Abstract
One of the potential benefits of using data-independent acquisition (DIA) proteomics protocols is that information not originally targeted by the study may be present and discovered by subsequent analysis. Herein we reanalyzed DIA data originally recorded for global proteomic analysis to look for isomerized peptides, which occur as a result of spontaneous chemical modifications to long-lived proteins. Examination of a large set of human brain samples revealed a striking relationship between Alzheimer’s disease (AD) status and isomerization of aspartic acid in a peptide from tau. Relative to controls, a surprising increase in isomer abundance was found in both autosomal dominant and sporadic AD samples. To explore potential mechanisms that might account for these observations, quantitative analysis of proteins related to isomerization repair and autophagy was performed. Differences consistent with reduced autophagic flux in AD-related samples relative to controls were found for numerous proteins, including most notably p62, a recognized indicator of autophagic inhibition. These results suggest, but do not conclusively demonstrate, that lower autophagic flux may be strongly associated with loss of function in AD brains. This study illustrates that DIA data may contain unforeseen results of interest, and may be particularly useful for pilot studies investigating new research directions. In this case, a promising target for future investigations into the therapy and prevention of AD has been identified.
Keywords: proteomics, age-related neurodegenerative disease, proteostasis, lysosome, aspartic acid, amyloid, neurofibrillary tangle, amyloid-beta, post-translational modification, hippocampus
Graphical Abstract
Introduction
A frequently mentioned advantage of data-independent acquisition (DIA) proteomics approaches is the potential capture of information not originally sought when then data was acquired. This advantage is anticipated because DIA experiments are designed to collect information about all species eluting from chromatographic separation, without selection as occurs in data-dependent acquisition (DDA). Despite the potential for re-analysis of DIA data, examples are limited.1,2 Recent work has posited a connection between Alzheimer’s disease (AD) and isomerization of amino acids in long-lived proteins, which may interfere with lysosomal digestion.3 Mass spectrometric analysis of peptide isomers (where typically L-Asp has been swapped for D/L-isoAsp) and epimers (where a single residue has been flipped from the L to D configuration) is typically stymied by several complicating factors including: no mass change following modification, absence of unique or characteristic fragments, and similarity of fragmentation patterns in general. An example of isomerization at Asp for the tryptic peptide 386TDHGAEIVYK395 from tau is illustrated in Scheme 1. Identification of isomers in DDA is further complicated by dynamic exclusion, which precludes examination of the same mass for a defined period of time. In theory, DIA data should contain information about isomers, but this capability has not been demonstrated because typical DIA analysis pipelines only identify the best peak for any given sequence.4 Some progress has been made with identification of phosphorylation isomers, though these isomers can be distinguished by many unique mass fragments.5
Scheme 1.
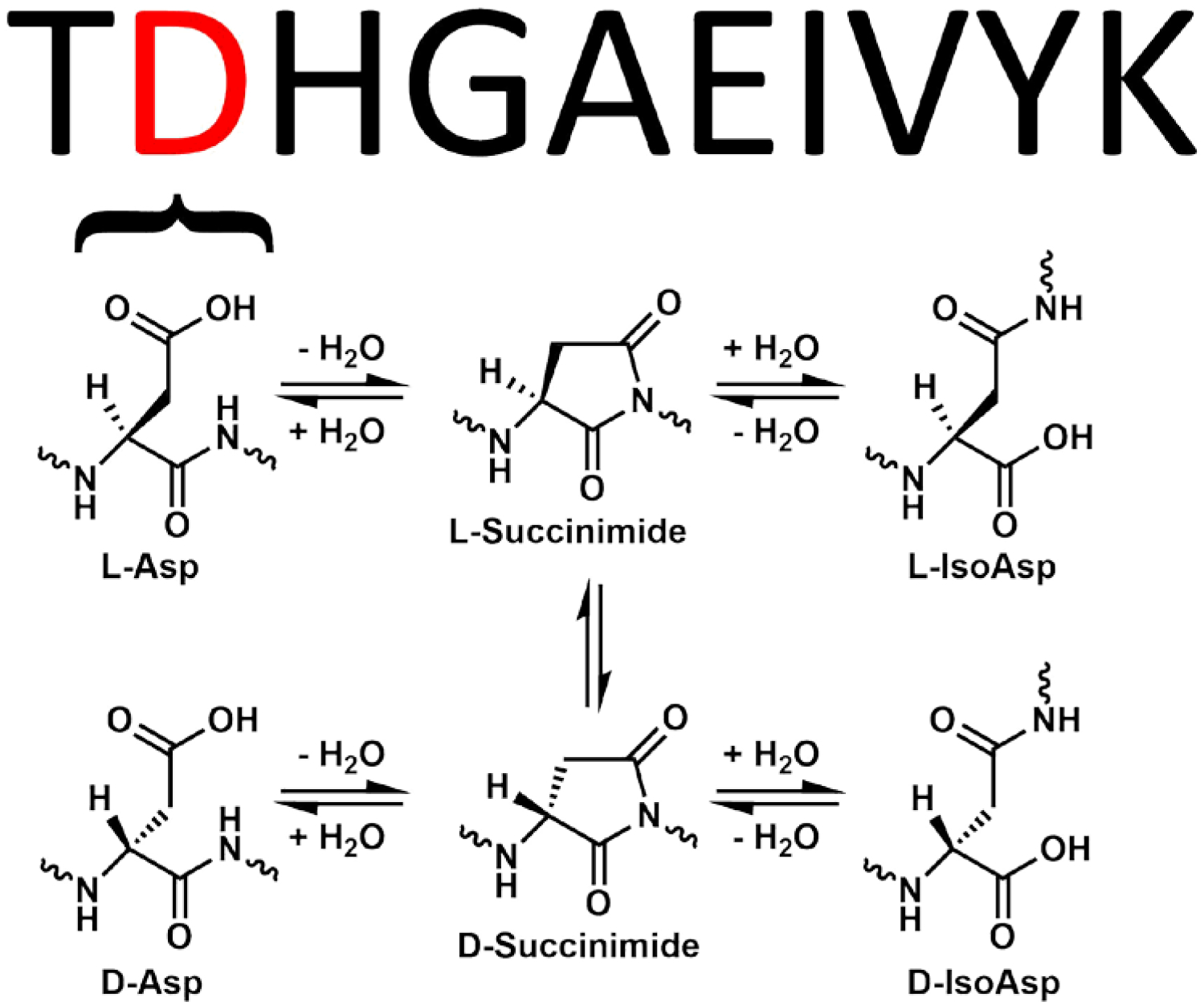
Chemical pathways leading to Asp isomerization
If the extent of amino acid isomerization in biological samples could be easily determined by DIA, then connections to the origin of AD and other age-related diseases could be investigated more easily. Despite decades of intense investigation, the underlying causes of AD remain incompletely understood. Many hypotheses have focused on aggregation of Aβ peptides, which form toxic oligomers as well as larger fibrils and ultimately extracellular deposits in multiple brain regions of people with AD.6 Abnormally phosphorylated and otherwise post-translationally modified tau has also received significant attention, as it is a primary constituent of neurofibrillary tangles (NFTs) and neuropil threads that are observed within neurons in partially overlapping brain regions of people with AD.7 The accumulation of pathologic tau is a better predictor of dementia,8 and recent work has shown that phosphorylated tau at positions 217, 181, and 205 indicate staging and development of AD9 with translation to reliable plasma markers that also correlate with degree of dementia.10,11 In addition, there are many other pathologic changes that accompany AD, including other aggregated deposits such as Lewy bodies,12 vascular disease,13 energy deficiency,14 and lysosomal storage.15,16 The relationships between all of these observations and their underlying causes remain to be fully elucidated, but it is clear that absent genetic predisposition, increasing age is the primary risk factor for AD.17
One aspect of aging that remains underexplored is the spontaneous chemical modification (SCM) of long-lived proteins. These modifications are not enzymatically catalyzed, and some fall largely outside the purview of biological control. Although many SCMs are easily detected and have been well documented, including oxidation,18 truncation,19 and deamidation,20,21 others are more pernicious and typically evade notice, such as the isomerization mentioned above.22,23 At the level of an individual residue, isomerization leads to dramatic structural changes, but these alterations are masked in the context of a protein where all other residues remain unmodified. Aspartic acid is the amino acid most prone to isomerize, producing four distinct structures by way of a succinimide intermediate.24 Although isomerization does not result in a change in mass, recently reported methods have enabled isomer detection in targeted proteomics-like experiments.25,26
In the more specific context of AD, isomerization has also received limited attention.27 Aβ extracted from amyloid plaques contains many sites of isomerization.28,29,30 This is perhaps not surprising, as extracellular plaques may persist for years, allowing ample opportunity for spontaneous modifications to occur. Isomerization of Aβ has been examined primarily within the context of its influence on aggregation, although recent results have illustrated that preventing lysosomal degradation may be more injurious.3 In the case of tau, isomerization has only been studied in a handful of reports. Initial work relied on methylation of L-isoAsp by protein isoaspartyl methyl-transferase (PIMT) to quantify isomerization of tau at the protein level.31 These results found greater isomerization in AD samples, but sites of modification and their relative contributions were not identified or quantified. Later work identified some sites of isomerization32 and examined qualitative spatial distributions by imaging select NFTs.33 However, despite the promising nature of these early results, many aspects of tau isomerization and its potential connection with AD remain unexplored.
In this study, we developed a novel analysis protocol and applied it to existing DIA data to explore the extent of isomerization in proteins extracted from human brain samples. We demonstrate that DIA data does contain information about isomerized peptides, and that this information can be easily extracted. Furthermore, our analysis revealed striking differences in the amount of isomerization of tau between AD and control samples. Isomerization of tau can also differentiate sporadic versus autosomal dominant AD, with isomerization being greater in autosomal dominant. To explore potential connections with lysosomal impairment, the data was examined for additional correlations with other autophagic markers, including p62. The results are consistent with decreased autophagic flux as the underlying cause of the observed isomerization, although this explanation cannot be conclusively demonstrated by analysis of the existing dataset. Discussion regarding the possibilities and limitations of DIA data re-analysis and the implications of our findings on future directions in AD research are detailed.
Methods.
Sample Stratification Description.
Brain tissue samples were stratified into 4 groups based on clinical, pathological and genetic data (Table S2A and S2B). Cognitive status was determined as dementia (AD/ADAD) or no dementia (controls) by DSM-IVR criteria; controls were from the Adult Changes in Thought (ACT) study and were included only if the last evaluation was within 2 years of death and the last CASI score was >90 (upper quartile for controls in the ACT cohort). Controls with no or low Alzheimer’s disease neuropathologic change (ADNC) were designated ‘control-low’ and those with intermediate or high ADNC were designated ‘control-high’. All cases with dementia had intermediate or high level ADNC and were classified as AD, and further subclassified as sporadic AD or autosomal Dominant AD (ADAD) when a dominantly inherited mutation in PSEN1 or PSEN2 was found. Sporadic AD cases came from the ACT and the University of Washington (UW) AD Research Center (ADRC), whereas ADAD cases came from the UW ADRC and the Dominantly Inherited Alzheimer Network (DIAN). Cases with Lewy body disease (other than those involving only the amygdala), territorial infarcts, more than 2 cerebral microinfarcts, or hippocampal sclerosis were excluded. Time from death to cryopreservation of tissue, post mortem interval (PMI), was <8 hr in all cases except for those in the ADAD cohort. Details of sample stratification for the 2 brain regions (SMTG and Hippocampus) are provided in Tables S2A and S2B in the supporting information.
