Abstract
Recapitulation of complex tissues signifies a remarkable challenge and, to date, only few approaches have emerged that can efficiently reconstruct necessary gradients in three-dimensional (3D) constructs. This is true even though mimicry of these gradients is of great importance to establish the functionality of engineered tissues and devices. Here, a composable-gradient digital light processing (DLP)-based (bio)printing system is developed for the first time, utilizing the unprecedented integration of a microfluidic mixer for the generation of either continual or discrete gradients of desired (bio)inks in real time. Notably, the precisely controlled gradients are composable on-the-fly by facilely adjusting the (bio)ink flow ratios. In addition, this setup is designed in such a way that no washing steps are needed when exchanging the gradient (bio)inks, further enhancing our time- and (bio)ink-saving strategy. Various planar and 3D structures exhibiting continual gradients of materials, of cell densities, of growth factor concentrations, of hydrogel stiffness, and of porosities in horizontal and/or vertical direction, are exemplified. The composable fabrication of multifunctional gradients strongly support the potential of our unique bioprinting system in numerous biomedical applications.
Keywords: gradient, digital light processing, vat polymerization, microfluidic mixer, bioprinting
Gradient structures in biological tissues play a pivotal role in organ development, physiology, and disease conditions, which are widely existing from the polarization of the neural tube to the architecture of the osteochondral interface.[1] Among a set of gradient classes, cellular gradients are the transitions of cell densities and cell types. The differences of cell densities and/or cell types influence cell-cell interactions, being relevant to the functions of related tissues.[2] In addition, the chemical gradients in extracellular matrix, such as cytokines, serve as microenvironmental cues for cell proliferation, migration, and differentiation, among other behaviors.[3] For example, mechanical gradients, involving the variation of compressive, tensile, or shear features, are crucial factors to influence cell functions and tissue interfaces such as the tendon entheses.[4] Porosity gradient in the radial direction is found in the bone structure, where the averaged porosity is increased from the cortical bone towards the trabecular bone.[5] Given the importance of various gradients in natural tissues, recapitulating continual gradient properties in the engineered graft tissues cannot be overlooked in either tissue regeneration or organ modeling applications.
Early studies on the fabrication of gradient structures such as sequential layering, convective stretching, and magnetic field manipulation, show efficacy to formulate gradients of different types.[3, 6, 7] However, the gradient controllability, in particular that in volumetric configuration, is generally insufficient and limited by the requirements of specialized, oftentimes manual or semi-manual procedures. The emergence of three-dimensional (3D) (bio)printing that incorporates (bio)inks, takes advantages of the controllable patterning to produce various gradients within complex tissue-engineered structures with biomimicry.[3] For instance, inkjet printing, depositing single droplets to build up a structure, has been applied to generate gradients of biologically active macromolecules or cells on substrate surfaces, as well as to achieve the graded components of hydrogels and polymers.[8, 9] Another printing technology, extrusion printing, successfully builds gradient constructs through altering the inks or the assistance of microfluidic printheads.[10, 11] However, both of these technologies hold intriguing features with inevitable bottlenecks as well. The inkjet approach displays the tradeoffs between printing resolution and material type-limitation, while extrusion printing is confined by the slow printing speed, and the insufficient complexity that can be achieved.[12, 13]
The stereolithography (SLA)-/digital light processing (DLP)-based 3D printing rises as a potential solution to obtain good printing resolutions and intricate architectures with a satisfactorily wide selection of inks.[14–16] In SLA, photopolymerization is performed by the focused laser beam scanning on the surface of the liquid ink, resulting in a high spatial resolution.[17] DLP uses a digital micromirror device (DMD) or a liquid crystal display, where the designed patterns are projected onto the ink reservoir for the ink photopolymerization leading to the formation of sophisticated structures.[18] Compared to extrusion printing, a higher x-y resolution of 25–50 μm can be achieved by the DLP approach, which is determined by parameters such as the pixel size of the DMD projecting the light patterns and the magnification of the projecting optics.[19] Meanwhile, DLP-based 3D printing generally exhibits faster printing speeds than the classical extrusion printing, especially for volumetric structures, because the fabrication relies on the layer-by-layer method instead of the linear filament deposition.[17, 20] The printing speed of the DLP-based method is primarily dependent on the layer thickness and influenced by the photocrosslinking efficiency of the ink, which can be on the scale of mm3 s-1.[21]
Furthermore, in junction with SLA-/DLP-based 3D printing, several studies investigated to incorporate transitions or multi-materials in desired sections of the 3D-printed constructs. For example, a 3D multi-material structure was patterned using the SLA approach, where bioinks encapsulating multiple cells were bioprinted by exchanging the bioinks within the vat.[22] Additionally, in our previous study, a multi-material bioprinting platform, which combined the DLP bioprinting system with a microfluidic device to achieve the convenient exchange of multiple bioinks, was reported to fabricate the 3D multi-material constructs at high speeds.[23] Indeed, this bioprinting platform enabled a fast switching between multiple materials, yet the multiple materials were only discretely distributed within the 3D-bioprinted constructs. As a result, there was limited capacity for composing these materials or achieving continual gradients in real time during bioprinting.
There is no denying that microfluidic devices have enabled the gradient formulation of continually multi-material patterns.[11] However, limitations of this method have been identified in effectively mixing multiple materials suggesting that there is room for improvement. An important question with regard to the fluid flow-mixing lies in the slow diffusion of laminar and uniaxial flows that requires long path lengths to ensure complete mixing.[24] The chaotic mixer has been proven promising for the rapid mixing of fluids by the geometric complexity within a microchannel, such as the slanted rib and barrier.[25, 26] By utilizing the inks mixed with the microfluidic chaotic mixer built into the DLP 3D bioprinting, we hypothesized that sophisticated, composable gradient structures could be fabricated in a single bioprinting session on-the-fly in a precisely controllable way and at desirable resolutions.
Here we introduce, for the very first time, a gradient DLP 3D (bio)printing system combining a DMD-based (bio)printer and a microfluidic chaotic mixer-linked vat to build functionally graded scaffolds and tissue constructs. After mixing a small volume of (bio)ink with the chaotic flow, sophisticated structures featuring real-time composable gradients, either continual or discrete, are fabricated in a precisely predictable manner. The diverse proof-of-concept biological applications of this bioprinting platform are demonstrated by generating cell gradients, chemical gradients, mechanical property gradients, porosity gradients, as well as dual gradients using poly(ethylene glycol) diacrylate (PEGDA) or gelatin methacryloyl (GelMA)-based (bio)inks.
