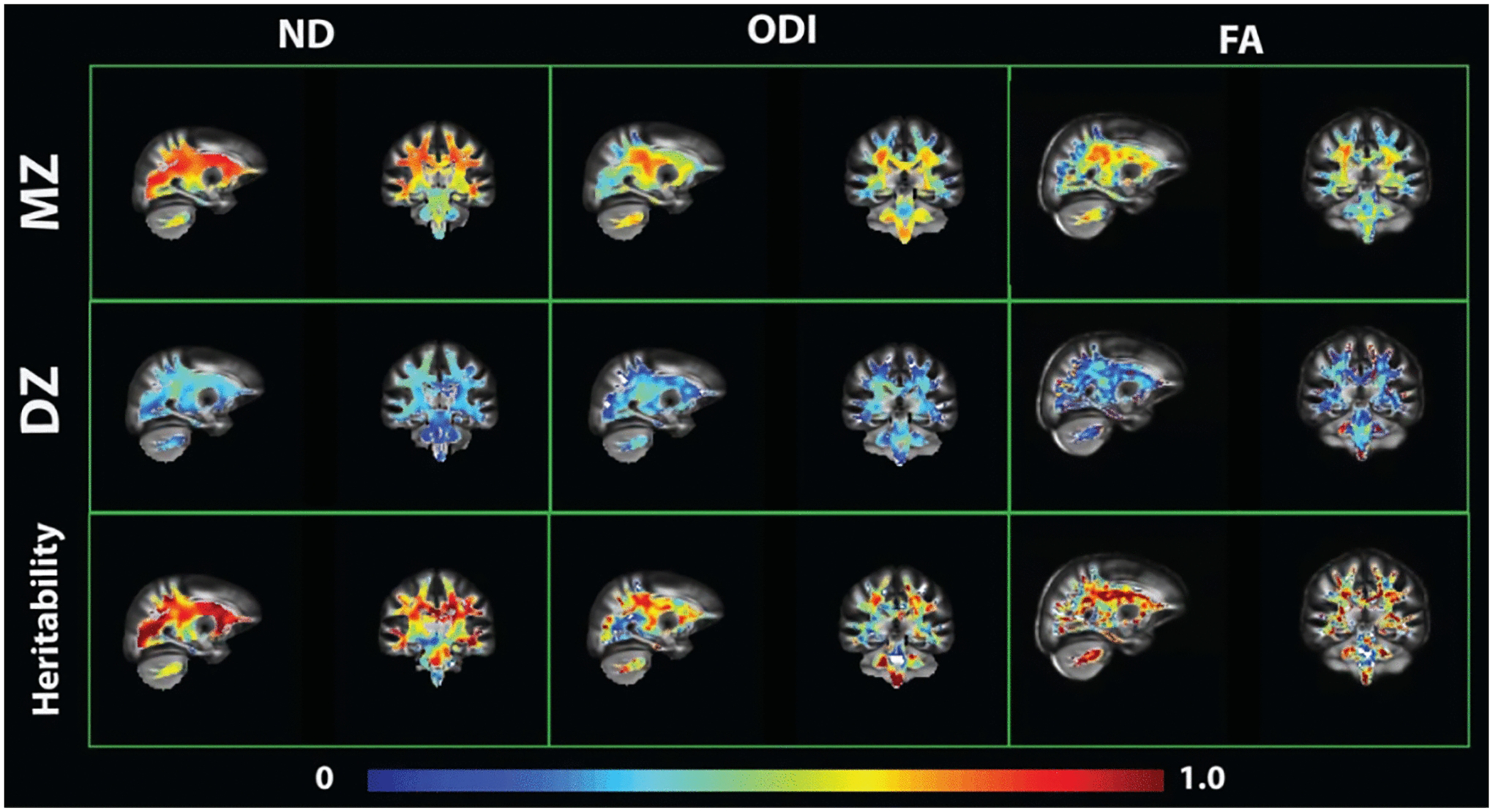
Voxel-wise ICC and Falconer’s heritability maps
Biometric ACE modeling for DTI and NODDI measures
Compared with Falconer’s heritability, the structural equation ACE modeling approach optimally weights estimates for MZ and DZ sample sizes, constrains estimates according to polygenic theory, provides standard errors for parameter estimates, and allows for tests of absolute model fit and comparative fit of nested models (Neale and Cardon, 1992). Model fitting output parameters are detailed in Supplement Tables S1–S6, including maximum likelihood variance components parameter estimates, 95% CIs, and statistical significance tests for of A and C. Heritability estimates of DTI and NODDI measures of JHU template ROIs are shown in Tables 3 and 4 and Figure 5.
