Abstract
Traumatic brain injury (TBI), often induced by sports, car accidents, falls, or other daily occurrences, is a primary non-genetically related risk factor for the development of subsequent neurodegeneration and neuronal cell death. However, the molecular mechanisms underlying neurodegeneration, cell death, and neurobehavioral dysfunction following TBI remain unclear. Here, we found that poly(ADP-ribose) polymerase-1 (PARP-1) was hyperactivated following TBI and its inhibition reduced TBI-induced brain injury. Macrophage migration inhibitory factor (MIF), a newly identified nuclease involved in PARP-1-dependent cell death, was translocated from the cytosol to the nucleus in cortical neurons following TBI and promoted neuronal cell death in vivo. Genetic deletion of MIF protected neurons from TBI-induced dendritic spine loss, morphological complexity degeneration, and subsequent neuronal cell death in mice. Moreover, MIF knockout reduced the brain injury volume and improved long-term animal behavioral rehabilitation. These neuroprotective effects in MIF knockout mice were reversed by the expression of wild-type MIF but not nuclease-deficient MIF mutant. In contrast, genetic deletion of MIF did not alter TBI-induced neuroinflammation. These findings reveal that MIF mediates TBI-induced neurodegeneration, neuronal cell death and neurobehavioral dysfunction through its nuclease activity, but not its pro-inflammatory role. Targeting MIF’s nuclease activity may offer a novel strategy to protect neurons from TBI.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04037-9.
Keywords: MIF, Traumatic brain injury, Neurodegeneration, Cell death
Introduction
Traumatic brain injury (TBI) frequently caused by sports or car accidents is a leading cause of death and long-term disability in the United States. Approximately, 1.7 million Americans suffer TBI annually, and about 5,400,000 TBI survivors are currently living with permanent deficits and require daily assistance, which has imposed an enormous economic burden on individuals and society (https://treatnow.org/knowledgebase/car-accidents-and-brain-injury-statistics-2020/) [1, 2]. Unfortunately, so far there is a lack of effective therapies. TBI is a devastating occurrence causing neurodegeneration and neuronal cell death that contributes to subsequent neurological dysfunction and an increased risk of neurodegenerative diseases including dementia, Alzheimer’s disease (AD), and Parkinson’s disease (PD) [3, 4]. The secondary brain injury following the initial TBI plays a significant role in neurodegeneration and neuronal cell death [1]. However, our understanding on the processes and mechanisms of neurodegeneration and neuronal cell death contributing to the secondary brain injury following TBI is limited.
Previous studies have implicated an important role of poly(ADP-ribose) polymerase-1 (PARP-1) in TBI as systemic inhibition of PARP-1 attenuates neuronal cell loss, reduces the lesion volume, and significantly improves the motor function recovery in the mouse model of TBI [5, 6]. PARP-1 belongs to a PARP superfamily consisting of 17 members, and is activated by oxidative stress or DNA damage to generate poly(ADP-ribose) (PAR), which functions as a death signal to trigger an intrinsic cell death program termed PARthanatos (PARP-1-dependent cell death) [7–9]. PARthanatos is widely involved in neurological diseases including ischemic stroke and neurodegenerative diseases like AD and PD [10–14]. The molecular mechanism underlying PARthanatos involves apoptosis-inducing factor (AIF) release from the mitochondria and translocation to the nucleus [8, 15]. AIF then recruits macrophage migration inhibitory factor (MIF), a previously known pro-inflammatory cytokine, to the nucleus, where MIF functions as a novel nuclease and induces chromatinolysis and PARthanatic cell death [7]. MIF is upregulated in the brain of AD patients and AD mouse model [16, 17]. High MIF levels are correlated with the worse outcome of neuroinflammation and cognitive deficits in a mouse model of sporadic AD and human patients with AD [18]. Similarly, elevated MIF levels have been observed in human stroke patients and mouse ischemic stroke models, leading to an increased injury volume following ischemic stroke [19–21], which is independent of its effect on alteration of pro-inflammatory cytokine levels [20, 21]. In contrast, inhibition of MIF nuclease activity reduces ischemic brain injury [7]. Although PARP-1/MIF-mediated PARthanotos plays an important role in glutamate cytotoxicity and ischemic brain injury [7, 8, 15, 22], it remains unknown whether MIF and its nuclease activity contribute to neurodegeneration and neuronal cell death following TBI.
In this study, we found that PARP-1 is hyperactivated and PARP-1 knockout (KO) reduces injury volume following TBI. MIF expression is translocated from the cytosol to the nucleus in neurons following TBI, which promotes neurodegeneration and neuronal cell death. MIF KO or nuclease-deficient MIF mutant preserves dendritic spine density and neuronal morphological complexity, reduces the injury volume, and improves long-term animal behavior rehabilitation in the TBI mouse model. Thus, targeting MIF’s nuclease activity may offer a novel strategy to protect neurons from TBI.
Materials and methods
Animals
The UT Southwestern Medical Center is fully accredited by the American Association for the Accreditation of Laboratory Animal Care (AAALAC). All research procedures performed in this study were approved the UT Southwestern Medical Center Animal Care and Use Committee (IACUC) in compliance with the Animal Welfare Act regulations and Public Health Service (PHS) Policy. Male mice (n = 3–28 per group) were used in our studies in a blinded fashion. Mouse genotype was determined by J.E.W. or other lab members. TBI surgery was performed by Z.R., Q.L. or J.E.W. Mouse genotypes were decoded after TBI, mouse behavior tests and data analysis. Based on their genotype, mice were grouped as WT, KO, KO-WT, and KO-E22A. Within each group, mice were randomly assigned to subgroups including sham, 1, 3, 7, 14 or 28 days post TBI.
The controlled cortical impact (CCI)
The open-head CCI has been well developed as a TBI mouse model and was performed as described previously using Impact One Stereotaxic Impactor (39,463,920) from Leica [23]. Briefly, mice were first anesthetized with 5% isoflurane, then placed in a stereotaxic frame and maintained with 2% isoflurane. After a midline skin incision, a 3.5 mm craniotomy was made in the right parietal bone midway between bregma and lambda with medial edge 1 mm lateral to the midline using a motorized drill to remove bone but leaving the dura intact. Mice were impacted for one time at 5 m/s with a 20 ms dwell time and 2 mm depression using a 3 mm diameter convex tip. Mice were expected to receive moderate brain injury. Mice in the sham group underwent the identical surgical procedures but were not impacted. After injury, skin was sutured and allowed to recover in a heated incubator. Body temperature was maintained at 37 °C using a small animal temperature controller throughout all procedures during the surgery. All surgical tools were sterilized for the surgery. 1 day, 3 days, 7 days, 14 days or 28 days after TBI, mice were perfused with PBS and stained with triphenyl tetrazolium chloride (TTC). The brains were further fixed with 4% paraformaldehyde (PFA) and sliced for Nissl or immunohistochemical staining.
Golgi staining and neuronal morphometric analysis
Golgi staining of adult MIF WT and KO mouse brain (3 months old) without TBI and 1 day or 28 days post TBI was performed using FD Rapid Golgi Stain Kit PK401A (FD NeuroTechnologies) as described previously [24]. All brains were blocked and embedded in OCT embedding medium (Tissue-Tek, catalog #4583). Coronal sections (150 µm) were cut with a Leica cryostat (CM3050S) and mounted on gelatin-coated microscopes slides (FD NeuroTechnologies). After natural dry at room temperature in the dark for at least 1 day, slides were stained with the staining solution freshly prepared from the kit, and then hydrated in distilled water (1 min) and gradually dehydrated in 50%, 75%, 95%, and 100% ethanol. After clearance in xylene, slides were mounted with Permount. For analysis of spine density and dendritic length and branching, z-stack images of layer V pyramidal neurons from the medial somatosensory cortex were taken with a motorized Axio Observer Z1 microscope (Carl Zeiss). 3D reconstruction of the entire dendritic processes in each neuron was made from the z-series stacks of images. 2D projection images were then traced using the NIH ImageJ program with NeuronJ plugin. Total dendritic length and branch number per neuron were analyzed as described previously [25]. Sholl analysis was performed using the NIH ImageJ Sholl analysis plugin as described previously [25]. The spine density in each cell was also measured from one apical dendritic branch in the terminal tuft, from an oblique branch running off the main apical dendritic shaft approximately half-way up the shaft, and from a secondary branch proximal to the cell body for one basilar branch using high magnification images [26].
TTC staining
The brain was cut into five equal pieces of coronal sections. Contralateral and ipsilateral hemisphere and injury areas were traced using ImageJ software. The volume of brain injury was calculated as the product of cross-sectional area for all sections and distance between each sections (1.5–2 mm), and corrected for edema using the formula: corrected injury volume (mm3) = injury volume × (contralateral volume/ipsilateral volume) [27, 28]. The data were presented as injury% = corrected injury volume/corrected hemisphere volume × 100%. Total 138 mice were used for TTC staining experiments and detailed mouse numbers used in the experiments have been indicated in the figure legends.
Nissl staining
Nissl staining was performed using standard methods as previously described [29]. Briefly, mice were anesthetized with 5% isoflurane and perfused with 4% paraformaldehyde (Sigma). Brains were extracted, postfixed and cryoprotected in 30% sucrose. Brain was then cut into 30 μm sections and free-floating sections from each mouse were divided into 4 equal series. Each series contains about 24–32 sections separated successively by 90 μm and is representative of all parts of mouse brain except cerebellum. A series of 24–32 brain sections from each mouse were mounted on the slide for Nissl staining. Sections were immersed for 2 min in each of the following solutions: 95% ethanol, 95% ethanol, 100% ethanol, 100% xylene, 100% ethanol, 95% ethanol, 95% ethanol and H2O. Then, sections were stained in 0.1% Thionin for 7 min followed by 3 times washing with H2O for 2 min each. After incubation in the Formalin Acetic Solution containing 1% glacial acetic acid and 0.39% formaldehyde for 40 s, sections were immediately washed with 3 times H2O for 2 min each. Next, sections were dehydrated through 95% ethanol, 100% ethanol, and cleared in 2 times 100% xylene for 10 min each before mounting the coverslip with Permount (Fisher, SP15-100) for image analysis. Brain injury volume (mm3) = injury volume × (contralateral volume/ipsilateral volume) [27, 28]. The data were presented as injury% = corrected injury volume/corrected hemisphere volume × 100%. Detailed mouse numbers used in the experiments have been indicated in the figure legends.