Sample Metadata, Batch Design and References.
Each region of human brain tissue was divided into batches of 14 individual samples and 2 pooled references for a total of 16. The first batch of each region was also used to create a region-specific reference pool to be used as a “common reference” and/or single point calibrant, which was homogenized, aliquoted, frozen, and used to compare between batches within a brain region. Human cerebellum and occipital lobe tissue was homogenized, pooled, aliquoted and frozen to be used as a “batch reference” for comparison between batches and other brain regions. Batch design was randomly balanced based on group ratios. For example, batches from the SMTG brain region contained 5 “Sporadic AD”, 4 “Autosomal Dominant AD”, 2 “Control Low Neuropathology”, and 3 “Control High Neuropathology” samples.
Lysis/Digestion.
Two 25 μm cryo slices (“curls”) of brain tissue were resuspended in 120 μl of lysis buffer of 5% SDS, 50mM Triethylammonium bicarbonate (TEAB), 2mM MgCl2, 1X HALT phosphatase and protease inhibitors, vortexed, and briefly sonicated at setting 3 for 10 seconds with a Fisher sonic dismembrator model 100. A microtube was loaded with 30 μl of lysate and capped with a micropestle for homogenization with a Barocycler 2320EXT (Pressure Biosciences Inc.) for a total of 20 minutes at 35°C with 30 cycles of 20 seconds at 45,000 psi followed by 10 seconds at atmospheric pressure. Protein concentration was measured with a BCA assay. Homogenate of 50 μg was added to a process control of 800 ng of yeast enolase protein (Sigma) which was then reduced with 20 mM DTT and alkylated with 40 mM IAA. Lysates were then prepared for S-trap column (Protifi) binding by the addition of 1.2% phosphoric acid and 350 μl of binding buffer (90% Methanol, 100 mM TEAB). The acidified lysate was bound to column incrementally, followed by 3 wash steps with binding buffer to remove SDS and 3 wash steps with 50:50 methanol:chloroform to remove lipids and a final wash step with binding buffer. Trypsin (1:10) in 50mM TEAB was then added to the S-trap column for digestion at 47°C for one hour. Hydrophilic peptides were then eluted with 50 mM TEAB and hydrophobic peptides were eluted with a solution of 50% acetonitrile in 0.2% formic acid. Elutions were pooled, speed vacuumed and resuspended in 0.1% formic acid.
Injection of samples consisted of one μg of total protein (16 ng of enolase process control) and 150 fmol of a heavy labeled Peptide Retention Time Calibrant (PRTC) mixture (Pierce). The PRTC was used as a peptide process control. Library pools contained an equivalent amount of every sample (including references) in the batch. For example, a batch library pool consisted of the 14 samples from the batch and two references. System suitability (QC) injections comprised 150 fmol of PRTC and BSA.
Liquid Chromatography and Mass Spectrometry.
One μg of each sample with 150 femtomole of PRTC was loaded onto a 30 cm fused silica picofrit (New Objective) 75 μm column and 3.5 cm 150 μm fused silica Kasil1 (PQ Corporation) frit trap loaded with 3 μm Reprosil-Pur C18 (Dr. Maisch) reverse-phase resin analyzed with a Thermo Easy-nLC 1200. The PRTC mixture was used to assess system suitability before and during analysis. Four of these system suitability runs were analyzed prior to any sample analysis and then after every six sample runs another system suitability run was analyzed. The 110-minute sample LC gradient consists of a 2 to 7% for 1 minutes, 7 to 14% B in 35 minutes, 14 to 40% B in 55 minutes, 40 to 60% B in 5 minutes, 60 to 98% B in 5 minutes, followed by a 9 minute wash and a 30 minute column equilibration. Peptides were eluted from the column with a 50°C heated source (CorSolutions) and electrosprayed into a Thermo Orbitrap Fusion Lumos Mass Spectrometer with the application of a distal 3 kV spray voltage. For the sample digest, first a chromatogram library of 6 independent injections was analyzed from a pool of all samples within a batch. For each injection a cycle, one 120,000 resolution full-scan mass spectrum was acquired with a mass range of 100 m/z (400–500 m/z, 500–600 m/z…900–1000 m/z) followed by data-independent MS/MS spectra on the loop count of 26 at 30,000 resolution, AGC target of 4e5, 60 sec maximum injection time, 33% normalized collision energy with a 4 m/z overlapping isolation window.34,35,36 The chromatogram library data was used to quantify proteins from individual sample runs. These individual runs consisted of a cycle of one 120,000 resolution full-scan mass spectrum with a mass range of 350–2000 m/z, AGC target of 4e5, 100 ms maximum injection time followed by a data-independent MS/MS spectra on the loop count of 76 at 15,000 resolution, AGC target of 4e5, 20 sec maximum injection time, 33% normalized collision energy with an overlapping 8 m/z isolation window. Application of the mass spectrometer and LC solvent gradients were controlled by the ThermoFisher XCalibur (version 3.1.2412.24) data system.
Data Analysis.
Thermo RAW files were converted to mzML format using Proteowizard (version 3.0.20064) using vendor peak picking and demultiplexing.37 Chromatogram spectral libraries were created using default settings (10 ppm tolerances, trypsin digestion, HCD b- and y-ions) of EncyclopeDIA (version 0.9.5) using a Prosit predicted spectra library based the Uniprot human canonical FASTA.38 Quantitative spectral libraries were created by mapping spectra to the chromatogram spectral library using EncyclopeDIA39,40,41 requiring a minimum of 3 quantitative ions and filtering peptides at a 1% FDR using Percolator 3.01.42,43 The quantitative spectral library is imported into Skyline (daily version 20.1.1.83) with the human Uniprot FASTA as the background proteome to map peptides to proteins.44,45 Transition retention time filtering settings set to “use only scans within 4 minutes of MS/MS IDs” for all batches and then removed all peptides/proteins from Skyline document except APP (Uniprot Accession P05067) and Tau (Uniprot Accession P10636).
Automated identification of potential isomers.
Raw files collected with 4-m/z staggered isolation windows were demultiplexed and converted to mzMLs using MSConvert (Proteowizard, v 3.0.20239).46 The resulting mzMLs were searched in Crux Tide (v 3.2) and the top 10,000 precursors per spectrum were reported so that the results file contained every possible spectrum/ peptide combination.47 To limit search size, two searches were performed. The first search covered the entire tryptic human proteome (UP000005640) with no allowed missed cleavages or modifications. The second search covered a small subset of relevant proteins allowing semi-tryptic peptides with up to two missed cleavages, N-terminal acetylation, and methionine oxidation. All peptides and spectra from each 2-m/z isolation window were converted into matrices with Tailor calibrated XCorr as the values where each row is a potential peptide and each column is a spectrum.48,49 All candidate peptides were filtered so they had at least one score above a given noise threshold, here we used a cutoff of 1.15 for the full proteome search and 1.1 for the smaller semi-tryptic search. Scores were baseline adjusted with simple subtraction so that the minimum score for each peptide was 0. Peaks were defined as multiple consecutive spectra scoring above 80% of the maximum baseline-subtracted score. The remaining candidates were filtered for peptides with multiple peaks separated by 4 or more scans below the 80% threshold. With the remaining list of peaks, overlapping peaks that were generated by two different peptides were identified. In each of these cases, angle cosine was calculated between the lower scoring of the two peptides and 1000 random decoys as well as the lower scoring and higher scoring peptide. If the angle cosine between the two candidate peptides was greater than the maximum angle cosine between the decoys, it is likely that these peaks were caused by the same sets of ions. Therefore, the lower scoring of the two peaks was removed and the process was repeated for all identified peaks. All peptides with two or more peaks remaining after this process were identified as likely isomers.
Quantification of isomerization and protein biomarkers.
Isomer quantification was determined using peak area values extracted from Skyline (Skyline-daily 20.9.234). To calculate peak areas, the boundaries for each individual isomer peak were adjusted manually to account for differences in retention times between chromatograms. Within this window, the total area of both the precursor and fragment peaks corresponding the peptide of interest were then summed to produce a “total isomer area” value for each individual isomer. Fragments or precursors not found in all datasets were not used to calculate area values to prevent skewing the results with marginally detectable ions. Using total isomer area values, the “percent isomerization” was calculated by dividing the sum of isomer peak areas by the total peak area (where the canonical form was assumed to be dominant), see eq. 1. The ionization efficiencies of isomeric peptides are expected to be very similar due to identical size, functional group composition, and similar structure. Furthermore, any differences in ionization would be reproduced between sample sets, meaning that comparisons of differences between samples will accurately reflect differences in the amounts of isomers present. Means, two-tailed p-values, and correlations were calculated in Microsoft Excel.
| Eq. 1. |
Protein quantification was determined for each sample by using the sum of all detected peptide areas assigned within the chromatogram by Skyline (eq. 2). The reported values therefore represent relative abundance normalized to the total ion count. Peptides not found in all datasets were not used to calculate protein area values. It was assumed that differences in fragment ions between datasets would not be significant, due to dilution across the large number of discovered peptides for each protein. Means, two-tailed p-values, correlations, and z-score conversions were calculated in Excel.
| Eq. 2. |
Searching for and quantifying deamidation.
The extent of deamidation in tau was determined by reanalysis of the mzML data created for isomer analysis (see data analysis). Chromatogram spectral libraries were built using DIA-Umpire under DIA workflow settings with asparagine and glutamine deamidation modifications included in the search. Ambiguous matches were included so peptides with multiple possible deamidation sites would be discovered despite sharing a precursor mass. MS1 and MS/MS filtering were set to 20ppm mass accuracy and retention time filtering was set to include scans within 5 minutes of MS/MS IDs. To automatically train the mProphet model, a FASTA file including Tau, amyloid beta precursor protein, and all other isomerized proteins (from Table S1) was imported. Decoys were generated by shuffling sequences with one decoy per target. MS Amanda search settings used an MS1 tolerance of 5ppm and an MS/MS tolerance of 10ppm. Discovered deamidation sites were considered real when the associated chromatogram peak was clear, included a variety of precursor and fragment ions, and had a retention time shift relative to the non-deamidated form. Oftentimes there were several deamidated peaks, which is to be expected considering the asparagine deamidation mechanism involves a succinimide ring, and therefore facilitates isomerization.24 Because only relative deamidation is of interest, for each deamidated peptide a single peak was selected across all samples, then its component ions were summed to yield a total peak area. This value was compared across all samples. Means, two-tailed p-values, correlations, and z-score conversions were calculated in Excel.
Peptide synthesis and isomer verification.