Table 3:
Twin ACE modeling of fractional anisotropy (FA) and neurite orientation dispersion and density imaging (NODDI) measures ND and ODI in specific Johns Hopkins University template white matter regions. N=438
| Fractional anisotropy (FA) | Neurite Density (ND) | Orientation Dispersion Index (ODI) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| a2 | c2 | e2 | a2 | c2 | e2 | a2 | c2 | e2 | |
| Projection fibers: | |||||||||
| Anterior corona radiata-L | 0.73 | 0.00 | 0.27 | 0.90 | 0.00 | 0.10 | 0.64 | 0.00 | 0.36 |
| Anterior corona radiata-R | 0.70 | 0.00 | 0.30 | 0.88 | 0.00 | 0.12 | 0.65 | 0.00 | 0.35 |
| Superior corona radiata-L | 0.67 | 0.00 | 0.33 | 0.86 | 0.00 | 0.14 | 0.61 | 0.00 | 0.39 |
| Superior corona radiata-R | 0.71 | 0.09 | 0.20 | 0.84 | 0.00 | 0.16 | 0.71 | 0.05 | 0.24 |
| Posterior corona radiata-L | 0.74 | 0.00 | 0.26 | 0.84 | 0.01 | 0.15 | 0.44 | 0.31 | 0.25 |
| Posterior corona radiata-R | 0.78 | 0.00 | 0.22 | 0.89 | 0.00 | 0.11 | 0.43 | 0.31 | 0.26 |
| Anterior limb of internal capsule-L | 0.73 | 0.00 | 0.27 | 0.79 | 0.00 | 0.21 | 0.33 | 0.07 | 0.60 |
| Anterior limb of internal capsule-R | 0.65 | 0.00 | 0.35 | 0.81 | 0.00 | 0.19 | 0.49 | 0.00 | 0.51 |
| Posterior limb of internal capsule-L | 0.64 | 0.04 | 0.32 | 0.41 | 0.18 | 0.41 | 0.76 | 0.00 | 0.24 |
| Posterior limb of internal capsule-R | 0.68 | 0.04 | 0.28 | 0.69 | 0.00 | 0.31 | 0.76 | 0.01 | 0.23 |
| Posterior thalamic radiation-L | 0.73 | 0.00 | 0.27 | 0.86 | 0.00 | 0.14 | 0.44 | 0.13 | 0.43 |
| Posterior thalamic radiation-R | 0.62 | 0.02 | 0.36 | 0.85 | 0.00 | 0.15 | 0.16 | 0.40 | 0.44 |
| Association fibers: | |||||||||
| Superior longitudinal fasciculus-L | 0.81 | 0.00 | 0.19 | 0.84 | 0.00 | 0.16 | 0.72 | 0.00 | 0.28 |
| Superior longitudinal fasciculus-R | 0.82 | 0.00 | 0.18 | 0.89 | 0.00 | 0.11 | 0.71 | 0.01 | 0.28 |
| Superior fronto-occipital fasciculus-L | 0.38 | 0.00 | 0.62 | 0.58 | 0.00 | 0.42 | 0.40 | 0.00 | 0.60 |
| Superior fronto-occipital fasciculus-R | 0.45 | 0.00 | 0.55 | 0.48 | 0.09 | 0.43 | 0.37 | 0.00 | 0.63 |
| Cingulum-L | 0.67 | 0.00 | 0.33 | 0.70 | 0.09 | 0.21 | 0.49 | 0.00 | 0.51 |
| Cingulum-R | 0.69 | 0.00 | 0.31 | 0.83 | 0.00 | 0.17 | 0.60 | 0.00 | 0.40 |
| Uncinate fasciculus-L | 0.62 | 0.00 | 0.38 | 0.50 | 0.00 | 0.50 | 0.64 | 0.00 | 0.36 |
| Uncinate fasciculus-R | 0.46 | 0.00 | 0.54 | 0.60 | 0.00 | 0.40 | 0.41 | 0.00 | 0.59 |
| Sagittal Stratum-L | 0.57 | 0.00 | 0.43 | 0.82 | 0.00 | 0.18 | 0.50 | 0.00 | 0.50 |
| Sagittal Stratum-R | 0.63 | 0.00 | 0.37 | 0.63 | 0.16 | 0.21 | 0.23 | 0.27 | 0.50 |
| Commissural fibers: | |||||||||
| Genu of corpus callosum | 0.75 | 0 | 0.25 | 0.88 | 0 | 0.12 | 0.47 | 0.18 | 0.35 |
| Body of corpus callosum | 0.64 | 0 | 0.36 | 0.74 | 0.07 | 0.19 | 0.56 | 0.05 | 0.39 |
| Splenium of corpus callosum | 0.59 | 0.06 | 0.35 | 0.81 | 0 | 0.19 | 0.58 | 0.19 | 0.23 |
| Tracts in the brainstem: | |||||||||
| Corticospinal tracts-L | 0.59 | 0.00 | 0.41 | 0.40 | 0.15 | 0.45 | 0.43 | 0.04 | 0.53 |
| Corticospinal tracts-R | 0.54 | 0.00 | 0.46 | 0.55 | 0.00 | 0.45 | 0.40 | 0.00 | 0.60 |
| Cerebral peduncle-L | 0.55 | 0.00 | 0.45 | 0.54 | 0.00 | 0.46 | 0.68 | 0.00 | 0.32 |
| Cerebral peduncle-R | 0.36 | 0.08 | 0.56 | 0.45 | 0.00 | 0.55 | 0.61 | 0.00 | 0.39 |
| Middle cerebellar peduncle | 0.72 | 0 | 0.28 | 0.55 | 0 | 0.45 | 0.73 | 0 | 0.27 |
| Global Measure: | 0.83 | 0 | 0.17 | 0.91 | 0 | 0.09 | 0.65 | 0.16 | 0.19 |