Fluoro-Jade C (FJC) staining
FJC staining of MIF WT and KO mouse sections (30 μm) post TBI was performed as previously described [30, 31]. Briefly, brain sections from each mouse were organized into a series of 24–32 sections separated successively by 90 μm. Each series is representative of all parts of mouse brain except cerebellum. A series of 24–32 brain sections from each mouse were mounted onto Superfrost Plus microscope slides (Fisher Scientific, 12-550-15). After dry, slides were immersed in 0.01% NaOH prepared in 80% ethanol solution for 5 min following by 70% alcohol for 2 min and distilled water for 2 min. Slides were then transferred to 0.06% potassium permanganate and shaked for 10 min in the dark. After rinsing with distilled water, slides were incubated in 0.0001% Fluoro-Jade C working solution (Sigma, AG325) with 0.1% acetic acid for 10 min in the dark. After staining, the sections were rinsed with distilled water 3 times for 1 min each. After dry, slides were immersed in 50%, 70% and 100% ethanol for 2 min each and then cleared in xylene. Lastly, slides were mounted with DPX (Sigma, 317616) for image analysis. All brain sections were checked under the microscope and 5–10 representative images were taken from the cortex of each single mouse and neurodegeneration was quantified as FJC+ cell number/image field from 15 to 30 images of WT and KO mouse following TBI.
TUNEL staining
DNA damage was assessed by TUNEL assay according to manufacturer instructions (In Situ Cell Death Detection Kit, TMR red, Millipore Sigma). Briefly, dehydrated mouse brain slices were mounted on slides and dried. After fixation with 4% PFA and permeabilization with 0.1% Triton X-100 solution, slides were incubated with reaction mixture containing terminal deoxynucleotidyl transferase and fluorescein-conjugated nucleotides for 1 h at 37 °C. Slides were counterstained with DAPI and quantified under a fluorescent microscope.
Immunofluorescence staining and microscopy images
Brains were fixed with 4% PFA, cryoprotected in 30% sucrose and cut into 30 μm sections, and free-floating sections were divided into 4 equal series. Each series contains about 24–32 sections separated successively by 90 μm and is representative of all parts of mouse brain except cerebellum. A series of 24–32 brain sections from each mouse were used for floating immunohistochemical staining. First, the sections were permeabilized with 0.1% Triton X-100 and blocked with 10% normal donkey serum in PBS. Brain sections were then incubated with the antibodies against MIF (Abcam, ab36146), glial fibrillary acidic protein (GFAP) (Agilent Technologies, Z033401-2), cleaved caspase-3 (Cell Signaling, 9661), iba1 (Fisher Scientific-Wako, NC1469672) and NeuN (Sigma, ABN90) followed by Cy2 affinipure donkey anti-goat IgG or Cy3 affinipure donkey anti-rabbit IgG. Immunofluorescent images were carried out using a motorized Axio Observer Z1 microscope (Carl Zeiss). 3–10 representative images were taken from the cortex of each single mouse and neurodegeneration was quantified as NeuN+ cell number/image field from identical cortex regions of WT and KO mice following TBI.
RNA sequencing
RNA sequencing was performed as descripted previously [32]. Total RNA was isolated from WT and MIF KO mouse cortex with or without TBI (n = 3–5 per group) using the RNeasy mini kit (Qiagen) and treated with DNase (Qiagen). The quality of total RNA was confirmed with a RIN score 8.5 or higher by the Agilent Tapestation 4200. mRNAs were used for library preparation with the TruSeq Stranded mRNA Library Prep Kit (Illumina), and sequenced on the Illumina NextSeq 500. Reads were mapped to the hg19) (UCSC version from igenomes) using Tophat and annotated using a custom R script using the UCSC known Genes table as a reference. Read counts were generated using feature Counts and the differential expression analysis was performed using edgeR. The differential expression results with a false discovery rate (FDR) < 0.05 and mRNA fold change > 1.5 as a cutoff were used for further downstream analysis. Gene ontology was analyzed using DAVID (version 6.8). Total 8 MIF mice and 6 MIF KO mice with or without TBI were tested in two independent RNA sequencing experiments.
Intracerebroventricular (ICV) injection
One microliter trypan blue, AAV2-MIF WT or E22A (1 × 1013 GC/ml, Vector BioLabs) was injected intracerebroventricularly into both sides of newborn MIF KO mice [7]. The efficiency and expansion pattern of ICV injection was visualized by trypan blue (1 µl/side). The expression of MIF and its mutant was confirmed by immunostaining without TBI surgery at 12 weeks of age. Each individual mouse after TBI treatment and TTC staining for the brain injury volume measurement was also verified whether ICV injection was performed successfully by immunostaining. Only those successfully injected mice were included in the study.
Open field tests
The open field tests were performed to evaluate the locomotor activity and anxiety in a rectangular plastic apparatus (25 cm × 25 cm × 25 cm) using the video-tracking software system (SMART 3.0) from PanLab. The field was divided into peripheral zone (5 cm from the edge of walls) and central zone (36% of the total surface of the arena). The time spent in and entries into the center were measured as an anxiolytic indicator [33]. Mice were first transported to the behavior room and left undisturbed to get acclimated to the environment for at least 30 min. Then, each mouse was placed into the open field facing the center and allowed to explore freely for 8 min. After each experiment, the arena was thoroughly wiped with 70% ethanol solution to remove any feces, urine and odor. Finally, animal travel distance, walk speed and time in each zone for the first 5 min were analyzed using the SMART 3 software (Harvard Apparatus-Panlab, Holliston, MA, USA).
Corner test
The corner test was performed before TBI as day 0 and at days 1, 3, 7, 14, and 28 after TBI to assess sensory and motor deficits following both cortical and striatal injury [7]. A video camera was placed on the top of the cage to record the activity of a mouse in the cage for 5 min. Mice were placed between two pieces of cardboard each with a dimension of 30 cm × 20 cm × 0.5 mm attached to each other from the edges with an angle of 30°. Once in the corner, mice usually rear and then turn either left or right. Prior to TBI, mice do not show a side preference. Mice with sensory and motor deficits following TBI turn toward the non-impaired side (right). The percentage of right turn = right turns/total turns × 100% was calculated and compared. Recordings were evaluated by observers who were blinded to the treatment and genotype of the animals.
Cylinder test
The cylinder test was performed to assess the forelimb asymmetry performance in mice using a transparent glass cylinder (9 cm in diameter and 15 cm in height) equipped with a video camera on the top [7]. A mouse was placed in the cylinder and forelimb use of the mouse was recorded for 5 min and analyzed according to the following parameters: (1) number of right forelimb touch to the cylinder wall, (2) number of left forelimb touch to the cylinder wall and (3) simultaneous use of both limbs. VLC media player was used to slow down the video to count the forelimb touches. The percent use of the right limb touch was quantified by right limb touches divided by the total number of touches during the period of observation. Recordings were evaluated by an observer who was blinded to the treatment and genotype of the animals.
Beam test
Fine motor coordination and balance were assessed by the beam walking assay using a modified method described previously [34]. The goal of this test is for the mouse to stay upright and walk across an elevated narrow beam to a safe platform. The mice were trained for 3 days before test. The beam apparatus consists of 1 m beam with a round surface of 7 mm diameter) resting 20 cm above the tabletop on two poles. The time to cross the center 60 cm was measured. Sponges were applied below the beam, ~ 7.5 cm above the tabletop, to cushion any falls.
Treadmill test
Mice were first placed on a two-lane horizontal treadmill (Panlab/Harvard Apparatus, Holliston, MA, USA) and trained/warmed up at a speed of 3 cm/s for 1 min, 6 cm/s for 1 min and 12 cm/s for 1 min daily for 3 days before TBI surgery. The test was performed at 12 cm/s for 2 min immediately after the daily warm-up procedures before TBI as day 0 and at days 1, 3, 7, 14, and 28 after TBI. The shock times during the test were recorded and compared.
Lentiviral and adeno-associated viral (AAV) constructs and virus production
Full-length mouse MIF-WT-Flag (NM_010798) or MIF-E22A-Flag cDNA was subcloned into a lentiviral cFugw vector by AgeI and EcoRI restriction sites, and its expression was driven by the human ubiquitin C promoter as described previously [7]. The lentivirus was produced by transient transfection of the recombinant cFugw vector into 293FT cells together with three packaging vectors: pLP1, pLP2, and pVSV-G (1.3:1.5:1:1.5). The viral supernatants were collected at 48 and 72 h after transfection and concentrated by ultracentrifuge for 2 h at 50,000g. MIF-WT-Flag or MIF-E22A-Flag cDNA was subcloned into an AAV-WPRE-bGH (044 AM/CBA-pI-WPRE-bGH) vector at BamHI and EcoRI restriction sites, and their expression was driven by chicken β-actin (CBA) promoter. All AAV2 viruses were produced by the Vector BioLabs. Other plasmid constructs have been described previously [7].
Immunoblotting assay
The proteins were separated on denaturing polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. The membrane was blocked and incubated overnight with primary antibodies: rabbit anti-PAR (Trevigen, 4336-BPC-100), mouse anti-PARP-1 (Fisher, BDB11039), mouse anti-Flag (Sigma, F3165), Goat anti-MIF (Abcam, ab36146), or mouse anti-actin (Proteintech, 66009-1-AP) at 4 °C, followed by donkey anti-mouse, donkey anti-goat or goat anti-rabbit IgG conjugated to HRP for 1 h at room temperature. After washing, the immune complexes were detected by the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Statistical analysis
The data were expressed as mean ± SEM (standard error of the mean). Statistical evaluation was performed by unpaired two-tailed Student’s t test between two groups and by one-way ANOVA with Dunnett’s multiple comparisons or two-way ANOVA with Tukey’s multiple comparisons or Sidak’s multiple comparisons within multiple groups as indicated using GraphPad Prism 8.0 software. For neuron intersection numbers quantified from the Sholl analysis, two-way ANOVA with Sidak’s multiple comparisons within groups at the identical distance from soma were performed. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 are considered significant and ns represents not significant. The precise sample number (n) was provided to indicate the number of independent biological samples in each experiment. All immunoblots were repeated at least three times independently with similar results.