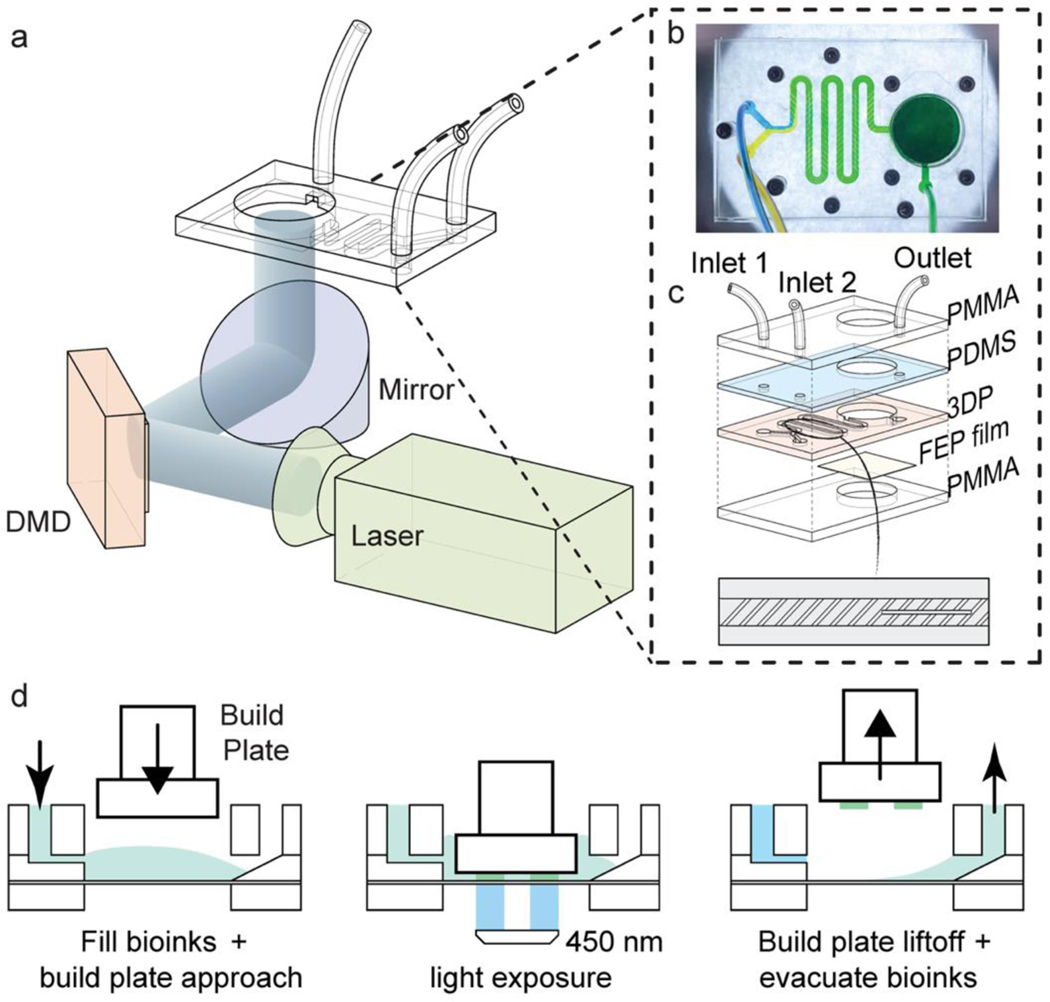
The (bio)printing system consisted of a microfluidic mixer chip to generate homogenous graded (bio)inks and a DLP-based 3D (bio)printer (Figure 1a). The DMD device with the display resolution of 1920 × 1080 pixels was applied, delivering a 38-μm resolution in the x-y plane on the build plate. To obtain the gradient (bio)inks in a controllable and predictable manner, we designed and fabricated a microfluidic mixer chip based on the chaotic mixing, as shown in Figures 1b, 1c, and S1. The microfluidic mixer chip was composited of multiple inlets, a microchannel, and a (bio)ink vat. In general, the fabrication and assembly of the microfluidic chip were extremely convenient and allowed for rapid modification of the inlet numbers as well as the (bio)ink vat size. The maximum size of the (bio)ink vat, also indicated the size of the projecting area at the vat, relied on the projecting capacity of the DMD device and the magnification of the optical system. In our setup, after the 5 times of expansion of the optical system, the total size of projected area at the (bio)ink vat was enlarged to 72.96 × 41.04 mm2, which was used to design our vat dimensions. Nevertheless, the vat size can be easily reduced or enlarged according to different printer setups.[27]
Figure 1.
The setup of the composable-gradient DLP (bio)printing platform and the (bio)printing workflow. (a) Schematic of the gradient DLP (bio)printer, combining the DLP (bio)printing system and the microfluidic mixing chip. (b) Photograph of the microfluidic mixing chip showing the blue and yellow colors mixed into the green, which is composed of a chaotic mixing microchannel and a (bio)ink vat. (c) Illustration showing the assembly of the microfluidic mixing chip bearing two inlets and one outlet. The bottom design presents the chaotic mixing microchannel containing slanted ribs and barriers. (d) The gradient DLP (bio)printing process for the continuous filling of the mixed (bio)ink, photocrosslinking, and evacuating the extra (bio)ink post-(bio)printing with each (bio)ink gradient.
The microchannel segment, as the key element in our gradient (bio)printing strategy, was designed by modifying the barrier-embedded chaotic micromixer reported by others.[25, 28] Relying on the theoretical, experimental, and numerical analyses, the mixing performance of the barrier-embedded chaotic micromixer can be optimized by controlling several geometric parameters, namely the rib angle, the ratio of rib width to rib height, the ratio of rib height to microchannel height, and the ratio of rib spacing to rib height. Among these, the ratio of rib height to microchannel height and the rib angle have been found to play dominant roles in the mixing outcomes.[25] The larger height ratio of rib to microchannel is related to the increased helical flow and the better mixing, resulting in the reduced microchannel length required for the complete mixing. Additionally, it has been demonstrated that the helical flow is maximized when the rib angle of 35.6° is applied,[29] which was also used in our study. In detail, our chaotic mixer was 350 μm in microchannel height and 1.5-mm-wide, with the slanted ribs (35.6°) being 100-μm-high and 250 μm in width, leaving a space of 250 μm between each other. The zigzag shape of the entire mixer was used to save space with a functional length of 99 mm, fitting 5 mixing cycles (a mixing cycle is composed of a sequential region of slanted ribs), and induced the helical type of flow. By inserting barriers parallel to the flow direction, the mixing was changed from the elliptic pattern to the hyperbolic pattern.[25] This type of mixer can work well with the fluids at the Reynolds number ranging from 1 to 100,[28] indicating the wide application of the chaotic mixer on the (bio)ink mixing for 3D (bio)printing.
The process of gradient (bio)printing is illustrated in Figure 1d. Once the mixed (bio)ink entered and filled the vat, the build platform was approached at the (bio)printing level. By projecting the desired patterns with the 450-nm blue light (0.5 mW cm−2), an optimal (bio)printing procedure was performed with the layer height of 300 or 200 μm and printed in 60 or 30 s for each layer, respectively. Tris(2,2-bipyridyl)dichlororuthenium(II) hexahydrate (Ru)/sodium persulfate (SPS) was chosen as the photoinitiator since its cytocompatibility and high photocrosslinking performance have been reported in previous reports involving DLP-based bioprinting.[30] After the photocrosslinking of each layer, the build platform was lifted, and the uncured (bio)ink was evacuated from the outlet. To avoid the contamination between the different gradient (bio)inks, the fluorinated ethylene propylene (FEP) film at the bottom of the vat was treated with a hydrophobic spray except for the outlet area, enabling the evacuation of the (bio)ink enriched to the outlet where it was relatively hydrophilic (Video S1). This unique setup largely overcame the cross-contamination issue between the (bio)ink exchanges and the challenge associated with significant bioink wasting observed in the other multi-material DLP bioprinting methods.[31, 32] Further, it would benefit the fabrication speed, considering that there is no need for vat-washing and the ability of efficient (bio)ink-mixing in real time.
