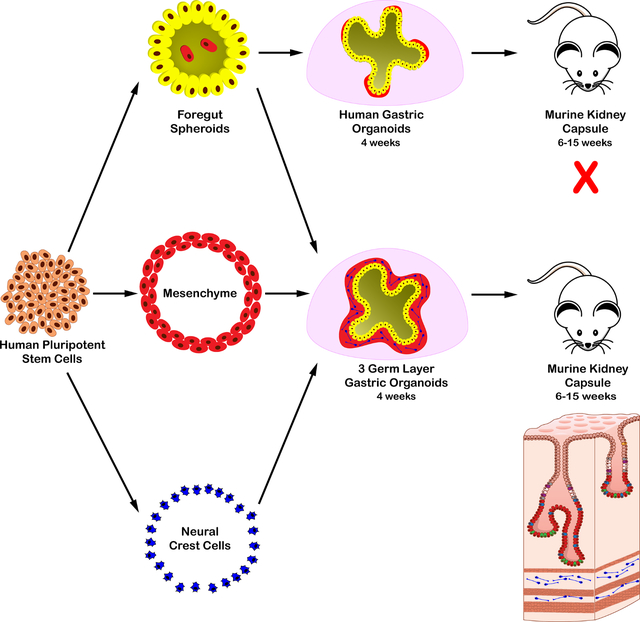
SUMMARY
Human organoid model systems all lack important cell types that, in the embryo, get incorporated into organ tissues during development. We developed an organoid assembly approach starting with cells from the three primary germ layers; enteric neuroglial, mesenchymal, and epithelial precursors, that were separately derived from human pluripotent stem cells (PSCs). From these three cell-types we generated human antral and fundic gastric tissue containing differentiated glands, surrounded by layers of smooth muscle containing functional enteric neurons that controlled contractions of the engineered antral tissue. Using this experimental system, we show that human enteric neural crest cells (ENCCs) promote mesenchyme development and glandular morphogenesis of antral stomach organoids. Moreover, ENCCs can act directly on the foregut to promote a posterior fate, resulting in organoids with a Brunner’s gland phenotype. Thus, germ layer components that are separately derived from PSCs can be used for tissue engineering to generate complex human organoids.
Keywords: tissue engineering, gastric, enteric nervous system, mesenchyme, human pluripotent stem cells
Graphical Abstract
eTOC Statement
Eicher et al. have generated complex three germ layer human gastric organoids from pluripotent stem cells. These three germ layer organoids contain glandular epithelium surrounded by multiple, organized layers of functionally innervated smooth muscle. Manipulating this system revealed key developmental roles of neural crest cells on gastric epithelium and mesenchyme.
INTRODUCTION
All organs of the gastrointestinal (GI) tract are assembled from cells derived from the three primary germ layers during embryonic development. These diverse cell types are required for the proper execution of the GI tract’s complex functions. For example, key functions of the stomach to chemically and mechanically breakdown orally ingested nutrients depend on a complex interaction of the epithelium to produce acid and proteases (Furness et al., 2020; Zhao et al., 2008; Norlen et al., 2005; Rydning et al., 2002), the smooth muscle to contract and relax, and the enteric nerves to coordinate both of these processes (Sung et al., 2018; Beckett, Sanders and Ward, 2017; Shaylor et al., 2016; Li et al., 2011). These three main components of the stomach develop separately from the three primary germ layers but then assemble into a tissue that is functional by birth, with the endoderm forming the epithelial lining, the mesoderm contributing to the stromal cells and smooth muscle layers, and the ectoderm giving rise to the enteric nervous system (ENS).
During organ development, the ENS, mesenchyme, and epithelium communicate with each other to regulate regional identity, assembly, morphogenesis, and differentiation into specific cell types (Le Guen et al., 2015). For example, neural progenitors of the ENS, enteric neural crest cells (ENCCs), migrate to the foregut in response to signals from the surrounding mesenchyme as this mesenchyme is differentiating into multiple, organized layers of smooth muscle. Reciprocal signaling between germ layers is observed across species. For example, sonic hedgehog (Shh) is secreted by the epithelium of several GI organs and regulates expression of bone morphogenetic protein (BMP) in adjacent mesenchyme. BMP activation in the hindgut promotes posterior mesenchyme patterning (Roberts et al., 1995; Roberts et al., 1998; Faure et al., 2002; De Santa Barbara et al., 2005) where in the stomach it regulates formation of the pyloric sphincter (Smith and Tabin, 1999; Smith et al., 2000; Moniot et al., 2004; Theodosiou and Tabin, 2005). Epithelial Shh is also known to indirectly regulate ENCC proliferation and differentiation through manipulation of key extracellular matrix proteins within the gut mesenchyme (Nagy et al., 2016). Reciprocal signaling from ENCCs regulate the growth, pattering, and maturation of developing stomach mesenchyme (Faure et al., 2015). Finally, epithelial Shh-induced BMP signaling and ENCC-produced BMP antagonists work in concert to regulate the radial position of the gut’s smooth muscle layers (Huycke et al., 2019).
Congenital and acquired disorders of the proximal GI tract include esophageal atresia, gastroparesis, and gastroesophageal reflux disease, which can arise from improper epithelial, ENS or smooth muscle development (Westfal and Goldstein, 2017; Brosens et al., 2016; Pesce et al., 2018; Perin et al., 2017). While animal models have been invaluable to study gastroesophageal development and disease (de Santa Barbara, van den Brink and Roberts, 2002), there are major structural and functional differences in this organ between different species. For example, rodents have a forestomach that does not exist in humans. The avian stomach contains a proventriculus, which is a proximal glandular compartment, somewhat paralleling the human fundus, and a gizzard, which is a more distal grinding compartment that is vastly different than the human antrum (Kim and Shivdasani, 2016). There are also developmental differences at the molecular level; Hedgehog signaling have different roles in the development of GI smooth muscle between chick and mouse embryos (Huycke et al., 2019). Each of the existing model systems have experimental strengths and weaknesses; with frog and chick embryos being easy to manipulate in vitro whereas mouse embryos being a strong genetic model. However, creation of genetically tractable, complex human GI tissue models that can be manipulated in vitro could accelerate studies of development and discovery of drugs that improve gastroesophageal function.
Current gastric and esophageal organoid models are limited in that they only contain epithelium (McCracken et al., 2014; McCracken et al., 2017; Trisno et al., 2018). We reasoned that we could engineer added complexity into gastroesophageal organoids by incorporating progenitor cells from the other 2 germ layers in vitro. In so doing we could both construct a more complex organoid and study human organ development in a way not previously possible. In this study, we incorporated human pluripotent stem cell (hPSC)-derived splanchnic mesenchyme and ENCCs into developing human antral and fundic gastric organoids (hAGOs and hFGO, respectively) to recapitulate normal gastric development. The resulting gastric organoids were composed of epithelial glands surrounded by multiple layers of functionally innervated smooth muscle. The technology was readily transferrable to other types of organoids and was used to engineer esophageal organoids containing all 3 germ layers. The tractability of this approach allowed us to study germ layer communication during stomach development. We found that human ENCCs promote gastric mesenchymal development and glandular morphogenesis and that the presence of adequate amount of gastric mesenchyme is essential for maintaining gastric regional identity.
RESULTS
Deriving mesenchyme from hPSCs and incorporation into gastric organoids
One of the first and most critical steps in GI development is the assembly of epithelium and mesenchyme into a primitive gut tube. While PSC-derived human gastric organoids have a full complement of epithelial cell types (McCracken et al., 2014; McCracken et al., 2017), they do not intrinsically develop a robust mesenchyme. To develop an approach to generate GI mesenchyme we used a method recently identified to direct the differentiation of hPSCs into splanchnic mesenchyme (SM), the source of gastric mesenchyme (Han et al., 2020). This method was based on the signaling pathways that drive the normal development of GI mesenchyme mouse. Briefly, hPSCs were differentiated into lateral plate mesoderm (LPM) with TGFβ inhibition, WNT inhibition, and BMP activation as previously published (Figure 1A) (Han et al., 2020; Loh et al., 2016). As LPM can give rise to both cardiac and SM, the LPM was treated with retinoic acid (RA) to induce a SM fate, resulting in loss of cardiac markers like ISL1 and, thus, an expansion of a FOXF1+/ISL1- SM cellular population (Figure 1B–C) (Han et al., 2020).
Figure 1.
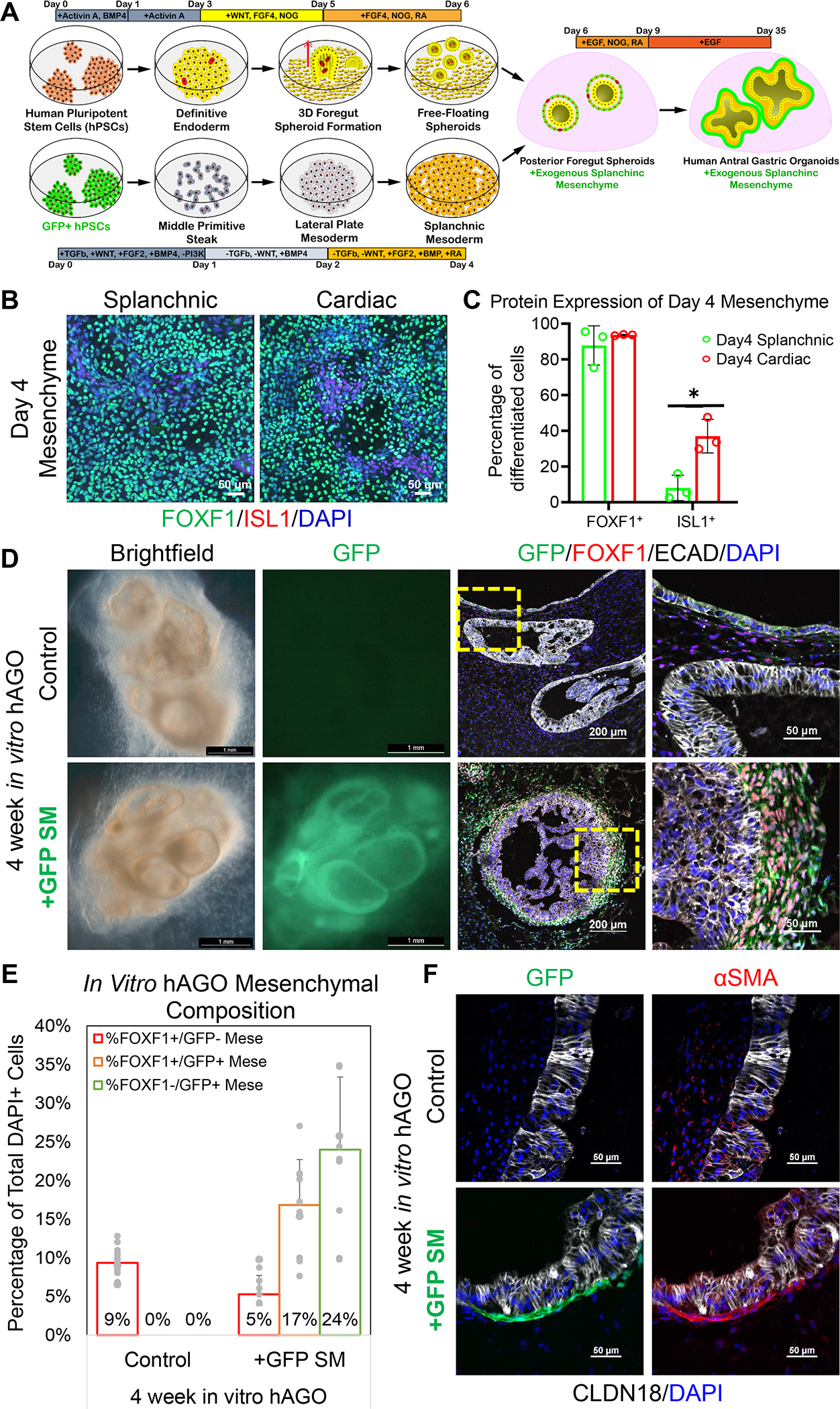
Incorporation of hPSC-derived splanchnic mesenchyme into hAGOs. (A) Schematic depicting the method of deriving and incorporating GFP+ splanchnic mesenchyme (SM) into hAGOs. SM was derived from an hPSC line that constitutively expresses GFP. (B) Representative immunofluorescent images of day 4 splanchnic (left) and cardiac (right) mesenchymal monolayers costained with FOXF1 (green) and ISL1 (red); images are 2×2 composite of 20x images stitched by Nikon NIS Elements. (C) Quantification of FOXF1+ (left) and ISL1+ (right) cells within day 4 splanchnic (green bar) and cardiac (red bar) mesenchymal monolayers (n=3 fields from one differentiation, *p<0.05, Student’s t-test). (D) Brightfield images of hAGOs grown for four weeks in vitro with and without recombination with exogenous GFP+ SM (green) costained with mesenchymal marker FOXF1 (red). Higher magnification images are shown to the right. (E) Quantification of FOXF1+ mesenchymal contribution (n=11–18 fields from at least 3 organoids per condition from one differentiation, same trend seen across more than two individually seeded differentiations). (F) Representative images of four week in vitro hAGOs with and without recombined SM (green) stained with smooth muscle marker αSMA (red) and gastric epithelial markers CLDN18 (white). See also Figure S1.
We investigated several approaches to incorporate mesenchyme into developing gastric organoids, including combining mesenchyme and epithelium at different epithelial developmental stages (i.e., at either day 6 or day 9 of the hAGO protocol), testing a single cell aggregation method versus using intact epithelial organoids, and utilizing differently patterned mesenchymal populations, including splanchnic, cardiac, septum transversum, and gastric-esophageal mesenchyme. We initially compared incorporation of varying concentrations of SM and septum transversum (STM) mesenchyme expressing GFP on day 6 and found that 50,000 cells/well at a 2:1 ratio of SM cells to foregut epithelial cells resulted in hAGO with a robust layer of mesenchyme surrounding the epithelium (Fig. S1A). We then repeated this with regionally patterned gastric-esophageal mesenchyme (GEM) on day 9 of hAGO protocol (Figure 1D, S1B). In all cases, SM formed an organized layer of GFP+ mesenchymal cells expressing the early SM marker FOXF1 surrounding the gastric epithelium (Figure 1D). Between a third to a half of all cells in hAGO +SM were mesenchyme, representing a 3–5-fold increase over control organoids (Figure 1E, S1C). In contrast, STM or GEM did not form a uniform layer of mesenchyme (Figure S1B) with less than a fourth of all cells being mesenchyme (Figure S1C). Finally, we observed GFP+ mesenchymal cells that expressed αSMA+, suggesting the potential for differentiation into smooth muscle (Figure 1F).