Results
PARP-1 is hyperactivated and mediates TBI-induced neuronal cell death
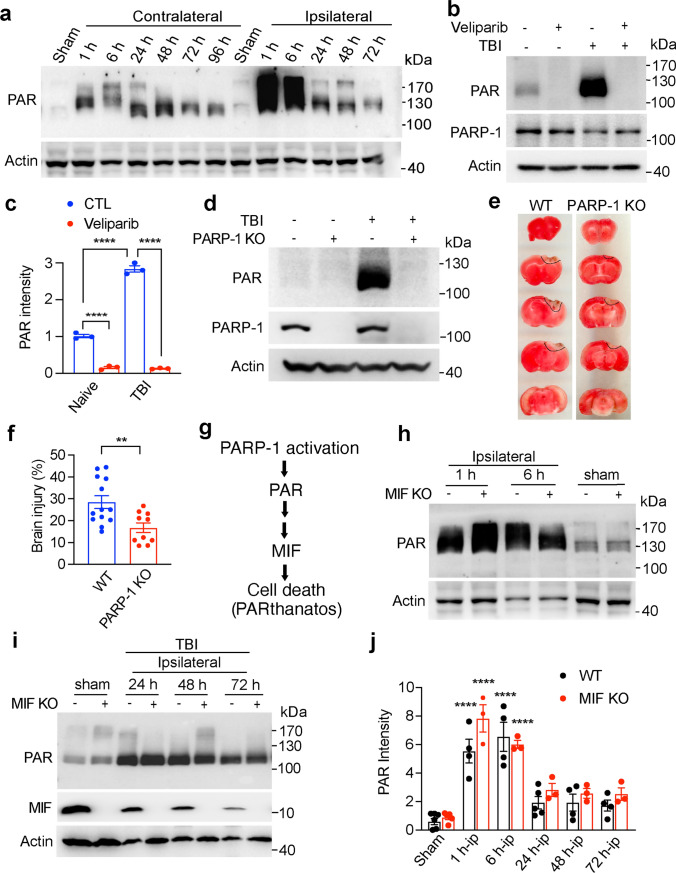
PAR, which is a bioproduct of PARP-1 activation, is a cell death signal and triggers PARthanatos contributing to ischemic neuronal cell death [7, 8]. To explore whether PARP-1 is hyperactivated and PARthanatos is involved in TBI, the PARP-1 activity was examined at 1 h, 6 h, 24 h, 48 h, and 72 h after CCI. We found that PARP-1 was hyperactivated at 1 h and 6 h in the ipsilateral side after TBI and then gradually decreased during 24 h–72 h post TBI (Fig. 1a). However, its PAR levels were still higher than those of the sham group during the first 24–72 h after TBI (Fig. 1a). The smear bands represent proteins covalently or non-covalently labeled with different length of linear or side chains of PAR during 1–72 h post TBI. Intraperitoneal (IP) injection of PARP inhibitor Veliparib (100 mg/kg every 8 h) completely blocked TBI-induced PAR accumulation at day 1 post TBI (Fig. 1b, c). In line with that, genetic deletion of PARP-1 (Fig. 1d) significantly reduced PAR accumulation and subsequent brain injury at day 3 post TBI (Fig. 1e, f). PARP-1 KO mice preserved more NeuN+ neurons in the injured cortex and reduced the number of degenerating/dying neurons indicated by Fluoro-Jade C (FJC) and TUNEL staining (Supplemental Fig. 1a–g). However, the cleaved caspase 3 staining did not reveal obvious difference between PARP-1 WT and KO mice 3 days following TBI (Supplemental Fig. 1a, b), which supports the previous findings that PARthanatos is independent of caspase-3 [22]. MIF was recently identified as an executor of PARthanatos, which cleaved DNA into large DNA fragmentation and caused cell death following ischemic stroke [7]. Therefore, MIF functions as a downstream factor of PARP-1 hyperactivation (Fig. 1g). As expected, MIF KO did not obviously affect PARP-1 activity in the ipsilateral side (Fig. 1h–j). The peak of PARP-1 activation was observed at 1 h and 6 h after TBI in the ipsilateral side of both WT and MIF KO mice (Fig. 1a, h–j). These data indicate that PARP-1 is hyperactivated in the ipsilateral side following TBI and PARthanatos contributes to TBI-induced neuronal cell death.
Fig. 1.
Effects of PARP-1 and MIF on PAR formation and brain injury following TBI. a PARP-1 activation following TBI on the ipsilateral and contralateral brain. b PARP inhibitor Veliparib (100 mg/kg, IP injection every 8 h) blocked PAR accumulation at 24 h following TBI. c Quantification of TBI-induced PAR accumulation with or without Veliparib treatment shown in b. Data are shown as mean ± SEM and analyzed for statistical significance by one-way ANOVA. ****p < 0.0001. d Expression of PAR and PARP-1 in PARP-1 WT and KO mice at 24 h post TBI. e Representative TTC images of WT and PARP-1 KO mice 3 days post TBI. f Quantification of the effects of PARP-1 KO on brain injury 3 days post TBI. The brain injury volume was calculated as % of hemisphere. n = 12 (WT) and n = 10 (PARP-1 KO). Data are shown as mean ± SEM and analyzed for statistical significance by two-tailed Student’s t test. **p < 0.01. g The cascade of PARthanatos pathway. PARP-1 activation functions as an upstream factor of MIF involved in PARthanatos. h–j Relative PAR intensity in the ipsilateral side at different time points following TBI in both WT and MIF KO mice (n = 3–5/group). Data are shown as mean ± SEM and analyzed for statistical significance by one-way ANOVA. ****p < 0.0001 vs. sham
MIF KO protects dendritic spine density and morphological complexity in neurons and reduces neuronal cell death following TBI
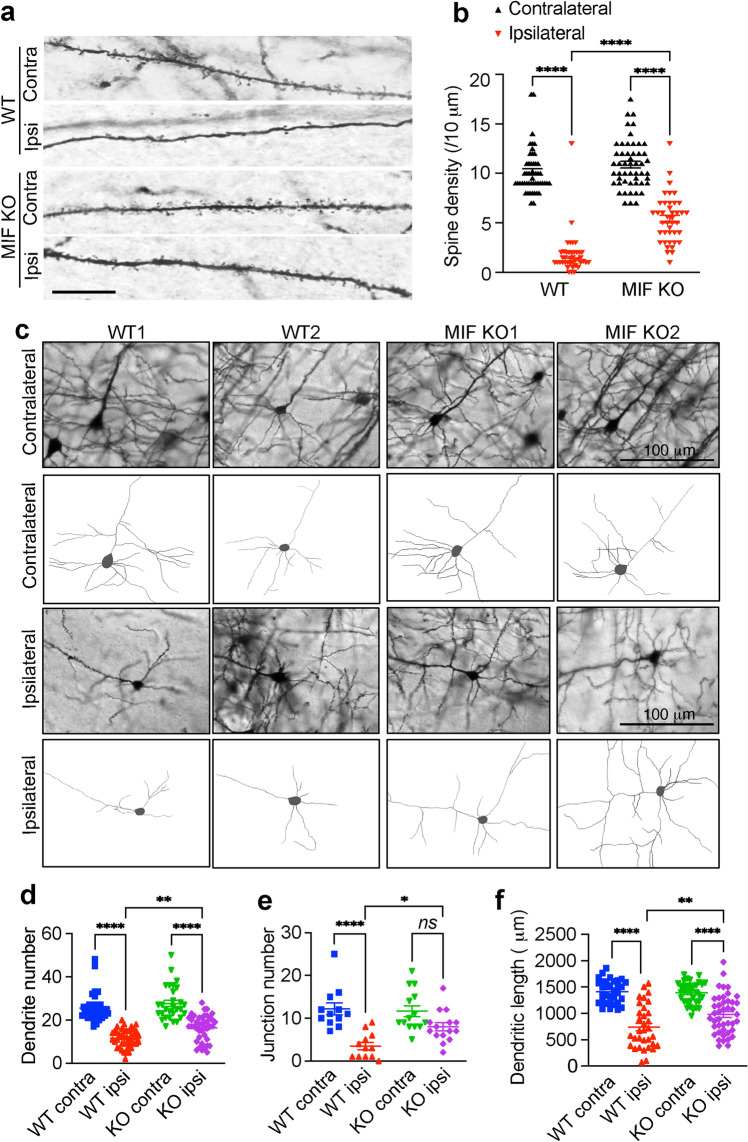
To study the role of MIF in TBI-induced neurodegeneration and neuronal cell death, MIF WT and KO mice were impacted for one time at 5 m/s speed using a 3 mm diameter convex tip with a 20 ms dwell time and 2.0 mm depression. Dendritic spine loss and neuronal morphological alteration were first examined 1 day post TBI. TBI significantly reduced dendritic spine density of MIF WT as well as KO neurons in the layer V cortex of the ipsilateral side compared to its contralateral side, whereas the spine density of MIF KO neurons was significantly higher than that of WT neurons (Fig. 2a, b). By tracing neurons in the ipsilateral side of WT and KO, we found that the dendrite number, junction number, and total dendritic length were significantly reduced in MIF WT neurons but were protected in MIF KO neurons (Fig. 2c–f).
Fig. 2.