Table 4:
Twin ACE modeling of diffusion tensor imaging (DTI) measures of AD, RD and MD in specific Johns Hopkins University template white matter regions. N=438
| AD | RD | MD | |||||||
|---|---|---|---|---|---|---|---|---|---|
| a2 | c2 | e2 | a2 | c2 | e2 | a2 | c2 | e2 | |
| Projection fibers: | |||||||||
| Anterior corona radiata-L | 0.73 | 0.00 | 0.27 | 0.81 | 0.00 | 0.19 | 0.82 | 0.00 | 0.18 |
| Anterior corona radiata-R | 0.74 | 0.00 | 0.26 | 0.83 | 0.00 | 0.17 | 0.87 | 0.00 | 0.13 |
| Superior corona radiata-L | 0.67 | 0.00 | 0.33 | 0.80 | 0.00 | 0.20 | 0.82 | 0.00 | 0.18 |
| Superior corona radiata-R | 0.82 | 0.00 | 0.18 | 0.83 | 0.00 | 0.17 | 0.85 | 0.00 | 0.15 |
| Posterior corona radiata-L | 0.52 | 0.28 | 0.20 | 0.81 | 0.00 | 0.19 | 0.72 | 0.14 | 0.14 |
| Posterior corona radiata-R | 0.40 | 0.43 | 0.17 | 0.83 | 0.00 | 0.17 | 0.88 | 0.00 | 0.12 |
| Anterior limb of internal capsule-L | 0.64 | 0.01 | 0.35 | 0.71 | 0.00 | 0.29 | 0.62 | 0.00 | 0.38 |
| Anterior limb of internal capsule-R | 0.62 | 0.00 | 0.38 | 0.70 | 0.00 | 0.30 | 0.66 | 0.00 | 0.34 |
| Posterior limb of internal capsule-L | 0.71 | 0.00 | 0.29 | 0.40 | 0.28 | 0.32 | 0.24 | 0.38 | 0.38 |
| Posterior limb of internal capsule-R | 0.55 | 0.07 | 0.38 | 0.70 | 0.00 | 0.30 | 0.56 | 0.00 | 0.44 |
| Posterior thalamic radiation-L | 0.32 | 0.39 | 0.29 | 0.71 | 0.05 | 0.23 | 0.43 | 0.35 | 0.22 |
| Posterior thalamic radiation-R | 0.41 | 0.36 | 0.23 | 0.75 | 0.00 | 0.25 | 0.71 | 0.08 | 0.21 |
| Association fibers: | |||||||||
| Superior longitudinal fasciculus-L | 0.47 | 0.28 | 0.25 | 0.87 | 0.00 | 0.13 | 0.85 | 0.02 | 0.13 |
| Superior longitudinal fasciculus-R | 0.56 | 0.20 | 0.24 | 0.86 | 0.00 | 0.14 | 0.87 | 0.00 | 0.13 |
| Superior fronto-occipital fasciculus-L | 0.40 | 0.00 | 0.60 | 0.54 | 0.00 | 0.46 | 0.62 | 0.00 | 0.38 |
| Superior fronto-occipital fasciculus-R | 0.42 | 0.00 | 0.58 | 0.62 | 0.00 | 0.38 | 0.62 | 0.00 | 0.38 |
| Cingulum-L | 0.51 | 0.08 | 0.41 | 0.73 | 0.00 | 0.27 | 0.73 | 0.01 | 0.26 |
| Cingulum-L | 0.66 | 0.00 | 0.34 | 0.76 | 0.00 | 0.24 | 0.78 | 0.00 | 0.22 |
| Uncinate fasciculus-L | 0.48 | 0.00 | 0.52 | 0.48 | 0.00 | 0.52 | 0.11 | 0.21 | 0.68 |
| Uncinate fasciculus-R | 0.22 | 0.23 | 0.55 | 0.39 | 0.00 | 0.61 | 0.39 | 0.05 | 0.56 |
| Sagittal Stratum-L | 0.53 | 0.13 | 0.34 | 0.73 | 0.00 | 0.27 | 0.78 | 0.00 | 0.22 |
| Sagittal Stratum-R | 0.33 | 0.30 | 0.37 | 0.75 | 0.00 | 0.25 | 0.61 | 0.14 | 0.25 |
| Commissural fibers: | |||||||||
| Genu of corpus callosum | 0.71 | 0 | 0.29 | 0.81 | 0 | 0.19 | 0.82 | 0 | 0.18 |
| Body of corpus callosum | 0.60 | 0 | 0.40 | 0.72 | 0 | 0.28 | 0.74 | 0 | 0.26 |
| Splenium of corpus callosum | 0.45 | 0.26 | 0.29 | 0.67 | 0 | 0.33 | 0.68 | 0 | 0.32 |
| Tracts in the brainstem: | |||||||||
| Corticospinal tracts-L | 0.56 | 0.00 | 0.44 | 0.65 | 0.00 | 0.35 | 0.60 | 0.00 | 0.40 |
| Corticospinal tracts-R | 0.37 | 0.01 | 0.62 | 0.59 | 0.00 | 0.41 | 0.51 | 0.00 | 0.49 |
| Cerebral peduncle-L | 0.29 | 0.16 | 0.55 | 0.54 | 0.00 | 0.46 | 0.13 | 0.13 | 0.74 |
| Cerebral peduncle-R | 0.44 | 0.04 | 0.52 | 0.37 | 0.03 | 0.60 | 0.30 | 0.09 | 0.61 |
| Middle cerebellar peduncle | 0.60 | 0.00 | 0.40 | 0.71 | 0 | 0.29 | 0.63 | 0.00 | 0.37 |
| Global Measure: | 0.65 | 0.15 | 0.20 | 0.73 | 0.00 | 0.27 | 0.67 | 0.00 | 0.33 |
Note: Full ACE model fit is shown. Estimates of zero for c2 were empirically driven to their boundary at zero in these ACE models.