Peptide standards for each isomeric form were synthesized following a standard solid phase peptide synthesis protocol.50 The synthesized standards were then lyophilized to dryness, dissolved in 1mL 50:50 ACN:H2O, and frozen in a -20C freezer. Sequence was confirmed by electrospraying 5uM peptide in H2O with 0.1% formic acid to verify exact mass and fragmentation patterns consistent with the desired peptide. An isomer mixture was then run through a setup equivalent to that used for all DIA data collection.
Results.
DIA isomer analysis.
It has been previously established that some peptide isomers/epimers can easily be separated with traditional chromatographic methods used in proteomics experiments.25 Therefore, DIA data should contain instances where the same peptide appears to elute at multiple retention times, with the same precursor and fragment masses in both peaks. Typical DIA analysis is engineered to select the highest scoring peak and ignore any others in such instances, which means that presence of isomers would not typically be detected. To overcome this limitation, we created a protocol to intentionally scan the data for instances where two or more peaks eluted at different chromatographic retention times yet corresponded to the same peptide sequence based on high confidence matches of both precursor mass and numerous fragment ions. An example of such data is illustrated for the peptide WDGQETTLVR in Fig. 1a. To eliminate false positives from partial matches of fragment ions that derived from nonequivalent precursors (see for example, the data in Fig. 1b), the results for each peak were ranked against all potential precursors coeluting across both peaks. For non-isomeric peaks, this yields a higher score for different precursors for each peak whereas isomers yield similar scores for both peaks. Applying this method to our brain dataset, we found 9 peptides from unique proteins meeting these criteria, which were assigned as potential sites of spontaneous isomerization, see Table S1. Peptides that exist in more than two isomeric forms are also possible and can be identified by our method. An example is illustrated in Fig. 1c for the peptide 386TDHGAEIVYK395 from tau.
Figure 1.
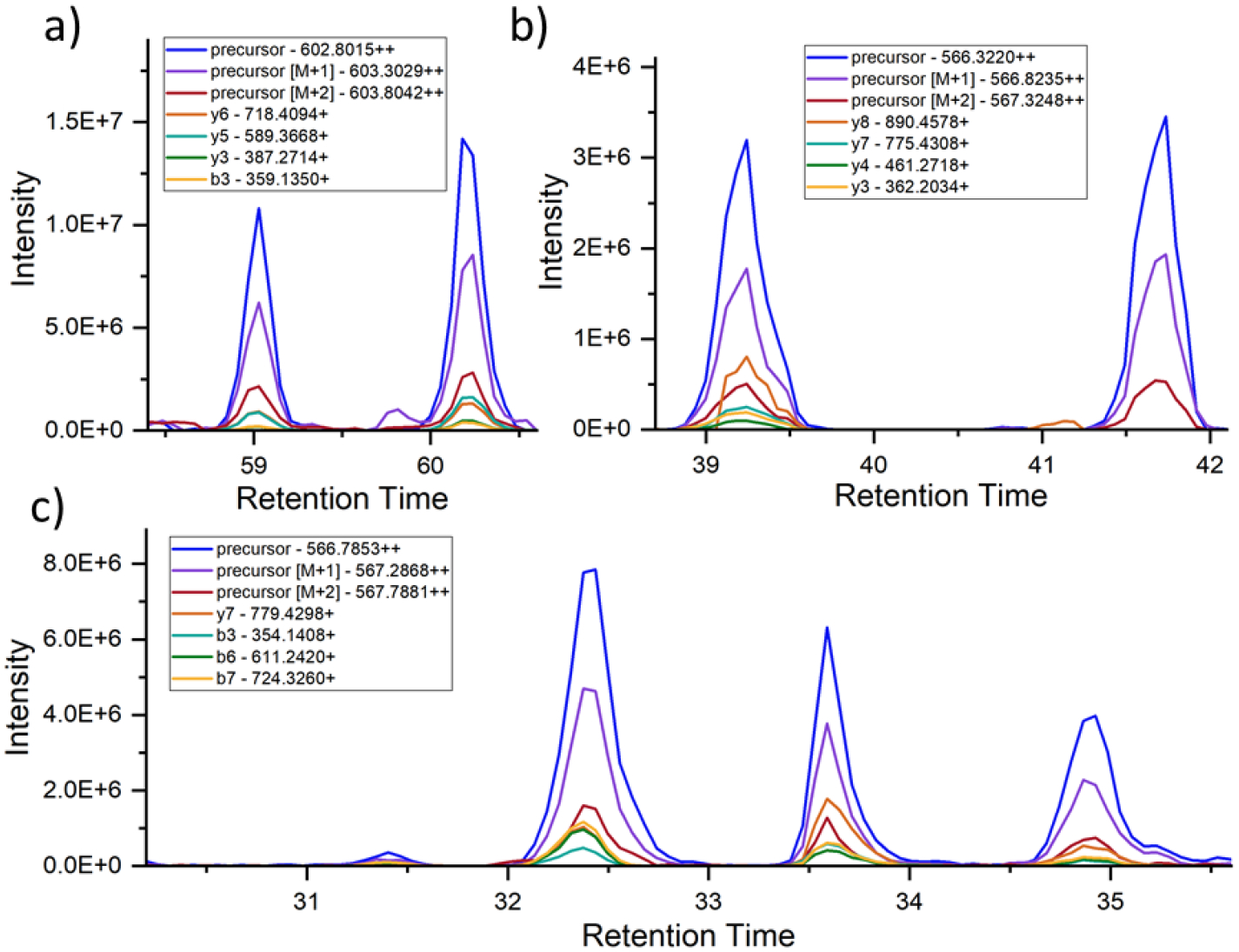
Skyline chromatograms for several peptides. The blue, purple, and red lines typically highest in intensity represent the major precursor isotopes. The other lines correspond to various fragment ions associated with the precursor in the spectral library. a) Fatty Acid Binding Protein, Heart peptide WDGQETTLVR [97–106]. There are two distinct peptide peaks in the chromatogram that are identical in their precursor and fragment ions but elute separately. This indicates two isomeric forms of the same sequence. Note that Skyline does not typically recognize the lower-intensity peptide peak. b) Microtubule-associated protein tau peptide KLDLSNVQSK [281–290]. Although two distinct peaks appear, the lack of fragment ion peaks in the later-eluting peak indicates a peptide with a similar precursor mass but dissimilar sequence. c) Tau peptide TDHGAEIVYK [386–395]. An example where multiple peaks share precursor and fragment ions in common, suggesting the existence of multiple isomers.
Alternative explanations for the existence of such data were also considered but deemed unlikely. For example, for different retention times to result from switching the order of two residues by genetic mutation, the same mutation would have to be present in all of the different individuals who were sampled. Furthermore, although the precursor mass would be unchanged in such a circumstance, some fraction of the fragment ions would be able to distinguish such isomers. The peptides do not contain mass-shifting post-translational modifications (PTMs), so the results are not due to alternating PTM locations. Finally, exchange of Leu/IIe or vice versa is not possible in most cases as the peptides do not contain these residues. Therefore, the peptides eluting at multiple times most likely represent products of spontaneous isomerization. The degree of isomerization for each peptide can be calculated by comparing the relative abundances of the separate peaks, with the assumption that the most abundant peak (particularly in samples where one form is dominant) is the native form.
Isomerization of Tau.
As shown for a single sample in Fig. 1, tau is one of the isomer-containing proteins identified from our search. The same protocol was repeated with identical amounts of total protein extracted from brain samples of 66 individuals. In Fig. 2a, the percent isomerization for the peptide 386TDHGAEIVYK395 is shown for 21 samples for sporadic AD and 2 samples for autosomal dominant AD (or ADAD) extracted from the hippocampus. The results are sorted by disease status and age, which is provided in the x-axis label. Although the percent isomerization values vary, it is clear that a considerable fraction of tau is isomerized in most AD/ADAD samples. In contrast, the % isomerization found in 22 hippocampal control samples was minimal (Fig. 2b). The AD and controls are easily distinguished by statistical analysis (Fig. 2c), with averages of 18.3% and 2.4%, respectively. Note, the controls constitute two groups, control-high (12 samples) and control-low (10 samples), where control-high brains exhibited pathological features typically associated with AD but the person remained cognitively normal (additional detail is provided in the Discussion and Supporting Information). Differences are more notable for samples extracted from the superior and middle temporal gyri (SMTG). In addition, the SMTG cohort contains 23 ADAD samples and 19 sporadic AD samples, allowing for statistical analysis of both cohorts. Most AD/ADAD samples exhibit some level of isomerization (Fig. 2d) while control samples (11 high, 9 low) are nearly devoid of isomerization (Fig. 2e). The magnitudes of the differences in the SMTG are also greater, yielding averages of 25.6%, 9.94%, and 0.571% isomerization for ADAD, sporadic AD, and control, respectively. The degree of isomerization in ADAD samples is distinguishable from both sporadic AD and controls (Fig. 2f). Notably, the majority of AD/ADAD samples exhibiting <5% isomerization in both the hippocampal and SMTG datasets are found in individuals over the age of 75 within the sporadic AD cohort. The degree of isomerization for the hippocampus and SMTG samples extracted from the same brains show strong correlation (Pearson’s 0.82). In contrast, other proteins identified in Table S1 do not exhibit different degrees of isomerization between AD and control groups (see Fig. S2 for an illustrative example). To place the results in the larger context, if compared with p-values based on protein abundances between AD/ADAD and control samples, the % isomerization ranks 4th lowest among 6050 p-values.
Figure 2.