MIF KO preserves dendritic spine density and neuronal morphology following TBI. Dendritic spine density (a) and quantification (b) in WT and MIF KO mouse cortex 1 day after TBI. Images (n = 50 and n = 41, respectively) in the contralateral and ipsilateral sides of WT and KO mice for quantification. Contra, contralateral; Ipsi, Ipsilateral. Scale bar, 10 µm. c Golgi staining and neuron tracing in WT and KO mice 1 day after TBI. Quantification of dendrite number (d, n = 38, n = 42, n = 29, n = 42 images in order for the contralateral and ipsilateral sides of WT and KO neurons), junction number (e, n = 12, n = 12, n = 14, n = 15 images in order for the contralateral and ipsilateral sides of WT and KO neurons) and total dendritic length (f, n = 33, n = 33, n = 35, n = 43 images in order for the contralateral and ipsilateral sides of WT and KO neurons) of WT and KO neurons after TBI. Data are shown as mean ± SEM and analyzed for statistical significance by one-way ANOVA. ****p < 0.0001, **p < 0.01, *p < 0.05
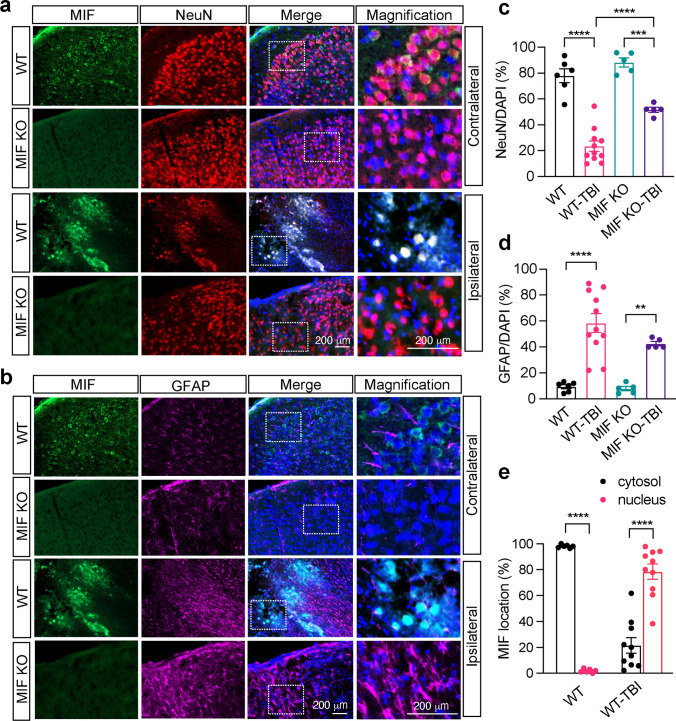
To determine which types of cells express MIF in brain and whether MIF localization is altered in these cells to promote neuronal cell death following TBI, MIF immunostaining was performed and overlayed with a neuron marker NeuN (Fig. 3a) as well as an astrocyte marker GFAP (Fig. 3b). In the contralateral side, MIF was mainly expressed in the cytosol of NeuN+ neurons, but not GFAP+ astrocytes (Fig. 3a, b). There were comparable levels of NeuN+ neurons observed in the contralateral cortex in both MIF WT and KO mice (Fig. 3a, c). Similar results were also observed in sham animals (Supplemental Fig. 2). In the ipsilateral side, NeuN+ cells were significantly reduced after TBI in WT MIF mice (Fig. 3a, c). However, MIF KO mice had much more NeuN+ cells than MIF WT mice, indicating that MIF KO protects neurons from TBI-induced brain injury (Fig. 3a, c). Few GFAP+ astrocytes were in the contralateral side, but these cells were clearly activated in the ipsilateral sides in both WT and MIF KO mouse brain (Fig. 3b, d). No obvious difference of GFAP+ cells was observed between MIF WT and KO mice. In line with the robust neuron loss in the ipsilateral side of WT mice, MIF was clearly translocated from the cytosol to the nucleus following TBI and overlayed with NeuN in the ipsilateral side next to the damage core, but not with GFAP (Fig. 3a, e). These results indicate that MIF nuclear translocation might be an early indicator of neuronal cell death.
Fig. 3.
Expression of MIF in the cortex of mouse brain 1 day after TBI. Co-immunostaining of MIF/NeuN (a) and MIF/GFAP (b) in the contralateral (non-TBI) and ipsilateral brain cortex of MIF WT and KO mice next to the damage core 1 day after TBI. Quantification of relative NeuN+ neurons/DAPI per field (c) and GFAP+ astrocytes/DAPI per field (d) in the contralateral (non-TBI) and ipsilateral brain cortex of MIF WT and KO mice 1 day after TBI. Images (n = 6, n = 11, n = 5, n = 5 in order) in the contralateral and ipsilateral sides of WT and KO mice for quantification. e Quantification of the percentage of MIF localization in the cytosol and nucleus per field in the contralateral (non-TBI) and ipsilateral brain cortex of MIF WT mice 1 day after TBI. Images (n = 6 and n = 10, respectively) in the contralateral and ipsilateral sides of WT mice for quantification. Data in c–e are shown as mean ± SEM and analyzed for statistical significance by one-way ANOVA. ****p < 0.0001, ***p < 0.001, **p < 0.01
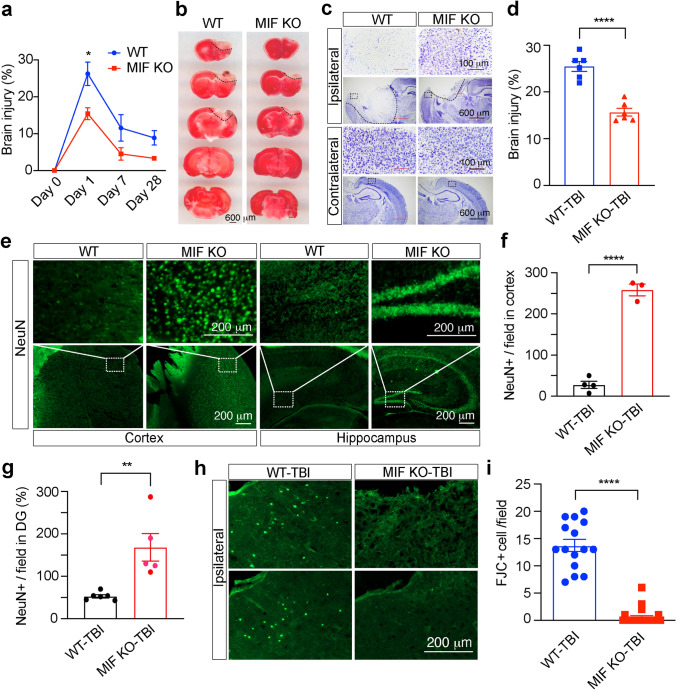
Our recent in vitro cortical neuronal culture studies showed that MIF knockdown by three different shRNAs or genetic deletion of MIF blocked N-methyl-d-aspartate (NMDA)-induced large DNA fragmentation and neuronal cell death, which can be reversed by expression of WT MIF, but not nuclease-deficient E22A MIF [7], supporting a critical role of MIF in neuronal cell death. To further explore the role of MIF in TBI-induced neuronal cell death, brain injury was examined 1, 7 and 28 days after TBI. TTC staining showed the brain injury was reached maximum at day 1 after TBI and then gradually reduced at days 7 and 28 after TBI in WT mice (Fig. 4a). In contrast to WT mice, MIF KO mice had much smaller brain injury volume after TBI (Fig. 4a, b). Consistently, Nissl staining revealed more neuronal cell loss in the cortex of the ipsilateral side of WT mouse brain than that of MIF KO mouse brain (Fig. 4c, d), which was confirmed by NeuN staining in the cortex next to the mechanical injury site as well as hippocampus in the ipsilateral side (Fig. 4e–g). FJC staining further supported that MIF promoted neurodegeneration in the ipsilateral cortex of WT mice, which was clearly reduced by MIF KO (Fig. 4h, i). These results reveal that MIF KO protects neurons from TBI-induced neurodegeneration and cell death.
Fig. 4.
MIF KO reduced the TBI volume. a Brain injury volume at day 0 before TBI (n = 3/group for WT and KO), day 1 (n = 6/group for WT and KO), day 7 (n = 6/group for WT and KO) and day 28 (n = 18 for WT and n = 8 for KO) following TBI determined by TTC staining. Data are shown as mean ± SEM. *p < 0.05 by two-way ANOVA Sidak’s multiple comparisons test. b Representative images of TTC staining of MIF WT and KO mice 1 day after TBI. Representative images (c) and injury quantification (d) of Nissl staining with MIF WT and KO mice 1 day after TBI. n = 6 mice per group. ****p < 0.0001. NeuN staining (e) and quantification (f, g) in the cortex next to the TBI site of MIF WT and KO mice (e, left two panels) and hippocampus (e, right two panels). Images [n = 4 (WT-TBI in f), n = 3 (KO-TBI in f), n = 6 (WT-TBI, g), n = 5 (KO-TBI, g)] from 3 pairs of MIF WT and KO mice for quantification. Fluoro-Jade C (FJC) staining (h) and quantification (i) in the cortex of MIF WT and KO mice 7 days following TBI. Images [n = 15 (WT-TBI), n = 23 (KO-TBI)] of MIF WT and KO mice for quantification. Data in a, d, f, g, i are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA (a) and unpaired two-tailed Student’s t test (d, f, g, i). ****p < 0.0001, **p < 0.01, *p < 0.05
Taken together, our findings indicate that MIF is mainly expressed in neurons and contributes to dendritic spine loss, morphological alteration and neuronal cell death following TBI. Moreover, MIF nuclear translocation provides a strong prediction of neuron loss following TBI.