Fig. 5:
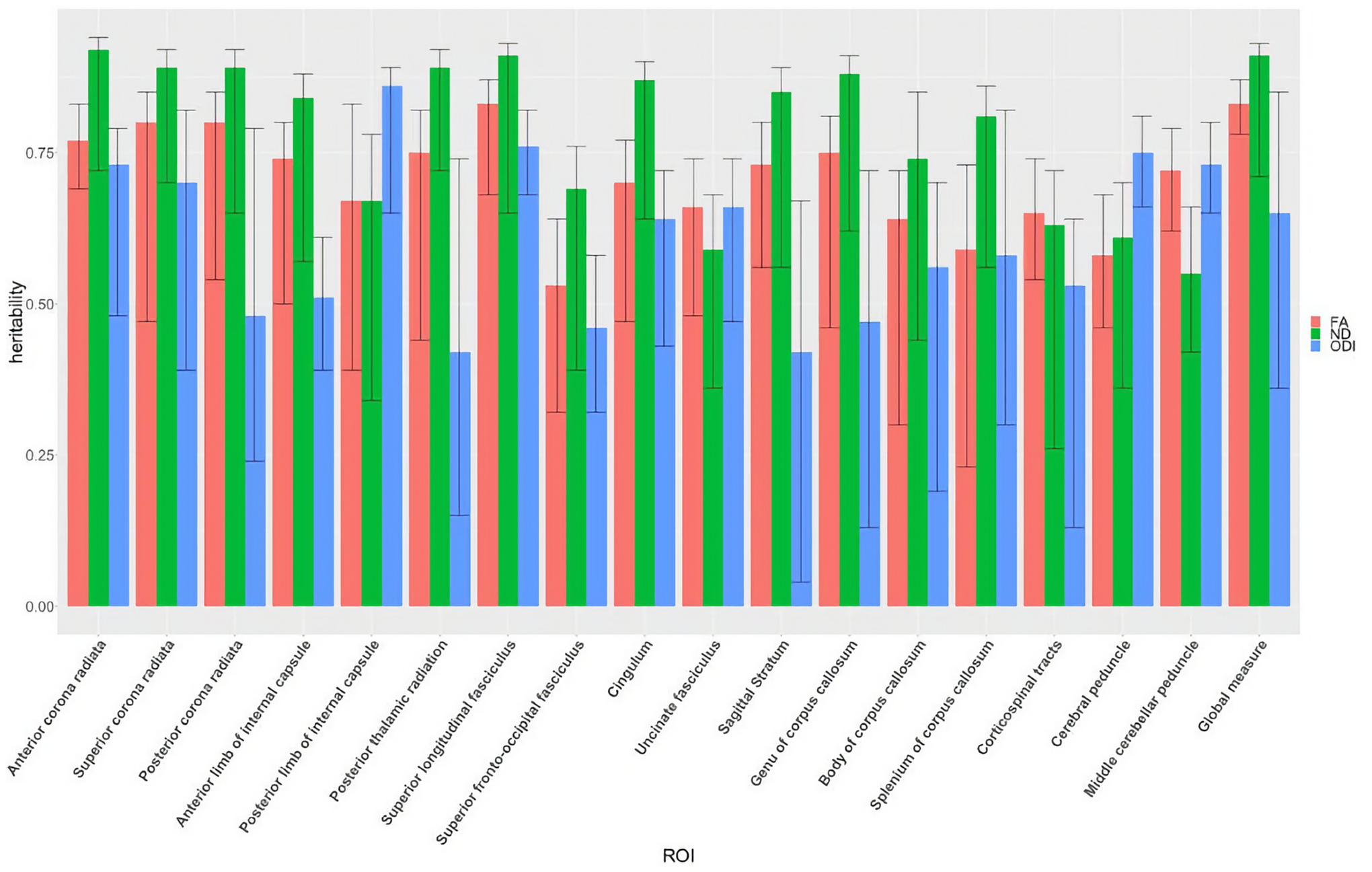
Comparative heritability estimates extracted from biometric ACE model fitting for fractional anisotropy (FA), neurite density (ND), and orientation dispersion index (ODI). Error bars provide 95% confidence intervals for each estimate
Global heritability estimates were 0.83 [CI 0.78, 0.87], 0.91 [CI 0.71, 0.93], and 0.65 [CI 0.36, 0.85] for FA, ND, and ODI, respectively (see Table 3). Additive genetic factors explained 83% of the variance in global FA, with individual tract heritability ranging from 0.53– 0.83. Major projection and association fibers had the highest levels of FA heritability compared with the other pathways. The superior longitudinal fasciculus, anterior limb of internal capsule, and anterior corona radiata had the highest FA heritability. The splenium of the corpus collosum, superior frontal-occipital fasciculus, and the cerebral peduncle had the lowest FA heritability. When compared with FA, heritability estimates of ND are higher in the majority of fiber tracts investigated, with the exception of tracts in the brain stem, such as the middle cerebellar peduncle and corticospinal tract, and in the uncinate fasciculus (Table 3, Figure 5). ODI estimates were generally lower than those for ND and FA, with the exception of posterior limb of internal capsule and cerebral peduncle (Table 3, Figure 5). Using the Wilcoxon signed rank test for the group of 30 ROIs, ND heritabilities are significantly higher than those for FA (z=2.85, p=0.04) and ODI (z=3.46, p=0.01).
Nearly 88% of the WM tracts showed heritability estimates for AD, RD, and MD of .40 or higher (ranging to .89), as shown in Table 4. Projection fibers had both the highest and lowest levels of AD and MD heritability, compared with the other pathways. Right anterior and superior corona radiata had the highest AD heritability of 0.74 and 0.82. The left posterior thalamic radiation, right corticospinal tracts, and left cerebral peduncle have the lowest heritabilities for AD (0.32, 0.37, and 0.29, respectively) and MD (0.43, 0.51, and 0.13, respectively).
Shared environmental contributions (c2) were generally small and close to 0 for FA, ND, and RD of all individual tracts. Removing shared environmental factors from the above-mentioned ACE models did not significantly reduce the model fit compared with the full model (see Supplement Table S1–6). On the other hand, AD, MD, and ODI measures of several tracts showed shared environmental contributions; these tracts included the posterior corona radiata, posterior thalamic radiation, and sagittal stratum (Tables 3 and 4).
Age as Moderator
Additional analyses accounted for the potential effect of age. At the phenotypic level, no significant linear relationship between age at the time of scan and DTI or NODDI measures emerged (p>0.05). Due to the non-normal distribution of age in our sample, with age clustering around 17 and 22, we subdivided our twins into an adolescent group (13–20) and an early adult group (21–24) (see Supplement Figure S1 for age distribution). Biometric modeling showed comparable heritability and variance components between groups, and thus age groups were pooled in the final analysis.
Discussion
This study first replicated previous findings that genetic factors explain a substantial portion of individual differences in FA (Brouwer et al., 2010; Chiang et al., 2011; Jahanshad et al., 2013; Kochunov et al., 2015; Lee et al., 2015). Further, our study suggests that NODDI ND and ODI measures are both highly heritable (with global heritability of 0.91 [CI 0.71, 0.93] and 0.65 [CI 0.36, 0.85], respectively). To our knowledge, this is the first report that systematically examines whole brain NODDI measures of WM in a large twin sample and establishes NODDI as a promising phenotype for future imaging genetics research.