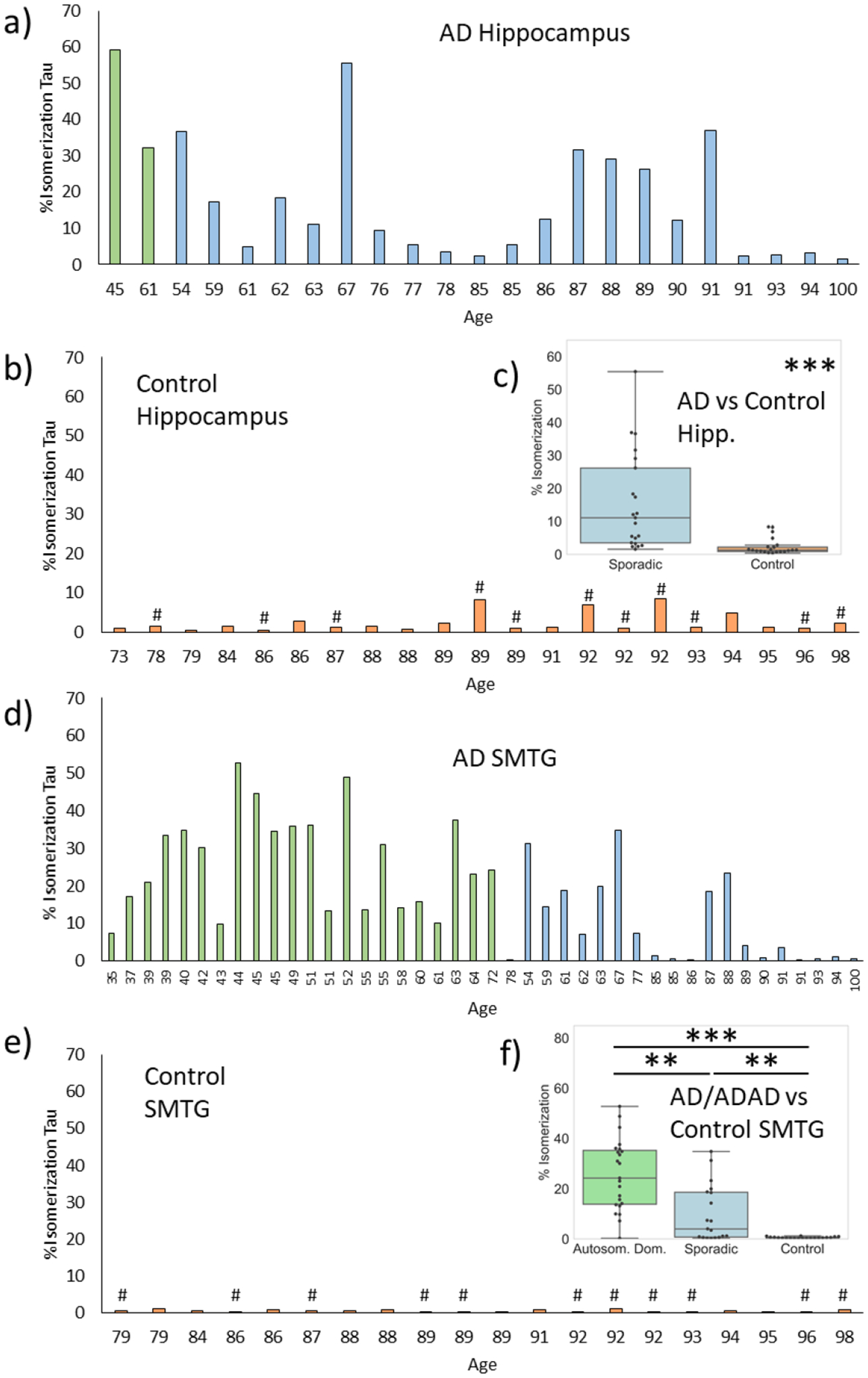
% isomerization of Asp387 in tau for a) AD (blue) and autosomal dominant AD (ADAD, green) samples, and b) control samples (orange) extracted from hippocampus. c) Comparison of %isomerization in AD versus control (note: ADAD values are not included). d)-f) analogous plots for superior and middle temporal gyri (SMTG). # indicates control-high. P-values are indicated by: * <0.05, ** <0.01, ***, <0.001.
To confirm the identity of isomers in the tissue samples, we collected LC-MS/MS data for a set of known synthetic tau isomer standards. Comparison of relative elution times confirms that 386TDHGAEIVYK395 from tau is isomerized at Asp387, in agreement with known amino acid propensities for isomerization and previous examination of tau, see Fig. S3. Synthetic standards also confirm the initial assignment of the native L-Asp form as the most abundant peak, followed by L-isoAsp. This observation is in agreement with previous results showing that L-isoAsp is the primary product of Asp isomerization.51
Autophagy and Repair.
In addition to our analysis of isomerization, we also examined differences in the abundance of proteins connected to autophagy using traditional DIA methodology. Seven (MPCP, MLP3A(LC3), Hsc70, Mtp70, p62, progranulin, and mTOR) exhibited some differences between AD and control groups, and six could not be distinguished between AD and control groups (Lamp2, CatB, Rack1, Hsp70, BiP, and 28S-RP-S27). Most strikingly, the chaperone p62 (a known marker for autophagy),52 shares the closest relationship with isomerization of tau, see Fig. 3. The relative abundance of p62 (represented by a z-score) correlates with the degree of isomerization of tau in both the hippocampus and SMTG AD/ADAD samples, yielding Pearson correlations of 0.83 for the hippocampus and 0.82 for the SMTG. Among all 6049 proteins examined, p62 shows the highest correlation with % isomerization for AD/ADAD samples. In contrast, p62 levels do not correlate with isomerization in controls. The total amount of p62 is also higher in AD/ADAD samples, particularly in the SMTG (Fig. 3c). The levels of several other proteins connected with autophagy (MPCP, MLP3A, and Hsc70) were also evaluated and found to be reduced by modest amounts in AD (Fig. 3c).
Figure 3.
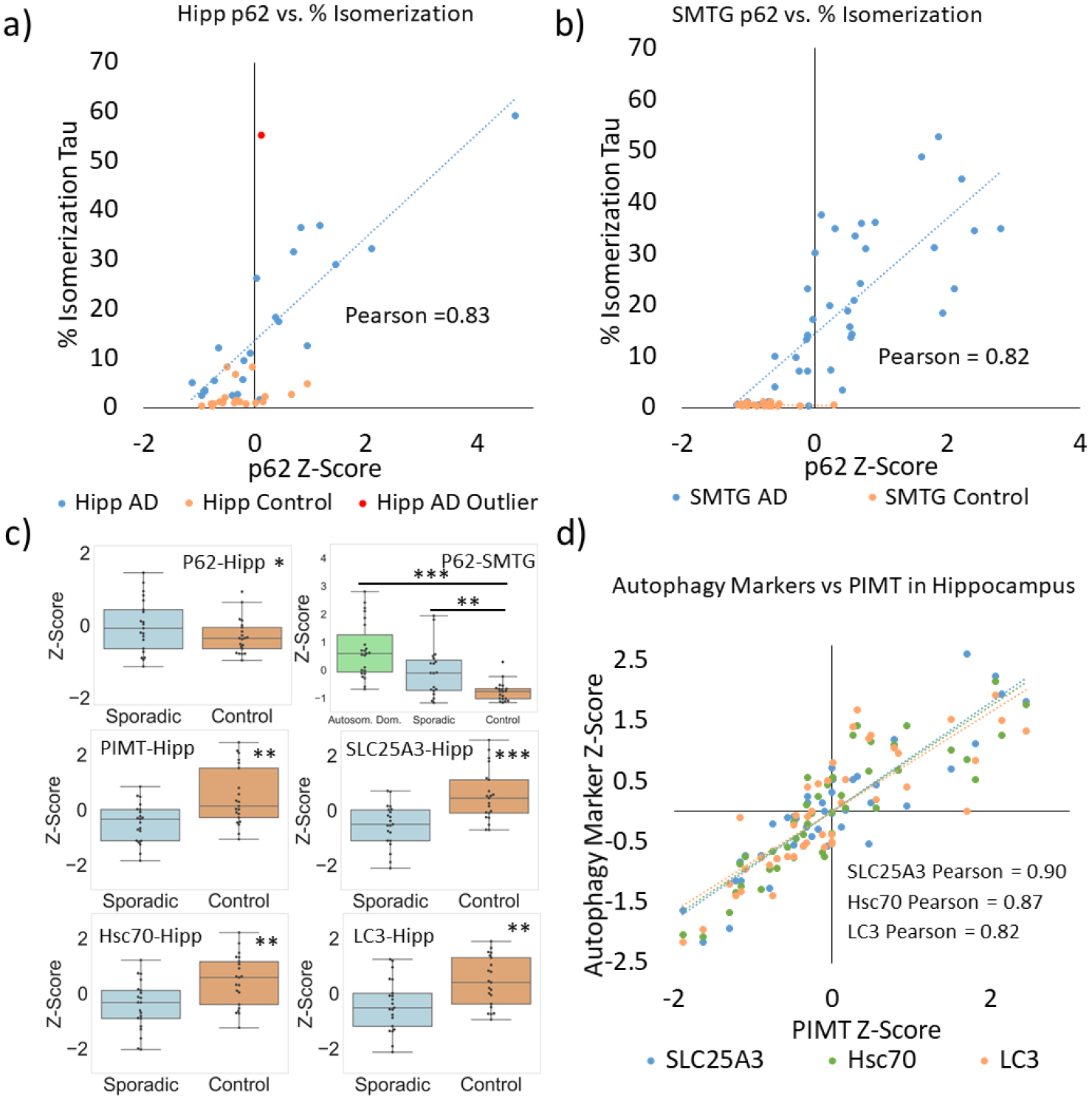
Correlation of tau %isomerization with p62 levels in the a) hippocampus and b) SMTG. c) Statistical comparisons of protein levels present in AD and control samples. Hippocampal samples do not include ADAD results. d) Correlation between various autophagy markers and PIMT levels.
Protein isoaspartyl methyl transferase (PIMT) is a repair enzyme that partially restores L-isoAsp to L-Asp. PIMT was detected in our dataset, and quantitative analysis revealed lower amounts of PIMT in hippocampal AD samples (Fig. 3c). Correlation between PIMT levels and isomerization of tau in the hippocampal dataset was low (Pearson’s −0.10) and modest for the SMTG (−0.41). Interestingly, the levels of MPCP, MLP3A, and Hsc70 correlate more strongly with PIMT levels in the hippocampus (Fig. 3d), with Pearson values above 0.80.
Deamidation in Tau.
Similar to isomerization, deamidation is largely an SCM that should therefore increase as a function of molecular lifetime. To establish the relationship between deamidation and isomerization in Tau, the extent of deamidation of Asn was measured for VQIINKK, LDLSNVQSK, and LTFRENAK in the SMTG samples. The results are shown in Fig. 4. Deamidation of all three peptides correlates strongly with isomerization of TDHGAEIVYK (Pearson values 0.88 VQIINKK, 0.78 LDLSNVQSK, and 0.86 LTFRENAK). Statistical comparison of deamidation between cohorts (Fig. 4) also shows a similar pattern to that observed for isomerization of TDHGAEIVYK. Differences in the extent of deamidation observed for each peptide likely originate from variations in the underlying rates of deamidation.
Figure 4.
