Abstract
To determine the phylogenetic position of two new rickettsial strains isolated from ticks in China, 16S ribosomal DNA, gltA, and ompA (apart from the tandem repeat units) genes were amplified by PCR and sequenced. The phylogenetic relationships between these strains and other rickettsiae were inferred from the comparison of sequences of the three genes by the parsimony, neighbor-joining, and maximum-likelihood methods. The results demonstrated that the 054 strain, a rickettsia pathogenic in humans, and the HL-93 strain were related and clustered together with Rickettsia japonica. Significant statistical bootstrap values (100 and 92%) supported the nodes in this cluster. Based on previous genotypic and antigenic data and the phylogenetic analysis presented here, the 054 and HL-93 strains should be considered as new species, and we formally propose that they be named “Rickettsia heilongjiangii” and “Rickettsia hulinii,” respectively.
The advent of new culture techniques, such as the shell vial-centrifugation technique (16), and the detection of rickettsial DNA have dramatically increased the number of rickettsial species detected (20). Before 1984, only six spotted fever group (SFG) rickettsioses were recognized in the world (20): Rocky Mountain spotted fever caused by Rickettsia rickettsii, Mediterranean spotted fever caused by Rickettsia conorii, Siberian tick typhus caused by Rickettsia sibirica, Queensland tick typhus caused by Rickettsia australis, rickettsialpox caused by Rickettsia akari, and Israeli spotted fever caused by Israeli tick typhus rickettsia. Over the last 12 years, eight new rickettsioses have been reported, including Japanese spotted fever caused by Rickettsia japonica, Flinders Island spotted fever caused by Rickettsia honei, Astrakhan fever caused by Astrakhan fever rickettsia, African tick bite fever caused by Rickettsia africae, California flea typhus caused by the ELB agent, and three unnamed spotted fevers caused by “Rickettsia mongolotimonae,” Rickettsia slovaca, and Rickettsia helvetica. In China, 20 strains of tick-associated rickettsiae, belonging to at least five serotypes, have been isolated over the last few years, including R. sibirica; the BJ-90 isolate (42); the Heilongjiang isolate (strain 054, also called “Rickettsia heilongjiangii”); the Inner Mongolian isolate HA-91, or “R. mongolotimonae,” also found in France; and the Hulin isolate (strain HL-93, also called “Rickettsia hulinii”) (9, 19, 33). Three of these rickettsiae, R. sibirica, “R. mongolotimonae,” and “R. heilongjiangii,” are human pathogens. “R. heilongjiangii” was first isolated in 1982 from Dermacentor silvarum ticks collected in Suifenhe, a city of Heilongjiang Province in China (Fig. 1). Between May and July 1988, 12 patients in Chunhua, Huichun County, Jilin Province, were reported to be infected by this strain on the basis of serological results. All patients had a history of fever, headache, rash, eschar, regional lymphadenopathy, and conjunctivitis following a tick bite, with fourfold rises in complement fixation antibodies in convalescent-phase sera (14). Between May and June 1996, seven patients with clinical manifestations of SFG rickettsial infections were found in Suifenhe and Dongning, in Heilongjiang Province. Isolates from these patients were found to be identical to the 054 strain by microimmunofluorescence and PCR plus restriction fragment length polymorphism (30). The “R. hulinii” strain was first isolated in 1993 from Haemaphysalis concinna ticks collected in Hulin County, Heilongjiang Province (Fig. 1), and was considered to be a unique SFG rickettsia (38). To date, the pathogenicity of this strain has been experimentally demonstrated in animals (38), but not in humans. In order to estimate the phylogenetic classification of strains 054 and HL-93, we sequenced their 16S rRNA (16S ribosomal DNA [rDNA])-, citrate synthase (gltA)-, and rOmpA (ompA)-encoding genes and compared these sequences with those of other SFG rickettsiae.
FIG. 1.
Map of China showing the areas of collection of ticks from which strains 054 and HL-93 were isolated.