We have previously shown that transplantation of PSC-derived intestinal and colonic organoids under the mouse kidney capsule allowed for continued growth and development. To identify if hAGOs with ENS and mesenchyme can grow and form tissue more comparable to human stomach, all organoids were transplanted into mice and grown for an additional 10–12 weeks (Table S1) and resulting tissues were compared to sections from a 38 week fetal and adult human stomach. At the end of 4 weeks in vitro, most hAGOs averaged between 1–2mm in diameter, had a glandular epithelial morphology and a single lumen. For growth in vivo, one organoid per mouse was transplanted under the kidney capsule and grown for 8–12 weeks. Most hAGOs without added mesenchyme did not survive (~60%) and those that did were small containing only a simple, one-cell layer thick cuboidal-like epithelium that did not fully differentiate (Figure 2B). In contrast, hAGOs with exogenous mesenchyme had a high survival rate (100%) and grew from a few millimeters to 0.5–1.5 cm in diameter, exhibiting up to 1000x increase in volume in some cases. The mesenchyme differentiated into layers of poorly organized αSMA+ smooth muscle that surround a simple layer of hAGO epithelium (Figure 2B). This data suggests that the incorporation of exogenous mesenchyme promotes engraftment and growth of hAGOs but does not result in normal gastric tissue with glandular units of simple columnar epithelium.
Figure 2.
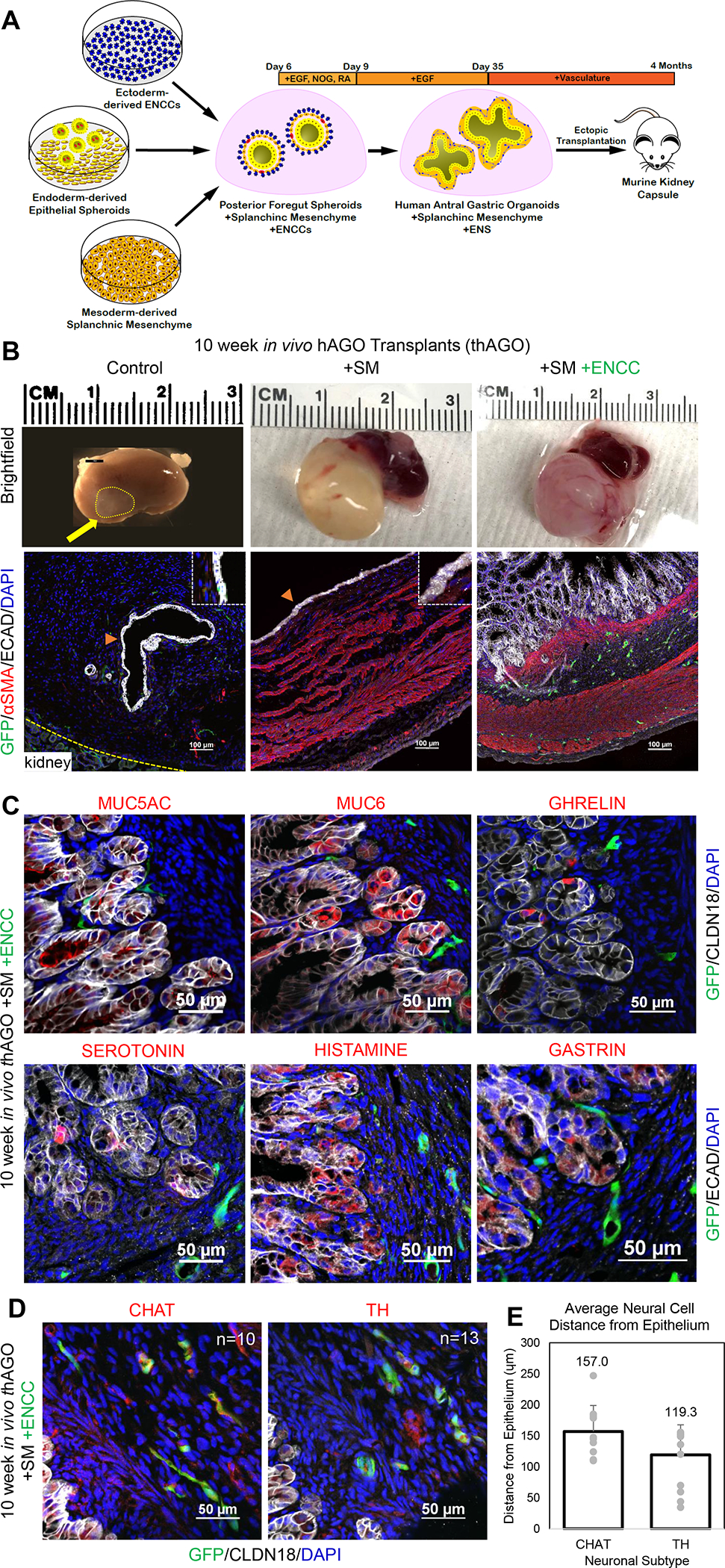
3 germ layer recombinants form human gastric tissue with innervated layers of smooth muscle and glandular epithelium. (A) Schematic depicting the generation of 3 germ layer recombinants using foregut endoderm, SM and ENCCs. (B) Morphological comparison between hAGO transplants with and without SM and ENCCs (top) and representative images of 10 week in vivo hAGOs stained with αSMA mesenchyme (red) (bottom). ENCCs are labeled with GFP (green) and counterstained with epithelial marker ECAD (white). (C) Gastric epithelial marker analysis of patterning and differentiated cell types in 3 germ layer thAGOs. MUC5AC (red, top left) and MUC6 (red, top middle) mark surface pit and gland mucous cells, respectfully. Hormone producing endocrine cells marked with ghrelin (red, top right), serotonin (red, bottom left), histamine (red, bottom middle), and gastrin (red, right bottom). GFP labels the recombined ENCC (green) and the epithelium is labeled with CLDN18 (white, top) and ECAD (white, bottom). (D) Marker analysis of neuronal differentiation in 3 germ layer thAGOs. GFP+ ENCC (green) is costained with choline acetyltransferase (CHAT, red, left) and tyrosine hydroxylase (TH, red, right). (E) Quantification of distance between epithelium of 3 germ layer thAGO and acetyltransferase (CHAT, left) and tyrosine hydroxylase (TH, right) neuronal subtypes (n=10–13 individual cells from two fields from one differentiation). See also Table S1 and Movie S1.
Engineering 3 germ layer human gastric tissue
While SM improved the growth of hAGOs, the smooth muscle was poorly organized and there was no evidence of epithelial development into the glandular structures similar to human stomach. We therefore investigated if incorporation of the ectodermally-derived ENCCs, in combination with SM, might result in more normal stomach development (Figure 2A). Although there are several published protocols to derived NCC aggregates from hPSCs (Barber, Studer and Fattahi, 2019), we used a protocol that allowed us to harvest migrating ENCCs (Workman et al., 2017; Bajpai et al., 2010) with the rationale that in the embryo, migrating ENCCs colonize the gut. RFP-expressing ENCCs were generated, harvested, and were recombined with or without GFP-expressing SM with hAGO spheroids. Recombinants were grown for 4 weeks in vitro (Figures S2A, S3G) and then transplanted into mice for an additional 10–12 weeks. While both hAGOs +SM and hAGO +SM +ENCC transplants grew to over 1 cm in diameter, only hAGO +SM +ENCC recombinants formed the stereotypic glandular structures found in the human stomach (Figures 2B, 3A–B). In addition, hAGO +SM +ENCC transplants had distinct layers of smooth muscle that were organized like the 38 week human fetal stomach that contains a muscularis mucosa and muscularis externa, comprising of inner circular and outer longitudinal layers of smooth muscle (Figures 2B, 3A–B, S2E–F). Quantitative comparison showed the muscle layers of hAGO +SM +ENCC transplants formed at a similar distance from the epithelium as the 38wk human stomach (Figure S2E–F). Embedded within the smooth muscle fibers was a network of enteric neurons arranged in characteristic plexi (Figure 2B, Movie S1). One plexus layer arranged in the muscularis mucosa-like muscle layer and the other arranged in the muscularis externa-like muscle layer. The organization of different ENS layers in hAGOs is spatially similar to the neuronal plexi of human stomach, with ENS neurons embedded in submucosa and muscularis externa-like muscle layers. While the hAGOs that contained all 3 germ layers were similar to the fetal stomach, they are not as architecturally mature as the adult human stomach (Figures 3A–B, S2E–F).
Figure 3.
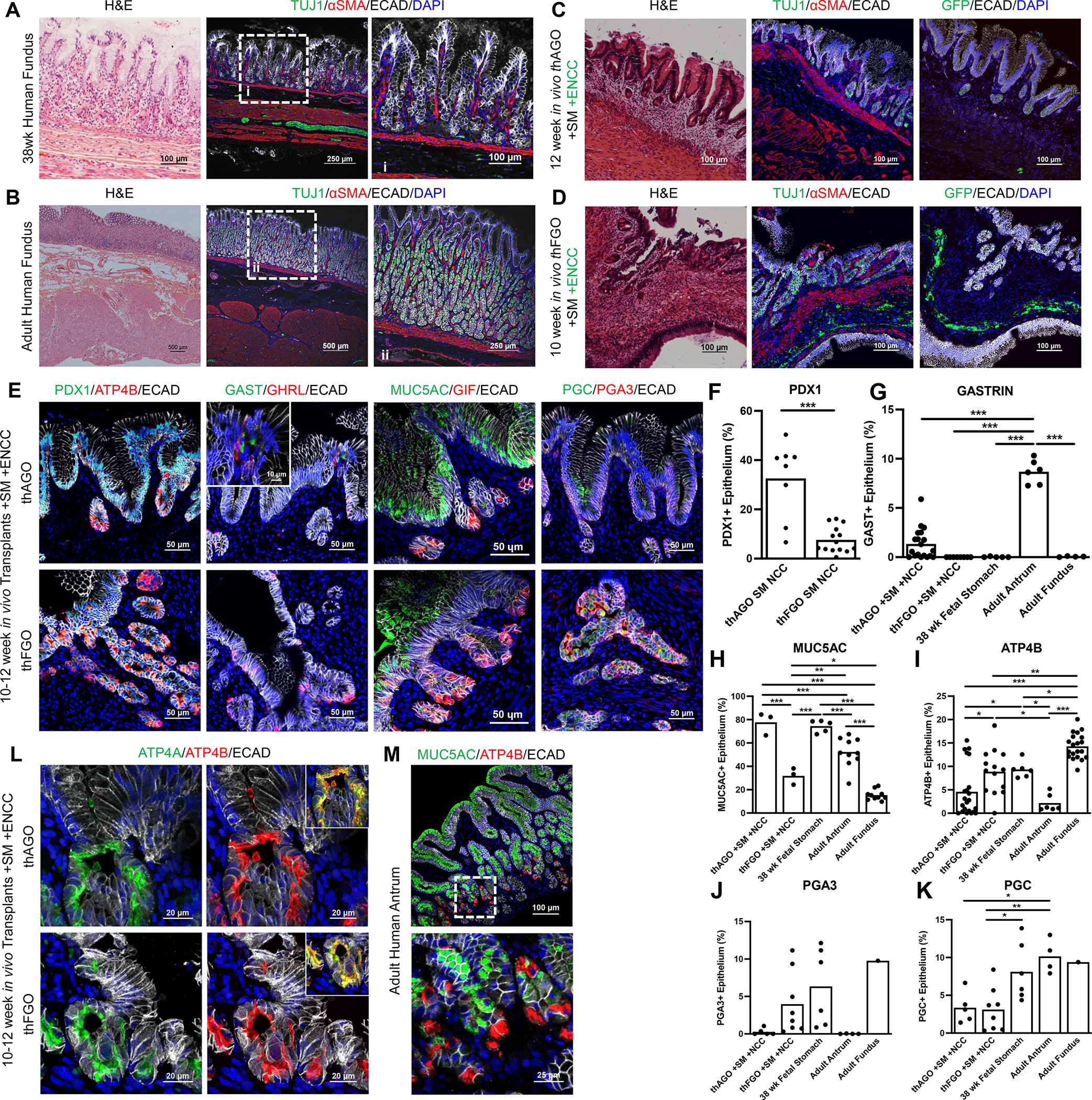
A comparison of engineered antral and fundic organoid tissue with the human stomach. Histological and immunofluorescence analysis of whole thickness gastric tissue from 38 week (A) and adult (B) human stomach taken from the distal fundus and 3 germ layer (C) thAGO and (D) thFGO. The 3 germ layers are labeled with neuronal marker TUJ1 or GFP (green), smooth muscle αSMA (red), and epithelial marker ECAD (white). (E) Representative images of gastric epithelial patterning and cell differentiation of 3 germ layer thAGOs (top) and hFGOs (bottom). PDX1 (green, left), endocrine cells expressing gastrin (GAST, green, middle), and the surface mucin (MUC5AC, green, middle) are (E) qualitatively and (F-H) qualitatively enriched in hAGOs and human antral biopsies. Parietal cells (ATP4B, red, left; and GIF, red, middle), and chief cells expressing fundic-specific pepsinogen A3 (PGA3, red, right) are (E) qualitatively and (I-J) quantitatively enriched in the hFGOs and human fundic biopsies. MUC5AC/GIF staining is representative of a maximum intensity projection (maxIP) rendered from a z-stack of images by Nikon NIS Elements. Endocrine cells expressing ghrelin (GHRL, red, middle) and chief cell marker pepsinogen C (PGC, green, right) are observed at relatively similar levels in both thAGOs and thFGOs (E,K). (L) Representative images of parietal cells in 3 germ layer thAGOs (top) and thFGOs (bottom) with colocalized apical expression of ATP4A (green) and ATP4B (red) H+/K+ ATPase subunits. (E, L, M) Epithelium is labeled with ECAD (white). Significance denoted as *p<0.05, **p<0.01, and ***p<0.001 determined by (F) Student t-test or (G-K) one-way ANOVA with Tukey’s Multiple Comparison (n=3–20 20x fields from 5 thAGO TXPs from two differentiations, 8–14 20x fields from 2 thFGO TXPs from one differentiation, 5–6 20x fields from fetal stomach from 1 patient, 1–20 20x fields from adult fundus from 3 patients, and 4–10 20x fields from adult antrum from 2 patients). (M) Representative human adult antral biopsy labeled with MUC5AC (green) and ATP4B (red); staining is representative of a maxIP rendered from a z-stack of images by Nikon NIS Elements. See also Figures S2 and S3.
The epithelium of hAGO +SM +ENCC transplants expressed the gastric marker CLDN18 and lacked the intestinal epithelial marker CDH17 (Figure 2C, data not shown) and contained the expected antral cell types including surface mucous cells (MUC5AC), gland mucous cells (MUC6), and endocrine cells expressing ghrelin, serotonin, histamine, and gastrin (Figures 2C, 3E). The GFP+ neurons formed a plexus-like structure that was embedded within the layers of smooth muscle. We also observed GFP+ choline acetyltransferase+ (ChAT+) and dopaminergic (TH+) neurons. The spatial organization of the neurons relative to the epithelium in the hAGO tissue was similar to the 38 week human stomach, where they were approximately 120–160μm away from the glands and in close proximity to the endocrine cells such as ghrelin and gastrin (Figure 2C–E). This association in vivo is important as neurotransmitters control secretion of a variety of stomach hormones including ghrelin and gastrin (Breit et al., 2018).
Generating fundic tissues containing 3 germ layers
One of the most prominent domains of the human stomach is the corpus, which contains fundic (oxyntic) glands with acid producing parietal cells and digestive enzyme secreting chief cells. The glands are also in close proximity to enteric neurons that, along with gastric endocrine cells, help to regulate acid production. We investigated if the 3 germ layer recombinant approach could also be used to engineer human fundic tissue with the above properties. We generated early stage hFGOs as previously described (McCracken et al., 2017) and recombined them with SM and GFP+ ENCCs. After four weeks we observed both SM and ENCCs incorporated into hFGOs (Figure S2H), similar to hAGOs (Figures S2A–D), and confirmed fundic identity by the presence of ATP4B+ parietal cells and very low expression of PDX1 (Figure S2H and data not shown).