Characterization of MIF function in mouse brain under physiological conditions
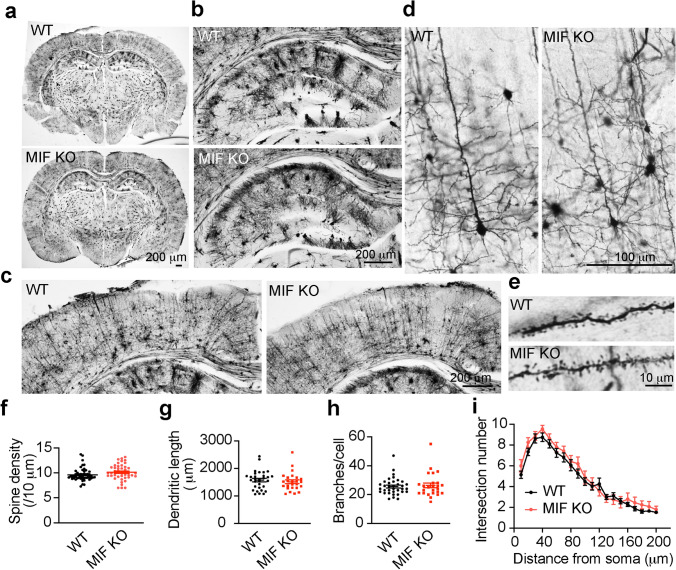
Given the importance of MIF in promoting neurodegeneration and neuronal cell death following TBI, it is essential to study whether MIF can be a safe target for TBI treatment. Therefore, we examined whether MIF KO has any adverse effect on spine density and neuronal morphology and whether MIF KO itself causes neurodegeneration in mouse brain under physiological conditions. We found that MIF KO mice were born at the expected Mendelian frequency and grew normally. These mice lived even longer [35]. By Golgi staining (Fig. 5a), MIF KO mice showed a similar brain size and neuronal cell density in the hippocampus (Fig. 5b) and cortex (Fig. 5c). No obvious cell loss was observed in MIF KO mouse brain. Further analysis of neuronal morphological complexity, dendritic spine density, and dendritic length, branches and intersection at different distance from soma revealed no obvious difference between WT and KO mice (Fig. 5d–i). These data indicate that MIF KO itself does not obviously alter neuronal morphology or cause neuronal injury in mouse brain, supporting the notion that targeting MIF might be a safe strategy to prevent neurodegeneration and neuronal cell death post TBI.
Fig. 5.
Characterization of dendritic spine density and neuronal morphology in MIF WT and KO mice under physiological conditions by Golgi staining. a Overview of whole brain. b Overview of hippocampus. c Overview of cortex. d Golgi staining of neurons in cortex. e Zoom in images of spines. f–i Quantification of dendritic spine density (f, n = 48 for WT and n = 43 for KO), dendritic length (g, n = 31 for WT and n = 25 for KO), branches (h, n = 35 for WT and n = 25 for KO) and intersection number at different distance from soma (i, n = 36 for WT and n = 25 for KO)
MIF-mediated neuronal cell death following TBI is independent of its proinflammatory effect on neuroinflammation
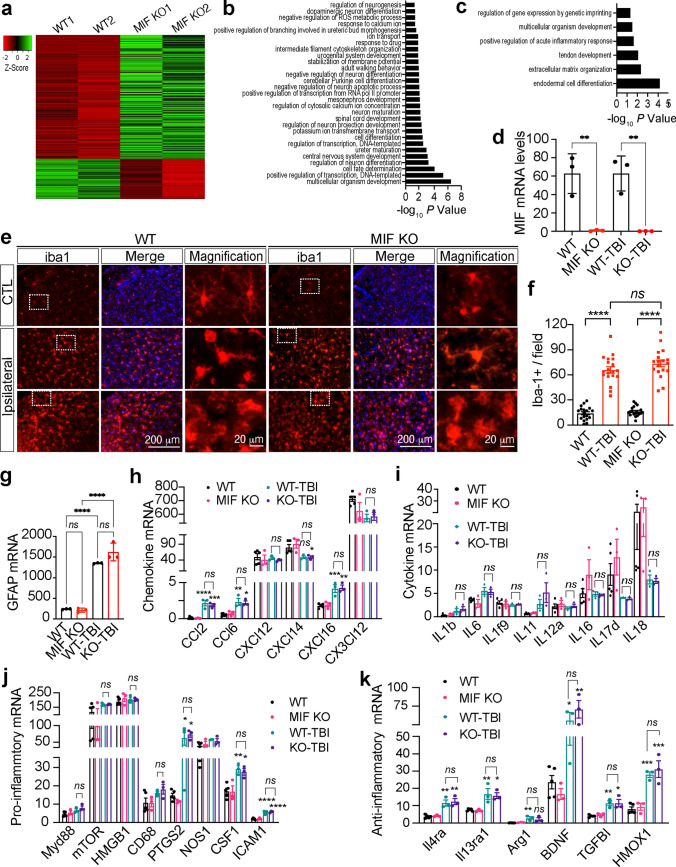
To study whether MIF mediates TBI-induced neurodegeneration and neuronal cell death via altering the expression of proinflammatory cytokines and chemokines, we first performed RNA-sequencing analysis in mouse ipsilateral forebrain. Total RNAs were extracted from both MIF WT and KO mice 1 day after TBI and mice (Balb/C background) without TBI were used as controls. According to the mRNA profiling, MIF KO upregulated 279 genes and downregulated 78 genes under physiological conditions (FDR > 0.05, LogCPM > 0, and fold change > 1.5) (Fig. 6a, Source Data-Table 1). Gene ontology analysis showed that MIF KO-upregulated genes (p < 0.05) were involved in positive regulation of neurogenesis, branching morphogenesis, neuron differentiation and maturation, animal behavior, and negative regulation of reactive oxygen species (ROS) metabolic and neuron apoptotic processes (Fig. 6b). MIF KO-downregulated genes were involved in processes including acute inflammatory response, extracellular matrix organization and organism development (Fig. 6c). At day 1 post TBI, MIF KO increased expression of 57 genes mainly involved in animal behavior regulation and decreased expression of 42 genes involved in inflammation or immune response (Supplemental Fig. 3a–c, Source Data-Table 2). However, the number of differentially expressed genes by MIF was minimal as TBI significantly upregulated 643 genes but downregulated 216 genes (fold change > 1.5) in both MIF WT and KO mouse brain (Supplemental Fig. 3d, e, Source Data-Table 3), suggesting that MIF has very little if any effect on TBI-induced gene expression. The majority of TBI-upregulated genes were involved in three pathways, including (1) defense response/wounding response/inflammatory response/leukocyte activation; (2) metabolic response/biosynthetic process; and (3) cell death regulation (Supplemental Fig. 3d, e). MIF KO in each mouse was confirmed at mRNA and protein levels (Figs. 1i, 6d).
Fig. 6.
Effects of MIF on gene expression and its role in neuroinflammation under physiological and TBI conditions. a Representative heatmap of MIF KO-up- and down-regulated gene expression under physiological conditions (fold change > 1.5) from a single RNA-sequencing dataset. Total 2 independent RNA-sequencing experiments were performed with n = 5 (WT mice) and n = 3 (KO mice). b, c Pathway analysis of MIF KO-up- and down-regulated gene expression under physiological conditions. d. MIF mRNA expression in MIF WT and KO mice without TBI and 1 day post TBI. n = 3 mice per group. e, f Expression and quantification of iba1+ cells in the contralateral (non-TBI) and ipsilateral (TBI) sides of MIF WT and KO mouse brains 1 day post TBI. Images (n = 20 per group) obtained from MIF WT and KO mice for quantification. g GFAP mRNA expression in MIF WT and KO mice without TBI and 1 day post TBI. n = 3 per group. h mRNA expression of chemokines in MIF WT and KO mice without TBI and 1 day post TBI. n = 3 mice per group. i mRNA expression of cytokines in MIF WT and KO mice without TBI and 1 day post TBI. n = 3 mice per group. mRNA expression of pro-inflammatory (j) and anti-inflammatory (k) genes in MIF WT and KO mice without TBI and 1 day post TBI. n = 3 mice per group. Data in d, f–k are shown as mean ± SEM and analyzed for statistical significance by one-way ANOVA. ****p < 0.0001, **p < 0.01. ns not significant
Since MIF was previously recognized as a secreted proinflammatory factor, we next studied whether MIF KO alters microglial activation following TBI, which is a hallmark of neuroinflammation. By iba1 staining, we found that microglial cells displayed a hyper-ramified morphology on the contralateral side of both MIF WT and KO mouse brain (Fig. 6e). However, the number of microglia was significantly increased in the ipsilateral side of MIF WT and KO mouse brain even 7 days post TBI (Fig. 6e, f). The shape of microglial cells was also altered significantly. Both bushy- and amoeboid-shaped microglial cells were observed in the cortex of MIF WT and KO mice 7 days post TBI, indicating hyperactivation of microglia at least during the first 7 days following TBI. Overall, no differences of microglial morphology and number were found between MIF WT and KO mice (Fig. 6e, f). Similar to microglia, astrocytes were activated following TBI as GFAP was increased at both mRNA and protein levels (Figs. 3b, 6g). MIF KO did not interfere with TBI-induced astrocyte activation (Figs. 3b, 6g).
Activated microglia are often classified into M1 and M2 microglia. M1 microglia produce pro-inflammatory cytokines/chemokines including interleukin (IL)1b, IL6, IL12, C–X–C motif chemokine ligand 8 (CXCL8), C–C motif chemokine ligand 2 (CCL2), and C–X3–C motif chemokine ligand 1 (CX3CL1), whereas M2 microglia produce anti-inflammatory cytokines/chemokines/factors including IL-4, IL10, IL13, transforming growth factor beta induced (TGFBI), brain-derived neurotrophic factor (BDNF), insulin-like growth factor (IGF-1) and heme oxygenase 1 (HMOX-1) [36, 37]. By RNA-seq, we found that TBI indeed significantly increased the expression of some chemokines including CCL2, CCL6, CXCL14, CXCL16 (Fig. 6h, i), but failed to regulate other cytokines or chemokines detected in MIF WT and KO mice. MIF KO did not alter the expression of any detectable chemokines (CCL2, CCL6, CXCL12, CXCL14, CXCL16 and CX3CL12) or cytokines (IL1b, IL6, IL1f9, IL11, IL12a, IL16, IL17d, and IL18) in mouse brain under both physiological and TBI conditions (Fig. 6h, i). In addition to cytokines/chemokines, we also studied the effect of MIF on the expression of the associated markers for pro-inflammatory response including MyD88, mTOR and high mobility group box 1 (HMGB1). We found that TBI did not regulate Myd88, mTOR, HMGB1, CD68 or nitric oxide synthase 1 (NOS1). In contrast, TBI significantly increased the expression of prostaglandin-endoperoxide synthase 2 (PTGS2), colony stimulating factor 1 (CSF1), and intercellular adhesion molecular 1 (ICAM1) (Fig. 6j). Interestingly, TBI also significantly increased a group of anti-inflammatory gene expression, including IL4ra, IL13ra1, Arg1, BDNF, TGFBI, HMOX1, indicating that both M1 and M2 microglia are activated following TBI (Fig. 6k). Again, MIF KO had no influence on the expression of pro- or anti-inflammatory genes (Fig. 6k). Taken together, these findings indicate that MIF-mediated neurodegeneration and neuronal cell death following TBI is independent of its effect on proinflammatory gene expression.