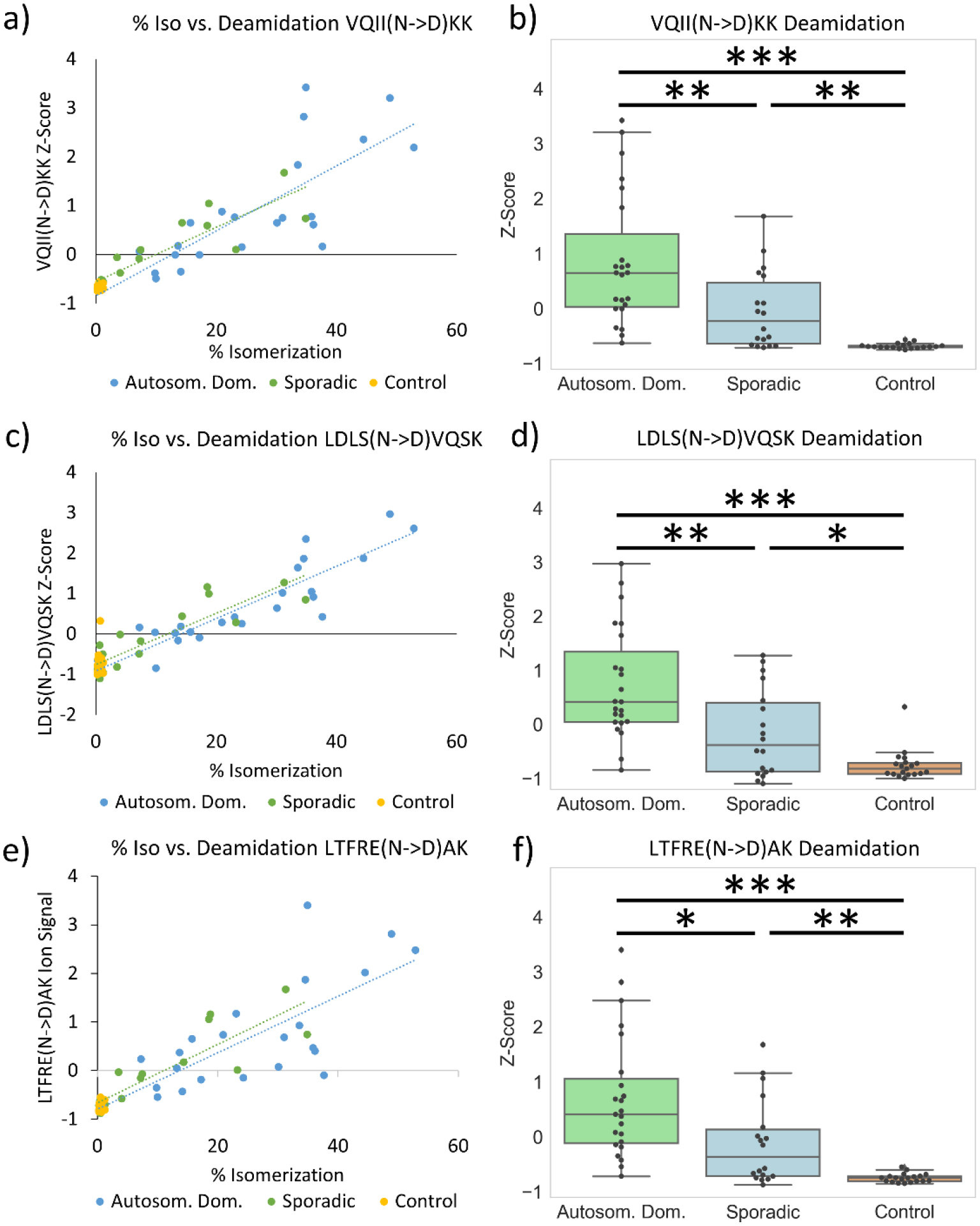
Degree of Asn deamidation in the SMTG for three peptides from Tau, a) VQIINKK, c) LDLSNVQSK, e) LTFRENAK. Strong correlation is observed between Asn deamidation and Asp isomerization in TDHGAEIVYK. b), d), f) Statistical analysis of deamidation extent for each cohort. In LTFRE(N->D)AK the +1 Da and +2 Da isotope precursor peaks are excluded due to overlapping precursor interference.
Isomerization of Aβ.
The % isomerization for HDSGYEVHHQK from Aβ is shown in Fig. 5. Isomerization of Aβ was not detected by our automated search because the isomeric forms are not baseline separated. However, given that Aβ isomerization has been documented previously and is a molecule of interest for AD, we examined Aβ in detail and found partial separation of isomeric forms. In addition to complicating identification, partial separation also interferes with quantitation because the amount of Aβ is not consistent between control and AD samples (see Fig. S4). At low intensity, distinguishing partially separated peaks is not possible. This issue is particularly problematic for control-low samples where HDSGYEVHHQK is essentially not detected. Within the confines of samples where HDSGYEVHHQK was detected and produced clear separation of isomers, the % isomerization in the hippocampus did not vary between AD and control, see Fig. 5a and 5b. However, in the SMTG samples, the % isomerization was greater in AD samples (means of 56.6%, 53.7%, and 44.4% for ADAD, sporadic AD, and control, respectively), although the difference was less than that observed for either isomerization or deamidation of tau.
Figure 5.
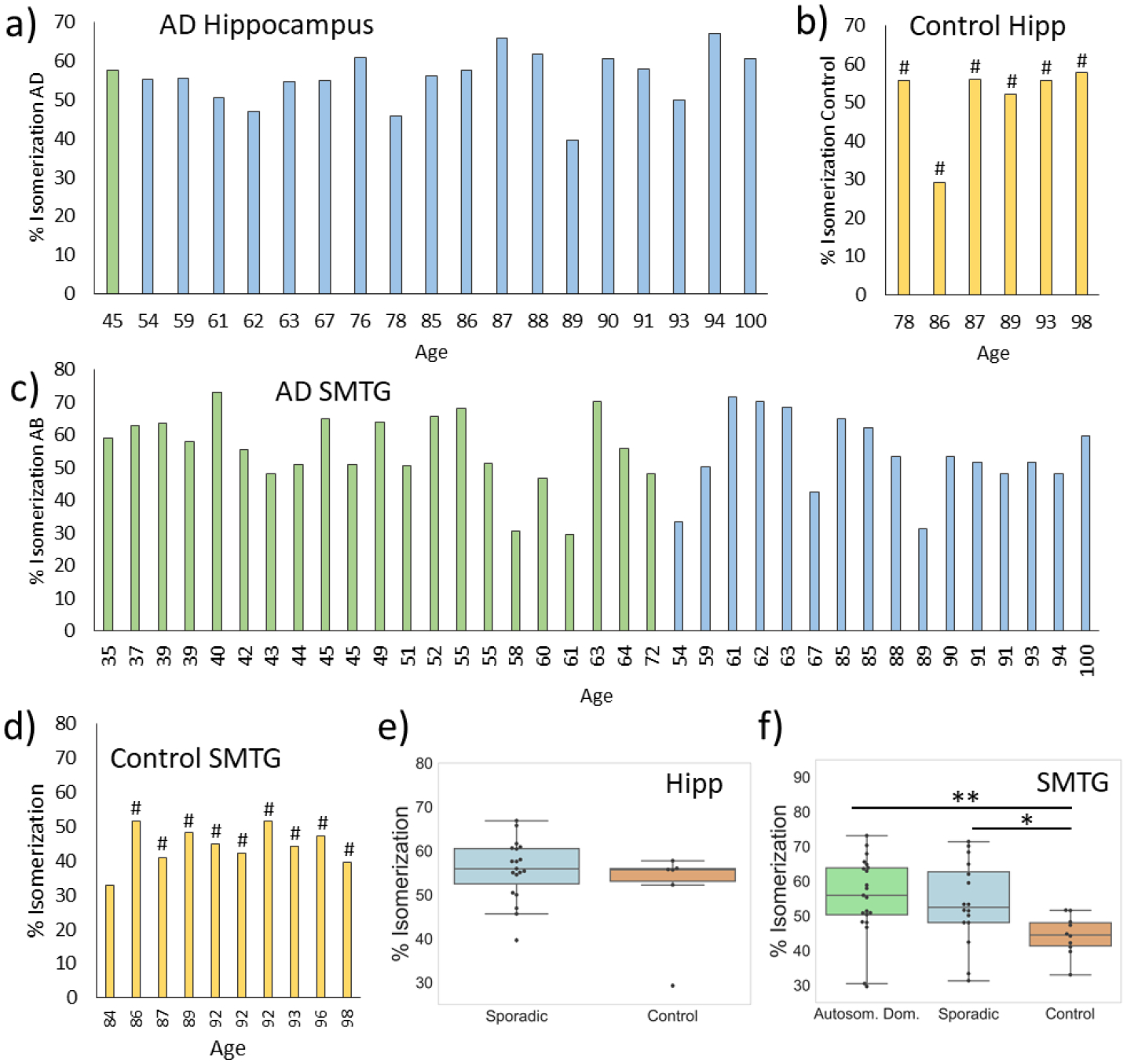
Percent isomerization of Aβ for AD (blue), ADAD (green), and control samples (orange) from the regions indicated (a-d). Statistical analysis of the data, revealing no difference in the hippocampus and minor difference in the SMTG (e-f). # indicates control-high. P-values are indicated by: * <0.05, ** <0.01, ***, <0.001.
Discussion.
It is clear that new information can be extracted from reanalysis of DIA data, as illustrated by the results in Fig. 2. Although isomerization of tau had been reported for this peptide previously in experiments on very limited samples,32 DIA enabled large-scale examination of many samples, including different cohorts, while enabling quantitative comparison between them. The results in Fig. 2 clearly illustrate that the % isomerization of tau is a strong indicator of AD/ADAD status. In the hippocampus samples, tau isomerization is 8x greater in AD relative to controls. In the SMTG dataset, tau isomerization is 45x greater in ADAD and 17x greater in sporadic AD relative to controls. One important difference between the hippocampus and SMTG results is the near absence of isomerization in SMTG control samples. Although the overall trend is the same in both the hippocampus and SMTG, it is clear that different regions of the brain do not behave identically with regard to tau isomerization. Importantly, there are two separate control groups in our analysis (denoted as control-high and control-low). The control-low group consists of individuals with limited observable pathology (B scores of 2 or lower and C scores of 0) who scored >90 on the CASI (Cognitive Abilities Screening Instrument) test within two years of death. In contrast, the control-high group is unusual and constitutes individuals with B scores of 2/3 and C scores of 2/3, yet also scored >90 on CASI within two years of death. In other words, in terms of B and C scores, the control-high group is very similar to the AD and ADAD cohorts, yet they did not exhibit signs of dementia within two years of death. Strikingly, isomerization of tau is not abundant in either control group (or distinguishable between control-low and control-high), meaning that isomerization can clearly distinguish control-high from AD/ADAD where B/C scores cannot. This suggests that tau isomerization occurs independently from aberrant protein aggregation, yet it is related in some fashion to the processes leading to cognitive and functional impairment.
Isomerization of amino acid residues in proteins is an SCM that requires no enzymatic intervention to proceed.53 The isomerization we observe in tau occurs primarily at Asp387, consistent with previous studies showing that Asp is the most readily isomerized amino acid in long-lived proteins,24,25,28 and that DH motifs are prone to isomerize.51 Because the process is spontaneous, for significant amounts of isomerization to accumulate in the brain, protein turnover must be sufficiently slow to allow isomerization to occur. The timeframe required to allow the observed extent of isomerization at Asp387 is not known, but in vitro experiments suggest a timeframe between months and years.54 Furthermore, the measured rates of Asp isomerization in other model peptides were found to proceed at rates typically less than ~1% per day.3 It is also important to recognize that Asp isomerization will trend towards an equilibrium, where all four potential isomers will be populated to some extent. Based on distributions observed previously in synthetic model peptides, the percent isomerization attained for Asp at equilibrium should be approximately 80%.51 This value is largely driven by the observation that L/D-isoAsp isomers are typically favored by ~3:1 over the L/D-Asp isomers.51 Although some values over 60% isomerization are observed in our dataset, none achieve the equilibrium value, indicating that tau isomerization on average was incomplete for all individuals that we examined.