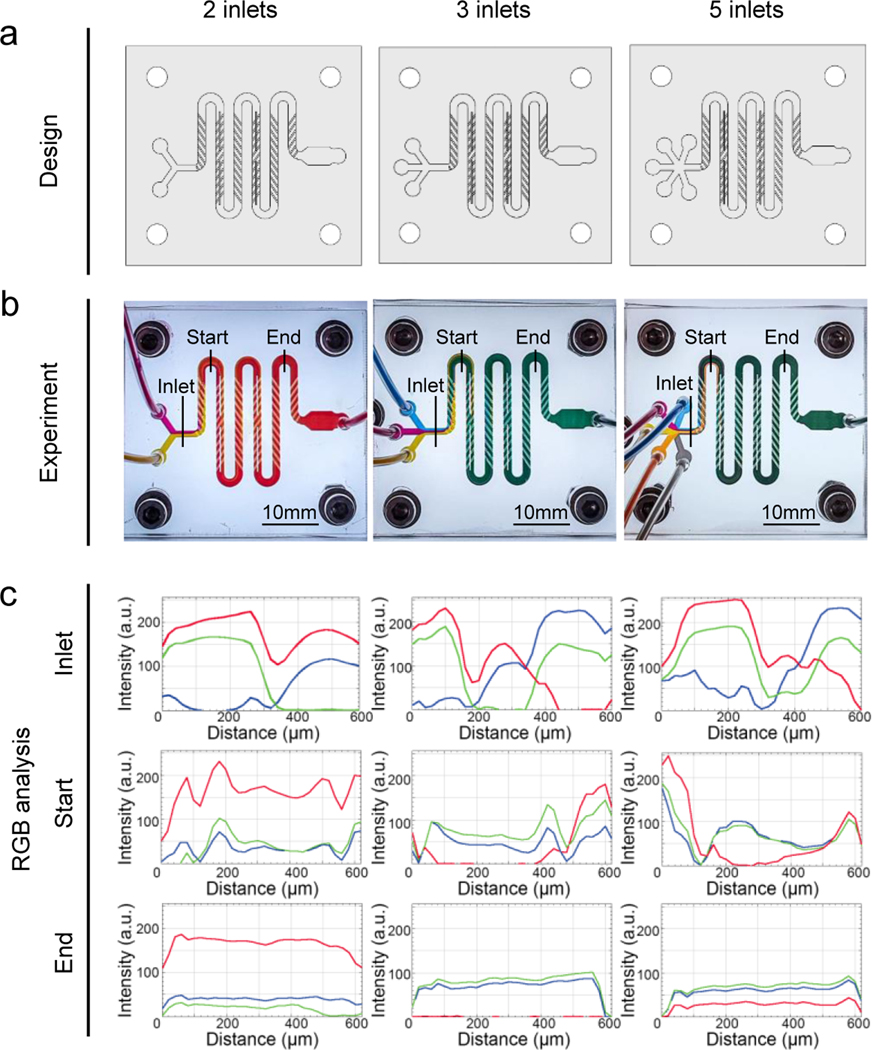
The mixing performance of the designed chaotic mixer was characterized by monitoring the mixing of 2, 3, or 5 fluid streams. On the basis of the designed mixers in Figure 2a, 40 wt.% PEGDA (molecular weight, Mw = 575 Da) colored with different dyes were delivered at a speed of 750 μL min-1. The mixing colors in the microchannels were observed over time and the results showed that the streams entered the junctions of the microchannels and quickly reached the steady state within a second. The optical images of the 2-, 3-, and 5-inlet mixers were captured once the steady flows were obtained, as shown in Figure 2b. The streamlines of the colors revealed the helical flow along the direction of the mixing liquid. Notably, the individual streamlines started to rotate due to the transverse velocity component induced by the slanted ribs from the beginning of the microchannels, and were mixed rapidly when they were crossing the first half cycle. Finally, they became fully mixed at the ending zones, confirming that the homogenous solutions at the desired gradient levels were obtained by the use of these chaotic mixers. The image of each mixer filled with different colors at steady state was applied to analyze the RGB (red, green, blue) components at the inlet, start (approximately 0.5 of the mixing circles), and end (approximately 4.5 of the mixing circles) positions. In each case, significant difference of RGB intensity distributions was observed at the inlet area whereas uniformly distributed intensities of RGB at the end were apparently realized (Figure 2c).
Figure 2.
Evaluations of the microfluidic chaotic mixers over 2, 3, and 5 different inks. (a) Designs of the mixers with 2, 3, and 5 inlets showing the zigzag shape of the microchannel (350 μm in height, 1.5 mm in width, and 99 mm in total length) containing 58 slanted ribs (100 μm in height, 250 μm in width, and leaving a lateral space of 250 μm between two ribs) and 4 barriers. (b) Experimental mixing observed by flow streamlines with PEGDA colored with different colors in the microfluidic chaotic mixers. (c) RGB analyses of the color intensities of the flow streamlines in (b) at the inlet, start (after 0.5 of the mixing cycle), and end (after 4.5 of the mixing cycles) positions.
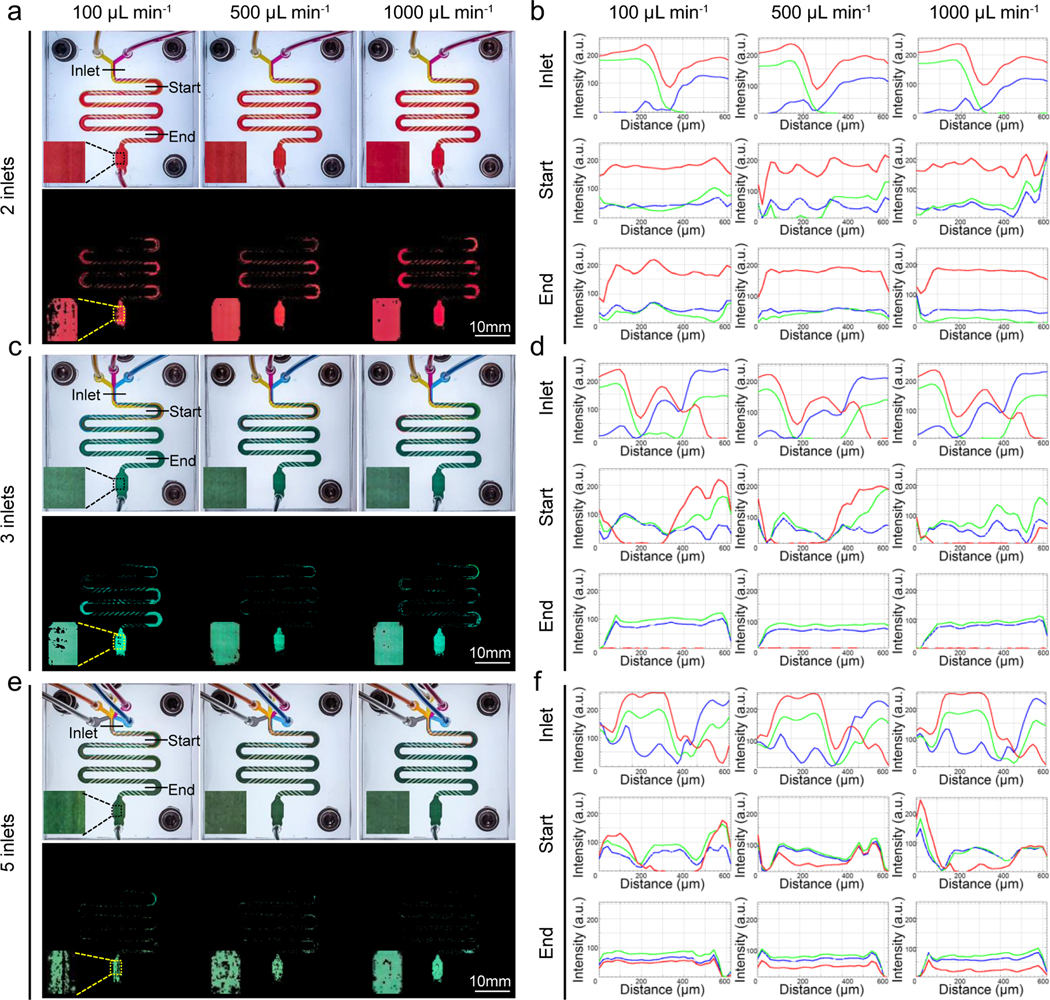
The chaotic mixers were also supplied with additional different flow rates to investigate the influence of fluid velocity on the mixing efficiency. The results of RGB intensity-analyses revealed that when mixing the same number of fluids, the colors were all evenly distributed at the ending regimes of mixers at the flow rates from 100 μL min−1 and 500 μL min−1 to 1000 μL min−1 (Figure 3). To confirm, we provided another analyzing method to quantify the degree of mixing by the term ‘mixing index (MI)’, which was acquired by quantifying the number of final mixed pixels compared to the number of total pixels. The well-mixed colors extracted from the ending zones of the mixers illustrated in Figure S2 implied that the mixing was quite thorough in all scenarios, consistent with the line profiles shown in Figure 3. The slight differences in results among the different flow rates could be seen from Figures 3 and S2. The colors in the ending area of 2-inlet mixer showed that the MI was increased from 79.0 ± 3.6% to 90.8 ± 3.7% and 97.1 ± 0.8% as the flow rate was elevated from 100 μL min−1 to 500 μL min−1 and 1000 μL min−1, indicating better mixing was achieved by increasing the fluid velocity. The same trends between flow rate and mixing performance could be observed in the 3- and 5-inlet mixers as well. The possible reason of the promoted mixing efficiency at the higher flow rate is the changes of Reynolds number. Reynolds number is the ratio of inertial to viscous forces, which is related to the flow speed, microchannel dimension, and viscosity of the fluid.[33] The number was higher with the flow rate increasing from 100 μL min−1 to 500 μL min−1 and 1000 μL min−1, resulting in the more chaotic advection of fluids and the better mixing result.[34] Overall, the developed chaotic mixer was capable of rapidly mixing multiple numbers of fluids exhibiting no less than 60% of completely mixing at the flow rates ranging from 100 μL min−1 to 1000 μL min-1. As the best mixing performance was achieved at 1000 μL min−1, it was used as the flow rate for the mixing in other subsequent experiments of this study. By far, numerous applications applying mixing strategies to extrusion 3D (bio)printing or other fabrication modalities have been carried out, yet few examples have reported characterizations of the mixing efficiencies. Here, we evaluated the mixing performance of the mixers with multiple inlets at a set of flow rates. These results extended our knowledge regarding the mixing process as well as integration of various components in the gradient-generation system where mixing of more than two types of (bio)inks are required.
Figure 3.
Mixing efficiencies of the microfluidic chaotic mixers over multiple inks at different collective flow rates. (a, c, e) Photos showing the mixing of 2, 3, and 5 inks at flow rates of (a) 100 μL min−1, (c) 500 μL min−1, and (e) 1000 μL min−1, and their corresponding images of extracted mixed colors. Magnified photographs of the end chambers are shown at the bottom left of each panel. (b, d, f) RGB profiles of the inlet, start, and end regions analyzed from photos in (a, c, e).