Our results show higher heritability for ND than FA and ODI in 25 out of 30 ROI measures, suggesting ND is more sensitive to underlying genetic sources of variation. The heritability of ODI, which is a measure of spatial coherence, is lower than that of FA and ND, with the exception of the cerebral peduncle, middle cerebellar peduncle, and posterior limb of internal capsule (Table 3, Figure 5). In a previous NODDI heritability study using an Amish family pedigree, Kochunov et al. (2016) found ND heritability of 0.70 and ODI heritability of 0.42 in the corpus collosum (CC), values that align reasonably well with our CC heritability estimates. Our study applied white matter ROI analysis and found heritability estimates of ODI ranging from 0.16 to 0.86 in all of the WM tracts investigated. Using the Wilcoxon signed rank test, we showed the ND heritabilities, which ranged from 0.45–0.90, are higher than those of FA and ODI. Our finding indicates a strong genetic basis for both NODDI parameters, suggesting genetic factors contribute to the dispersion of axons as well as their density.
Our heritability estimates of traditional DTI measures (Tables 3 & 4) are also consistent with the range of estimates in other independent cohorts and consortium analyses (Jahanshad et al., 2013; Vuoksimaa et al., 2017). Our global AD heritability of 0.65 is lowest among all the metrics we investigated. This finding is consistent with results from recent twin samples composed of infants and middle-aged adults (Brouwer et al., 2010; Lee et al., 2015; Vuoksimaa et al., 2017). One study with a mixed family design also showed levels of AD heritability similar to ours (Kochunov et al., 2010). RD and MD heritability estimates are higher than AD estimates, with global heritability of 0.73 [CI 0.51, 0.92] and 0.67 [CI 0.37, 0.85], respectively. The similar RD and MD heritabilities agree with recent twin studies where complete DTI metrics—not just FA—are reported (Gustavson et al., 2019; Vuoksimaa et al., 2017). Thus, our results support and expand trends in the literature concerning genetic effects within the DTI framework.
When examining voxel-level heritability maps, we note that the patterns across FA and ODI are similar, whereas ND heritability appears more homogenous across the brain (Figure 4). In our investigations of specific WM regions, we found projection fiber regions to generally have the highest heritability in ND and DTI measures, followed by commissural fiber regions, association fiber regions, and lastly regions of the brain stem (see Tables 3 and 4). Among the association fiber regions that connect different brain regions of the same hemisphere, the superior longitudinal fasciculus has the highest heritability, compared with more inferior regions such as uncinate fasciculus or sagittal stratum. This trend among association fibers is also observed in other studies (Hatton et al., 2018; Shen et al., 2014). The lower heritability estimates in midbrain regions (Tables 3 and 4), particularly for small regions such as the cortical spinal tract (CST), may be more influenced by errors in registration with the region template that contribute to the variation in the measurements. Alternatively, measures from the CST may be more influenced by environmental factors.
In our study, homologous left and right hemisphere WM tracts were examined separately. Although we did not statistically test for differences in heritability, the heritability estimates for left and right hemispheric ROIs were comparable, with more than 80% of regions showing left and right hemisphere heritability differences smaller than 10%. Regions indicating high asymmetry in heritability include several association fibers such as uncinate fasciculus, cingulum, and sagittal stratum (Tables 3 and 4), which is also shown in other studies (Geng et al., 2012; Lee et al., 2015; Vuoksimaa et al., 2017). However, the consistency and the effect size of the left versus right differences in heritability should be investigated in future studies.
The absence of shared environmental influence in the majority of tracts that we investigated is consistent with the twin DTI literature (Gustavson et al., 2019; Vuoksimaa et al., 2017) and other structural brain measurements (Giedd et al., 2007; Jansen et al., 2015). However, some regions with non-zero shared environmental contribution were observed, especially among ODI indices. For instance, the posterior thalamic radiation showed environmental contribution in AD, ODI, and MD (see Tables 3 and 4). Sources of shared environments may first arise during the prenatal period when neuronal migration, neurulation, and myelination are occurring. Subsequent stressors and other experiential factors may similarly affect MZ and DZ twins, thus contributing to shared environment estimates. Unique environmental influences (i.e., unique to one member within a pair) account for the remaining variance.
Although age-related changes in brain structure have been observed in WM microstructure (Lebel & Beaulieu, 2011), most imaging genetics studies are limited to cross-sectional study designs (Chiang et al., 2011), with the exception of infant twin studies (Lee et al., 2018). In a recent longitudinal study that investigated the age-related development of NODDI measures among singletons from infancy through early childhood, Lynch et al. (2020) found that age is positively correlated with ND, but that age showed little or no correlation with ODI (Lynch et al., 2020). Longitudinal twin imaging studies spanning adolescence and adulthood may better illuminate age-related changes in genetic and environmental influences than cross-sectional dichotomization of younger and older age groups. The limited age range in the current study may partly explain why we did not observe significant age-related differences in heritability (Supplemental Figure 1).
Despite histological validation studies (Sepehrband et al., 2015), we should be wary of overly strong biological interpretation of NODDI measures. While restricted intra-axonal diffusion is likely to be a major factor in the neurite signal compartment, the model is still vastly over-simplified, given the complex microstructure of white matter. In addition, recent studies highlight some potential issues in the NODDI model assumptions (Lampinen et al., 2017) and default diffusivity values (Guerrero et al., 2019), which may reduce model performance.
With such limitations recognized, recent work aims to improve NODDI’s formalism and its performance (Tariq et al., 2016). Moreover, the improved tissue specificity from the existing NODDI model, when combined with traditional DTI measures, has proved beneficial in studying typical brain development (Beck et al., 2021; Lynch et al., 2020) and neurological disorders such as metabolic diseases and multiple sclerosis (Grussu et al., 2017; Timmers et al., 2016). Confidence in our results is strengthened when we note that the phenotypic correlations between DTI and NODDI measures also align with published studies, where FA is negatively correlated ODI and positively correlated with ND (Table 2, Figure S2) (Zhang et al., 2012).
The differences in the model frameworks of Gaussian diffusion for the DTI model and multi-compartmental modeling of NODDI in our study provide added evidence of the genetic basis of white matter microstructure. Our findings contribute to the current imaging genetics literature by providing evidence that within the limited domain of the NODDI framework, (1) additive genetic factors contribute to individual differences of both ND and ODI in white matter; and (2) significant heritabilities are also observed in brain regions with more complex fiber organization, such as the corona radiata.