The quantitative differences in isomerization between AD, ADAD, and controls suggests that some underlying biological process or processes likely differ between these groups. Again, the unbiased data collection approach inherent with DIA is advantageous, as it allows the testing of possible hypotheses. Due to the previously established connections between lysosomal malfunction and isomerization, we examined proteins associated with autophagy in each cohort. A variety of autophagy-related proteins are present in lower amounts in the AD cohorts, see Fig. 3c.55,56,57 Although the differences are statistically significant, the magnitudes of the changes are not particularly large. However, we did discover greater differences in one autophagic marker, p62, which is known to accumulate when autophagy is impaired.52 The amount of p62 is greater in AD by a factor of 1.6x in the hippocampus and 2.2x in the SMTG. Although previous studies of the frontal cortex using Western blot found lower levels of p62 in AD brains relative to controls, those results may represent levels of cytosolic p62.58 Consistent with this possibility, other studies have demonstrated strong co-localization of p62 within tau inclusions.59 In our studies, we found strong correlation between the amount of p62 and degree of isomerization in samples for both the hippocampal and SMTG datasets (Fig. 3a and 3b), although isomerization more clearly distinguished AD/ADAD samples from controls. The buildup of p62 suggests that isomerization is greater in AD/ADAD due to slowed autophagic flux, which is known to cause p62 accumulation.60 Since Asp isomerization is an SCM, longer protein lifetime due to reduced autophagic flux would be expected to increase the degree of isomerization. Furthermore, soluble tau is intrinsically disordered,61 which is known to facilitate isomerization of Asp residues in general. Although there are known limitations to the use of p62 to monitor autophagic flux,62 our findings also support other connections that have been reported between autophagy, p62, and AD.63,64 The strong correlation between isomerization of TDHGAEIVYK and deamidation in several peptides from tau, as illustrated in Fig. 4, also supports the possibility for reduced autophagic flux in ADAD and sporadic AD cohorts because the underlying chemistries that lead to deamidation and isomerization are closely related.
It has been suggested previously that tau isomerization may simply be a consequence of the long half-life of tau present in neurofibrillary tangles (NFTs),32,33 but our data is not consistent with this interpretation. For example, isomerization is nearly absent in the control-high group and some AD samples despite the presence of abundant NFTs in both. Furthermore, comparison of tau isomerization vs age within the hippocampal sporadic AD cohort revealed poor correlation (Pearson −0.35, note the trend is downward with age). To be consistent with isomerization representing NFT half-life, this would require NFTs in older people to have been present for less time. Furthermore, continued isomerization may not be possible for tau trapped in NFTs, in which case NFT age would not correlate with degree of isomerization. Indeed, structures for the fibrils constituting neurofibrillary tangles have recently been determined, and Asp387 lies just outside the highly ordered domain in a partially ordered region.65 It is likely difficult to accommodate succinimide formation (Scheme 1) in such close proximity to densely packed fibrils because the structural flexibility required for proper backbone reorientation should be hindered. Furthermore, recent results have demonstrated that tau exchanges dynamically in and out of NFTs.66 Collectively, these observations suggest that the degree of isomerization in NFTs may simply reflect the amount of tau isomerization already present at the time of incorporation into the NFT. This idea offers the most straightforward explanation for the control-high samples and select AD samples that exhibited low isomerization despite the extensive presence of NFTs. Alternatively, the structures of the NFTs could theoretically differ between these groups, allowing for isomerization in one case and not in the other. If this is the underlying cause, the relevant structural differences in the NFTs have thus far eluded detection but isomerization could be used to identify and separate samples for further investigation. Finally, isomerization represents another difference between the behavior of tau/NFTs and Aβ/amyloid plaque. Aβ is extensively isomerized in the AD brain.67 However, isomerization of Aβ is known to continue in the fibril state,3 and the total degree of isomerization therefore reflects a combination of the initial and subsequent modification from before and after incorporation into fibrils. This may account for the reduced difference in isomerization of Aβ observed within our cohorts (see Fig. 5).
To ascertain the extent to which other important factors might influence the degree of tau isomerization, we examined the amount of PIMT in each sample. PIMT can repair some products of Asp isomerization by methylation, which occurs primarily at L-isoAsp.68,69 Comparison between AD and controls reveals that the levels of PIMT are lower in AD samples, although the effect is modest and unlikely to fully account for the observed differences in isomerization. In addition, there is weak correlation between PIMT levels and isomerization of tau, further suggesting that PIMT is not a dominant controlling factor. However, levels of some of the autophagic markers in Fig. 3 do show strong correlation with the level of PIMT (Fig. 3d), suggesting that expression of PIMT is connected with (or perhaps regulated by) autophagy.
ADAD and autophagy.
The disease-causing mutations within our ADAD cohort occur exclusively at PSEN1 and PSEN2. The associated proteins, presenilin 1 (PS1) and presenilin 2 (PS2), function as components of γ-secretase complex, which is responsible for enzymatic cleavage of amyloid precursor protein.70 Although mutant presenilins are often discussed in the context of Aβ cleavage,71 the same modifications can also impact autophagy.72,73 For example, mutant presenilins are associated with increased lysosomal malfunction that may occur by disruption of lysosomal acidification or autophagosome formation.74,75 In other cases, evidence implicates disruption of calcium homeostasis as the source of lysosomal impairment.76,77 Regardless of the precise underlying cause, there is a clear potential for disrupted autophagy resulting from the mutations in PSEN1 and PSEN2. Our data reveals that tau isomerization is greatest in ADAD (Fig. 2f), suggesting that autophagic flux is reduced most in ADAD. The lesser (yet still greater than control) degree of isomerization in sporadic AD suggests a more subtle diminution of autophagic flux. Consistent with this thought, numerous studies have shown that autophagy declines with age in general.78,79 Furthermore, PS1 expression naturally declines with increasing age.73 Decreased autophagic flux is therefore consistent with age being the greatest risk factor in AD.
Conclusion.
This study highlights some of the strengths and weaknesses of retrospective DIA data analysis. Clearly, it is possible to identify new meaningful information, but this may be most likely in situations similar to that demonstrated here, i.e. where the target falls outside the traditional proteomics envelope. Furthermore, interesting results obtained by retrospective analysis are unlikely to match the expectations associated with a purpose-built study, but rather may point towards interesting directions for future investigation. For example, our observations identified some dramatic differences in the isomerization of tau when AD, ADAD, and two unique control groups were examined. Subsequent re-examination of the data also revealed that deamidation likewise differs between these groups. Consideration of other information from proteins related to autophagy also points towards a connection between autophagic flux and the isomerization of tau, but the constraints imposed by examination of an existing dataset impede conclusive confirmation of this hypothesis. On the other hand, once new targets (such as isomerization) have been identified, analysis of other existing DIA datasets is straightforward and could reveal other interesting, unanticipated results. For example, one of the more striking observations highlighted in this study is that isomerization can easily distinguish between AD and control-high groups even though both exhibit substantial aberrant protein aggregation. This observation can be rationalized if protein aggregation occurs independently of and more rapidly than isomerization, in which case aggregation would indicate one level of slow-down in autophagic flux while isomerization would point towards greater (and perhaps more pathological) slowdown. Additional research will be required to investigate these possibilities. In summary, existing DIA data may be an excellent resource for obtaining ‘preliminary results’ to guide future studies.
Supplementary Material
Figure S1. Isomerization compared in hippocampus and SMTG of samples from the same brain.
Figure S2. Quantification of isomerization in peptide WDGQETTLVR from fatty acid binding protein, heart
Figure S3. Chromatograms of synthesized TDHGAEIVYK isomers compared to a Skyline chromatogram from SMTG containing all isomers
Figure S4. Quantification and chromatogram from isomerized amyloid beta peptide HDSGYEVHHQK
Table S1. List of proteins found to contain isomerization via automated search
Table S2. Brain Tissue Stratification Description for SMTG
Table S3. Brain Tissue Stratification Description for Hippocampus
Acknowledgements.
The authors gratefully acknowledge funding from the NIH (R01 AG066626 for RRJ, RF1 AG053959 for MM and TJM, U01 AG006781 (ACT study) and P30 AG066509 (UW ADRC), and the Nancy and Buster Alvord Endowment (CDK), P30 AG062421 for MPF, U19 AG032438 (DIAN) for RB. Data collection and sharing for this project was supported by The Dominantly Inherited Alzheimer’s Network (DIAN, UF1AG032438, see ListS1 in supporting information) funded by the National Institute on Aging (NIA), the German Center for Neurodegenerative Diseases (DZNE), Raul Carrea Institute for Neurological Research (FLENI), Partial support by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI). This manuscript has been reviewed by DIAN Study investigators for scientific content and consistency of data interpretation with previous DIAN Study publications. We acknowledge the altruism of the participants and their families and contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study.
Footnotes
Declaration of Interests. Dr. Chhatwal has served on a medical advisory board for Otsuka Pharmaceuticals. This work is unrelated to the content of this manuscript.
Supporting Information: The following supporting information is available free of charge at ACS website http://pubs.acs.org
Data Availability.