To obtain a better understanding of how the gradient-patterning in the printing processes can be controlled, the mixing was modulated by adjusting the ratio of the different inks on-the-fly. By controlling the inlet flow rates, the desired volume of each ink was precisely injected into the mixing microchannel resulting in a continual or discrete gradient of mixed inks at the ending zone of the mixer (also the ink vat of the printer). In our experiments, the collective flow rate was fixed at 1000 μL min−1 in the microchannel, no matter how many inlets the mixer had. The continual gradient in the slices of a hydrogel pie shown in Figure 4a was achieved by using this microfluidic mixer chip with 3 inlets, in combination with 40 wt.% PEGDA (Mw = 575 Da) inks colored with magenta, green, and blue containing 2-mM/20-mM Ru/SPS, with all the adjustments facilely achieved on-the-fly in real time during a single printing session. As such, it was also not necessary to prepare each of the inks beforehand prior to printing as needed in the previous setup with the ink-exchange mechanism, the latter of which could be labor-intensive especially when the required number of gradients is increased.[35] The pie, which was divided into 12 equal sizes with gradient colors, showed almost continuous color distributions at each slice, which we term as ‘continual’. The color distributions of the printed structure were in a good agreement with the corresponding mixed inks directly collected at the end of pumps without printing. Our findings led us to conclude that the microfluidic mixer chip enabled to gain predictable mixing of multiple inks in a precisely controllable manner. It offered unprecedented opportunities for fabricating highly complex gradients that can be further adjusted in real time, which was difficult to obtain before in the other DLP-based methods.
Figure 4.
Illustrations of 2D and 3D structures produced by the composable-gradient DLP printing technique. (a) The continual gradient of colors in the slices of a hydrogel pie printed by mixing merely three colored PEGDA inks. The inks in the Eppendorf tubes represent the colors collected after mixing before printing. (b) The 2D structures including a circle with 4 color gradients, a heptagon with 7 color gradients, and a vascular network with 4 color gradients, generated by mixing two differently colored PEGDA inks at different ratios followed by printing. The corresponding designed patterns are shown at the left, while the actual printed results are presented at the right. (c) The 3D constructs generated using PEGDA inks showing shapes of a cube, a pyramid, and a twisting hollow vase, featuring color gradients in the vertical direction. (d) The 2D maple leaf with horizontal gradients and the 3D maple leaf with vertical gradients printed with the same composable gradient colors mixed from green to red in real time during the printing sessions. (e) The Rubik’s cubes consisted of 2-by-2-by-2, 3-by-3-by-3, and 6-by-6-by-6 units presenting color gradients from discrete to continual.
To assess the possibility of generating gradients in both vertical and horizontal directions, we conducted 2D and 3D printing sessions using PEGDA inks as demonstrations. The composable gradient colors from yellow to green in printed planar constructs were formed by mixing yellow- and blue-colored inks through simply modulating their volumetric ratios. The printed structures in Figure 4b illustrated a circle (diameter: 9 mm) with 4 gradient colors in the radial direction, a heptagon (side length of each triangle: 2 mm) possessing 7 continual gradients, and a capillary-like network structure (horizontal length: 10 mm) containing 4 gradient color variants, all in the planar direction. The homogenous color distribution in each gradient bioink again indicated the efficient mixing obtained by the microfluidic mixer. The consistent colors between the designs and experimental results suggested the successful prediction of gradients by controlling the volumetric ratios of the inlet inks. We also printed 3D structures with varying colors to demonstrate the powerful printing capacity with vertical gradients. Figure 4c presents a cube (6 × 4 × 4 mm3) with a continual yellow-to-green gradient from the top to the bottom, which was achieved in 8 min of printing including 2 min of total ink-filling sessions and 6 min of total light exposures. The printed cube showed the gradually reduced size from top to bottom, in the same direction of dark green color-addition. The result explained that the darker color absorbed more projected light and therefore the photopolymerized area was slightly reduced than that with lighter color in the same crosslinking time.
We further fabricated a pyramid construct, in which color transitions occurred along the vertical direction. In addition, a more complex vase architecture with inner hollow and twisted orientation also exhibited the continual color gradients in the z-direction. By utilizing the same composable gradient colors mixed from green to red again in real time during the printing sessions, a 2D maple leaf with horizontal gradients and a 3D maple leaf with vertical gradient, both continuous in desired regions, were printed (Figure 4d). These printed spatially and material-wise heterogenous constructs highlighted the potential of our unique strategy to precisely control the gradient properties within sophisticated objects in-plane and throughout the volume. It should be noted that the discrete or continual of gradient relies on the numbers by which the printing pattern is divided. The more segments the pattern is divided into, the finer gradient can be obtained. For example, the printing model of the Rubik’s cube was divided into 2-by-2-by-2, 3-by-3-by-3, and 6-by-6-by-6 blocks (Figure 4e). Through printing with inks obtained by mixing 2, 3, and 6 sets of fluid ratios adjusted in real time, the discrete to continual (or near-continuous) gradients could be identified in the printed cubes.