We next investigated if three germ layer hFGOs engrafted under the murine kidney capsule would form more mature fundic tissues (Figure S2G). After 10–12 weeks, hFGOs grew up to a centimeter in size (Figure S2C and S2G) and had a similar histological architecture to that of 38 week human fundic tissues, with glandular epithelium surrounded by multiple layers of innervated smooth muscle (Figure 3A–D). Both hAGOs and hFGOs maintained their regional identity after transplantation and the proportions of cell types that normally distinguish the human corpus/fundic from the antrum also distinguished hAGOs from hFGOs. Specifically, hAGOs expressed higher levels of PDX1, antral-specific gastrin-expressing endocrine cells, and MUC5AC+ surface mucus cells compared to hFGOs (Figure 3E–H). Conversely, hFGOs contained more ATP4B+/GIF+ parietal cells than the hAGOs, and hFGOs had fundic-specific PGA3+ chief cells, which were absent in hAGOs (Figure 3E, 3I–J). Ghrelin-expressing endocrine cells and PGC+ cells at the bases of the glands were observed in both regions of the stomach, with higher ghrelin expression observed in hFGOs (Figures 3E, 3K). In general, the extent of glandular morphogenesis of transplanted hFGOs was less than that of hAGOs (Figure 3C–E).
We previously demonstrated that differentiation of parietal cells in vitro required BMP signaling and MEK inhibition (Figure S2H) (McCracken et al., 2017), however transplanted hFGOs required no additional factors for robust parietal cell differentiation (Figures 3E, 3I, and 3L–M) demonstrating that the signaling processes that control gastric cell type specification occur normally in engineered tissue. As observed in human stomach biopsies, engineered antral tissue does contain parietal cells, but at lower numbers than are found in fundic glands (Figures 3E, 3I, and 3L–M) (Choi et al., 2014). Furthermore, parietal cells in hFGOs in vitro only expressed the ATP4B+ subunit of the H+/K+ ATPase and the cellular localization is primarily cytoplasmic (Figure S2H) while hFGOs matured in vivo expressed much higher levels of both the ATP4A and ATP4B subunits which colocalize on the apical membrane (Figure 3L) suggesting that these parietal cells are more mature than their in vitro counterparts. Together these data show that engineering mesenchyme and ENS cells into hAGOs and hFGOs results in the formation of gastric tissues that more closely resemble human stomach.
Antral 3 germ layer organoids exhibit functional muscle contraction
The stomach plays an essential role in the mechanical breakdown of food and in emptying it into the duodenum. This gastric motility involves the ENS, which functionally controls smooth muscle contractions. To investigate if the ENS and smooth muscle in the 3 germ layer hAGOs formed a functional neuromuscular unit, we isolated tissue strips from transplanted hAGOs and placed them in an organ bath chamber system to monitor contractility. After an equilibration period, spontaneous contractile oscillations were observed from tissues derived from 1 hAGO +SM and 3 separate hAGO +SM +ENCC transplants (Figure 4A). The presence of phasic contractions indicated that intramuscular interstitial cells of Cajal (ICCs) were present within both the 2 and 3 germ layer organoids. However, the contractile activity was irregular in hAGO +SM tissue, in contrast to the highly regular contractions in hAGO +SM +ENCC. This suggested that ENCCs may interact with ICCs, an idea that was further supported by the close association with TUJ1+ neuroglia cells with clusters of mesenchymal cells expressing the ICC marker c-KIT (Figure 4B). These mesenchymal clusters are ~7% of total cells present (Figure 4C) and are arranged within the muscularis externa-like muscle layers of the 3 germ layer hAGO transplants comparable to the human stomach. The force of smooth muscle contractions increased in response to the muscarinic receptor agonist bethanechol and muscle relaxation was triggered with addition of scopolamine, a muscarinic antagonist (Figure 4D and E). These findings demonstrate the presence of functional smooth muscle in both hAGO +SM and hAGO +SM +ENCC.
Figure 4.
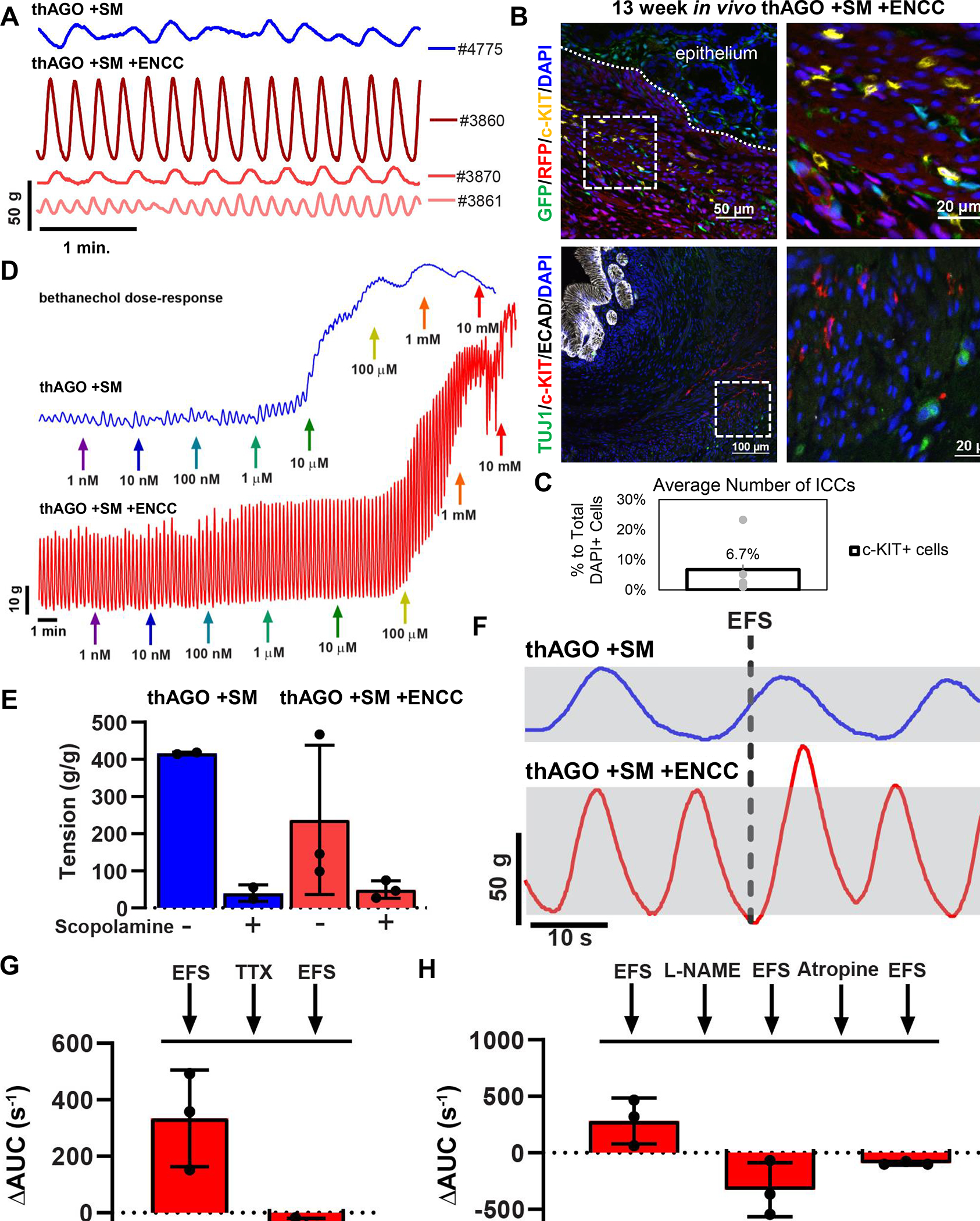
Antral 3 germ layer organoids have a functional ENS that regulates gastric tissue contractions. (A) Isometric force contractions in tissues isolated from one individual thAGO +SM (blue) and three individual thAGO +SM +ENCC (red) after an equilibrium period. Contractile activity was triggered using electrical field stimulation (EFS). (B) Neuronal (GFP, green, top; and TUJ1, green, bottom) and interstitial cells of Cajal (ICC) (c-KIT, yellow, top; and red, bottom) stained in 13 week in vivo hAGO +SM +ENCC grafts. RFP (top) marks SM. (C) Quantification of the average number of ICCs in in vivo hAGO +SM +ENCC grafts (n=7 fields from 3 organoids from 2 differentiations). (D) Activation of muscarinic receptors induced contractions in tissues isolated from a thAGO +SM (blue) and hAGO +SM +ENCC (red). Increasing doses of bethanechol were added to the tissues at times indicated by the colored arrows. (E) Inhibition of the muscarinic receptor with scopolamine induced muscle relaxation. Calculated maximal and minimal tissue tension of tissues from hAGO +SM (blue) and hAGO +SM +ENCC (red). (F) Representative tracings of contractile activity in response to electrical field stimulation (EFS) in thAGO +SM (blue) and hAGO +SM +ENCC (red). Dashed line indicates timing of EFS application and gray rectangles highlight pre-EFS contractile amplitude. (G) Inhibition of ENS activation with the neurotoxin tetrodotoxin (TTX) abrogates EFS-mediated contractions. Change in area under the curve following a control EFS stimulation measured for one minute after stimulation, followed by TTX treatment, and a final EFS stimulation in hAGO +SM +ENCC. (H) Functionally testing the role of nitrergic and cholinergic neuronal activity in smooth muscle contractions. Change in area under the curve induced by EFS stimulation and following treatment with the nitrergic inhibitor L-NAME and the cholinergic inhibitor Atropine. All data was normalized to tissue mass; n=1 hAGO +SM; n=3 hAGO +SM +ENCC from two differentiations.
The close proximity of neurons with c-KIT clusters in hAGO +SM +ENCC tissues suggests that the ENS may functionally regulate smooth muscle contractions. To test this, we exposed tissue strips to an electrical field stimulation (EFS) at a level that would trigger neuronal firing, but would not directly cause smooth muscle contractions. EFS pulses were administered to 2 and 3 germ layer hAGO muscle strips and only resulted in an increase in contractile activity in hAGO +SM +ENCC, indicating that the ENS was regulating smooth muscle (Figure 4F). We then blocked ENS function using the neurotoxin tetrodotoxin (TTX), which abolished the ability of EFS to stimulate contractile activity (Figure 4G). Lastly, we investigated the involvement of nitrergic and cholinergic neuronal activity in regulating smooth muscle contractions. We inhibited nitric oxide synthetase (nNOS)-expressing neurons with NG-nitro-l-arginine methyl ester (L-NAME), a nitric oxide synthesis inhibitor, and inhibited cholinergic neurons using atropine, an acetylcholine (Ach) receptor antagonist. Contractile activity was measured following control stimulation and stimulation after compound exposure and was expressed as the change in the area under the curve (AUC) immediately before and after each EFS stimulation (Figure 4H). These data provide insight into the proportions of nitrergic and cholinergic neuronal activity compared to the total ENS activity (control EFS) and show that gastric tissue contractions involved both nitrergic and cholinergic neuronal activities.
3 germ layer esophageal organoids
To test if our approach of combining tissue from 3 germ layers was broadly applicable to engineering other organs, we attempted to incorporate SM and ENCCs into developing human esophageal organoids (HEOs) (Trisno et al., 2018). Like hAGOs and hFGOs, we started with HEOs that were largely epithelial and added GFP+ SM (Figure S3A–B). After four weeks in vitro HEOs +SM had a robust layer of GFP+ mesenchyme surrounding the epithelium (Figure S3A) with a high percent of these co-expressing FOXF1 or the more differentiated marker vimentin (VIM) (Figure S3A). Quantification showed that control HEOs only contain ~1% of endogenous mesenchyme while HEOs +SM contain ~25% mesenchymal cells (Figure S3B).
We next incorporated ENCCs into HEOs with or without SM (Figure S3C–H). After 1 month in culture, the ENCCs in HEOs without SM had differentiated into TUJ1/MAP2/NESTIN+ enteric neurons that aggregated tightly around the epithelium but did not organize into a neuronal plexus, likely due to the sparsity (less than 1%) of endogenous mesenchyme present (Figure S3D–E). In contrast when both ENCCs and SM were both recombined with HEO epithelium we observed a TUJ1+ neuronal plexus associated within a FOXF1+ mesenchymal layer (Figure S3F–I). Overall, these finding show that different human GI organ tissues can be engineered by combining progenitors from all 3 germ layers and emphasize the importance of reciprocal cell-cell communication between the epithelial, mesenchymal, and ENCCs for proper assembly and function of embryonic organs.
ENCCs differentiation into ENS neuroglial cell types does not require the addition of exogenous mesenchyme
One powerful use of this system is the capability to study interactions between different germ layers during organogenesis. The above results suggest that the presence of ENCCs is important for the development of both smooth muscle and gastric epithelium, but it was not clear if ENCCs act directly on either of these germ layers. We therefore took advantage of the ability to add or remove germ layers to interrogate how ENCCs impact the development of other germ layers. ENCCs were generated, quality tested by expression of NCC markers (Figure S4), and recombined with hAGOs with and without exogenous mesenchyme at day 6 of hAGO development (Figure 5A). We also recombined ENCCs at day 9 to avoid exposing ENCCs to retinoic acid (RA) and noggin (NOG) that are applied to hAGO cultures between days 6–9 as it is known that RA can directly impact the patterning and differentiation of ENCCs (Figure S4E) (Simoes-Costa and Bronner, 2015; Workman et al., 2017). Surprisingly at either time point, ENCCs incorporated well into hAGOs and formed TUJ1+ neurons and S100b+ glial cells adjacent to gastric epithelium (Figure S4B,F–H and Movie S2). Even without exogenous mesenchyme, ENCCs differentiated into a diverse array of neuroglial subtypes, including inhibitory (nNOS), interneurons (Synaptophysin), dopaminergic (TH), sensory (Calbindin) neurons, and glial cells (GFAP were 2% of the total cells) (Figure S4I,J and Table S2). However, without exogenous mesenchyme, ENS development was abnormal, with disorganized neurons being immediately adjacent to the epithelium (Figure S4K–L). There were, however, a comparable number of nNOS+ inhibitory neurons present in hAGOs +ENCC compared to mouse E13.5 stomach (Figure S4M–N). These data show that ENCCs incorporated into hAGOs differentiated into neuroglial subtypes without the addition of exogenous mesenchyme, but that ENS plexus development likely requires a robust population of mesenchyme.
Figure 5.