The Skyline documents and raw files for mass spectrometry data are available at Panorama Public (PXD025668, https://panoramaweb.org/ADiso.url).80,81
References
- 1.Rosenberger G; Liu Y; Röst HL; Ludwig C; Buil A; Bensimon A; Soste M; Spector TD; Dermitzakis ET; Collins BC; Malmström L; Aebersold R Inference and Quantification of Peptidoforms in Large Sample Cohorts by SWATH-MS. Nat. Biotechnol 2017, 35 (8), 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye Z; Mao Y; Clausen H; Vakhrushev SY Glyco-DIA: A Method for Quantitative O-Glycoproteomics with in Silico-Boosted Glycopeptide Libraries. Nat. Methods 2019, 16 (9), 902–910. [DOI] [PubMed] [Google Scholar]
- 3.Lambeth TR; Riggs DL; Talbert LE; Tang J; Coburn E; Kang AS; Noll J; Augello C; Ford BD; Julian RR Spontaneous Isomerization of Long-Lived Proteins Provides a Molecular Mechanism for the Lysosomal Failure Observed in Alzheimer’s Disease. ACS Cent. Sci 2019, 5(8), 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting YS; Egertson JD; Payne SH; Kim S; MacLean B; Käll L; Aebersold R; Smith RD; Noble WS; MacCoss MJ Peptide-Centric Proteome Analysis: An Alternative Strategy for the Analysis of Tandem Mass Spectrometry Data. Mol. Cell. Proteomics 2015, 14 (9), 2301–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Searle BC; Lawrence RT; MacCoss MJ; Villén J Thesaurus: Quantifying Phosphopeptide Positional Isomers. Nat. Methods 2019, 16 (8), 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selkoe DJ; Hardy J The Amyloid Hypothesis of Alzheimer’s Disease at 25 Years. EMBO Mol. Med 2016, 8 (6), 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballatore C; Lee VMY; Trojanowski JQ Tau-Mediated Neurodegeneration in Alzheimer’s Disease and Related Disorders. Nat. Rev. Neurosci 2007, 8 (9), 663–672. [DOI] [PubMed] [Google Scholar]
- 8.Gordon BA; Blazey TM; Christensen J; Dincer A; Flores S; Keefe S; Chen C; Su Y; McDade EM; Wang G; Li Y; Hassenstab J; Aschenbrenner A; Hornbeck R; Jack CR; Ances BM; Berman SB; Brosch JR; Galasko D; Gauthier S; Lah JJ; Masellis M; van Dyck CH; Mintun MA; Klein G; Ristic S; Cairns NJ; Marcus DS; Xiong C; Holtzman DM; Raichle ME; Morris JC; Bateman RJ; Benzinger TLS Tau PET in Autosomal Dominant Alzheimer’s Disease: Relationship with Cognition, Dementia and Other Biomarkers. Brain 2019, 142 (4), 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barthélemy NR; Li Y; Joseph-Mathurin N; Gordon BA; Hassenstab J; Benzinger TLS; Buckles V; Fagan AM; Perrin RJ; Goate AM; Morris JC; Karch CM; Xiong C; Allegri R; Mendez PC; Berman SB; Ikeuchi T; Mori H; Shimada H; Shoji M; Suzuki K; Noble J; Farlow M; Chhatwal J; Graff-Radford NR; Salloway S; Schofield PR; Masters CL; Martins RN; O’Connor A; Fox NC; Levin J; Jucker M; Gabelle A; Lehmann S; Sato C; Bateman RJ; McDade E A Soluble Phosphorylated Tau Signature Links Tau, Amyloid and the Evolution of Stages of Dominantly Inherited Alzheimer’s Disease. Nat. Med 2020, 26 (3), 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmqvist S; Janelidze S; Quiroz YT; Zetterberg H; Lopera F; Stomrud E; Su Y; Chen Y; Serrano GE; Leuzy A; Mattsson-Carlgren N; Strandberg O; Smith R; Villegas A; Sepulveda-Falla D; Chai X; Proctor NK; Beach TG; Blennow K; Dage JL; Reiman EM; Hansson O Discriminative Accuracy of Plasma Phospho-Tau217 for Alzheimer Disease vs Other Neurodegenerative Disorders. JAMA 2020, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barthélemy NR; Horie K; Sato C; Bateman RJ Blood Plasma Phosphorylated-Tau Isoforms Track CNS Change in Alzheimer’s Disease. J. Exp. Med 2020, 217 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotzbauer PT; Tojanowski JQ; Lee VMV Lewy Body Pathology in Alzheimer’s Disease. J. Mol. Neurosci 2001, 17 (2), 225–232. [DOI] [PubMed] [Google Scholar]
- 13.Govindpani K; McNamara LG; Smith NR; Vinnakota C; Waldvogel HJ; Faull RL; Kwakowsky A Vascular Dysfunction in Alzheimer’s Disease: A Prelude to the Pathological Process or a Consequence of It? J. Clin. Med 2019, 8 (5), 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freiherr J; Hallschmid M; Frey WH; Brünner YF; Chapman CD; Hölscher C; Craft S; De Felice FG; Benedict C Intranasal Insulin as a Treatment for Alzheimer’s Disease: A Review of Basic Research and Clinical Evidence. CNS Drugs 2013, 27 (7), 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Acker ZP; Bretou M; Annaert W Endo-Lysosomal Dysregulations and Late-Onset Alzheimer’s Disease: Impact of Genetic Risk Factors. Mol. Neurodegener 2019, 14 (1), 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nixon RA The Lysosome in Aging-Related Neurodegenerative Diseases. In Lysosomes: Biology, Diseases, and Therapeutics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp 137–179. [Google Scholar]
- 17.Carr DB; Goate A; Phil D; Morris JC Current Concepts in the Pathogenesis of Alzheimer’s Disease. Am. J. Med 1997, 103 (3), 3S–10S. [DOI] [PubMed] [Google Scholar]
- 18.Truscott RJW Age-Related Nuclear Cataract-Oxidation Is the Key. Exp. Eye Res 2005, 80 (5), 709–725. [DOI] [PubMed] [Google Scholar]
- 19.Kapphahn RJ; Ethen CM; Peters E A; Higgins, L.; Ferrington, D. A. Modified Alpha A Crystallin in the Retina: Altered Expression and Truncation with Aging. Biochemistry 2003, 42 (51), 15310–15325. [DOI] [PubMed] [Google Scholar]
- 20.Tonie Wright H; Urry DW Nonenzymatic Deamidation of Asparaginyl and Glutaminyl Residues in Protein. Crit. Rev. Biochem. Mol. Biol 1991, 26 (1), 1–52. [DOI] [PubMed] [Google Scholar]
- 21.Cournoyer JJ; Pittman JL; Ivleva VB; Fallows E; Waskell L; Costello CE; O’Connor PB Deamidation: Differentiation of Aspartyl from Isoaspartyl Products in Peptides by Electron Capture Dissociation. Protein Sci 2005, 14 (2), 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H; Fung EYM; Zubarev AR; Zubarev RA Toward Proteome-Scale Identification and Quantification of Isoaspartyl Residues in Biological Samples. J. Proteome Res 2009, 8 (10), 4615–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansson ET Strategies for Analysis of Isomeric Peptides. J. Sep. Sci 2018, 41 (1), 385–397. [DOI] [PubMed] [Google Scholar]
- 24.Geiger T; Clarke S Deamidation, Isomerization, and Racemization at Asparaginyl and Aspartyl Residues in Peptides - Succinimide-Linked Reactions That Contribute to Protein-Degradation. J. Biol. Chem 1987, 262 (2), 785–794. [PubMed] [Google Scholar]
- 25.Lyon YA; Sabbah GM; Julian RR Identification of Sequence Similarities among Isomerization Hotspots in Crystallin Proteins. J. Proteome Res 2017, 16 (4), 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia C; Lietz CB; Yu Q; Li L Site-Specific Characterization of D -Amino Acid Containing Peptide Epimers by Ion Mobility Spectrometry. Anal. Chem 2014, 86 (6), 2972–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H; Lyutvinskiy Y; Soininen H; Zubarev RA Alzheimer’s Disease and Mild Cognitive Impairment Are Associated with Elevated Levels of Isoaspartyl Residues in Blood Plasma Proteins. J. Alzheimer’s Dis 2011, 27 (1), 113–118. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu T; Watanabe A; Ogawara M; Mori H; Shirasawa T Isoaspartate Formation and Neurodegeneration in Alzheimer’s Disease. Arch. Biochem. Biophys 2000, 381 (2), 225–234. [DOI] [PubMed] [Google Scholar]
- 29.Roher AE; Lowenson JD; Clarke S; Wolkow C; Wang R; Cotter RJ; Reardon IM; Zurcherneely HA; Heinrikson RL; Ball MJ; et al. Structural alterations in the peptide backbone of beta-amyloid core protein may account for its deposition and stability in Alzheimer’s-disease. J. Biol. Chem 1993, 268 (5), 3072–3083. [PubMed] [Google Scholar]
- 30.Shapira R; Austin GE; Mirra SS Neuritic Plaque Amyloid in Alzheimer’s Disease Is Highly Racemized. J. Neurochem 1988, 50 (1), 69–74. [DOI] [PubMed] [Google Scholar]
- 31.Payan IL; Chou S-J; Fisher GH; Man EH; Emory C; Frey WH Altered Aspartate in Alzheimer Neurofibrillary Tangles. Neurochem. Res 1992, 17 (2), 187–191. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe A; Takio K; Ihara Y Deamidation and Isoaspartate Formation in Smeared Tau in Paired Helical Filaments. Unusual Properties of the Microtubule-Binding Domain of Tau. J. Biol. Chem 1999, 274 (11), 7368–7378. [DOI] [PubMed] [Google Scholar]
- 33.Miyasaka T; Watanabe A; Saito Y; Murayama S; Mann DMA; Yamazaki M; Ravid R; Morishima-Kawashima M; Nagashima K; Ihara Y Visualization of Newly Deposited Tau in Neurofibrillary Tangles and Neuropil Threads. J. Neuropathol. Exp. Neurol 2005, 64 (8), 665–674. [DOI] [PubMed] [Google Scholar]
- 34.Egertson JD; MacLean B; Johnson R; Xuan Y; MacCoss MJ Multiplexed Peptide Analysis Using Data-Independent Acquisition and Skyline. Nat. Protoc 2015, 10 (6), 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amodei D; Egertson J; MacLean BX; Johnson R; Merrihew GE; Keller A; Marsh D; Vitek O; Mallick P; MacCoss MJ Improving Precursor Selectivity in Data-Independent Acquisition Using Overlapping Windows. J. Am. Soc. Mass Spectrom 2019, 30 (4), 669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egertson JD; Kuehn A; Merrihew GE; Bateman NW; MacLean BX; Ting YS; Canterbury JD; Marsh DM; Kellmann M; Zabrouskov V; Wu CC; MacCoss MJ Multiplexed MS/MS for Improved Data-Independent Acquisition. Nat. Methods 2013, 10 (8), 744–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adusumilli R; Mallick P Data Conversion with ProteoWizard MsConvert. Methods Mol. Biol 2017, 339–368. [DOI] [PubMed] [Google Scholar]
- 38.Gessulat S; Schmidt T; Zolg DP; Samaras P; Schnatbaum K; Zerweck J; Knaute T; Rechenberger J; Delanghe B; Huhmer A; Reimer U; Ehrlich H-C; Aiche S; Kuster B; Wilhelm M Prosit: Proteome-Wide Prediction of Peptide Tandem Mass Spectra by Deep Learning. Nat. Methods 2019, 16 (6), 509–518. [DOI] [PubMed] [Google Scholar]
- 39.Searle BC; Pino LK; Egertson JD; Ting YS; Lawrence RT; MacLean BX; Villén Judit; MacCoss MJ Chromatogram Libraries Improve Peptide Detection and Quantification by Data Independent Acquisition Mass Spectrometry. Nat. Commun 2018, 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ting YS; Egertson JD; Bollinger JG; Searle BC; Payne SH; Noble WS; MacCoss MJ PECAN: Library-Free Peptide Detection for Data-Independent Acquisition Tandem Mass Spectrometry Data. Nat. Methods 2017, 14 (9), 903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pino LK; Just SC; MacCoss MJ; Searle BC Acquiring and Analyzing Data Independent Acquisition Proteomics Experiments without Spectrum Libraries. Mol. Cell. Proteomics 2020, 19 (7), 1088–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Käll L; Canterbury JD; Weston J; Noble WS; MacCoss MJ Semi-Supervised Learning for Peptide Identification from Shotgun Proteomics Datasets. Nat. Methods 2007, 4 (11), 923–925. [DOI] [PubMed] [Google Scholar]