We further illustrated biological applications of our gradient DLP-bioprinter in fabricating tissue constructs with multifunctional gradients, including that of cell density, of matrix stiffness, of growth factor concentration, and of porosity. We first evaluated the cytocompatibility of our gradient bioprinting method, where bioinks were mixed in the microchannel before bioprinting under the different fluid ratios. The results of live/dead staining in Figure S3 presented that a high proportion of C2C12 myoblasts encapsulated in 10 wt.% GelMA was viable after the mixing and bioprinting processes. Through the quantifications of live cell numbers, the comparable cell numbers between the group without passing through the mixer (i.e., mixed by pipetting; Ctrl) and the group mixed by the microfluidic mixing chip (Mix) were observed immediately after bioprinting and over 7 days of culture. Meanwhile, with the ratios of GelMA and GelMA encapsulating C2C12 increasing, the total cell numbers were elevated. The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was also conducted to confirm the enhanced metabolic activities of C2C12 cells measured at days 0, day 3, and 7 after bioprinting (Figure S4).
For the cell density gradient-fabrication, the green fluorescent protein (GFP)-human umbilical vein endothelial cells (HUVECs) were bioprinted via arbitrarily adjusting the velocity ratios of 10 wt.% GelMA bioink containing HUVECs and 10 wt.% GelMA ink without cells on-the-fly, which were altered between 0.20 and 9.50 in a randomly designed manner (Figure 5a). The green fluorescent intensity profiles of the bioprinted samples, reflecting the cell densities, displayed the consistent variations with the flow velocity ratios that were arbitrarily designed as the inputs during the corresponding bioprinting sessions. Therefore, we established a bioprinting method of achieving arbitrarily adjustable gradients almost in real time in a highly predictable manner.
Figure 5.
Biomedical applications of gradient-bioprinted tissue-like constructs. (a) Two examples showing cell density gradients of GFP-HUVECs bioprinted in an arbitrarily controllable manner at any desired density in any given single printing session. The intensity analysis for each example shows the consistency of output intensity pattern (green) and the velocity ratios of input bioink flows (red). (b-d) Gradient bioprinting of cells within the 10 wt.% GelMA hydrogels. (b) A circle pattern with MDA-MB-231 cells (blue) showing the cell density gradient in the radial direction and the quantitative analysis of the fluorescence intensities. (c) The vascular network structure with the cell density gradient in the horizontal direction encapsulating HUVECs (green) and the quantitative analysis of the fluorescence intensities. (d) A construct containing RFP-HUVECs and GFP-HUVECs showing the positive gradient of red cells and the negative gradient of green cells from left to right, and the quantitative analyses of the fluorescence intensities. (e) The photograph captured on EDD 14 of the CAM assay implanted with the printed VEGF-gradient scaffold and its corresponding image processed with the traced vessels. Bar graphs showing the quantitative values of the total vessel lengths and vessel numbers. (f) C2C12 myoblasts stained with F-actin/nucleus (red/blue) cultured on a bioprinted stiffness-gradient hydrogel presenting various morphologies. The quantifications of single-cell areas and cell aspect ratios (major axis to the minor axis) of C2C12, as well as the compressive modulus of 10–30 wt.% GelMA hydrogel. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA (e, compared with the control group of 0 μg mL−1 VEGF; f, compared with the group of 10 wt.% GelMA), mean ± s.d. (e, n = 3; f, n = 10).
Many natural tissues are composed of multiple cell types that are distributed heterogeneously, playing an essential role in the establishment of cell functions and cell-cell communications.[36] To replicate the cellular gradient structure, a circle with three continual cell densities was bioprinted through mixing the GelMA solution with different volumetric ratio of MDA-MB-231 breast cancer cells. This multiple-zone concentric circular pattern shown in Figure 5b presented the reduced cell density in the radical direction from the central area to the peripheries. The gradient in the number of cells was supported by the quantification result of fluorescence intensities along the direction, decreasing from 70 at the center (0 μm) to only 9 at the edge (380 μm). In addition, bioprinting with GFP-HUVECs in the gradient density allowed to fabricate a vascular network with the minimum vessel diameter of 50 μm (Figure 5c). The visualized image exhibited the decreasing cellular fluorescence from the left to the right in the network. Identical conclusion was derived from quantified result that the average intensity was approximately 50 in the left end but was reduced to 20 at the right.
To achieve multiple gradients of multiple cells at the same time, two bioinks of 10 wt.% GelMA containing GFP-HUVECs and red fluorescent protein (RFP)-HUVECs were loaded into the two inlets of the microfluidic mixing chip. By dynamically changing the flow velocities of the two bioinks during the same bioprinting session, one increasing whereas another decreasing, the construct containing two cell gradients in reverse directions was obtained. The result presented that in the bioprinted construct, the intensities of the RFP cells graded from 0 to 450 in one direction, while the GFP cell intensities graded from 8000 to 30 conversely (Figure 5d). This cell-type or cell-density gradient could potentially be applied to mimicking various biological tissues such as the cartilage, where chondrocytes, hypertrophic chondrocytes, and osteoblasts are graded in their distributions.[37, 38] The similar patterns of results were demonstrated in previous studies using the extrusion-based gradient bioprinting, in which the chondrocytes were bioprinted with the density gradient, or, mesenchymal stem cells (MSCs) and chondrocytes were deposited at the graded ratio.[10, 39] The microfluidic mixing system combined with the DLP technique continues to be a great impetus to embed the cell gradient within the more complex tissue architectures than typically attainable with extrusion bioprinting.
For the functional tissue engineering, the chemical gradients in the matrix or induced by the surrounding cells serve as cues for cell guidance, migration, and fate-determining.[40] Take vascular endothelial growth factor (VEGF) as an example, the soluble VEGF gradient has been demonstrated to enhance vessel branching during angiogenesis and vasculogenesis.[41] More importantly, the gradient of substrate-conjugated VEGF plays an essential role in increasing the sprouting of vessel branches.[42] Therefore, we selected heparin methacryloyl (HepMA) as a macromolecular host of VEGF to enable the sustained VEGF release.[43] The HepMA-GelMA hydrogel was mixed with VEGF and formulated the latter into the horizontal gradient from 0 ng mL−1 to 100 ng mL−1 (Figure 5e). The ex ovo chorioallantoic membrane (CAM) assay was performed to evaluate the angiogenic capacity of the printed VEGF-gradient constructs. At embryonic development day (EDD) 14, the photographs of the printed construct and the surrounding CAM regions suggested that new microvessels developed towards the implanted hydrogel. The total lengths and numbers of blood vessels growing on the printed construct increased in the areas printed with higher concentrations of VEGF. The successful patterning of the growth factor gradient induced sufficient and differential vascularization, offering a great potential as an efficient tool in tissue regeneration.