Other DWI reconstruction techniques also help elucidate complex underlying microstructural features. In a twin study using high angular resolution imaging (HARDI), Shen et al. (2014) found that the WM fiber modeled by the fiber orientation distribution (FOD) function is more heritable than FA. The fiber orientation distribution function may be a more accurate representation of axonal fiber organization and possibly more sensitive to underlying genetic factors in areas of large populations of crossing fibers. Similarly, our adoption and validation of NODDI techniques within the classic twin modeling research framework should augment the current literature on the heritability of DTI indices.
Our analyses focused on standardized ROI measurements that may not capture heritability changes along the full length of WM pathways. For instance, Kochunov et al. (2015) showed that regions closer to the thalamus (the center of the brain in MNI space) had higher FA heritability compared with distal regions. Our qualitative observation that Falconer’s heritability in FA decreases at the edge of WM and cortical GM (Figure 4) likely supports this prior finding. Moreover, preliminary voxel-based Falconer’s heritability estimates in our study largely corroborate our ROI-based findings. Future studies will perform ACE modeling at the voxel level and investigate any regional changes in heritability. Additionally, twin modeling of NODDI measures can be extended to the whole brain to study the genetic basis of cortical and subcortical grey matter microstructure (Elman et al., 2017). For instance, orientation dispersion should capture the fanning and complexity of neurite fiber orientation (Fukutomi et al., 2018), which are important characteristics for brain development and neurodegenerative disorders.
In conclusion, by incorporating NODDI’s estimate of neurite density (ND) and orientation dispersion (ODI) index measures in a twin sample, we estimated the magnitude of genetic influences on variation in WM microstructure with a multi-compartment model of water diffusion, which previously has not been explored. The high and significant degree of heritability seen in many brain WM tracts suggests that genetic factors play an important role in the development of neurite density and dispersion measures estimated via water diffusion within white matter and highlights NODDI’s potential in the future of imaging genetics research.
Supplementary Material
ACKNOWLEDGEMENTS:
We dedicate this manuscript in memory of our friend and colleague, first author Zhan “Ross” Luo, who contributed the vast majority of this research and manuscript preparation but unexpectedly passed away during the review of the manuscript. Publication of this manuscript is supported by Ross’s next of kin. We thank Michael J. Anderle, Scott Mikkelson, Ronald Fisher, and Ian Carroll for assistance with data collection; Jaeyoon Choi, Amer Marzuki, Carol Van Hulle, and Jeanette Mumford for assistance with analyses; and, Steven Kecskemeti, Nicole Schmidt, and Elizabeth M. Planalp for technical and administrative support.
Funding:
Our research was supported by NIH grants R01 MH101504, P50 MH100031; and grants to the Waisman Center (P30 HD003352 and U54 HD090256-01). Z.L. is partly supported by National Research Service Award (NRSA) GM007507. N.A. is partially supported by the BRAIN Initiative R01-EB022883, the Center for Predictive and Computational Phenotyping (CPCP) U54 AI117924, the Alzheimer’s Disease Connectome Project (ADCP) UF1 AG051216 and R01 AG037639 grants. D.D. is partially supported by a Pathways to Independence Award, R00 MH110596.
Footnotes
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher’s embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. See here for Springer Nature’s terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
Conflicts of Interest: None of the authors have any conflicts of interest to declare.
Code availability: Genetic analyses with structural equation modeling were conducted with the publicly available R package umx (Bates et al., 2019)
Ethics Approval: Institutional Review Board (IRB) at the University of Wisconsin–Madison approved all study protocols.
Consent to Participate: Both twins and their legal guardian provided informed assent and consent, respectively before research participation.
Availability of Data and Material:
Multi-model neuroimaging data, including diffusion weighted imaging, are publicly available via the National Data Archive (Validating RDoC for Children and Adolescents: A Twin Study with Neuroimaging, collection #2105).
References
- Alexander AL, Lee JE, Lazar M, Field AS, 2007. Diffusion tensor imaging of the brain. Neurotherapeutics 4, 316–29. 10.1016/j.nurt.2007.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JLR, Sotiropoulos SN, 2016. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage 125, 1063–1078. 10.1016/j.neuroimage.2015.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y, Basser PJ, 2005. Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. NeuroImage 27, 48–58. 10.1016/j.neuroimage.2005.03.042 [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12, 26–41. 10.1016/j.media.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TC, Maes H, Neale MC, 2019. umx: Twin and Path-Based Structural Equation Modeling in R. Twin Res Hum Genet 22, 27–41. 10.1017/thg.2019.2 [DOI] [PubMed] [Google Scholar]
- Beck D, de Lange A-MG, Maximov II, Richard G, Andreassen OA, Nordvik JE, Westlye LT, 2021. White matter microstructure across the adult lifespan: A mixed longitudinal and cross-sectional study using advanced diffusion models and brain-age prediction. NeuroImage 224, 117441. 10.1016/j.neuroimage.2020.117441 [DOI] [PubMed] [Google Scholar]
- Bigos KL, Hariri AR, Weinberger DR (Eds.), 2016. Neuroimaging genetics: principles and practices. Oxford University Press, Oxford ; New York. [Google Scholar]