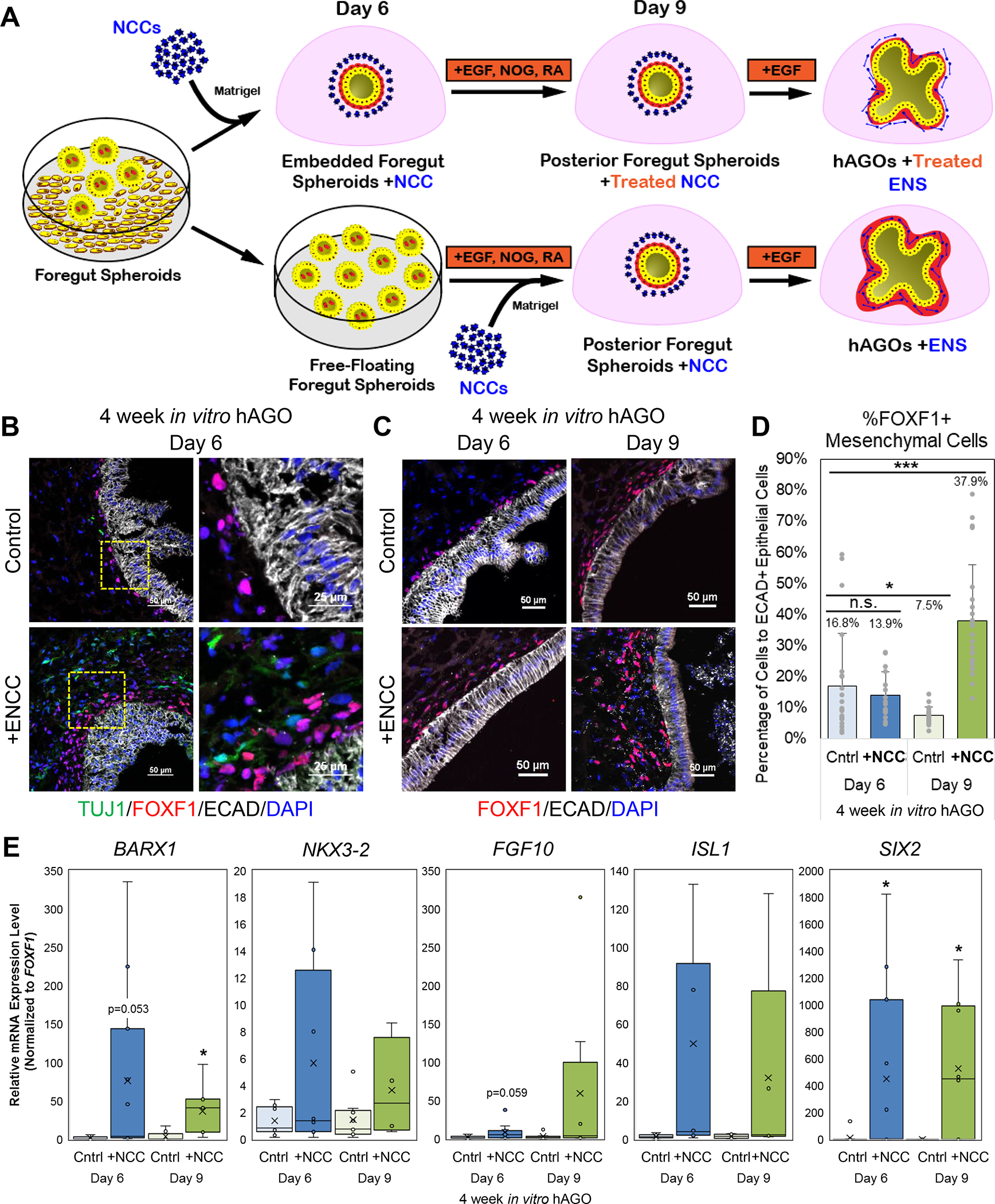
ENS cells promote in vitro growth and patterning of gastric mesenchyme. (A) Schematic illustrating the method of recombining hAGOs with ENCCs at day 6 and day 9 of hAGO protocol. (B) Representative images of four week in vitro hAGOs with (bottom) and without (top) ENCCs recombined on day 6 of hAGO protocol stained with TUJ1 neurons (green) and FOXF1 mesenchyme (red) and epithelial ECAD (white). Higher magnification images are shown to the right. (C) Representative images of four week in vitro hAGOs with (bottom) and without (top) ENCCs recombined on either day 6 (left) or day 9 (right) of hAGO protocol, demonstrate an increase in FOXF1+ mesenchyme (red). (D) Quantification of FOXF1+ mesenchymal contribution (n=16–24 fields from at least 3 organoids from one differentiation, same trend seen across at least two individually seeded differentiations, *p<0.05, ***p<0.001, Student’s t-test). (E) Relative expression of key gastric mesenchymal genes (BARX1, NKX3–2, FGF10, ISL1, SIX2) in hAGOs +ENCC when compared to control hAGOs -ENCC. (n=4–12 wells, with a minimum of 3 organoids per well, from 5 individual differentiations, *p<0.05, Student’s t-test). See also Figure S4, Movie S2, and Table S1.
ENS cells promote the growth and gastric identity of mesenchyme
Previous studies in developing chicken embryos suggest that ENCCs are involved in gastric mesenchyme development (Faure et al., 2015). We therefore analyzed the impact of ENCCs on the development of the small amount of endogenous mesenchyme present in hAGOs. Addition of ENCCs at day 6 of hAGO development had little effect on the number of FOXF1+ mesenchyme cells; in contrast, addition of ENCCs at day 9 resulted in 2–4 times more FOXF1+ mesenchyme surrounding the epithelium (Figure 5A–D). Addition of ENCCs at either day 6 or day 9 also correlated with increased levels of gastric mesenchymal genes BARX1, NKX3–2, FGF10, ISL1, and SIX2 (Figure 5E) (Faure et al., 2013). This suggested that the enteric neurons not only encourage the growth of mesenchyme in vitro, but also support its proper regional patterning into gastric-specific mesenchyme. It is interesting that addition of ENCCs at day 9 promotes the expansion of mesenchyme whereas addition at day 6 does not. The main difference is ENCCs recombined with hAGOs at day 6 are exposed to the BMP inhibitor NOG and RA from day 6–9 as part of the normal hAGO protocol. The impact of this treatment on ENCCs will be discussed below.
ENCC cells support the growth and morphogenesis of organoid epithelium in vivo
We described above that addition of exogenous mesenchyme alone was not sufficient to promote growth and morphogenesis of organoid epithelium (Figure 2). However, we did not investigate how the addition of ENCCs alone without exogenous mesenchyme might impact epithelial development. Therefore, hAGOs with ENCCs recombined at day 6 and 9 were transplanted into mice and grown for 6–15 weeks at which time they were scored for graft survival, overall growth, and epithelial morphogenesis (Figure S5A–B). The presence of an ENS, even in the absence of exogenous mesenchyme, improved both number and epithelial growth of hAGO +ENCC grafts (Figures 6A–B and S5B–D). In most cases the epithelium of the grafts was a simple gastric epithelium with gastric hormonal cells, such as gastrin, ghrelin, somatostatin, and serotonin, as well as surface mucous cells marked by MUC5AC (Figure S5E). However, in 5/21 hAGO +ENCC grafts recombined at day 9, we observed pronounced glandular epithelial morphogenesis as compared to 0/19 hAGO -ENCC (control) grafts (Figure 6A–B, right). Analysis of grafts 4, 10, and 14 weeks following transplantation showed examples of differentiated smooth muscle (Figure S5F) and neuroglial cells expressing TUJ1, S100b, peripherin, nNOS, and GFAP (Figure S6A–B). GFAP cells represent approximately 1% of the total cells present in engrafted hAGOs +ENCC (Figure S6C). These neurons are arranged near the epithelium (Figure S6D and Movie S3) and capable of effluxing calcium as measured using a GCaMP reporter as previously shown (Figure S6E–F and Movie S4) (Workman et al., 2017; Chen et al., 2013). From these findings we conclude that ENCCs promote survival and engraftment of hAGOs and the development of glandular tissue in subset of grafts. However, without a sufficient amount of mesenchyme, addition of ENCCs alone did not result in the development of normal gastric tissue.
Figure 6.
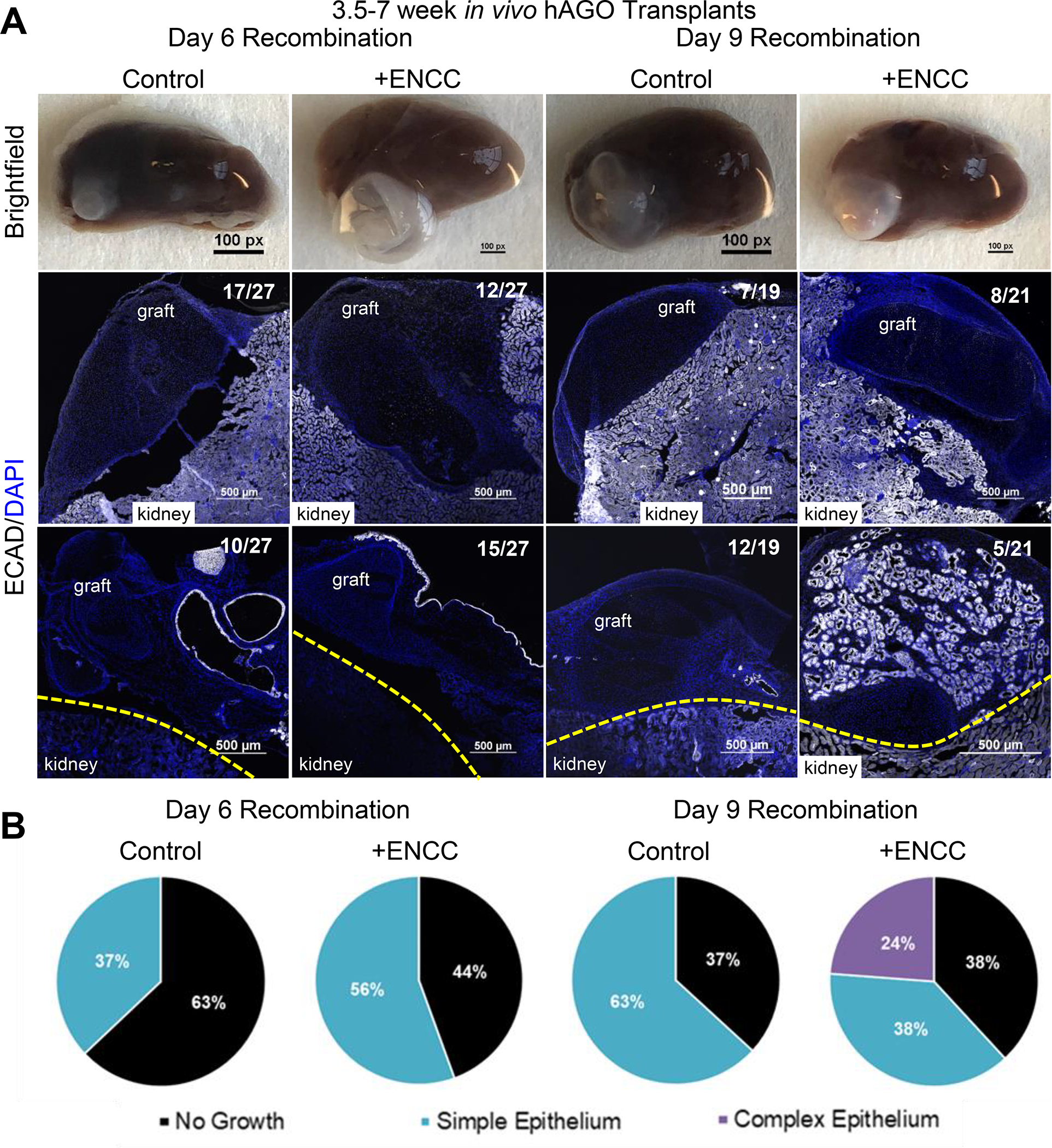
ENCCs promote hAGO engraftment and epithelial growth. (A) Representative low magnification images of gross organoids of ECAD+ epithelium (white) from thAGOs with and without ENCC following recombination at day 6 or day 9. (B) Quantification of organoid engraftment and epithelial growth from thAGOs with and without ENCCs. 24% (5/21) of hAGO +ENCC recombined at day 9 had complex glandular epithelium. See also Figures S5 and S6 and Movies S3 and S4.
The epithelium of hAGOs +ENCCs is morphologically and molecularly similar to Brunner’s glands
A number of hAGO +ENCC grafts displayed a complex glandular epithelial morphology (Figures 6A–B and S7C–D), expressed PDX1 and GATA4 indicative of correct gastrointestinal regional identity, and had gastric hormone-expressing cells such as serotonin, ghrelin, histamine, and somatostatin (Figure 7A, data not shown). However, they did not express key gastric-specific epithelial markers CLDN18 or SOX2 (Figure 7A–B) or have characteristic gastric cell types like MUC5AC-expressing mucous cells (Figure 7D). The glandular epithelium was also negative for intestinal epithelial markers CDX2 and CDH17 (Figure 7A–B). These tissues were human and not a contaminant mouse tissue from the host (Figure S7C). Given the glandular epithelium of the grafts was neither gastric nor intestinal, we explored the possibility that these were Brunner’s glands. Brunner’s glands are glandular structures found within the submucosa of the proximal part of the duodenum, near the pyloric junction. They serve to secrete sodium bicarbonate to neutralize any escaping gastric acids. While some protein markers exist, most analyses have been done on biopsies from pathologic samples, like adenomas (Wang et al., 2015). Given the lack of definitive markers for human Brunner’s glands, we established a combinatorial marker profile for Brunner’s glands using patient biopsies (Figure S7A–B) and published reports (Figure 7C). Human Brunner’s glands were negative for gastric markers CLDN18, SOX2, and MUC5AC and intestinal markers CDH17 and MUC2 and had low levels of CDX2 compared to adjacent duodenal epithelium (Figure S7). Human Brunner’s glands were positive for glucagon-like peptide-1 receptor (GLP-1R) (Holst, Andersen and Grunddal, 2021) and MUC6 and co-expression of these markers occurs only in Brunner’s glands. Using this combinatorial expression profile of 9 different markers on hAGO +ENCC grafts allowed us to conclude that they were most similar to Brunner’s glands (Figures 7C and S7A–B) (Tan et al., 2020; Wang et al., 2015; Balbinot et al., 2017).
Figure 7.