Apart from the chemical gradients, the stiffness of extracellular matrix is of great interest for determining cell morphology, migration, and differentiation of multipotential cells.[40, 44] Mechanical gradients exist in the interfaces between different tissues, and meanwhile the heterogenous mechanical properties can be presented within the same tissues.[45, 46] Considering that the stiffness of GelMA hydrogel is tunable by the macromolecule concentration, the degree of methacryloyl-substitution, and the crosslinking conditions,[47, 48] continually formulating GelMA hydrogels with different concentrations would likely guarantee stiffness gradients through our gradient DLP-based printing (Figure 5f). Indeed, C2C12 myoblasts cultured on the surface of a printed GelMA matrix featuring a stiffness gradient presented various morphologies after 1 day of incubation. The cells located at the stiffer side (30 wt.% GelMA with a compressive modulus of 232.8 ± 14.9 kPa) exhibited a spindle-shaped morphology, with the largest cell area and aspect ratio. On the contrary, the cells on the softer and intermediate areas (10 wt.%, 11.9 ± 1.7 kPa; and 20 wt.%, 80.4 ± 4.7 kPa) appeared to be less elongated, consistent with the prior reports.[49] In addition to fabricating the biomimicry structures, our method for creating the stiffness gradients in bioprinted constructs allows the investigation of cell responses to substrates potentially in a more predictable and controllable manner.
Engineered scaffolds with porous features provide several advantages in tissue regeneration.[50] For instance, porous structures promote the transport of nutrients and wastes, offer large surface areas and spaces for the cell attachment and spreading, and allow the close interactions between the implants and the adjacent tissues. The aqueous two-phase system is formed by blending a polymer and salt or two polymers and water, which has been mainly used as the liquid-liquid fractionation approach to extract, purify, and enrich of biomolecules. Recent developments in producing hydrogel microspheres and porous hydrogels utilizing the aqueous two-phase system have attracted much attention.[51–53] To fabricate the 3D porous gradient architecture, 10 wt.% GelMA bioink containing human MSCs was mixed with the 10 wt.% GelMA solution containing the porogen (porogen, Mw = 2,000,000 Da). By adjusting the flow ratios of the two bioinks, we formulated the 10 wt.% GelMA bioinks with 3.0 wt.%, 1.5 wt.%, and 0.5 wt.% of the porogen, resulting in three gradual zones featuring different pore sizes as observed from the optical micrographs (Figure S5a–b). The average pore sizes within the different hydrogel sections made of these mixed bioinks changed from 65 μm and 29 μm to 12 μm (Figure S5c). The MSCs encapsulated within the bioprinted construct exhibited enhanced spreading in the portions with larger pore sizes at 7 days after the bioprinting (Figure S5d). Compared to the hydrogel segment mixed with 0.5 wt.% porogen, the sizes of cell cluster was promoted to 2.5-fold and 4-fold within the hydrogel regions containing 1.5 wt.% and 3.0 wt.% porogen, respectively. The existent of porogen effectively formed pores in the bioprinted hydrogel constructs, facilitating the diffusion of nutrients and oxygen and the removal of wastes, thus providing an environment to promote cell spreading and proliferation.[51–53]
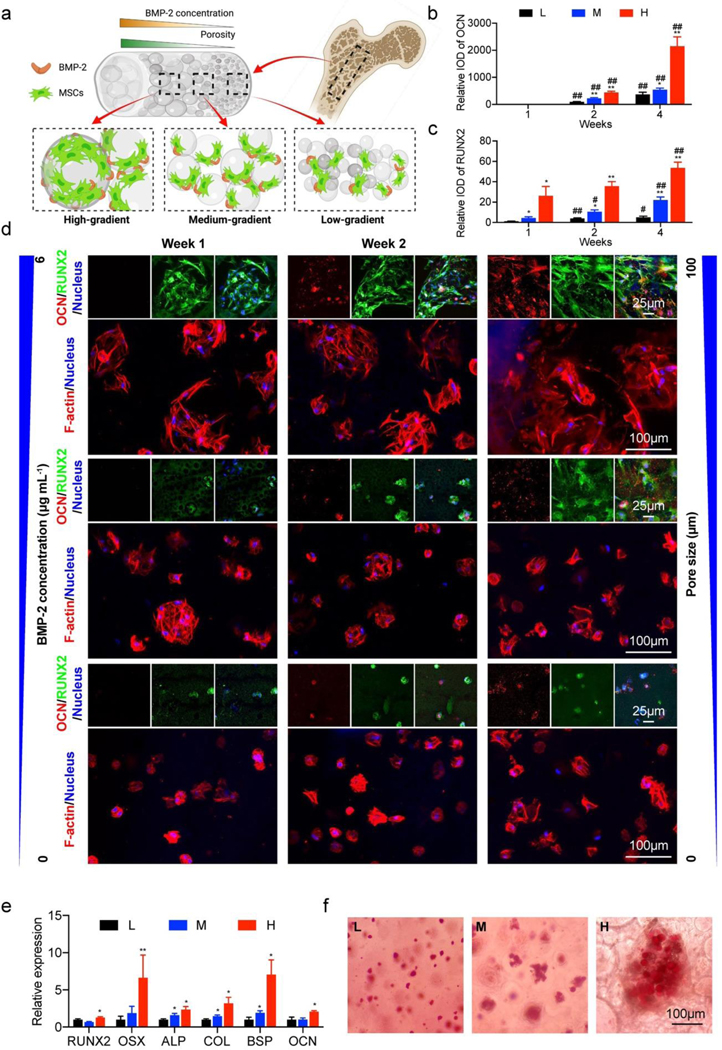
The native bone tissue has a complex heterogenous architecture that possesses regional differences in structures and compositions.[54–56] Dual or multiple gradients in the fabrication of tissue-mimicking constructs are therefore required to achieve a better recapitulation of the complex heterogenous tissue patterns. Utilizing our porosity-gradient construct, we further integrated the second biological gradient of bone morphogenetic protein 2 (BMP-2) to enhance osteogenesis of MSCs. BMP-2, as a crucial growth factor that can induce the osteogenic differentiation of MSCs,[57, 58] was first evaluated by mixing in our microfluidic mixer with the 10 wt.% GelMA ink into the final concentrations of 0, 1, 2, and 6 μg mL-1. The MSCs seeded onto the surface of the printed BMP-2-graded scaffold exhibited a more fibroblastic morphology when the BMP-2 concentration was increased (Figure S6).
In our 4-week osteogenesis study, the fabrication started by mixing the pore-forming GelMA-porogen/BMP-2/MSC bioink with the GelMA/MSC bioink under three sets of different flow ratios. We were able to bioprint the 3D constructs with dual gradients of BMP-2 (0, 3, and 6 μg mL−1) and porosity (where the concentrations of porogen were 0.5, 1.5, and 3.0 wt.%) (Figure 6a). The results of F-actin staining indicated that MSCs grew as cell clusters and filled in the pore areas of the hydrogels. Better cell spreading and proliferation could be observed in the portion with higher concentration of porogen.
Figure 6.
Osteogenesis induced by the dual gradients of porosity and BMP-2. (a) Schematic showing the bioprinted 3D construct encapsulated with MSCs for the investigation of osteogenic effects of the dual gradients of porosity and BMP-2. (b-d) Immunostaining results of RUNX2 (green) and OCN (red), as well as F-actin (red) of MSCs in the bioprinted bone-like structures at 1, 2, and 4 weeks following osteogenic differentiation. The quantifications show integrated optical densities (IODs) for RUNX2 and OCN in the regions of low-gradient (L), medium-gradient (M), and high-gradient (H). (e) Osteogenic gene expression levels of MSCs in the bioprinted dual-gradient hydrogels at 4 weeks following osteogenic differentiation in the regions of low-gradient (L), medium-gradient (M), and high-gradient (H). (f) ARS staining of bioprinted dual-gradient hydrogels at 4 weeks following osteogenic differentiation in the regions of low-gradient (L), medium-gradient (M), and high-gradient (H). *P < 0.05, **P < 0.01; #P < 0.05, ##P < 0.01; one-way ANOVA (b and c, *, compared with the group of low-gradient, #, compared with the group of 1 week; e, compared with the group of low-gradient at 1 week), mean ± s.d. (n = 3).