MIF nuclease activity is required for TBI-induced neurodegeneration and neuronal cell death
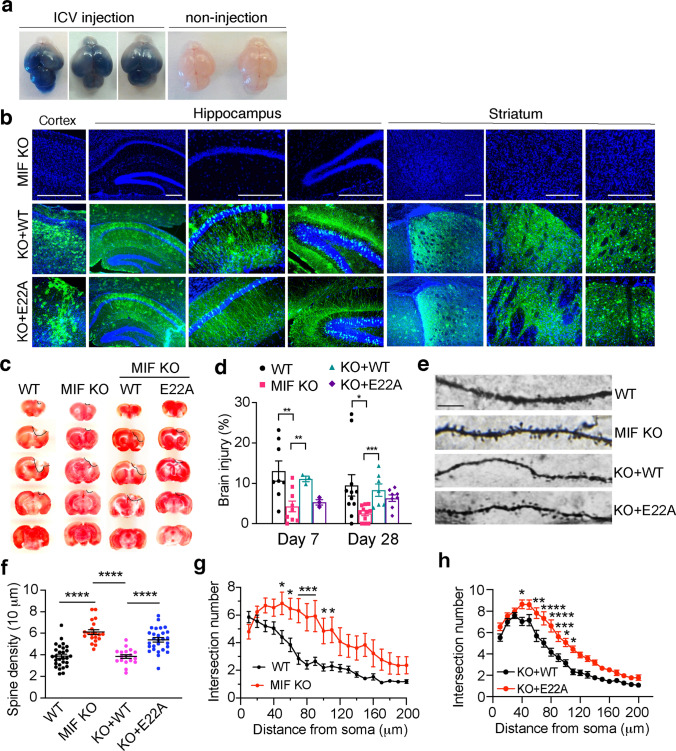
Recently, MIF has been identified as a novel nuclease that cleaves DNA into large fragments and causes neuronal cell death [7]. To examine whether MIF’s nuclease activity plays a role in TBI-induced neuronal cell death, we performed ICV injection with AAV encoding WT-MIF or nuclease-deficient E22A MIF mutant in newborn MIF KO pups (P0). The efficiency of ICV injection at P0 was consistent and widespread in the entire brain as indicated by trypan blue (Fig. 7a). The expression of WT and E22A MIF in the cortex, hippocampus, and striatum was also confirmed in 3-month-old mice without brain injury by immunostaining (Fig. 7b), which is highly consistent with our previous tests [7]. Mice after TBI treatment and TTC staining for the brain injury volume measurement were also verified for the success of ICV injection. Only those successfully injected mice were included in the study. TBI-induced brain injury was compared at days 7 and 28 after TBI. MIF KO significantly reduced TBI-induced injury volume at day 7 as well as day 28 after TBI. Expression of WT-MIF in MIF KO mouse brain restored TBI-induced brain injury at both day 7 and day 28 post TBI, whereas expression of E22A MIF did not obviously reverse the phenotype (Fig. 7c, d). In parallel with this experiment, the effects of MIF’s nuclease activity on long-term dendritic spine loss and neuronal morphological alteration were also examined at day 28 post TBI. MIF KO clearly suppressed TBI-induced reduction of dendritic spine density and intersection number (Fig. 7e–g). Overexpression of WT MIF, but not nuclease-deficient E22A MIF in MIF KO mice, significantly reduced dendritic spine density as well as dendritic intersection number (Fig. 7e, f, h). These findings reveal that MIF and its nuclease activity are required for TBI-induced neurodegeneration and neuronal cell death.
Fig. 7.
MIF nuclease activity is required for inducing brain injury and altering dendritic spine density and neuronal morphology following TBI. a Representative images of the efficiency and expansion pattern of ICV injection at P0 indicated by trypan blue. 10 mice with ICV injection and 5 mice with non-injection were from 3 independent experiments. b Representative expression of WT and nuclease-deficient mutant E22A-MIF in cortex, hippocampus and striatum of MIF KO mice 3 months after ICV injection without brain injury. c TTC staining of MIF WT, KO, and KO mice expressing WT or nuclease-deficient MIF at day 28 post TBI. d Quantification of TBI-induced injury at day 7 and day 28 in MIF WT (n = 8 and n = 11, respectively), KO (n = 9 and n = 11, respectively), and KO mice expressing WT (n = 3 and n = 7, respectively) or nuclease-deficient E22A MIF (n = 3 and n = 8, respectively). Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Sidak’s or Tukey’s multiple comparisons test. ***p < 0.001, **p < 0.01, *p < 0.05. e Representative images of dendritic spine in WT, KO, and KO mice expressing WT and nuclease-deficient mutant E22A-MIF at day 28 following TBI. f Quantification of the spine density in WT, KO, and KO mice expressing WT and nuclease-deficient mutant E22A-MIF at day 28 post TBI. n = 28 (WT); n = 20 (KO); n = 18 (KO + WT); n = 28 (KO + E22A) images. Data are shown as mean ± SEM and analyzed for statistical significance by one-way ANOVA with Sidak’s multiple comparisons test. ****p < 0.0001. g–h Quantification of the intersection number of neurons in WT (n = 21), KO (n = 15), and KO mice expressing WT (n = 31) and nuclease-deficient mutant E22A-MIF (n = 32) at day 28 post TBI by Sholl analysis. Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Sidak’s multiple comparisons test. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05
MIF impairs behavioral recovery in mice with TBI through its nuclease activity
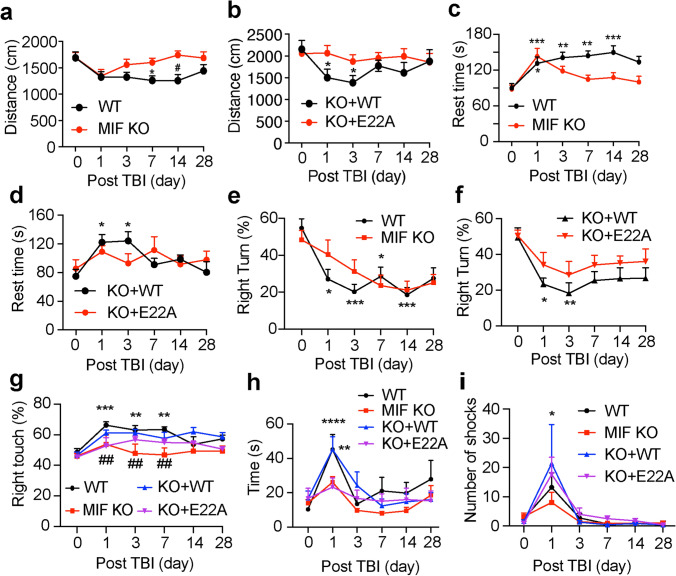
Finally, we studied the effects of MIF and its nuclease activity on animal rehabilitation following TBI. Animal behavioral tests including open field test, corner test, beam test, cylinder test, and treadmill test were performed to study animal’s spontaneous activity, sensory and motor functions before TBI as day 0 and at days 1, 3, 7, 14 and 28 following TBI. TBI was performed in the right hemisphere. The spontaneous locomotor activity was reduced in both WT and MIF KO mice at day 1 post TBI as indicated by the total distance of open field test (Fig. 8a). At day 3 post TBI, MIF KO mice started to recover and had better improvement than WT mice for the time period from day 3 to day 14 post TBI, but no obvious difference was observed between WT and MIF KO mice at day 28 post TBI (Fig. 8a). Overexpression of WT MIF in MIF KO mice significantly reduced the spontaneous activity at days 1 and 3 post TBI, when compared to overexpression of nuclease-deficient E22A MIF mutant, although the spontaneous activity was slowly restored in WT MIF rescued mice at days 7, 14 and 28 post TBI (Fig. 8b). In line with the total distance measurement, analysis of the rest time (speed < 2.5 cm/s) showed that TBI significantly increased the rest time in WT mice at day 1–day 28 post TBI as well as WT MIF rescued mice at day 1 and day 3 post TBI (Fig. 8c, d). In contrast, TBI did not obviously increase the rest time in MIF KO mice or MIF KO mice expressing nuclease-deficient E22A at day 3–day 28 post TBI (Fig. 8c, d).
Fig. 8.
Loss of MIF or its nuclease activity improved mouse neurobehavioral recovery following TBI. a–d Open field test of WT (n = 18), KO (n = 19), and KO mice expressing WT (n = 17) or E22A (n = 15) at days 0, 1, 3, 7, 14 and 28 post TBI. Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05 vs. day 0. e–f Corner test of WT (n = 28), KO (n = 23), and KO mice expressing WT (n = 14) or E22A (n = 14) at days 0, 1, 3, 7, 14 and 28 post TBI. Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Tukey’s multiple comparisons test. ***p < 0.001, **p < 0.01, *p < 0.05 vs. day 0. g Cylinder test of WT (n = 13), KO (n = 9), and KO mice expressing WT (n = 10) or E22A (n = 14) at days 0, 1, 3, 7, 14 and 28 post TBI. Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Tukey’s multiple comparisons test. ***p < 0.001, **p < 0.01 vs WT at day 0; ##p < 0.01 vs. WT at days 1, 3, 7 post TBI, respectively; h beam test of WT (n = 28), KO (n = 27), and KO mice expressing WT (n = 10) or E22A (n = 8) at days 0, 1, 3, 7, 14 and 28 post TBI. Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Tukey’s multiple comparisons test. ****p < 0.0001, **p < 0.01 vs. day 0. i Treadmill test of WT (n = 33), KO (n = 37), and KO mice expressing WT (n = 9) or E22A (n = 6) at days 0, 1, 3, 7, 14 and 28 post TBI. Data are shown as mean ± SEM and analyzed for statistical significance by two-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05 vs. day 0
The corner test was performed to assess sensory and motor deficits following both cortical and striatal injury [7]. Prior to TBI, both WT and KO mice showed a similar percentage of right and left turns in the corner test. However, the right turn was significantly reduced in WT mice, but not MIF KO mice, at days 1, 3, 7, 14 following TBI (Fig. 8e). Similarly, MIF KO mice expressing WT MIF showed a significant reduction of right turn at days 1 and 3 following TBI, whereas MIF KO mice expressing nuclease-deficient E22A MIF mutant showed slightly reduced right turn but without significance (Fig. 8f). On the other hand, the cylinder test was performed to assess the forelimb asymmetry performance by studying the preference of use right or left paw. TBI at the right hemisphere significantly increase the use of right paw of WT mice to stand up inside the cylinder at days 1, 3 and 7 post injury (Fig. 8g). MIF KO mice did not show any obvious preference of using right or left paw (Fig. 8g). Overexpression of WT MIF in MIF KO mice revealed a tendency to increase the preference of using right paw post TBI (Fig. 8g).