- Brouwer RM, Mandl RCW, Peper JS, van Baal GCM, Kahn RS, Boomsma DI, Hulshoff Pol HE, 2010. Heritability of DTI and MTR in nine-year-old children. NeuroImage 53, 1085–1092. 10.1016/j.neuroimage.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Barysheva M, Shattuck DW, Lee AD, Madsen SK, Avedissian C, Klunder AD, Toga AW, McMahon KL, De Zubicaray GI, Wright MJ, Srivastava A, Balov N, Thompson PM, 2009. Genetics of Brain Fiber Architecture and Intellectual Performance. J. Neurosci 29, 2212–2224. 10.1523/JNEUROSCI.4184-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M-C, McMahon KL, de Zubicaray GI, Martin NG, Hickie I, Toga AW, Wright MJ, Thompson PM, 2011. Genetics of white matter development: a DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage 54, 2308–2317. 10.1016/j.neuroimage.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan N, Siow B, O’Callaghan JM, Harrison IF, Wells JA, Holmes HE, Ismail O, Richardson S, Alexander DC, Collins EC, Fisher EM, Johnson R, Schwarz AJ, Ahmed Z, O’Neill MJ, Murray TK, Zhang H, Lythgoe MF, 2016. Application of neurite orientation dispersion and density imaging (NODDI) to a tau pathology model of Alzheimer’s disease. Neuroimage 125, 739–744. 10.1016/j.neuroimage.2015.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack R, Brett M, Osswald K, 2003. An Evaluation of the Use of Magnetic Field Maps to Undistort Echo-Planar Images. NeuroImage 18, 127–142. 10.1006/nimg.2002.1281 [DOI] [PubMed] [Google Scholar]
- Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran J-P, 2015. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage 105, 32–44. 10.1016/j.neuroimage.2014.10.026 [DOI] [PubMed] [Google Scholar]
- Elman JA, Panizzon MS, Hagler DJ, Fennema-Notestine C, Eyler LT, Gillespie NA, Neale MC, Lyons MJ, Franz CE, McEvoy LK, Dale AM, Kremen WS, 2017. Genetic and environmental influences on cortical mean diffusivity. Neuroimage 146, 90–99. 10.1016/j.neuroimage.2016.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutomi H, Glasser MF, Zhang H, Autio JA, Coalson TS, Okada T, Togashi K, Van Essen DC, Hayashi T, 2018. Neurite imaging reveals microstructural variations in human cerebral cortical gray matter. Neuroimage 182, 488–499. 10.1016/j.neuroimage.2018.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Prom-Wormley EC, Perez J, Kubarych T, Styner M, Lin W, Neale MC, Gilmore JH, 2012. White matter heritability using diffusion tensor imaging in neonatal brains. Twin Res Hum Genet 15, 336–350. 10.1017/thg.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Schmitt JE, Neale MC, 2007. Structural brain magnetic resonance imaging of pediatric twins. Hum Brain Mapp 28, 474–481. 10.1002/hbm.20403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grussu F, Schneider T, Tur C, Yates RL, Tachrount M, Ianuş A, Yiannakas MC, Newcombe J, Zhang H, Alexander DC, DeLuca GC, Gandini Wheeler-Kingshott CAM, 2017. Neurite dispersion: a new marker of multiple sclerosis spinal cord pathology? Ann Clin Transl Neurol 4, 663–679. 10.1002/acn3.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero JM, Adluru N, Bendlin BB, Goldsmith HH, Schaefer SM, Davidson RJ, Kecskemeti SR, Zhang H, Alexander AL, 2019. Optimizing the intrinsic parallel diffusivity in NODDI: An extensive empirical evaluation. PLoS ONE 14, e0217118. 10.1371/journal.pone.0217118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavson DE, Hatton SN, Elman JA, Panizzon MS, Franz CE, Hagler DJ, Fennema-Notestine C, Eyler LT, McEvoy LK, Neale MC, Gillespie N, Dale AM, Lyons MJ, Kremen WS, 2019. Predominantly global genetic influences on individual white matter tract microstructure. Neuroimage 184, 871–880. 10.1016/j.neuroimage.2018.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton SN, Panizzon MS, Vuoksimaa E, Hagler DJ, Fennema-Notestine C, Rinker D, Eyler LT, Franz CE, Lyons MJ, Neale MC, Tsuang MT, Dale AM, Kremen WS, 2018. Genetic relatedness of axial and radial diffusivity indices of cerebral white matter microstructure in late middle age. Hum Brain Mapp 39, 2235–2245. 10.1002/hbm.24002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov PV, Sprooten E, Mandl RC, Nichols TE, Almasy L, Blangero J, Brouwer RM, Curran JE, de Zubicaray GI, Duggirala R, Fox PT, Hong LE, Landman BA, Martin NG, McMahon KL, Medland SE, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Hulshoff Pol HE, Bastin ME, McIntosh AM, Deary IJ, Thompson PM, Glahn DC, 2013. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. Neuroimage 81, 455–469. 10.1016/j.neuroimage.2013.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AG, Mous SE, White T, Posthuma D, Polderman TJC, 2015. What Twin Studies Tell Us About the Heritability of Brain Development, Morphology, and Function: A Review. Neuropsychology Review 25, 27–46. 10.1007/s11065-015-9278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JH, Helpern JA, 2010. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed 23, 698–710. 10.1002/nbm.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezzard P, Balaban RS, 1995. Correction for geometric distortion in echo planar images from B0 field variations. Magnetic Resonance in Medicine 34, 65–73. 10.1002/mrm.1910340111 [DOI] [PubMed] [Google Scholar]
- Jones DK, Basser PJ, 2004. “Squashing peanuts and smashing pumpkins”: how noise distorts diffusion-weighted MR data. Magn Reson Med 52, 979–993. 10.1002/mrm.20283 [DOI] [PubMed] [Google Scholar]