To further investigate the impact of porosity and BMP-2 gradients on the osteogenic differentiation of MSCs, immunostainings of RUNX2 and osteocalcin (OCN) were carried out (Figure 6b–d). RUNX2 is a transcription factor that is related to the early stage of osteoblastic differentiation, while OCN is a late-stage bone marker, with the increased expression during osteoblast maturation.[59] Quantitative results confirmed that only RUNX2, as an osteoprogenitor at the early stage, was expressed at 1 week of differentiation, and its expression was enhanced within the areas with larger pores and higher BMP-2 concentrations. At 2 and 4 weeks of differentiation, the OCN signals were detected when the new bone was forming towards the higher dual-gradient sites. We also found the significantly increased intensities of both markers in the areas with larger pores and higher BMP-2 concentrations at week 2 and week 4. The quantitative reverse transcription-polymerase chain reaction (RT-PCR) analyses were preformed to further evaluate the expressions of specific osteogenic markers. Osteoprogenitor cell (RUNX2, osterix (OSX)), pre-osteoblast (alkaline phosphatase (ALP), type-I collagen (COL1A1)), and osteoblast (bone sialoprotein (BSP), OCN) gene expressions in the three gradient zones of the bioprinted constructs after 4 weeks of differentiation were compared and are shown in Figure 6e. Results implied that the expression levels of OSX, BSP, and COL were significantly upregulated at the medium gradient (1.5 wt.% of porogen with 3 μg mL−1 of BMP-2) and the high gradient (3.0 wt.% of porogen with 6 μg mL−1 of BMP-2), compared with their expressions in the low-gradient region (0.5 wt.% of porogen with 0 μg mL−1 of BMP-2). The expression levels of RUNX2, ALP, and OCN were slightly augmented from the low-gradient to medium-gradient and high-gradient regions.
Additionally, we verified the calcium-deposition in the bioprinted gradient scaffolds using Alizarin Red S (ARS) staining. The images of ARS exhibited no calcium deposition at 1 week, and disperse and limited calcium-deposition at 2 weeks (Figure S7). In comparison, the obviously improved calcium-deposition was visualized at 4 weeks within the MSC clusters, where the ARS-positive area was increased when the porosity and the BMP-2 concentration became higher (Figure 6f). Taken together, the immunofluorescence staining, gene-expression, and calcium-deposition data strongly suggested that the dual gradients of porosity and BMP-2 successfully stimulated osteogenesis of MSCs, as well as to a good extent recapitulated the architecture of the bone tissue.
In summary, a (bio)printing system able to produce composable gradients was established based on the DLP method for the first time, by taking advantage of a chaotic mixing chip connected to a specially designed vat. The gradients of (bio)inks, either continual or discrete, were conveniently formulated by mixing the desired amounts of (bio)inks at controllable volumes (determined by their infusion rates) on-the-fly in real time. We demonstrated the (bio)printing of sophisticated hydrogel constructs with both horizonal and vertical gradients in both 2D and 3D at reasonably high resolutions. More relevantly, the feasibility of our unique platform on functional (bio)printing was evaluated by (bio)printing (bio)inks with gradients in cell densities of single or multiple cell types, in substrate stiffness, in growth factor concentration, and in porosity and pore size. Overall, this DLP-based gradient (bio)printing system enables to recapitulate the gradient properties of natural tissues, at good resolution and with complex architectures, not demonstrated with DLP (bio)printing yet to date.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health ( R21EB025270, R00CA201603), the National Science Foundation (CBET-EBMS-123859), and the Brigham Research Institute. We thank the NeuroTechnology Studio at Brigham and Women’s Hospital for providing Zeiss LSM880 instrument access and consultation on data acquisition and data analysis.
Footnotes
Experimental section
The detailed experimental process is available in the Supporting Information.
Supporting information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Mian Wang, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Wanlu Li, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Luis S. Mille, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Terry Ching, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Pillar of Engineering Product Development, Singapore University of Technology and Design, Singapore 48737; Digital Manufacturing and Design Centre, Singapore University of Technology and Design, Singapore 487372; Department of Biomedical Engineering, National University of Singapore, Singapore 117583.
Zeyu Luo, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Guosheng Tang, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Carlos Ezio Garciamendez, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Ami Lesha, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Michinao Hashimoto, Pillar of Engineering Product Development, Singapore University of Technology and Design, Singapore 48737; Digital Manufacturing and Design Centre, Singapore University of Technology and Design, Singapore 487372.
Yu Shrike Zhang, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
References
- [1].Li C, Ouyang L, Armstrong JPK, Stevens MM, Trends Biotechnol 2020. [DOI] [PubMed] [Google Scholar]
- [2].Wu J, Chen Q, Liu W, Lin JM, Lab Chip 2013. 13(10) 1948–54. [DOI] [PubMed] [Google Scholar]
- [3].Bracaglia LG, Smith BT, Watson E, Arumugasaamy N, Mikos AG, Fisher JP, Acta Biomater 2017. 56 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Singh M, Berkland C, Detamore MS, Tissue Eng Part B Rev 2008. 14(4) 341–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cheng A, Schwartz Z, Kahn A, Li X, Shao Z, Sun M, Ao Y, Boyan BD, Chen H, Tissue Eng Part B Rev 2019. 25(1) 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang X, Liu Z, Pang Y, RSC advances 2017. 7(48) 29966–29984. [Google Scholar]
- [7].Seidi A, Sampathkumar K, Srivastava A, Ramakrishna S, Ramalingam M, J Nanosci Nanotechnol 2013. 13(7) 4647–55. [DOI] [PubMed] [Google Scholar]
- [8].Ilkhanizadeh S, Teixeira AI, Hermanson O, Biomaterials 2007. 28(27) 3936–43. [DOI] [PubMed] [Google Scholar]
- [9].de Gans BJ, Schubert US, Langmuir 2004. 20(18) 7789–93. [DOI] [PubMed] [Google Scholar]
- [10].Idaszek J, Costantini M, Karlsen TA, Jaroszewicz J, Colosi C, Testa S, Fornetti E, Bernardini S, Seta M, Kasarełło K, Biofabrication 2019. 11(4) 044101. [DOI] [PubMed] [Google Scholar]
- [11].Costantini M, Jaroszewicz J, Kozoń Ł, Szlązak K, Święszkowski W, Garstecki P, Stubenrauch C, Barbetta A, Guzowski J, Angewandte Chemie International Edition 2019. 58(23) 7620–7625. [DOI] [PubMed] [Google Scholar]
- [12].Singh M, Haverinen HM, Dhagat P, Jabbour GE, Adv Mater 2010. 22(6) 673–85. [DOI] [PubMed] [Google Scholar]