- 43.Käll L; Storey JD; MacCoss MJ; Noble WS Posterior Error Probabilities and False Discovery Rates: Two Sides of the Same Coin. J. Proteome Res 2008, 7 (1), 40–44. [DOI] [PubMed] [Google Scholar]
- 44.MacLean B; Tomazela DM; Shulman N; Chambers M; Finney GL; Frewen B; Kern R; Tabb DL; Liebler DC; MacCoss MJ Skyline: an Open Source Document Editor for Creating and Analyzing Targeted Proteomics Experiments. Bioinformatics 2010, 26 (7), 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pino LK; Searle BC; Bollinger JG; Nunn B; MacLean B; MacCoss MJ The Skyline Ecosystem: Informatics for Quantitative Mass Spectrometry Proteomics. Mass Spectrom. Rev 2020, 39 (3), 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chambers MC; Maclean B; Burke R; Amodei D; Ruderman DL; Neumann S; Gatto L; Fischer B; Pratt B; Egertson J; Hoff K; Kessner D; Tasman N; Shulman N; Frewen B; Baker TA; Brusniak M-Y; Paulse C; Creasy D; Flashner L; Kani K; Moulding C; Seymour SL; Nuwaysir LM; Lefebvre B; Kuhlmann F; Roark J; Rainer P; Detlev S; Hemenway T; Huhmer A; Langridge J; Connolly B; Chadick T; Holly K; Eckels J; Deutsch EW; Moritz RL; Katz JE; Agus DB; MacCoss M; Tabb DL; Mallick P A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nature Biotechnology 2012, 30 (10), 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park CY; Klammer AA; Käll Lukas; MacCoss MJ; Noble WS Rapid and Accurate Peptide Identification from Tandem Mass Spectra. J. Proteome Res 2008, 7 (7), 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eng JK; Fischer B; Grossmann J; MacCoss MJ A Fast SEQUEST Cross Correlation Algorithm. J. Proteome Res 2008, 7 (10), 4598–4602. [DOI] [PubMed] [Google Scholar]
- 49.Sulimov P; Kertész-Farkas A Tailor: A Nonparametric and Rapid Score Calibration Method for Database Search-Based Peptide Identification in Shotgun Proteomics. J. Proteome Res 2020, 19 (4), 1481–1490. [DOI] [PubMed] [Google Scholar]
- 50.Hood CA; Fuentes G; Patel H; Page K; Menakuru M; Park JH Fast Conventional Fmoc Solid-Phase Peptide Synthesis with HCTU. J. Pept. Sci 2008, 14, 97–101. [DOI] [PubMed] [Google Scholar]
- 51.Riggs DL; Gomez SV; Julian RR Sequence and Solution Effects on the Prevalence of D -Isomers Produced by Deamidation. ACS Chem. Biol 2017, 12 (11), 2875–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rusten TE; Stenmark H p62, an Autophagy Hero or Culprit? Nat. Cell Biol 2010, 12 (3), 207–209. [DOI] [PubMed] [Google Scholar]
- 53.Truscott RJW; Schey KL; Friedrich MG Old Proteins in Man: A Field in Its Infancy. Trends Biochem. Sci 2016, 41 (8), 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe A; Hong WK; Dohmae N; Takio K; Morishima-Kawashima M; Ihara Y Molecular Aging of Tau: Disulfide-Independent Aggregation and Non-Enzymatic Degradation in Vitro and in Vivo. J. Neurochem 2004, 90 (6), 1302–1311. [DOI] [PubMed] [Google Scholar]
- 55.Tanida I; Ueno T; Kominami E LC3 and Autophagy. Autophagosome and Phagosome 2008, 77–88. [DOI] [PubMed] [Google Scholar]
- 56.Majeski AE; Fred Dice J Mechanisms of Chaperone-Mediated Autophagy. Int. J. Biochem. Cell Biol 2004, 36 (12), 2435–2444. [DOI] [PubMed] [Google Scholar]
- 57.Wang J; Zhang J; Lee Y-M; Koh P-L; Ng S; Bao F; Lin Q; Shen H-M Quantitative Chemical Proteomics Profiling of De Novo Protein Synthesis during Starvation-Mediated Autophagy. Autophagy 2016, 12 (10), 1931–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du Y; Wooten MC; Gearing M; Wooten MW Age-Associated Oxidative Damage to the P62 Promoter: Implications for Alzheimer Disease. Free Radic. Biol. Med 2009, 46 (4), 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuusisto E; Salminen A; Alafuzoff I Ubiquitin-Binding Protein P62 Is Present in Neuronal and Glial Inclusions in Human Tauopathies and Synucleinopathies. Neuroreport 2001, 12 (10), 2085–2090. [DOI] [PubMed] [Google Scholar]
- 60.Lau A; Zheng Y; Tao S; Wang H; Whitman SA; White E; Zhang DD Arsenic Inhibits Autophagic Flux, Activating the Nrf2-Keap1 Pathway in a P62-Dependent Manner. Mol. Cell. Biol 2013, 33 (12), 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mandelkow E-M; Mandelkow E Biochemistry and Cell Biology of Tau Protein in Neurofibrillary Degeneration. Cold Spring Harb. Perspect. Med 2012, 2 (7), a006247–a006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu WJ; Ye L; Huang WF; Guo LJ; Xu ZG; Wu HL; Yang C; Liu HF P62 Links the Autophagy Pathway and the Ubiqutin–Proteasome System upon Ubiquitinated Protein Degradation. Cell. Mol. Biol. Lett 2016, 21 (1), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu H; Dai C; Fan Y; Guo B; Ren K; Sun T; Wang W From Autophagy to Mitophagy: The Roles of P62 in Neurodegenerative Diseases. J. Bioenerg. Biomembr 2017, 49 (5), 413–422. [DOI] [PubMed] [Google Scholar]
- 64.Zheng X; Wang W; Liu R; Huang H; Zhang R; Sun L Effect of P62 on Tau Hyperphosphorylation in a Rat Model of Alzheimer’s Disease. Neural Regen. Res 2012, 7 (17), 1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fitzpatrick AWP; Falcon B; He S; Murzin AG; Murshudov G; Garringer HJ; Crowther RA; Ghetti B; Goedert M; Scheres SHW Cryo-EM Structures of Tau Filaments from Alzheimer’s Disease. Nature 2017, 547 (7662), 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Croft CL; Goodwin MS; Ryu DH; Lessard CB; Tejeda G; Marrero M; Vause AR; Paterno G; Cruz PE; Lewis J; Giasson BI; Golde TE Photodynamic Studies Reveal Rapid Formation and Appreciable Turnover of Tau Inclusions. Acta Neuropathol 2021, 141 (3), 359–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukherjee S; Perez KA; Lago LC; Klatt S; McLean CA; Birchall IE; Barnham KJ; Masters CL; Roberts BR Quantification of N-Terminal Amyloid-β Isoforms Reveals Isomers Are the Most Abundant Form of the Amyloid-β Peptide in Sporadic Alzheimer’s Disease. Brain Commun 2021, 3 (2), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reissner KJ; Aswad DW Deamidation and Isoaspartate Formation in Proteins: Unwanted Alterations or Surreptitious Signals? Cell. Mol. Life Sci 2003, 60 (7), 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Connor CM; Aswad DW; Clarke S Mammalian Brain and Erythrocyte Carboxyl Methyltransferases Are Similar Enzymes That Recognize Both D-Aspartyl and L-Isoaspartyl Residues in Structurally Altered Protein Substrates. Proc. Natl. Acad. Sci 1984, 81 (24), 7757–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cacace R; Sleegers K; Van Broeckhoven C Molecular Genetics of Early-Onset Alzheimer’s Disease Revisited. Alzheimer’s Dement 2016, 12 (6), 733–748. [DOI] [PubMed] [Google Scholar]
- 71.Jankowsky JL; Fadale DJ; Anderson J; Xu GM; Gonzales V; Jenkins NA; Copeland NG; Lee MK; Younkin LH; Wagner SL; Younkin SG; Borchelt DR Mutant Presenilins Specifically Elevate the Levels of the 42 Residue β-Amyloid Peptide in Vivo: Evidence for Augmentation of a 42-Specific γ Secretase. Hum. Mol. Genet 2004, 13 (2), 159–170. [DOI] [PubMed] [Google Scholar]
- 72.Shen J; Kelleher RJ The Presenilin Hypothesis of Alzheimer’s Disease: Evidence for a Loss-of-Function Pathogenic Mechanism. Proc. Natl. Acad. Sci 2007, 104 (2), 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chong C-M; Ke M; Tan Y; Huang Z; Zhang K; Ai N; Ge W; Qin D; Lu J-H; Su H Presenilin 1 Deficiency Suppresses Autophagy in Human Neural Stem Cells through Reducing γ-Secretase-Independent ERK/CREB Signaling. Cell Death Dis 2018, 9 (9), 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neely KM; Green KN; LaFerla FM Presenilin Is Necessary for Efficient Proteolysis through the Autophagy-Lysosome System in a -Secretase-Independent Manner. J. Neurosci 2011, 31 (8), 2781–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J-H; Yu WH; Kumar A; Lee S; Mohan PS; Peterhoff CM; Wolfe DM; Martinez-Vicente M; Massey AC; Sovak G; Uchiyama Y; Westaway D; Cuervo AM; Nixon RA Lysosomal Proteolysis and Autophagy Require Presenilin 1 and Are Disrupted by Alzheimer-Related PS1 Mutations. Cell 2010, 141 (7), 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coen K, Flannagan RS, Baron S, et al. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo-lysosomal dysfunction in PSEN-deficient cells. J Cell Biol 2012, 198(1), 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fedeli C, Filadi R, Rossi A, Mammucari C, Pizzo P. PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca2+ homeostasis. Autophagy 2019, 15(12), 2044–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryazanov AG; Nefsky BS Protein Turnover Plays a Key Role in Aging. Mech. Ageing Dev 2002, 123 (2–3), 207–213. [DOI] [PubMed] [Google Scholar]
- 79.Cuervo AM; Bergamini E; Brunk UT; Dröge W; Ffrench M; Terman A Autophagy and Aging: The Importance of Maintaining “Clean” Cells. Autophagy 2005, 1 (3), 131–140. [DOI] [PubMed] [Google Scholar]
- 80.Sharma V; Eckels J; Taylor GK; Shulman NJ; Stergachis AB; Joyner SA; Yan P; Whiteaker JR; Halusa GN; Schilling B; Gibson BW; Colangelo CM; Paulovich AG; Carr SA; Jaffe JD; MacCoss MJ; MacLean B Panorama: A Targeted Proteomics Knowledge Base. J. Proteome Res 2014, 13 (9), 4205–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma V; Eckels J; Schilling B; Ludwig C; Jaffe JD; MacCoss MJ; MacLean B Panorama Public: A Public Repository for Quantitative Data Sets Processed in Skyline. Mol. Cell. Proteomics 2018, 17 (6), 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Isomerization compared in hippocampus and SMTG of samples from the same brain.
Figure S2. Quantification of isomerization in peptide WDGQETTLVR from fatty acid binding protein, heart
Figure S3. Chromatograms of synthesized TDHGAEIVYK isomers compared to a Skyline chromatogram from SMTG containing all isomers
Figure S4. Quantification and chromatogram from isomerized amyloid beta peptide HDSGYEVHHQK
Table S1. List of proteins found to contain isomerization via automated search
Table S2. Brain Tissue Stratification Description for SMTG
Table S3. Brain Tissue Stratification Description for Hippocampus
Data Availability Statement
The Skyline documents and raw files for mass spectrometry data are available at Panorama Public (PXD025668, https://panoramaweb.org/ADiso.url).80,81