- Kochunov P, Fu M, Nugent K, Wright SN, Du X, Muellerklein F, Morrissey M, Eskandar G, Shukla DK, Jahanshad N, Thompson PM, Patel B, Postolache TT, Strauss KA, Shuldiner AR, Mitchell BD, Hong LE, 2016. Heritability of complex white matter diffusion traits assessed in a population isolate. Hum Brain Mapp 37, 525–535. 10.1002/hbm.23047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Lancaster JL, Winkler AM, Smith S, Thompson PM, Almasy L, Duggirala R, Fox PT, Blangero J, 2010. Genetics of microstructure of cerebral white matter using diffusion tensor imaging. Neuroimage 53, 1109–1116. 10.1016/j.neuroimage.2010.01.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Marcus D, Winkler A, Sprooten E, Nichols TE, Wright SN, Hong LE, Patel B, Behrens T, Jbabdi S, Andersson J, Lenglet C, Yacoub E, Moeller S, Auerbach E, Ugurbil K, Sotiropoulos SN, Brouwer RM, Landman B, Lemaitre H, den Braber A, Zwiers MP, Ritchie S, van Hulzen K, Almasy L, Curran J, deZubicaray GI, Duggirala R, Fox P, Martin NG, McMahon KL, Mitchell B, Olvera RL, Peterson C, Starr J, Sussmann J, Wardlaw J, Wright M, Boomsma DI, Kahn R, de Geus EJC, Williamson DE, Hariri A, van ‘t Ent D, Bastin ME, McIntosh A, Deary IJ, Hulshoff pol HE, Blangero J, Thompson PM, Glahn DC, Van Essen DC, 2015. Heritability of fractional anisotropy in human white matter: A comparison of Human Connectome Project and ENIGMA-DTI data. NeuroImage 111, 300–311. 10.1016/j.neuroimage.2015.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampinen B, Szczepankiewicz F, Mårtensson J, van Westen D, Sundgren PC, Nilsson M, 2017. Neurite density imaging versus imaging of microscopic anisotropy in diffusion MRI: A model comparison using spherical tensor encoding. Neuroimage 147, 517–531. 10.1016/j.neuroimage.2016.11.053 [DOI] [PubMed] [Google Scholar]
- Lebel C, Beaulieu C, 2011. Longitudinal development of human brain wiring continues from childhood into adulthood. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 10937–47. 10.1523/JNEUROSCI.5302-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Steiner RJ, Luo S, Neale MC, Styner M, Zhu H, Gilmore JH, 2015. Quantitative tract-based white matter heritability in twin neonates. Neuroimage 111, 123–135. 10.1016/j.neuroimage.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Zhang J, Neale MC, Styner M, Zhu H, Gilmore JH, 2018. Quantitative tract-based white matter heritability in 1- and 2-year-old twins. Hum Brain Mapp. 10.1002/hbm.24436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KM, Cabeen RP, Toga AW, Clark KA, 2020. Magnitude and timing of major white matter tract maturation from infancy through adolescence with NODDI. Neuroimage 212, 116672. 10.1016/j.neuroimage.2020.116672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastropietro A, Rizzo G, Fontana L, Figini M, Bernardini B, Straffi L, Marcheselli S, Ghirmai S, Nuzzi NP, Malosio ML, Grimaldi M, 2019. Microstructural characterization of corticospinal tract in subacute and chronic stroke patients with distal lesions by means of advanced diffusion MRI. Neuroradiology 61, 1033–1045. 10.1007/s00234-019-02249-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J, 2008. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage 40, 570–582. 10.1016/j.neuroimage.2007.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M, Cardon L, 1992. Methodology for Genetic Studies of Twins and Families, Nato Science Series D: Springer; Netherlands. 10.1007/978-94-015-8018-2 [DOI] [Google Scholar]
- Reeder SB, Pineda AR, Wen Z, Shimakawa A, Yu H, Brittain JH, Gold GE, Beaulieu CH, Pelc NJ, 2005. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med 54, 636–644. 10.1002/mrm.20624 [DOI] [PubMed] [Google Scholar]
- Schmidt NL, Brooker RJ, Carroll IC, Gagne JR, Luo Z, Moore MN, Planalp EM, Sarkisian KL, Schmidt CK, Van Hulle CA, Lemery-Chalfant K, Goldsmith HH, 2019. Longitudinal Research at the Interface of Affective Neuroscience, Developmental Psychopathology, Health and Behavioral Genetics: Findings from the Wisconsin Twin Project. Twin Research and Human Genetics 22, 233–239. 10.1017/thg.2019.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NL, Van Hulle CA, Brooker RJ, Meyer LR, Lemery-Chalfant K, Goldsmith HH, 2013. Wisconsin Twin Research: Early Development, Childhood Psychopathology, Autism, and Sensory Over-responsivity. Twin Research and Human Genetics 16, 376–384. 10.1017/thg.2012.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepehrband F, Clark KA, Ullmann JFP, Kurniawan ND, Leanage G, Reutens DC, Yang Z, 2015. Brain tissue compartment density estimated using diffusion-weighted MRI yields tissue parameters consistent with histology. Hum Brain Mapp 36, 3687–3702. 10.1002/hbm.22872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K-K, Rose S, Fripp J, McMahon KL, de Zubicaray GI, Martin NG, Thompson PM, Wright MJ, Salvado O, 2014. Investigating brain connectivity heritability in a twin study using diffusion imaging data. NeuroImage 100, 628–641. 10.1016/j.neuroimage.2014.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Schneider T, Alexander DC, Gandini Wheeler-Kingshott CA, Zhang H, 2016. Bingham–NODDI: Mapping anisotropic orientation dispersion of neurites using diffusion MRI. NeuroImage 133, 207–223. 10.1016/j.neuroimage.2016.01.046 [DOI] [PubMed] [Google Scholar]
- Timmers I, Roebroeck A, Bastiani M, Jansma B, Rubio-Gozalbo E, Zhang H, 2016. Assessing Microstructural Substrates of White Matter Abnormalities: A Comparative Study Using DTI and NODDI. PLoS ONE 11, e0167884. 10.1371/journal.pone.0167884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuoksimaa E, Panizzon MS, Hagler DJ, Hatton SN, Fennema-Notestine C, Rinker D, Eyler LT, Franz CE, Lyons MJ, Neale MC, Tsuang MT, Dale AM, Kremen WS, 2017. Heritability of white matter microstructure in late middle age: A twin study of tract-based fractional anisotropy and absolute diffusivity indices. Hum Brain Mapp 38, 2026–2036. 10.1002/hbm.23502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC, 2012. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 61, 1000–1016. 10.1016/j.neuroimage.2012.03.072 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Multi-model neuroimaging data, including diffusion weighted imaging, are publicly available via the National Data Archive (Validating RDoC for Children and Adolescents: A Twin Study with Neuroimaging, collection #2105).