- [13].Placone JK, Engler AJ, Adv Healthc Mater 2018. 7(8) e1701161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Proceedings of the National Academy of Sciences 2016. 113(8) 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen S, Tuszynski MH, Nat Med 2019. 25(2) 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Science 2019. 364(6439) 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ligon SC, Liska R, Stampfl J, Gurr M, Mulhaupt R, Chem Rev 2017. 117(15) 10212–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li W, Mille LS, Robledo JA, Uribe T, Huerta V, Zhang YS, Advanced Healthcare Materials 2020. 2000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schreurs M, Suttorp CM, Mutsaers HAM, Kuijpers-Jagtman AM, Von den Hoff JW, Ongkosuwito EM, Carvajal Monroy PL, Wagener F, Med Res Rev 2020. 40(1) 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lim KS, Galarraga JH, Cui X, Lindberg GC, Burdick JA, Woodfield TB, Chemical reviews 2020. 120(19) 10662–10694. [DOI] [PubMed] [Google Scholar]
- [21].Zhang J, Hu Q, Wang S, Tao J, Gou M, International Journal of Bioprinting 2020. 6(1) 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R, Lab on a Chip 2010. 10(16) 2062–2070. [DOI] [PubMed] [Google Scholar]
- [23].Miri AK, Nieto D, Iglesias L, Goodarzi Hosseinabadi H, Maharjan S, Ruiz-Esparza GU, Khoshakhlagh P, Manbachi A, Dokmeci MR, Chen S, Shin SR, Zhang YS, Khademhosseini A, Adv Mater 2018. 30(27) e1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pfahler J, Harley J, Bau H, Zemel J, Sensors and Actuators A: Physical 1990. 22(1–3) 431–434. [Google Scholar]
- [25].Kim DS, Lee SW, Kwon TH, Lee SS, Journal of micromechanics and microengineering 2004. 14(6) 798. [Google Scholar]
- [26].Hessel V, Löwe H, Schönfeld F, Chemical Engineering Science 2005. 60(8–9) 2479–2501. [Google Scholar]
- [27].Li W, Wang M, Mille LS, Robledo Lara JA, Huerta V, Uribe Velazquez T, Cheng F, Li H, Gong J, Ching T, Murphy CA, Lesha A, Hassan S, Woodfield TBF, Lim KS, Zhang YS, Adv Mater 2021. 33(35) e2102153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM, Science 2002. 295(5555) 647–51. [DOI] [PubMed] [Google Scholar]
- [29].Kim BS, Kwak BS, Shin S, Lee S, Kim KM, Jung H-I, Cho HH, International journal of heat and mass transfer 2011. 54(1–3) 118–125. [Google Scholar]
- [30].Lim KS, Levato R, Costa PF, Castilho MD, Alcala-Orozco CR, van Dorenmalen KMA, Melchels FPW, Gawlitta D, Hooper GJ, Malda J, Woodfield TBF, Biofabrication 2018. 10(3) 034101. [DOI] [PubMed] [Google Scholar]
- [31].Lee J, Kim KE, Bang S, Noh I, Lee C, International journal of precision engineering and manufacturing 2017. 18(4) 605–612. [Google Scholar]
- [32].Han D, Yang C, Fang NX, Lee H, Additive Manufacturing 2019. 27 606–615. [Google Scholar]
- [33].Purcell EM, American journal of physics 1977. 45(1) 3–11. [Google Scholar]
- [34].Xia H, Wan S, Shu C, Chew Y, Lab on a Chip 2005. 5(7) 748–755. [DOI] [PubMed] [Google Scholar]
- [35].Miri AK, Nieto D, Iglesias L, Goodarzi Hosseinabadi H, Maharjan S, Ruiz-Esparza GU, Khoshakhlagh P, Manbachi A, Dokmeci MR, Chen S, Advanced Materials 2018. 30(27) 1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jadin KD, Wong BL, Bae WC, Li KW, Williamson AK, Schumacher BL, Price JH, Sah RL, J Histochem Cytochem 2005. 53(9) 1109–19. [DOI] [PubMed] [Google Scholar]
- [37].Zhu D, Tong X, Trinh P, Yang F, Tissue Engineering Part A 2018. 24(1–2) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sun Y, You Y, Jiang W, Wang B, Wu Q, Dai K, Sci Adv 2020. 6(37). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ren X, Wang F, Chen C, Gong X, Yin L, Yang L, BMC musculoskeletal disorders 2016. 17(1) 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sant S, Hancock MJ, Donnelly JP, Iyer D, Khademhosseini A, Can J Chem Eng 2010. 88(6) 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Byrne AM, Bouchier-Hayes DJ, Harmey JH, J Cell Mol Med 2005. 9(4) 777–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jeong GS, Han S, Shin Y, Kwon GH, Kamm RD, Lee SH, Chung S, Anal Chem 2011. 83(22) 8454–9. [DOI] [PubMed] [Google Scholar]
- [43].Li C, Ouyang L, Pence IJ, Moore AC, Lin Y, Winter CW, Armstrong JPK, Stevens MM, Adv Mater 2019. 31(17) e1900291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brandl F, Sommer F, Goepferich A, Biomaterials 2007. 28(2) 134–46. [DOI] [PubMed] [Google Scholar]
- [45].Seidi A, Ramalingam M, Elloumi-Hannachi I, Ostrovidov S, Khademhosseini A, Acta Biomater 2011. 7(4) 1441–51. [DOI] [PubMed] [Google Scholar]
- [46].Guimarães CF, Gasperini L, Marques AP, Reis RL, Nature Reviews Materials 2020. 1–20.33078077 [Google Scholar]
- [47].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A, Biomaterials 2015. 73 254–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gong J, Shuurmans C, Cao X, Van Genderen AM, Li W, Cheng F, He JJ, López A, Huerta V, Manríquez J, Li R, Li H, Delavaux C, Sebastian S, Wang H, Xie J, Yu M, Masereeuw R, Vermonden T, Zhang YS, Nature Communications 2020. 11 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Boontheekul T, Hill EE, Kong H-J, Mooney DJ, Tissue engineering 2007. 13(7) 1431–1442. [DOI] [PubMed] [Google Scholar]
- [50].Loh QL, Choong C, Tissue Eng Part B Rev 2013. 19(6) 485–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ying GL, Jiang N, Maharjan S, Yin YX, Chai RR, Cao X, Yang JZ, Miri AK, Hassan S, Zhang YS, Adv Mater 2018. 30(50) e1805460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ying G, Manriquez J, Wu D, Zhang J, Jiang N, Maharjan S, Hernandez Medina DH, Zhang YS, Mater Today Bio 2020. 8 100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ying G, Jiang N, Parra-Cantu C, Tang G, Zhang J, Wang H, Chen S, Huang NP, Xie J, Zhang YS, Advanced Functional Materials 2020. 30(46) 2003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li C, Ouyang L, Armstrong JPK, Stevens MM, Trends Biotechnol 2021. 39(2) 150–164. [DOI] [PubMed] [Google Scholar]
- [55].Bose S, Vahabzadeh S, Bandyopadhyay A, Materials today 2013. 16(12) 496–504. [Google Scholar]
- [56].Bose S, Roy M, Bandyopadhyay A, Trends Biotechnol 2012. 30(10) 546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ryoo HM, Lee MH, Kim YJ, Gene 2006. 366(1) 51–7. [DOI] [PubMed] [Google Scholar]
- [58].He X, Ma J, Jabbari E, Langmuir 2008. 24(21) 12508–16. [DOI] [PubMed] [Google Scholar]
- [59].Huang W, Yang S, Shao J, Li YP, Front Biosci 2007. 12 3068–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Liu W, Zhang YS, Heinrich MA, De Ferrari F, Jang HL, Bakht SM, Alvarez MM, Yang J, Li Y-C, Trujillo-de Santiago G, Miri AK, Zhu K, Khoshakhlagh P, Prakash G, Cheng H, Guan X, Zhong Z, Ju J, Zhu GH, Jin X, Shin SR, Dokmeci MR, Khademhosseini A, Adv. Mater. 2017. 29 1604630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lim KS, Schon BS, Mekhileri NV, Brown GCJ, Chia CM, Prabakar S, Hooper GJ, Woodfield TBF, ACS Biomaterials Science & Engineering 2016. 2(10) 1752–1762. [DOI] [PubMed] [Google Scholar]
- [62].Mangir N.i., Dikici S, Claeyssens F, MacNeil S, ACS Biomaterials Science & Engineering 2019. 5(7) 3190–3200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.