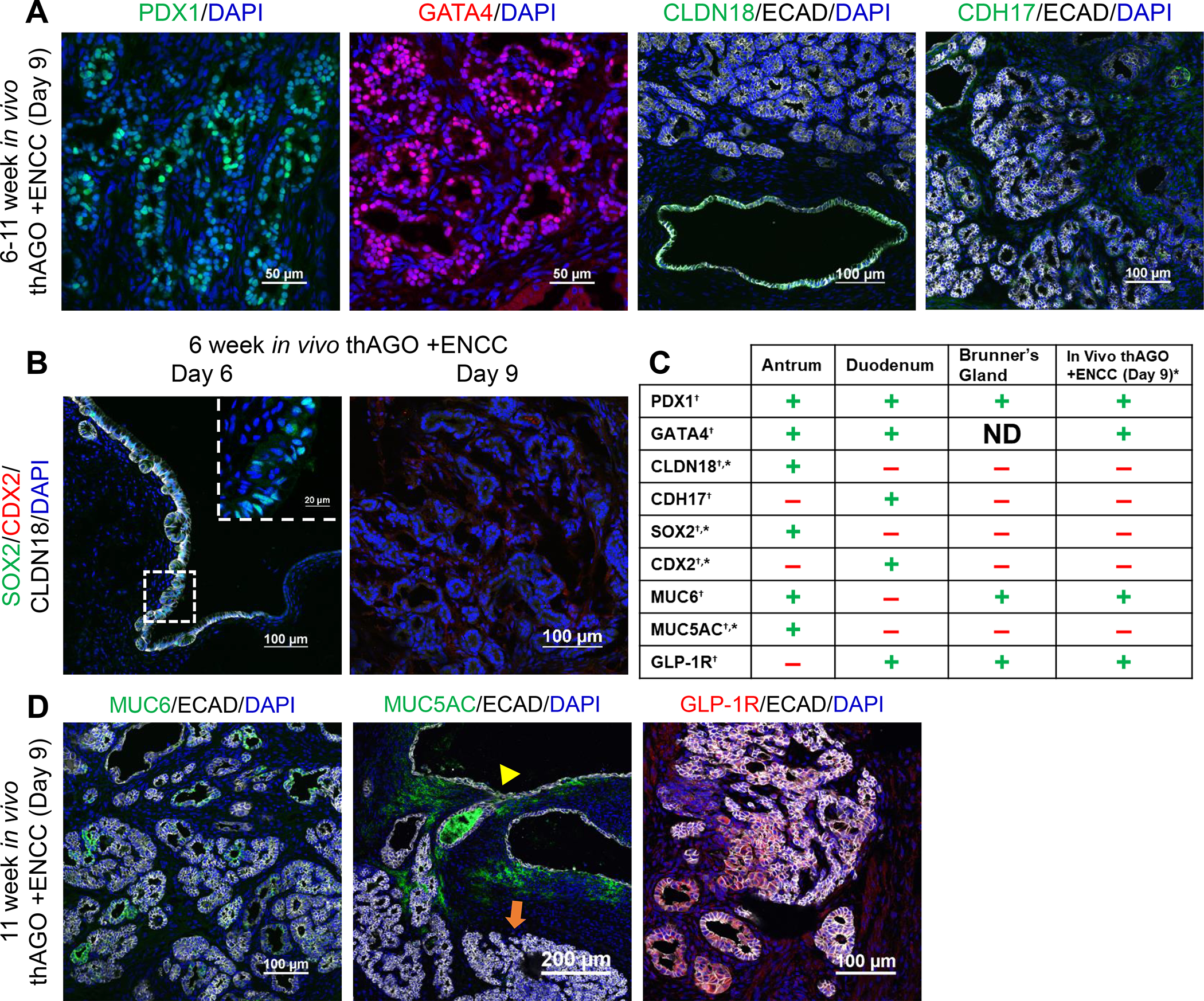
Identification of hAGO +ENCC glandular epithelium as Brunner’s glands. (A) Complex glandular epithelium expressed the pyloric junction marker PDX1 (green) and gastrointestinal columnar epithelial marker GATA4 (red) but did not express gastric epithelial marker CLDN18 or intestinal epithelial marker CDH17 in thAGOs recombined with ENCCs on day 9. (B-D) Marker analysis of organoid epithelium at different time points following transplantation. (B) After 6 weeks growth in vivo hAGOs with ENCCs recombined at day 6 had a simple epithelium expressing the gastric markers SOX2 (green, inset) and CLDN18 (white) but not the intestinal marker CDX2 (red). The glandular epithelium of from day 9 recombinants did not express these gastric or intestinal markers. (C) Comparison of antral and duodenal markers to known markers of Brunner’s glands and how these align with observed protein expression profile of complex epithelial growths from hAGOs +ENCC day 9 recombined grafts. †determined from previously published data; *determined experimentally on human tissue samples of Brunner’s glands (see Figure S7B) (D) At 11 week post-transplant, the glandular epithelium in organoids did express MUC6 (green, left) and GLP-1R (green, middle), similar to human Brunner’s glands. The simple epithelium (yellow triangle) expressed MUC5AC (green, right) while complex epithelium (orange arrow) the glandular epithelium did not. See also Figure S7.
This posterior shift from hAGO epithelium to Brunner’s Gland suggests that the added ENCCs may produce a factor(s) that posteriorizes gastric epithelium. One candidate pathway was BMP signaling, which is known to promote posterior fate at GI junctions (Smith and Tabin, 1999; Smith et al., 2000; Faure et al., 2002; Tiso et al., 2002; De Santa Barbara et al., 2005; Theodosiou and Tabin, 2005; Davenport et al., 2016; Stevens et al., 2017). Analysis of ENCCs shows high levels of both BMP4 and BMP7 transcripts (Figure S7D–E). To functionally investigate if BMP activity might mediate the ability of ENCCs to promote a Brunner’s gland fate, we analyzed hAGOs where ENCCs were incorporated at day 6 and then cultured with the BMP inhibitor NOG from day 6–9, along with RA, which are components of the normal hAGO protocol. None of the grafts (0/27) had Brunner’s gland epithelium following 3 days of NOG treatment as compared to hAGOs +ENCCs grafts that were not treated with NOG, where the 5/21 grafts contained Brunner’s gland epithelium (Figure S7F). Analysis of ENCCs showed that NOG treatment caused reduced levels of both BMP4 and 7 (Figure S7G), suggesting this may explain their reduced posteriorizing activity.
DISCUSSION
By understanding and applying the key signaling pathways known to regulate the development of different cell types, many protocols are now published to direct the differentiation of hPSCs into germ layer-specific fates, including endoderm, mesoderm, and ectoderm derivatives. However, we know that organs of the GI tract are assembled from all 3 germ layers in a step wise manner. This research combines our knowledge of organ assembly with the ability to differentiate single germ layers from hPSCs to develop an enhanced paradigm for tissue engineering complex organoids. Starting with 3-dimensional gut tube-like spheroids, we layered on separately derived SM and ENCCs, both having the intrinsic ability to self-organize around the epithelium. To our knowledge, this is the first demonstration of GI organoid generation from the three primary germ layers. Moreover, the engineered gastric tissue has many properties of bona fide human stomach antrum. The basic concept of assembling organoids from separately-derived germ layer progenitors was also applied to both fundus and esophagus, suggesting that this technology could be broadly applied to tissue engineer other organs, like lung, liver, and bladder.
Congenital diseases, due to impacts on multiple germ layers, often affects several organs. These 3 germ layer organoids represent improved model systems to study both the effects of patient-specific mutations on multiple organs and how gene mutations impact individual germ layers, similar to a cell specific Cre approaches in mice. This approach has been used to study the impact of patient mutations on human PSC-derived ENCCs (Workman et al., 2017) and PSC-derived epithelial cell types (Zhang et al., 2019), and could be used to study mutations that largely effect mesenchyme (Gilbert et al., 2020). Analyses of organoids could identify previously unappreciated patient pathologies thus improving clinical care. However, to effectively understand congenital disorders, we need to better understand the processes of epithelial-mesenchyme-ENS communication during normal organ development. The tractability of this system is ideally suited to interrogate such signaling pathways mediating this crosstalk. Our data suggests ENCCs impact the growth and patterning of endogenous mesenchyme. Moreover, without addition of exogenous SM, ENCCs re-pattern gastric epithelium to a more posterior identity similar to Brunner’s glands. However, when ENCCs are added simultaneously with a robust population of SM, together these germ layers maintain gastric patterning and promote gastric gland morphogenesis.
The ability to manipulate signaling pathways in a germ layer-specific manner in vitro is a powerful way to dissect the molecular basis of organ development. For example, it is known that WNT and BMP signaling pathways control the anterior-posterior and dorso-ventral patterning of the developing GI tract in many model organisms (Smith and Tabin, 1999; Smith et al., 2000; Faure et al., 2002; Tiso et al., 2002; De Santa Barbara et al., 2005; Theodosiou and Tabin, 2005; Davenport et al., 2016; Stevens et al., 2017) and in hPSC-derived colonic organoids (Munera and Wells, 2017; Múnera et al., 2017). In the gastro-duodenal boundary, WNT-mediated crosstalk between the epithelium and mesenchyme are essential for establishing and maintaining a molecular boundary between the gastric and duodenal epithelium (McCracken et al., 2017; Kim et al., 2005). Evidence from chick studies show that ENCCs regulate the anterior-posterior patterning of stomach (Faure et al., 2015), and our data now show that ENCCs express BMP ligands, can posteriorize human gastric epithelium, and that inhibiting BMP signaling prevents the posteriorizing effects of ENCCs.
Our findings highlight that the only context where we see formation of normal gastric tissue is when foregut spheroids are combined with a carefully controlled amount of SM and ENCCs. Adding SM alone results in organoids with poorly organized smooth muscle and a simple epithelium and adding ENCCs alone results in mispatterned epithelium and the formation of Brunner’s glands. However, recombining robust populations of SM and ENCCs results in well-organized smooth muscle, an organized neuroglial plexus, and the formation of properly patterned gastric glands with chief and parietal cells. Moreover, the neuroglial plexus forms a functional link with the smooth muscle to regulate rhythmic gastric contractions. We conclude that communication between all 3 germ layers is essential for proper assembly and morphogenesis of stomach tissue. While growth of more mature gastric tissue requires the transplantation of organoids into mice, we believe that the animals largely act as an incubator, providing a blood supply that allows for continued organoid growth and development (Watson et al., 2014).
We observed that posteriorizing effects of ENCCs correlated with the formation of Brunner’s gland-like structures following transplantation and growth in vivo. Since little is known about the embryonic development of these glands in any species, or what markers define them, we identified a marker profile of normal human Brunner’s glands, which will aid in studying the molecular pathways that drive development of these glands. For example, LRIG and ERB2 signaling play a role in Brunner’s gland development (Wang et al., 2015) and our data suggest that BMP patterning may play into this pathway. These markers could be used with hPSC-derived Brunner’s gland organoids to perform a systematic functional study of how these pathways may interact during Brunner’s gland development. Another application would be to study how ENCCs stimulate the formation of Brunner’s gland organoids. A role for ENCC-derived neurons in glandular morphogenesis has been established. Submandibular glands in mouse are responsive to neuronal signals like VIP and Ach during branching morphogenesis in vitro (Knox et al., 2010) and in vivo (Nedvetsky et al., 2014). By comparison, Brunner’s glands are also innervated by VIP and Ach neurons (Moore, Morris and Vanner, 2000; Moore, Kim and Vanner, 2000).
A possible mechanism to explain why gastric gland morphogenesis requires both ENS and mesenchyme comes from studies of intestinal and lung development. In the intestine of mice and chicks, mesenchymal clusters (Freddo et al., 2016; Walton et al., 2012) and smooth muscle (Shyer et al., 2013) regulate villus morphogenesis and lung branches (Goodwin et al., 2019). Additionally, in chick, BMP signaling from both epithelium and NCCs regulates the radial position of developing smooth muscle layers (Huycke et al., 2019). Thus, innervation of smooth muscle layers in human gastric antrum may promote development of organized smooth muscle and glandular morphogenesis. Our data suggest that ENCCs require a robust population of mesenchyme to promote gastric gland morphogenesis. However, addition of ENCCs alone can promote formation of Brunner’s gland-like epithelium, suggesting the addition of mesenchyme is important to maintain gastric identity and/or synergize with the signals coming from the ENCCs. This seems plausible considering the close proximity and physical connection of enteric nerves with both smooth muscle and epithelial cells n stomach glands that is necessary for proper stomach function.
Little is known about ENS development and the specifics of neuronal diversity within proximal GI tract, relative to the intestine and colon ENS (Kaelberer et al., 2018; Lasrado et al., 2017; Rakhilin et al., 2016; Bohorquez et al., 2015; Walsh and Zemper, 2019; Nagy and Goldstein, 2017; Brookes et al., 2013) and we have observed differences in ENS development between human gastric and intestinal organoids. Innervated intestinal organoids did not contain nNOS+ neurons in vitro (Workman et al., 2017) but these neurons did form in hAGOs +ENCCs. This implies regional differences exist in ENS development between proximal and distal GI organs that could be studied using human organoid systems. Additional differences in stomach and intestinal development, from orientation and innervation of smooth muscle to glandular morphogenesis and neuronal control of secretion, can be modeled in these systems. Moreover, human organoids are now being used to study human organ physiology. hPSC-derived human intestinal organoids were used as a model of malabsorption in humans and led to the discovery of a previously unknown mechanism by which enteroendocrine cells control nutrient absorption (McCauley et al., 2020).
In summary, we have generated 3 germ layer organoids that are morphologically, cellularly, and functionally similar to human stomach tissues. Engineered gastric tissue has glands with surface and pit mucous cells, as wells as chief and parietal cells. We observed oriented layers of smooth muscle innervated by functional enteric nerves and used this highly manipulable system to start defining cellular communications that occur during human stomach development. This technology is broadly translatable to other organs so it is possible that engineered tissue might be a source of material for reconstruction of congenital disorders and acute injuries of the upper GI tract.
LIMITATIONS OF THE STUDY
HPSC-derived gastric organoids can be variable between differentiations. Coordinating the timing of recombination of three simultaneously generated germ layers, as well as the requirement of kidney capsule transplantation into immunocompromised mice necessary to promote 3 germ layer organoid maturity is technically challenging, lengthy, and can limit protocol scalability and forward progression of developing transplantable material.
STAR METHODS
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James Wells (james.wells@cchmc.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Experimental Model and Subject Details
Animals
All mice used in kidney capsule transplantation experiments were housed in the animal facility at Cincinnati Children’s Hospital Medical Center (CCHMC) in accordance with NIH Guidelines for the Care and Use of Laboratory animals. Animals were maintained on a 12 hour light-dark cycle with access to water and standard chow ad libitum. Healthy male and female immune-deficient NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice, aged between 8 and 16 weeks old, were used in all experiments. These mice were obtained from the Comprehensive Mouse and Cancer Core Facility. All experiments were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of CCHMC.
Timed mattings of wildtype mice were used to generate e13.5 embryos for immunohistological analysis. The morning that the vaginal plug was observed was denoted as e0.5.
Human Biopsy Tissue
The use of human tissues was approved by an Institutional Review Board (IRB) at CCHMC (protocol number 2015–5056). Informed consent for the collection and use of tissues was obtained from all donors, parents, or legal guardians. Full-thickness fundic and antrum stomach tissue samples obtained from bariatric procedures came from the Helmrath Lab at CCHMC under IRB protocol number 2014–0427. Human surgical samples were collected from patients between the ages of 15 and 17, and included both males and females of Caucasian and African American backgrounds. Healthy human full-thickness stomach and duodenal tissue samples were obtained from the CCHMC Pathology Core.
Human ESC/iPSC lines and maintenance
Human embryonic stem cell (hESC) lines H1 (WA-01) and H9 (WA-09) were purchased from WiCell (NIH approval number NIHhESC-10–0043 and NIHhESC-10–0062). The H1 line is male and the H9 line is female. H9-GAPDH-GFP and H9-GAPDH-mCherry hESCs along with human induced pluripotent stem cell (iPSC) line 77.3-GFP were all generated and obtained from the CCHMC Pluripotent Stem Cell Facility (PSCF) and approved by the institutional review board (IRB) at CCHMC. Human iPSC line WTC11 AAVS1-CAG-GCaMP6f was obtained from Bruce Conklin’s laboratory at UCSF (Maddah et al., 2015). All hPSC lines were analyzed for pluripotency and the absence of karyotypic abnormalities and mycoplasma contamination by the CCHMC PSCF. Human iPSC line WTC11 was analyzed for karyotype by Cell Line Genetics. All human hPSCs were maintained in an undifferentiated state as colonies in feeder-free conditions. They were plated on human-ES-cell-qualified Matrigel (BD Biosciences) and maintained at 37°C with 5% CO2 with daily replacement of mTeSR1 media (STEMCELL Technologies). Cells were routinely passaged every 4 days with Dispase (STEMCELL Technologies) after a confluency of about 80–90% was reached.