Next, the beam test was performed to determine mouse balance and fine motor coordination functions before and post TBI. TBI clearly increased the time of WT mice climbing through the 60 cm-length of beam at day 1 post TBI and these mice recovered at day 3 post TBI (Fig. 8h). However, TBI did not significantly disturb MIF KO mice’s ability to climb through the beam (Fig. 8h). Overexpression of WT MIF, but not nuclease-deficient E22A MIF, in MIF KO mice restored the slow climbing phenotype as WT mice at day 1 post TBI. No obvious difference was observed at the late phase of recovery.
The treadmill test utilizes a shock grid, which forces mice to quickly learn and keep running to maintain certain distance from the shock grid. Before TBI, all groups of mice learned rapidly to avoid the shock while running at speed of 12 cm/s. At day 1 post TBI, injury clearly increased the shock numbers in all groups. Starting at day 3 post TBI, all groups of mice were again able to keep running to avoid the shock. No obvious difference was observed between WT-MIF and KO-MIF mice with the treadmill test (Fig. 8i).
Taken together, MIF KO or loss of MIF nuclease activity shows a beneficial improvement of early neurobehavioral recovery, especially the spontaneous activity and motor functions.
Discussion
In the present study, we provide direct evidence that PARP-1 is rapidly hyperactivated to produce PAR leading to neuronal cell death in the ipsilateral side following TBI and PARP-1 KO protects mice from TBI. PARP-1 activation occurs in a time-dependent manner in mouse brain after TBI. Previous pharmacological studies revealed a protective role of PARP-1 inhibition in brain damage and functional recovery in the murine model of TBI [38–40]. Our results suggest that PARthanatos plays a pivotal role in neuronal cell death following TBI and early interference with PARP-1 activation may provide maximal therapeutic benefits. Increased PARP-1 activation has been observed in many neurological diseases including stroke, AD, and PD. A recent study showed that α-synuclein accumulation activates PARP-1 to increase PAR formation and dopaminergic cell death through PARthanatos. PARP-1 inhibitors or genetic KO prevents neuronal cell death [11, 41, 42]. These findings support a pivotal role of PARP-1 and PARthanatos in neuronal cell death in the broad neurological disorders.
MIF is widely expressed in various cell types, including neurons, monocytes, macrophages, vascular smooth muscle cells, and cardiomyocytes [43, 44]. The expression of MIF was reported to be upregulated in both mouse neurons and astrocytes following ischemic stroke [20, 21]. Here, we found that MIF is primarily expressed in mouse neurons, but not astrocytes, under physiological conditions. Although the overall expression of MIF in the brain is not obviously altered or even slightly reduced following TBI due to the neuron loss, the expression pattern of MIF is clearly altered from the cytosol to the nucleus of neurons in the damage core and strongly associated with increased neuronal cell death. This notion is also supported by our recent findings that MIF is recruited by AIF and translocated to the nucleus of cortical neurons following the ischemic injury as shown by both immunostaining and subcellular fraction studies [7]. Interfering with MIF–AIF interaction to block MIF nuclear translocation prevents ischemic neuronal cell death [7]. These data together indicate that MIF nuclear translocation could be an early indicator of neuronal injury.
Here, we found that MIF KO protects neurons by preserving dendritic spine density and neuronal morphological complexity and attenuating TBI-induced neuron loss and injury volume in mice. MIF has been well appreciated to function as a pro-inflammatory mediator released from cells and regulate immune and inflammatory responses including neuroinflammation [18, 45]. Although TBI indeed activates both M1 and M2 microglia and upregulates a subset of pro-inflammatory and anti-inflammatory genes, MIF KO does not obviously alter the expression of any detectable cytokines and chemokines under physiological conditions and during the first 7 days post TBI. TBI-altered gene expression is likely to play a role in pathogenesis of brain injuries, however, we exclude a possible role of MIF in regulation of inflammatory genes following TBI. This observation is highly consistent with another previous study showing that MIF KO does not affect hallmarks of inflammatory responses during the first week after ischemic stroke [21]. On the other hand, although MIF KO has no effect on those TBI-sensitive genes, MIF KO does increase the expression of a group of genes involved in either positive regulation of neurogenesis, branching morphogenesis, neuron differentiation and maturation, animal behavior or negative regulation of ROS metabolic and neuron apoptotic processes under physiological conditions (Fig. 6b). These findings indicate that MIF contributes to TBI-induced neurodegeneration and subsequent neuronal cell death via its unappreciated functions rather than its role in pro-inflammatory response.
We recently identified MIF as a 3′ nuclease and the executor of PARthanatos in ischemic stroke. In this study, we showed that TBI-induced dendritic spine loss, neuronal morphological complexity alteration and neuronal cell death were blocked by MIF deletion. Moreover, the neuroprotective effects of MIF KO were reversed by the expression of WT MIF but not its nuclease-deficient MIF E22A mutant in mice, supporting that MIF’s nuclease activity is required for neurodegeneration, neuronal cell death and behavioral impairment in mice with TBI. Our study also provides evidence that PARP-1/MIF-mediated PARthanatos contributes to TBI in vivo.
Apart from studying neuronal morphological degeneration and neuronal cell death, we also performed motor functional related behavioral tests, which are critical to reflect mouse functional changes in vivo. Open field test, corner test, cylinder test, beam test, and treadmill test before TBI and 28 days after TBI allow us to determine the effect of MIF on long-term mouse spontaneous activity, sensory activity, balance ability and forced/life threaten movement ability. These behavioral results strongly support that MIF KO and nuclease-deficient mice have better and early recovery following TBI than MIF WT mice. However, MIF expression does not obviously affect the forced/life threaten movement ability.
Functions of PARP-1 and MIF are highly context dependent [9, 46]. In response to severe brain injury or DNA damage, PARP-1 hyperactivation and MIF nuclear translocation trigger PARthanatic neuronal cell death as shown in this study and our previous ischemic stroke study [7]. However, in response to mild stress or DNA damage, PARP-1 has been shown to play a role in DNA repair and MIF has been shown to facilitate DNA replication in cancer cells [47]. Therefore, the functions of PARP-1 and MIF in the contralateral side might be different from those in the ipsilateral side post TBI. Although MIF is not expected to alter its upstream PARP-1 activation in the ipsilateral side of TBI, we do not exclude the possibility that MIF might affect DNA damage response and PARP-1 activation in the contralateral side. Further studies are needed to test the precise functions of MIF and PARP-1 in the contralateral side.
TBI is often induced by sports, car accidents, falls, and other daily occurrences, in addition to blast explosions. Unfortunately, no effective treatment is currently available. MIF KO mice are normal in appearance and give birth as the expected Mendelian rate. Our studies here revealed that MIF KO does not affect murine neuronal morphology or its spontaneous neurobehavior under physiological conditions. Moreover, MIF impairs/delays recovery of the spontaneous activity and motor functions in mice following TBI, which requires its nuclease activity. These findings indicate that targeting MIF and its nuclease activity could be a potential strategy to prevent neurodegeneration and the secondary brain injury following TBI.
Taken together, we found that PARP-1 is hyperactivated and MIF plays a pivotal role in dendritic spine degeneration and neuronal cell death after TBI. MIF KO or nuclease-deficient MIF mutant suppresses neurodegeneration, reduces neuronal cell death caused by TBI and improves animal behavioral rehabilitation. Targeting MIF and its nuclease activity may offer a novel strategy to protect neurons from TBI.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. W. Lee Kraus for providing PARP-1 KO mice and the UT Southwestern Medical Center Next Generation Sequencing Core for assistance with RNA-Seq.
Author contributions
YW and WL conceived the idea, designed research, analyzed data and wrote the paper. ZR initiated the project and performed the majority of the experiments including TBI surgery, Nissl staining, Golgi staining, and neuron morphology tracing and analysis. QL continued the project and performed TBI surgery, Nissl staining, Golgi staining, and neuron morphology tracing and analysis. JEW maintained the mouse lines and performed mouse genotyping, TBI surgery on both MIF and PARP mice, and animal behavioral tests. MZ performed mRNA sequencing studies. SL performed FJC and microglial activation experiments. HZ performed PARP inhibitor injection study. AD contributed to the blind animal behavior data analysis. YJW performed MIF western blots. YNW performed PARP-1 KO western blots. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke R00NS078049, National Institute of General Medical Sciences R35GM124693, and National Institute on Aging R01AG066166, Darrell K Royal Research Fund (DKR), Welch Foundation (I-1939-20170325), CPRIT-HIHR RP170671, TIBIR pilot Grant, the University of Texas (UT) Southwestern Medical Center Startup funds and UT Rising Stars to Y.W., and the Welch Foundation I-1903-20190330 to W.L.
Availability of supporting data
The datasets used and/or analyzed during the current study are included in this article.
Declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhi Ruan, Qing Lu and Jennifer E. Wang contributed equally.
References
- 1.Fernandez-Gajardo R, Matamala JM, Carrasco R, Gutierrez R, Melo R, Rodrigo R. Novel therapeutic strategies for traumatic brain injury: acute antioxidant reinforcement. CNS Drugs. 2014;28(3):229–248. doi: 10.1007/s40263-013-0138-y. [DOI] [PubMed] [Google Scholar]
- 2.Meaney DF, Morrison B, Dale Bass C. The mechanics of traumatic brain injury: a review of what we know and what we need to know for reducing its societal burden. J Biomech Eng. 2014;136(2):021008. doi: 10.1115/1.4026364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham NS, Sharp DJ. Understanding neurodegeneration after traumatic brain injury: from mechanisms to clinical trials in dementia. J Neurol Neurosurg Psychiatry. 2019;90(11):1221–1233. doi: 10.1136/jnnp-2017-317557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R, Sen N. Traumatic brain injury: a risk factor for neurodegenerative diseases. Rev Neurosci. 2016;27(1):93–100. doi: 10.1515/revneuro-2015-0017. [DOI] [PubMed] [Google Scholar]
- 5.Barr TL, Conley YP. Poly(ADP-ribose) polymerase-1 and its clinical applications in brain injury. J Neurosci Nurs. 2007;39(5):278–284. doi: 10.1097/01376517-200710000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Komjati K, Besson VC, Szabo C. Poly (adp-ribose) polymerase inhibitors as potential therapeutic agents in stroke and neurotrauma. Curr Drug Targets CNS Neurol Disord. 2005;4(2):179–194. doi: 10.2174/1568007053544138. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, An R, Umanah GK, Park H, Nambiar K, Eacker SM, Kim B, Bao L, Harraz MM, Chang C, Chen R, Wang JE, Kam TI, Jeong JS, Xie Z, Neifert S, Qian J, Andrabi SA, Blackshaw S, Zhu H, Song H, Ming GL, Dawson VL, Dawson TM. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016 doi: 10.1126/science.aad6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218(2):193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Luo W, Wang Y. PARP-1 and its associated nucleases in DNA damage response. DNA Repair (Amst) 2019;81:102651. doi: 10.1016/j.dnarep.2019.102651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abeti R, Abramov AY, Duchen MR. Beta-amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain. 2011;134(Pt 6):1658–1672. doi: 10.1093/brain/awr104. [DOI] [PubMed] [Google Scholar]
- 11.Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, Qi C, Poirier GG, Pletnikova O, Troncoso JC, Bekris LM, Leverenz JB, Pantelyat A, Ko HS, Rosenthal LS, Dawson TM, Dawson VL. Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson's disease. Science. 2018 doi: 10.1126/science.aat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love S, Barber R, Wilcock GK. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer's disease. Brain. 1999;122(Pt 2):247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- 13.Paldino E, D'Angelo V, Laurenti D, Angeloni C, Sancesario G, Fusco FR. Modulation of inflammasome and pyroptosis by olaparib, a PARP-1 inhibitor, in the R6/2 mouse model of Huntington's disease. Cells. 2020 doi: 10.3390/cells9102286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F, Hafezparast M, Caldecott KW. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42(1):307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newell-Rogers MK, Rogers SK, Tobin RP, Mukherjee S, Shapiro LA. Antagonism of macrophage migration inhibitory factory (MIF) after traumatic brain injury ameliorates astrocytosis and peripheral lymphocyte activation and expansion. Int J Mol Sci. 2020 doi: 10.3390/ijms21207448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Zhao J, Zhang Y, Zhang Y, Cai F, Wang L, Song W. Upregulation of MIF as a defense mechanism and a biomarker of Alzheimer's disease. Alzheimers Res Ther. 2019;11(1):54. doi: 10.1186/s13195-019-0508-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasiri E, Sankowski R, Dietrich H, Oikonomidi A, Huerta PT, Popp J, Al-Abed Y, Bacher M. Key role of MIF-related neuroinflammation in neurodegeneration and cognitive impairment in Alzheimer's disease. Mol Med. 2020;26(1):34. doi: 10.1186/s10020-020-00163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu YC, Tsai YH, Tang SC, Liou HC, Kang KH, Liou HH, Jeng JS, Fu WM. Cytokine MIF enhances blood–brain barrier permeability: impact for therapy in ischemic stroke. Sci Rep. 2018;8(1):743. doi: 10.1038/s41598-017-16927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inacio AR, Ruscher K, Leng L, Bucala R, Deierborg T. Macrophage migration inhibitory factor promotes cell death and aggravates neurologic deficits after experimental stroke. J Cereb Blood Flow Metab. 2011;31(4):1093–1106. doi: 10.1038/jcbfm.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inacio AR, Bucala R, Deierborg T. Lack of macrophage migration inhibitory factor in mice does not affect hallmarks of the inflammatory/immune response during the first week after stroke. J Neuroinflammation. 2011;8:75. doi: 10.1186/1742-2094-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 23.Guo D, Zeng L, Brody DL, Wong M. Rapamycin attenuates the development of posttraumatic epilepsy in a mouse model of traumatic brain injury. PLoS One. 2013;8(5):e64078. doi: 10.1371/journal.pone.0064078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, Kolodkin AL. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462(7276):1065–1069. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130(6):1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woolley CS, Gould E, Frankfurt M, McEwen BS. Naturally occurring fluctuation in dendritic spine density on adult hippocampal pyramidal neurons. J Neurosci. 1990;10(12):4035–4039. doi: 10.1523/JNEUROSCI.10-12-04035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Bezanilla S, TeBay C, Nilsson M, Walker FR, Ong LK. Visual discrimination impairment after experimental stroke is associated with disturbances in the polarization of the astrocytic aquaporin-4 and increased accumulation of neurotoxic proteins. Exp Neurol. 2019;318:232–243. doi: 10.1016/j.expneurol.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Belayev L, Alonso OF, Busto R, Zhao W, Ginsberg MD. Middle cerebral artery occlusion in the rat by intraluminal suture. Neurological and pathological evaluation of an improved model. Stroke. 1996;27(9):1616–1622. doi: 10.1161/01.str.27.9.1616. [DOI] [PubMed] [Google Scholar]
- 29.Savitt JM, Jang SS, Mu W, Dawson VL, Dawson TM. Bcl-x is required for proper development of the mouse substantia nigra. J Neurosci. 2005;25(29):6721–6728. doi: 10.1523/JNEUROSCI.0760-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmued LC, Hopkins KJ. Fluoro-Jade: novel fluorochromes for detecting toxicant-induced neuronal degeneration. Toxicol Pathol. 2000;28(1):91–99. doi: 10.1177/019262330002800111. [DOI] [PubMed] [Google Scholar]
- 31.Simoes AP, Silva CG, Marques JM, Pochmann D, Porciuncula LO, Ferreira S, Oses JP, Beleza RO, Real JI, Kofalvi A, Bahr BA, Lerma J, Cunha RA, Rodrigues RJ. Glutamate-induced and NMDA receptor-mediated neurodegeneration entails P2Y1 receptor activation. Cell Death Dis. 2018;9(3):297. doi: 10.1038/s41419-018-0351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Zhang B, Bao L, Jin L, Yang M, Peng Y, Kumar A, Wang JE, Wang C, Zou X, Xing C, Wang Y, Luo W. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J Clin Investig. 2018;128(5):1937–1955. doi: 10.1172/JCI95089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata K, Jin L, Jha S. Lack of promoter IV-driven BDNF transcription results in depression-like behavior. Genes Brain Behav. 2010;9(7):712–721. doi: 10.1111/j.1601-183X.2010.00605.x. [DOI] [PubMed] [Google Scholar]
- 34.Luong TN, Carlisle HJ, Southwell A, Patterson PH. Assessment of motor balance and coordination in mice using the balance beam. J Vis Exp. 2011 doi: 10.3791/2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper JM, Wilkinson JE, Miller RA. Macrophage migration inhibitory factor-knockout mice are long lived and respond to caloric restriction. FASEB J. 2010;24(7):2436–2442. doi: 10.1096/fj.09-152223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das R, Chinnathambi S. Microglial priming of antigen presentation and adaptive stimulation in Alzheimer's disease. Cell Mol Life Sci. 2019;76(19):3681–3694. doi: 10.1007/s00018-019-03132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolos M, Perea JR, Avila J. Alzheimer's disease as an inflammatory disease. Biomol Concepts. 2017;8(1):37–43. doi: 10.1515/bmc-2016-0029. [DOI] [PubMed] [Google Scholar]
- 38.Stoica BA, Loane DJ, Zhao Z, Kabadi SV, Hanscom M, Byrnes KR, Faden AI. PARP-1 inhibition attenuates neuronal loss, microglia activation and neurological deficits after traumatic brain injury. J Neurotrauma. 2014;31(8):758–772. doi: 10.1089/neu.2013.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaPlaca MC, Zhang J, Raghupathi R, Li JH, Smith F, Bareyre FM, Snyder SH, Graham DI, McIntosh TK. Pharmacologic inhibition of poly(ADP-ribose) polymerase is neuroprotective following traumatic brain injury in rats. J Neurotrauma. 2001;18(4):369–376. doi: 10.1089/089771501750170912. [DOI] [PubMed] [Google Scholar]
- 40.Whalen MJ, Clark RS, Dixon CE, Robichaud P, Marion DW, Vagni V, Graham SH, Virag L, Hasko G, Stachlewitz R, Szabo C, Kochanek PM. Reduction of cognitive and motor deficits after traumatic brain injury in mice deficient in poly(ADP-ribose) polymerase. J Cereb Blood Flow Metab. 1999;19(8):835–842. doi: 10.1097/00004647-199908000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson TM, Snyder SH, Dawson VL. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3(10):1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263(5147):687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 43.Merk M, Mitchell RA, Endres S, Bucala R. D-dopachrome tautomerase (D-DT or MIF-2): doubling the MIF cytokine family. Cytokine. 2012;59(1):10–17. doi: 10.1016/j.cyto.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grieb G, Merk M, Bernhagen J, Bucala R. Macrophage migration inhibitory factor (MIF): a promising biomarker. Drug News Perspect. 2010;23(4):257–264. doi: 10.1358/dnp.2010.23.4.1453629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S, Luo W, Wang Y. Emerging role of PARP-1 and PARthanatos in ischemic stroke. J Neurochem. 2021 doi: 10.1111/jnc.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Chen Y, Wang C, Yang M, Wang Y, Bao L, Wang JE, Kim B, Chan KY, Xu W, Capota E, Ortega J, Nijhawan D, Li GM, Luo W, Wang Y. MIF is a 3′ flap nuclease that facilitates DNA replication and promotes tumor growth. Nat Commun. 2021;12(1):2954. doi: 10.1038/s41467-021-23264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are included in this article.