Differentiations of the following lineages for construction of three-germ layers organoids are not dependent or variable based on starting hPSC line. Please see the Key Resource Table under Experimental Modes: Cell Lines for minimum number of differentiations performed using each cell line.
Method Details
Differentiation of hPSCS into mesenchyme
Partially confluent hPSCs colonies were dissociated into single cells using Accutase (Thermo Fisher Scientific), resuspended in mTesR1 with thiazovivin (1 μM, Tocris), and passaged 1:20 onto new Geltrex-coated 24-well plates (Sigma Aldrich). The directed differentiation of hPSCs into lateral plate mesoderm has been previously described (Han et al., 2020; Loh et al., 2016). Briefly, hPSCs were exposed to Activin A (30 ng/ml, Cell Guidance Systems), BMP4 (40 ng/ml, R&D Systems), CHIR99021 (CHIR, 6 μM, ReproCell), FGF2 (20 ng/ml, ThermoFisher Scientific), and PIK90 (100 nM, EMD Millipore) for 24 hours. A basal media composed of Advanced DMEM/F12 (ThermoFisher Scientific) supplemented with B27 supplement (1X, ThermoFisher Scientific), N2 supplement (1X, ThermoFisher Scientific), HEPES (13 mM, ThermoFisher Scientific), L-Glutamine (2 mM ThermoFisher Scientific), and penicillin-streptomycin (1X, ThermoFisher Scientifics) was used for this and all subsequent differentiation steps. Cells were then exposed to A8301 (1 μM, Tocris), BMP4 (30 ng/ml), and C59 (1 μM, Cellagen Technology) for 24 hours. For splanchnic mesoderm generation, cells were cultured in A8301 (1 μM), BMP4 (30 ng/ml), C59 (1 μM), FGF2 (20 ng/ml), and RA (2 μM, Sigma-Aldrich) from Day 2 to Day 4. To further direct regional splanchnic mesoderm, RA (2 μM), PMA (2 μM, Tocris) was used for 2 days, and then RA (2 μM), PMA (2 μM), NOG (100 ng/ml, R&D Systems) was used at the last 1 day to promote esophageal/gastric mesenchyme fate. Medium was changed every day throughout protocol. Confluent cells were dissociated into single cells using Accutase (6–10 min), resuspended in 1–8mL DMEM/F-12 (Gibco), counted, and immediately combined with hAGOs, hFGOs, and hEOs (see below for recombination procedure).
Differentiation of hPSCS into ENCCs
The generation of hPSC-derived ENCCs has been previously published (Bajpai et al., 2010; Workman et al., 2017). Briefly for ENCC generation, confluent hPSCs were treated with collagenase IV (500 U/ml, Gibco) in mTeSR1 for 60–90 mins to detach colonies. Cells were diluted and washed with DMEM/F-12 (Gibco) and then gently triturated and resuspended in neural induction media, 1:1 ratio of DMEM/F12-GlutaMAX (Gibco) and Neurobasal Medium (Gibco) with B27 supplement (0.5x, Gibco), N2 supplement (0.5x, Gibco), pen-strep (1x, Gibco), insulin (5 μg/mL, Sigma-Aldrich), FGF2 (20 ng/mL, R&D Systems), and EGF (20 ng/mL, R&D Systems), on non-TC-treated petri dishes (6cm, Fisherbrand). Neural induction media was changed daily and all-trans RA (2 μM) was added on days 4 and 5 for posteriorization. Day 6 free-floating neurospheres were plated on human fibronectin (HFN, 3 μg/cm2, Corning) and fed neural induction media without RA for 4 days. Migrated cells were collected using a 90 sec Accutase treatment and passaged onto HFN. Passaged cells were allowed to grow to confluency for an additional 4 days and fed neural induction media without RA every day. Confluent cells were dissociated into single cells using Accutase (2–6 min), resuspended in 1–4mL DMEM/F-12 (Gibco), counted, and immediately combined with hAGOs, hFGOs, and hEOs (see below for recombination procedure).
Differentiation of hPSCS into human organoids
We utilized slightly modified previously published protocols to generate hAGOs, hFGOs and hEOs (McCracken et al. 2014, 2017, Trisno et al., 2018). For hAGO and hFGO generation, confluent hPSC cultures were treated with Accutase to resuspend as single cells in mTeSR1 with ROCK inhibitor Y-27632 (10 μM; Tocris) and plated onto a Matrigel-coated 24-well dish (Sigma Aldrich). To direct the differentiation into definitive endoderm (DE), the hPSCs were exposed to Activin A (100 ng/ml) and BMP4 (50 ng/ml) in RPMI 1640 media (Life Technologies). For the following two days, cells were exposed to only Activin A (100 ng/ml) in RPMI 1640 media containing increasing concentrations (0.2% and 2.0%, respectfully) of defined fetal bovine serum (dFBS; HyClone). To then pattern DE into posterior foregut endoderm spheroids, cells were treated with FGF4 (500 ng/ml, R&D systems), NOG (200 ng/ml), and CHIR (2 μM) for 3 days, with media changed daily, in RPMI 1640 with 2% dFBS. RA (2 μM) was added on the third day of FGF4/NOG/CHIR treatment.
Recombination and spheroid patterning
Single cell suspensions of mesenchymal cells and ENCCs were counted and appropriate volumes added to foregut spheroids collected in a 1.5mL tube at an approximate ratio of 1,000 ENCCs and 2,500 mesenchyme cells per spheroid. Cell mixtures were mixed via gentle pipetting, centrifuged at 300g for 3–5 minutes, gently resuspended into 50 μL of basement membrane Matrigel, and pipetted onto a TC dish to allow three-dimensional in vitro culture. Organoids were fed with a base media of Advanced DMEM/F12 supplemented with B27 supplement (1X), N2 supplement (1X), HEPES (13 mM), L-Glutamine (2 mM), penicillin-streptomycin (1X), and EGF (100 ng/mL). In addition to this base media, the first three days were supplemented with NOG (200 ng/mL) and RA (2 μM). In addition to EGF, hFGOs were supplemented with CHIR (2 μM) throughout the organoid outgrowth and also received a 48 hr. pulse of BMP4 (50 ng/mL) and PD0325901 (2 μM, Stem Cell Technologies) 96 hours prior to collection for parietal cell differentiation in vitro. Media was replaced every 3–4 days. Two weeks following spheroid embedding in Matrigel, the organoids were collected and re-plated in fresh Matrigel at a dilution of ~1:12.
In vivo transplantation of hAGOs and hFGOs
hAGO, hFGO, hAGO +ENCC, hAGO +SM, hAGO +SM +ENCC, and hFGO +SM +ENCC were all ectopically transplanted into the kidney capsule of NSG mice as previously described (Watson et al., 2014). Briefly, four week old hAGOs or hFGOs were removed from Matrigel and transplanted into the kidney subcapsular space. Engrafted organoids were harvested 6–15 weeks after transplantation and analyzed for neuroglial, epithelial, and mesenchymal maturation.
Ex vivo muscle contraction and ENS function
Muscle contraction was assayed as previously described (Poling et al., 2018) and ENS function and motility were assayed as previously described with slight modifications (Workman et al., 2017). Briefly, strips of tissue approximately 2 × 6 mm in size were dissected and the epithelium mechanically removed in a method similar to seromuscular stripping as previously described (Workman et al., 2017). No chelation buffer was used. Resulting strips of muscle from hAGO +SM +ENCC were mounted within an organ bath chamber system (Radnoti) to isometric force transducers (ADInstruments) and contractile activity continuously recorded using LabChart software (ADInstruments). After an equilibrium period, a logarithmic dose response to Carbamyl-β-methylcholine chloride (Bethanechol; Sigma-Aldrich) was obtained through the administration of exponential doses with concentrations of 1 nM to 10 mM at 2 min intervals before the administration of 10 μM scopolamine (Tocris Bioscience). Data are normalized to muscle strip mass. After another equilibrium period, muscle preparations were then stimulated with a control EFS pulse. NG-nitro-L-arginine methyl ester (L-NAME; 50 μM; Sigma) was applied 10 min before EFS stimulation to observe the effects of NOS inhibition. Without washing, Atropine (atropine sulfate salt monohydrate; 1 μM; Sigma) was the applied 10 min prior to a final EFS stimulation to observe the cumulative effect of NOS and Ach receptor inhibition. After several washes and an additional equilibrium period, another control EFS pulse was administered. Neurotoxin tetrodotoxin (TTX; 4 μM; Tocris) was administered 5 min before a final EFS stimulation. Analysis was performed by calculating the integral (expressed as area under the curve, AUC) immediately before and after stimulation for 60s. Data are normalized to muscle strip mass.
Ex vivo GCamP6f calcium imaging
Detection of calcium transients was performed using the above-mentioned human iPSC line WTC11 AAVS1-CAG-GCaMP6f. Transplanted hAGOs +ENCC were harvested and then cultured on 8-well micro-slide (Ibidi) for 24 hours prior to imaging. They were then imaged every 4–15 sec for 3–10 min using either a 10x or 20x objective on a Nikon Ti-E inverted A1 confocal microscope with NIS elements software to obtain background fluorescence level. Transplanted hAGOs +ENCC were then treated with 30 mM KCl. Experiments were carried out at RT.
Tissue Processing, IHC, and Microscopy
Cell monolayers, ENCCs, and day 0 spheroids were washed with 1x phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde (PFA) at room temperature (RT) for 15 min, washed, and stored in PBS at 4°C. Four week old in vitro organoids and in vivo transplants were washed with PBS, fixed in 4% PFA at 4°C overnight, washed, and then placed in either PBS, 30% sucrose in PBS, or 70% ethanol at 4°C overnight for downstream whole mount, cryogenic, or paraffin processing, respectively. Prior to fixation, whole mount tissues were extracted from Matrigel using manual pipetting in cold PBS and Cell Recovery Solution (Corning). Tissues were then embedded in either O.C.T. Compound (Tissue-Tek) or paraffin and were serially sectioned at a thickness of 7–8 μm onto Superfrost Plus glass slides (Fisherbrand). Cryosection slides and paraffin slides were stored at −80°C and RT, respectively. Routine Hematoxylin & Eosin (H&E) staining was performed by the Research Pathology Core at CCHMC.
Frozen slides were thawed to room temperature (RT) and rehydrated in PBS, while paraffin slides were deparaffinized, rehydrated, and subjected to heat- and pressure-induced antigen retrieval in citrate buffer (0.192% citric acid and 0.0005% Tween 20 in dH20 of pH 6.0 with NaOH) for 30 minutes and brought to RT on ice. All slides and cells were washed with PBS, permeabilized with 0.5% Triton X-100 in PBS (PBST) for 15 min at RT and then blocked with 5% normal donkey serum (NDS, Jackson ImmunoResearch) in PBS for one hour at RT. Tissue was incubated at 4°C overnight in primary antibodies diluted in 5% NDS in PBS. Specific antibody details are listed in the Key Resource Table. The following day, tissue was washed and incubated with secondary antibodies at RT for one hour, thoroughly washed, and cover slipped with Fluoromount-G (Southern Biotech).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat anti-H/K ATPase α Subunit (ATP4A) | Santa Cruz Biotechnology | #sc-167155 |
| rabbit anti-H/K ATPase β Subunit (D-18) (ATP4B) | Santa Cruz Biotechnology | #sc-84304 |
| mouse anti-αSmooth Muscle Actin (αSMA) | Sigma-Aldrich | #A2547 |
| rabbit anti-αSmooth Muscle Actin (αSMA) | GeneTex | #981303701 |
| rabbit anti-βcatenin (β-CAT) | Santa Cruz Biotechnology | #sc-7199 |
| rabbit anti-βIII-tubulin (TUJ1) | Abcam | #ab18207 |
| rabbit anti-Cadherin 17 (CDH17) | Sigma-Aldrich | #HPA023616 |
| mouse anti-Cadherin 17 (CDH17) | R&D Systems | #MAB1032 |
| rabbit anti-Calbindin (CLBN1) | Gift from Kenny Campbell | N/A |
| mouse anti-CDX2 | BioGenex | MU392A-UC |
| rabbit anti-CDX2 | Cell Marque | #EPR2764Y |
| mouse anti-Chromogranin A (CHGA) | DSHB | #CPTC-CHGA-1 |
| rabbit anti-Claudin 18 (CLDN18) | Sigma-Aldrich | HPA018446 |
| goat anti-E-Cadherin (ECAD) | R&D Systems | #AF648 |
| rat anti-E-Cadherin (ECAD) | R&D Systems | #MAB7481 |
| mouse anti-E-Cadherin (ECAD) | BD Transduction Laboratories | #610182 |
| goat anti-FOXF1 | R&D Systems | AF4798 |
| rabbit anti-gastric intrinsic factor (GIF) | Sigma-Aldrich | HPA040774 |
| mouse anti-Gastric Mucin (MUC6) | Abcam | #ab49462 |
| rabbit anti-Gastrin (GAST) | Dako | #A0568 |
| goat anti-GATA4 | Santa Cruz Biotechnology | #sc-1237 |
| chicken anti-GFP | Aves Labs | GFP-1010 |
| goat anti-Ghrelin (GHRL) | Santa Cruz Biotechnology | #sc-10368 |
| rabbit anti-Glucagon-like peptide-1 receptor (GLP-1R) | Novus Biologicals | #NLS1205 |
| rabbit anti-Histamine | Immunostar | #22939 |
| rat anti-Keratin 8 (KRT8) | Developmental Studies Hybridoma Bank | #TROMA1 |
| rabbit anti-mucin 2 (MUC2) | Abcam | #ab134119 |
| mouse anti-Mucin-5AC (MUC5AC) | Abcam | #ab3649 |
| rabbit anti-Nitric oxide synthase (nNOS) | Abcam | #ab76067 |
| goat anti-PDX1 | Abcam | #ab47383 |
| goat anti-Peripherin | Santa Cruz Biotechnology | #sc-7604 |
| mouse anti-Pepsinogen I (PGA3) | Abcam | #ab31466 |
| sheep anti-Pepsinogen II (PGC) | Abcam | #ab31464 |
| rabbit anti-S100β | Dako | #Z031129 |
| rabbit anti-Serotonin (5-HT) | Immunostar | #20080 |
| goat anti-Somatostatin | Santa Cruz Biotechnology | #sc-7819 |
| goat anti-SOX2 | Santa Cruz Biotechnology | #sc-17320 |
| goat anti-SOX2 | R&D Systems | AF2018 |
| rabbit anti-SOX2 | Abcam | #ab97959 |
| guinea pig anti-Synaptophysin | Synaptic Systems | #101004 |
| chicken anti-Tyrosine Hydroxylase (TH) | Aves Labs | #TYH |
| Bacterial and virus strains | ||
| N/A | N/A | N/A |
| Biological samples | ||
| Human Stomach and Duodenum Samples | Wells Lab CCHMC | GI-1B-9952 A B |
| Human Antral and Fundic Samples | Helmrath Lab CCHMC | 14-241, 14-242, 14-243, 14-244 |
| Human Stomach and Duodenum Samples | CCHMC Pathology Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 1× DMEM | Invitrogen | Cat #11965-084 |
| 50× B27 supplement | Thermo Fisher Scientific | Cat #17504044 |
| 50× B27 supplement w/o Vitamin A | Thermo Fisher Scientific | Cat #12587-010 |
| A8301 | Torcis | Cat #A83-01 |
| Activin A | Cell Guidance Systems | Cat #GFH6 |
| Accutase | Thermo Fisher Scientific | Cat #A11105-01 |
| Advanced DMEM:F12 | Thermo Fisher Scientfic | Cat #12634-010 |
| Atropine sulfate salt monohydrate | Sigma-Aldrich | Cat #A0257 |
| Bethanechol | Sigma-Aldrich | Cat # 1071009 |
| C59 | Cellagen Technology | Cat #C7641-2s |
| CHIR99021 | ReproCell | Cat #04-0004-10 |
| Collagen Type IV | Thermo Fisher Scientific | Cat #17104019 |
| Defined fetal bovine serum (dFBS) | Hyclone | Cat #SH30070.02 |
| Dispase | Thermo Fisher Scientific | Cat #17105-041 |
| DMEM/F12 | Thermo Fisher Scientific | Cat #11320033 |
| DMEM/F-12, GlutaMAX | Thermo Fisher Scientific | Cat #10565018 |
| EGF | Sigma-Aldrich | Cat #1257-0.1MG |
| F12 Media | Invitrogen | Cat #11765-054 |
| Fetal Bovine Serum (FBS) | HyClone | Cat #SH30071.03 |
| Fibronectin | Corning | Cat #354008 |
| Geltrex | Thermo Fisher Scientific | Cat #A1413201 |
| HEPES Buffer | Thermo Fisher Scientific | Cat #15630080 |
| hESC-qualified Matrigel | BD Biosciences | Cat #354277 |
| Insulin from bovine pancreas | Sigma-Aldrich | Cat #I4011-50 |
| L-glutamine (100×) | Thermo Fisher Scientific | Cat #25030-081 |
| L-NAME | Sigma-Aldrich | Cat #N5751 |
| Matrigel Matrix Basement Membrane | BD Biosciences | Cat #354234 |
| mTeSR1 media | Stem Cell Technologies | Cat #5850 |
| N2 Supplement | Thermo Fisher Scientific | Cat #17502-048 |
| Non-essential Amino Acids (100×) | Thermo Fisher Scientific | Cat #11140050 |
| Normal donkey serum | Jackson Immunoresearch Labs | Cat #017-000-121 |
| Neurobasal Medium | Thermo Fisher Scientific | Cat #21103049 |
| PD0325901 | Stem Cell Technologies | Cat #72184 |
| Pen/Strep (100×) | Thermo Fisher Scientific | Cat #15140-122 |
| PIK90 | EMD Millipore | Cat #528117 |
| PMA | Torcis | Cat #1201 |
| recombinant human BMP4 | R&D Systems | Cat #314-BP |
| recombinant human EGF | R&D Systems | Cat #236-EGF |
| recombinant human FGF2 | R&D Systems | Cat #233-FB |
| recombinant human FGF4 | R&D Systems | Cat #235-F4 |
| recombinant human FGF10 | R&D Systems | Cat #345-FG |
| recombinant human Noggin | R&D Systems | Cat #6057-NG |
| recombinant human WNT3A | R&D Systems | Cat #5036-WN |
| RNAlater | Thermo Fisher Scientific | Cat #AM7020 |
| Scopolamine | Tocris | Cat #1414 |
| Thiazovivin | Tocris | Cat #3845 |
| TTX | Tocris | Cat #1078 |
| Y-27632 dihydrochloride | Tocris | Cat #1254 |
| Critical commercial assays | ||
| Quantitect SYBR Green | Qiagen | Cat #204145 |
| Nucleospin RNA | Macherey-Nagel | Cat #740955 |
| SuperScript VILO cDNA Synthesis Kit | Thermo Fisher Scientific | Cat #11754250 |
| Deposited data | ||
| N/A | N/A | N/A |
| Experimental models: cell lines | ||
| Human: H1 ES cells; >28 hAGO, >18 NCC, and >10 SM differentiations | CCHMC Pluripotent Stem cell core / WiCell Research Institute | NIH hESC-10-0043 |
| Human: H9 ES cells | CCHMC Pluripotent Stem cell core / WiCell Research Institute | NIH hESC-10-0043 |
| Human: H9-GAPDH-GFP ES cells, >5 NCC differentiations | CCHMC Pluripotent Stem cell core | N/A |
| Human: H9 RFP ES cells, >4 SM differentiations | CCHMC Pluripotent Stem cell core | N/A |
| Human: H9 mCherry ES cells, >1 NCC differentiation and >3 SM differentiations | CCHMC Pluripotent Stem cell core | N/A |
| Human: iPSC77.3 GFP iPS cells, >6 NCC differentiations and >2 SM differentiations | CCHMC Pluripotent Stem cell core | N/A |
| Human: WTC11 AAVS1-CAG-GCaMP6f iPS cells, 2 NCC differentiations | Bruce Conklin’s Lab at UCSF | N/A |
| Experimental models: organisms/strains | ||
| Mouse: NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) | Comprehensive Mouse and Cancer Core Facility CCHMC | N/A |
| Oligonucleotides | ||
| Please see Table S3. | ||
| Recombinant DNA | ||
| N/A | N/A | N/A |
| Software and algorithms | ||
| IMARIS | Bitplane | N/A |
| NIS Elements | Nikon | N/A |
| LabChart software | ADInstruments | N/A |
| Other | ||
| 8-well micro-slide | Ibidi | N/A |
| Organ bath chamber | Radnoti | N/A |
| Isometric force transducers | ADInstruments | N/A |
For wholemount staining, organoids were washed at RT and then permeabilized with PBST at 4°C overnight. The next day, organoids were blocked in 5% NDS in PBST for 6–8 hours at RT and then incubated in primary antibodies at 4°C overnight on a rocking platform. Organoids were extensively washed in PBST and then incubated in secondary antibodies at 4°C overnight. Finally, organoids were washed with PBST, PBS and then serially dehydrated to 100% methanol. Organoids were then optically cleared with Murray’s Clear (2:1 benzyl benzoate: benzyl alcohol, Sigma) for at least 15 minutes prior to imaging.
Brightfield and GFP fluorescence images of live tissue samples were captured using either a Leica DMC5400 or DFC310 FX camera attached to a stereomicroscope. Whole mount and all immunofluorescent images were captured using a Nikon Ti-E inverted A1 confocal microscope. Images were processed, including any composite stitching and maxIP merging, and quantified using Nikon NIS Elements, Bitplane Imaris, Adobe Illustrator, and Microsoft PowerPoint software.
RNA isolation and qRT-PCR
Spheroids and organoids were harvested in RA1 Lysis Buffer and β-mercapethanol and stored at −80°C until total RNA was isolated using NucleoSpin RNA Isolation Kit (Macherey-Nagel) according to manufacturers’ instructions. Complementary DNA (cDNA) was reverse transcribed from 116 ng of RNA using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). qRT-PCR was performed using a QuantiTect SYBR Green PCR Kit (Qiagen) in MicroAmp EnduraPlate Optical 96-Well Fast Reaction Plates (Applied Biosystems) and run on a QuantStudio 6 Real-Time PCR Detection System (Applied Biosystems). Primer sequences are listed in the Key Resource Table. Analysis was performed using the ΔΔCt method by first normalizing all cycle threshold (Ct) values to a base housekeeping gene (GAPDH, PPIA, or FOXF1) and then to the control hAGO samples. Statistical analysis was performed using Student’s t-test
Quantification and Statistical Analysis
For analysis of organoid patterning, “n” represents the number of replicates performed in each experiment and each replicate is defined as 1 well of approx. 3–5 organoids in Matrigel culture. Any analysis presented from one individual experiment is representative of trends seen across at least two individually seeded experiments. All data are represented as mean ± s.d. Student’s t-tests with 2-tailed distribution and un-equal variance was completed using Microsoft Excel, where p ≤ 0.05 is symbolized by *, p ≤ 0.01 is symbolized by **, and p ≤ 0.001 is symbolized by ***. The determined significance cutoff was p ≤ 0.05. No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. No randomization was made.
Supplementary Material
Movie S1. 3-dimensional video image of wholemount immunofluorescence of 10 week in vivo 3 germ layer hAGO +SM +GFP ENCC (neurons-green, αSMA-red, ECAD-white, DAPI-blue, 10x magnification). Video corresponds to Figure 2A, bottom right panel.
Movie S2. 3-dimensional video image of wholemount immunofluorescence of 4 week in vitro hAGO +GFP ENCC demonstrates network morphology of TUJ1+ neurons (neurons-green, TUJ1-red, ECAD-white, DAPI-blue, 10x magnification). Video corresponds to Figure S4F, relating to Figure 5.
Movie S3. 3-dimensional video image of wholemount immunofluorescence of 8 week in vivo hAGO graft +GFP ENCC demonstrates network morphology of TUJ1+ neurons (neurons-green, TUJ1-red, ECAD-white, DAPI-blue, 10x magnification). Video corresponds to Figure S6D, relating to Figure 6.
Movie S4. Time-lapse video of live-imaged in vivo thAGO graft +ENCC where ENCC contain a GCaMP6f reporter. GFP fluorescent firing demonstrates Ca2+ flux of multiple individual neurons. hAGOs were generated with H1 cells, which do not have a Ca2+ indicator. Images were collected every 4 seconds for 3 minutes using a 20x objective. Video corresponds to Figure S6F, relating to Figure 6.
Highlights.
Human stem cell-derived lineages combine to generate multi-layer gastric organoids.
3 germ layer gastric organoids form glands and functionally innervated smooth muscle.
Neural crest cells promote growth, patterning, and morphogenesis of gastric tissues.
3 germ layer tissue engineering also generates complex fundic and esophageal organoids.
ACKNOWLEDGEMENTS
We would like to thank all the members of the Wells, Zorn, and Helmrath laboratories for reagents and feedback. We specifically thank Heather McCauley and Jacob Enriquez from the Wells lab for technical assistance with ectopic transplantation surgeries and harvests. We also thank Chris Mayhew and Amy Pitstick from the Pluripotent Stem Cell Facility as well as Matt Kofron and Evan Meyer from the Confocal Imaging Core at Cincinnati Children’s Hospital for constant support and guidance. This research was supported by the grants from the NIH, U18 EB021780 (JMW, MAH), U19 AI116491 (JMW), P01 HD093363 (JMW), UG3 DK119982 (JMW), U01 DK103117 (MAH), 1F31DK118823–01 (AKE), NIEHS 5T32-ES007250–29 (DOK), the Shipley Foundation (JMW), and the Allen Foundation (JMW). We also received support from the Digestive Disease Research Center (P30 DK078392).
Footnotes
DECLARATION OF INTERESTS
No competing interests declared related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T and Wysocka J (2010) ‘CHD7 cooperates with PBAF to control multipotent neural crest formation’, Nature, 463(7283), pp. 958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbinot C, Vanier M, Armant O, Nair A, Penichon J, Soret C, Martin E, Saandi T, Reimund JM, Deschamps J, Beck F, Domon-Dell C, Gross I, Duluc I and Freund JN (2017) ‘Fine-tuning and autoregulation of the intestinal determinant and tumor suppressor homeobox gene CDX2 by alternative splicing’, Cell Death Differ, 24(12), pp. 2173–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber K, Studer L and Fattahi F (2019) ‘Derivation of enteric neuron lineages from human pluripotent stem cells’, Nat Protoc, 14(4), pp. 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett EA, Sanders KM and Ward SM (2017) ‘Inhibitory responses mediated by vagal nerve stimulation are diminished in stomachs of mice with reduced intramuscular interstitial cells of Cajal’, Sci Rep, 7, pp. 44759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohorquez DV, Shahid RA, Erdmann A, Kreger AM, Wang Y, Calakos N, Wang F and Liddle RA (2015) ‘Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells’, J Clin Invest, 125(2), pp. 782–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breit S, Kupferberg A, Rogler G and Hasler G (2018) ‘Vagus Nerve as Modulator of the Brain-Gut Axis in Psychiatric and Inflammatory Disorders’, Front Psychiatry, 9, pp. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes SJ, Spencer NJ, Costa M and Zagorodnyuk VP (2013) ‘Extrinsic primary afferent signalling in the gut’, Nat Rev Gastroenterol Hepatol, 10(5), pp. 286–96. [DOI] [PubMed] [Google Scholar]
- Brosens E, Burns AJ, Brooks AS, Matera I, Borrego S, Ceccherini I, Tam PK, García-Barceló MM, Thapar N, Benninga MA, Hofstra RM and Alves MM (2016) ‘Genetics of enteric neuropathies’, Dev Biol, 417(2), pp. 198–208. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K and Kim DS (2013) ‘Ultrasensitive fluorescent proteins for imaging neuronal activity’, Nature, 499(7458), pp. 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Roland JT, Barlow BJ, O’Neal R, Rich AE, Nam KT, Shi C and Goldenring JR (2014) ‘Cell lineage distribution atlas of the human stomach reveals heterogeneous gland populations in the gastric antrum’, Gut, 63(11), pp. 1711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport C, Diekmann U, Budde I, Detering N and Naujok O (2016) ‘Anterior-Posterior Patterning of Definitive Endoderm Generated from Human Embryonic Stem Cells Depends on the Differential Signaling of Retinoic Acid, Wnt-, and BMP-Signaling’, Stem Cells, 34(11), pp. 2635–2647. [DOI] [PubMed] [Google Scholar]
- de Santa Barbara P, van den Brink GR and Roberts DJ (2002) ‘Molecular etiology of gut malformations and diseases’, Am J Med Genet, 115(4), pp. 221–30. [DOI] [PubMed] [Google Scholar]
- De Santa Barbara P, Williams J, Goldstein AM, Doyle AM, Nielsen C, Winfield S, Faure S and Roberts DJ (2005) ‘Bone morphogenetic protein signaling pathway plays multiple roles during gastrointestinal tract development’, Dev Dyn, 234(2), pp. 312–22. [DOI] [PubMed] [Google Scholar]
- Faure S, de Santa Barbara P, Roberts DJ and Whitman M (2002) ‘Endogenous patterns of BMP signaling during early chick development’, Dev Biol, 244(1), pp. 44–65. [DOI] [PubMed] [Google Scholar]
- Faure S, Georges M, McKey J, Sagnol S and de Santa Barbara P (2013) ‘Expression pattern of the homeotic gene Bapx1 during early chick gastrointestinal tract development’, Gene Expr Patterns, 13(8), pp. 287–92. [DOI] [PubMed] [Google Scholar]
- Faure S, McKey J, Sagnol S and de Santa Barbara P (2015) ‘Enteric neural crest cells regulate vertebrate stomach patterning and differentiation’, Development, 142(2), pp. 331–42. [DOI] [PubMed] [Google Scholar]
- Freddo AM, Shoffner SK, Shao Y, Taniguchi K, Grosse AS, Guysinger MN, Wang S, Rudraraju S, Margolis B, Garikipati K, Schnell S and Gumucio DL (2016) ‘Coordination of signaling and tissue mechanics during morphogenesis of murine intestinal villi: a role for mitotic cell rounding’, Integr Biol (Camb), 8(9), pp. 918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB, Di Natale M, Hunne B, Oparija-Rogenmozere L, Ward SM, Sasse KC, Powley TL, Stebbing MJ, Jaffey D and Fothergill LJ (2020) ‘The identification of neuronal control pathways supplying effector tissues in the stomach’, Cell Tissue Res, 382(3), pp. 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MA, Schultz-Rogers L, Rajagopalan R, Grochowski CM, Wilkins BJ, Biswas S, Conlin LK, Fiorino KN, Dhamija R, Pack MA, Klee EW, Piccoli DA and Spinner NB (2020) ‘Protein-elongating mutations in MYH11 are implicated in a dominantly inherited smooth muscle dysmotility syndrome with severe esophageal, gastric, and intestinal disease’, Hum Mutat, 41(5), pp. 973–982. [DOI] [PubMed] [Google Scholar]
- Goodwin K, Mao S, Guyomar T, Miller E, Radisky DC, Košmrlj A and Nelson CM (2019) ‘Smooth muscle differentiation shapes domain branches during mouse lung development’, Development, 146(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Chaturvedi P, Kishimoto K, Koike H, Nasr T, Iwasawa K, Giesbrecht K, Witcher PC, Eicher A, Haines L, Lee Y, Shannon JM, Morimoto M, Wells JM, Takebe T and Zorn AM (2020) ‘Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis’, Nature Communications, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst JJ, Andersen DB and Grunddal KV (2021) ‘Actions of GLP-1 receptor ligands in the gut’, Br J Pharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke TR, Miller BM, Gill HK, Nerurkar NL, Sprinzak D, Mahadevan L and Tabin CJ (2019) ‘Genetic and Mechanical Regulation of Intestinal Smooth Muscle Development’, Cell, 179(1), pp. 90–105.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelberer MM, Buchanan KL, Klein ME, Barth BB, Montoya MM, Shen X and Bohorquez DV (2018) ‘A gut-brain neural circuit for nutrient sensory transduction’, Science, 361(6408). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT and Shivdasani RA (2005) ‘The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling’, Dev Cell, 8(4), pp. 611–22. [DOI] [PubMed] [Google Scholar]
- Kim TH and Shivdasani RA (2016) ‘Stomach development, stem cells and disease’, Development, 143(4), pp. 554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS and Hoffman MP (2010) ‘Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis’, Science, 329(5999), pp. 1645–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasrado R, Boesmans W, Kleinjung J, Pin C, Bell D, Bhaw L, McCallum S, Zong H, Luo L, Clevers H, Vanden Berghe P and Pachnis V (2017) ‘Lineage-dependent spatial and functional organization of the mammalian enteric nervous system’, Science, 356(6339), pp. 722–726. [DOI] [PubMed] [Google Scholar]
- Le Guen L, Marchal S, Faure S and de Santa Barbara P (2015) ‘Mesenchymal-epithelial interactions during digestive tract development and epithelial stem cell regeneration’, Cell Mol Life Sci, 72(20), pp. 3883–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chalazonitis A, Huang YY, Mann JJ, Margolis KG, Yang QM, Kim DO, Côté F, Mallet J and Gershon MD (2011) ‘Essential roles of enteric neuronal serotonin in gastrointestinal motility and the development/survival of enteric dopaminergic neurons’, J Neurosci, 31(24), pp. 8998–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Chen A, Koh PW, Deng TZ, Sinha R, Tsai JM, Barkal AA, Shen KY, Jain R, Morganti RM, Shyh-Chang N, Fernhoff NB, George BM, Wernig G, Salomon REA, Chen Z, Vogel H, Epstein JA, Kundaje A, Talbot WS, Beachy PA, Ang LT and Weissman IL (2016) ‘Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types’, Cell, 166(2), pp. 451–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddah M, Heidmann JD, Mandegar MA, Walker CD, Bolouki S, Conklin BR and Loewke KE (2015) ‘A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing’, Stem Cell Reports, 4(4), pp. 621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley HA, Matthis AL, Enriquez JR, Nichol JT, Sanchez JG, Stone WJ, Sundaram N, Helmrath MA, Montrose MH, Aihara E and Wells JM (2020) ‘Enteroendocrine cells couple nutrient sensing to nutrient absorption by regulating ion transport’, Nat Commun, 11(1), pp. 4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Aihara E, Martin B, Crawford CM, Broda T, Treguier J, Zhang X, Shannon JM, Montrose MH and Wells JM (2017) ‘Wnt/beta-catenin promotes gastric fundus specification in mice and humans’, Nature, 541(7636), pp. 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y and Wells JM (2014) ‘Modelling human development and disease in pluripotent stem-cell-derived gastric organoids’, Nature, 516(7531), pp. 400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniot B, Biau S, Faure S, Nielsen CM, Berta P, Roberts DJ and de Santa Barbara P (2004) ‘SOX9 specifies the pyloric sphincter epithelium through mesenchymal-epithelial signals’, Development, 131(15), pp. 3795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BA, Kim D and Vanner S (2000) ‘Neural pathways regulating Brunner’s gland secretion in guinea pig duodenum in vitro’, Am J Physiol Gastrointest Liver Physiol, 279(5), pp. G910–7. [DOI] [PubMed] [Google Scholar]
- Moore BA, Morris GP and Vanner S (2000) ‘A novel in vitro model of Brunner’s gland secretion in the guinea pig duodenum’, Am J Physiol Gastrointest Liver Physiol, 278(3), pp. G477–85. [DOI] [PubMed] [Google Scholar]
- Munera JO and Wells JM (2017) ‘Generation of Gastrointestinal Organoids from Human Pluripotent Stem Cells’, Methods Mol Biol, 1597, pp. 167–177. [DOI] [PubMed] [Google Scholar]
- Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, Hudson JR, Howell JC, Chatuvedi P, Spence JR, Shannon JM, Zorn AM, Helmrath MA and Wells JM (2017) ‘Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling’, Cell Stem Cell, 21(1), pp. 51–64.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy N, Barad C, Graham HK, Hotta R, Cheng LS, Fejszak N and Goldstein AM (2016) ‘Sonic hedgehog controls enteric nervous system development by patterning the extracellular matrix’, Development, 143(2), pp. 264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy N and Goldstein AM (2017) ‘Enteric nervous system development: A crest cell’s journey from neural tube to colon’, Semin Cell Dev Biol, 66, pp. 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedvetsky PI, Emmerson E, Finley JK, Ettinger A, Cruz-Pacheco N, Prochazka J, Haddox CL, Northrup E, Hodges C, Mostov KE, Hoffman MP and Knox SM (2014) ‘Parasympathetic innervation regulates tubulogenesis in the developing salivary gland’, Dev Cell, 30(4), pp. 449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norlen P, Ericsson P, Kitano M, Ekelund M and Hakanson R (2005) ‘The vagus regulates histamine mobilization from rat stomach ECL cells by controlling their sensitivity to gastrin’, J Physiol, 564(Pt 3), pp. 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin S, McCann CJ, Borrelli O, De Coppi P and Thapar N (2017) ‘Update on Foregut Molecular Embryology and Role of Regenerative Medicine Therapies’, Front Pediatr, 5, pp. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, Borrelli O, Saliakellis E and Thapar N (2018) ‘Gastrointestinal Neuropathies: New Insights and Emerging Therapies’, Gastroenterol Clin North Am, 47(4), pp. 877–894. [DOI] [PubMed] [Google Scholar]
- Poling HM, Wu D, Brown N, Baker M, Hausfeld TA, Huynh N, Chaffron S, Dunn JCY, Hogan SP, Wells JM, Helmrath MA and Mahe MM (2018) ‘Mechanically induced development and maturation of human intestinal organoids in vivo’, Nat Biomed Eng, 2(6), pp. 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhilin N, Barth B, Choi J, Munoz NL, Kulkarni S, Jones JS, Small DM, Cheng YT, Cao Y, LaVinka C, Kan E, Dong X, Spencer M, Pasricha P, Nishimura N and Shen X (2016) ‘Simultaneous optical and electrical in vivo analysis of the enteric nervous system’, Nat Commun, 7, pp. 11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DJ, Johnson RL, Burke AC, Nelson CE, Morgan BA and Tabin C (1995) ‘Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut’, Development, 121(10), pp. 3163–74. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Smith DM, Goff DJ and Tabin CJ (1998) ‘Epithelial-mesenchymal signaling during the regionalization of the chick gut’, Development, 125(15), pp. 2791–801. [DOI] [PubMed] [Google Scholar]
- Rydning A, Lyng O, Falkmer S and Grønbech JE (2002) ‘Histamine is involved in gastric vasodilation during acid back diffusion via activation of sensory neurons’, Am J Physiol Gastrointest Liver Physiol, 283(3), pp. G603–11. [DOI] [PubMed] [Google Scholar]
- Shaylor LA, Hwang SJ, Sanders KM and Ward SM (2016) ‘Convergence of inhibitory neural inputs regulate motor activity in the murine and monkey stomach’, Am J Physiol Gastrointest Liver Physiol, 311(5), pp. G838–g851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ and Mahadevan L (2013) ‘Villification: how the gut gets its villi’, Science, 342(6155), pp. 212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M and Bronner ME (2015) ‘Establishing neural crest identity: a gene regulatory recipe’, Development, 142(2), pp. 242–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Nielsen C, Tabin CJ and Roberts DJ (2000) ‘Roles of BMP signaling and Nkx2.5 in patterning at the chick midgut-foregut boundary’, Development, 127(17), pp. 3671–81. [DOI] [PubMed] [Google Scholar]
- Smith DM and Tabin CJ (1999) ‘BMP signalling specifies the pyloric sphincter’, Nature, 402(6763), pp. 748–9. [DOI] [PubMed] [Google Scholar]
- Stevens ML, Chaturvedi P, Rankin SA, Macdonald M, Jagannathan S, Yukawa M, Barski A and Zorn AM (2017) ‘Genomic integration of Wnt/beta-catenin and BMP/Smad1 signaling coordinates foregut and hindgut transcriptional programs’, Development, 144(7), pp. 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung TS, Hwang SJ, Koh SD, Bayguinov Y, Peri LE, Blair PJ, Webb TI, Pardo DM, Rock JR, Sanders KM and Ward SM (2018) ‘The cells and conductance mediating cholinergic neurotransmission in the murine proximal stomach’, J Physiol, 596(9), pp. 1549–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SH, Swathi Y, Tan S, Goh J, Seishima R, Murakami K, Oshima M, Tsuji T, Phuah P, Tan LT, Wong E, Fatehullah A, Sheng T, Ho SWT, Grabsch HI, Srivastava S, Teh M, Denil S, Mustafah S, Tan P, Shabbir A, So J, Yeoh KG and Barker N (2020) ‘AQP5 enriches for stem cells and cancer origins in the distal stomach’, Nature, 578(7795), pp. 437–443. [DOI] [PubMed] [Google Scholar]
- Theodosiou NA and Tabin CJ (2005) ‘Sox9 and Nkx2.5 determine the pyloric sphincter epithelium under the control of BMP signaling’, Dev Biol, 279(2), pp. 481–90. [DOI] [PubMed] [Google Scholar]
- Tiso N, Filippi A, Pauls S, Bortolussi M and Argenton F (2002) ‘BMP signalling regulates anteroposterior endoderm patterning in zebrafish’, Mech Dev, 118(1–2), pp. 29–37. [DOI] [PubMed] [Google Scholar]
- Trisno SL, Philo KED, McCracken KW, Cata EM, Ruiz-Torres S, Rankin SA, Han L, Nasr T, Chaturvedi P, Rothenberg ME, Mandegar MA, Wells SI, Zorn AM and Wells JM (2018) ‘Esophageal Organoids from Human Pluripotent Stem Cells Delineate Sox2 Functions during Esophageal Specification’, Cell Stem Cell, 23(4), pp. 501–515.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KT and Zemper AE (2019) ‘The Enteric Nervous System for Epithelial Researchers: Basic Anatomy, Techniques, and Interactions With the Epithelium’, Cell Mol Gastroenterol Hepatol, 8(3), pp. 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S and Gumucio DL (2012) ‘Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi’, Proc Natl Acad Sci U S A, 109(39), pp. 15817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi C, Lu Y, Poulin EJ, Franklin JL and Coffey RJ (2015) ‘Loss of Lrig1 leads to expansion of Brunner glands followed by duodenal adenomas with gastric metaplasia’, Am J Pathol, 185(4), pp. 1123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CL, Mahe MM, Munera J, Howell JC, Sundaram N, Poling HM, Schweitzer JI, Vallance JE, Mayhew CN, Sun Y, Grabowski G, Finkbeiner SR, Spence JR, Shroyer NF, Wells JM and Helmrath MA (2014) ‘An in vivo model of human small intestine using pluripotent stem cells’, Nat Med, 20(11), pp. 1310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfal ML and Goldstein AM (2017) ‘Pediatric enteric neuropathies: diagnosis and current management’, Curr Opin Pediatr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, Chang CF, Schiesser J, Aubert P, Stanley EG, Elefanty AG, Miyaoka Y, Mandegar MA, Conklin BR, Neunlist M, Brugmann SA, Helmrath MA and Wells JM (2017) ‘Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system’, Nat Med, 23(1), pp. 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, McGrath PS, Salomone J, Rahal M, McCauley HA, Schweitzer J, Kovall R, Gebelein B and Wells JM (2019) ‘A Comprehensive Structure-Function Study of Neurogenin3 Disease-Causing Alleles during Human Pancreas and Intestinal Organoid Development’, Dev Cell, 50(3), pp. 367–380.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CM, Martinez V, Piqueras L, Wang L, Taché Y and Chen D (2008) ‘Control of gastric acid secretion in somatostatin receptor 2 deficient mice: shift from endocrine/paracrine to neurocrine pathways’, Endocrinology, 149(2), pp. 498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. 3-dimensional video image of wholemount immunofluorescence of 10 week in vivo 3 germ layer hAGO +SM +GFP ENCC (neurons-green, αSMA-red, ECAD-white, DAPI-blue, 10x magnification). Video corresponds to Figure 2A, bottom right panel.
Movie S2. 3-dimensional video image of wholemount immunofluorescence of 4 week in vitro hAGO +GFP ENCC demonstrates network morphology of TUJ1+ neurons (neurons-green, TUJ1-red, ECAD-white, DAPI-blue, 10x magnification). Video corresponds to Figure S4F, relating to Figure 5.
Movie S3. 3-dimensional video image of wholemount immunofluorescence of 8 week in vivo hAGO graft +GFP ENCC demonstrates network morphology of TUJ1+ neurons (neurons-green, TUJ1-red, ECAD-white, DAPI-blue, 10x magnification). Video corresponds to Figure S6D, relating to Figure 6.
Movie S4. Time-lapse video of live-imaged in vivo thAGO graft +ENCC where ENCC contain a GCaMP6f reporter. GFP fluorescent firing demonstrates Ca2+ flux of multiple individual neurons. hAGOs were generated with H1 cells, which do not have a Ca2+ indicator. Images were collected every 4 seconds for 3 minutes using a 20x objective. Video corresponds to Figure S6F, relating to Figure 6.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request