Strains 054 and HL-93 were cultivated and purified as previously described (12), and their DNA was extracted from purified organisms with the QIAmp tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. PCR amplification and sequencing reactions were performed with the oligonucleotide primers mentioned below. All primers were purchased from GIBCO BRL (Custom Primers, Life Technologies Sarl. B.P., Cergy Pontoise Cedex, France), and amplifications were carried out in a Peltier model PTC-200 DNA thermal cycler (MJ Research, Inc., San Francisco, Calif.) under conditions previously described (11, 22–24). R. japonica and distilled water were used as a positive and negative control, respectively. Each amplicon obtained was purified for sequencing with a QIAquick Spin PCR Purification kit (Qiagen, Courtaboeuf, France) according to the manufacturer's protocol. Sequencing reactions were carried out with the Dye Terminator kit (d-rhodamine terminator cycle DNA sequencing kit; Perkin-Elmer) as described by the manufacturer. Sequencing reaction products were resolved by electrophoresis with an ABI Prism 377 Sequencer (Perkin-Elmer). The results obtained were processed into sequence data by using the Sequence Navigator and AutoAssembler software. Each base position was established at least three times in both the forward and reverse directions. Sequences of the 16S rDNA, gltA, and ompA genes were aligned by using the multisequence alignment program CLUSTAL, within the BISANCE environment (7). Phylogenetic relationships between strains 054 and HL-93 and other SFG rickettsiae were inferred by using version 3.4 of the PHYLIP software package (10). The distance matrices generated by DNADIST were determined under the assumptions of Kimura (13) and were used to infer dendrograms by the neighbor-joining method (25). Two other dendrograms were constructed by data processing with the maximum-likelihood and parsimony programs in PHYLIP. A bootstrap analysis based on 100 randomly generated trees by using SEQBOOT and CONSENSE in PHYLIP was performed to estimate the node reliability of the trees obtained by the three phylogenetic methods (4).
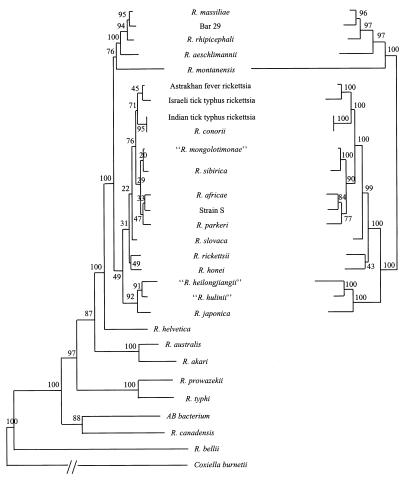
The primers fD1 and rP2 amplified 1,463- and 1,461-bp fragments of the 16S rDNA gene from strains 054 and HL-93, respectively. The gltA gene was amplified in two fragments. Overall, sequences of 1,238 and 1,237 bp were obtained from the O54 and HL-93 strains, respectively. PCR amplification and sequencing of the ompA gene excluded the central tandemly repeat region. The 5′-end fragment of the gene was 611 bp for both the 054 and HL-93 strains. The 3′ part of the gene was amplified in four fragments. Overall, sequences of 3,187 and 3,181 bp were obtained from strains 054 and HL-93, respectively. The sequenced fragments of the rOmpA gene were analyzed from base positions 91 to 680 and 3608 to 6789 with respect to the sequence published for R. rickettsii (2) and were therefore submitted to GenBank as two separate fragments. Strains 054 and HL-93 clustered together with R. japonica into a well-defined, strongly supported monophyletic group within the SFG rickettsiae, with bootstrap values of 91 and 100% for the node where strains and HL-93 branched together in the gltA and ompA trees, respectively, and values of 92 and 100% for the node where both strains clustered with R. japonica. Similar phylogenetic organizations were inferred from the analysis of all three genes by using the three phylogenetic analysis methods (Fig. 2).
FIG. 2.
Dendrogram representing phylogenetic relationships between Rickettsia species inferred from the comparison of gltA (left) and ompA (right) sequences by the neighbor-joining method. The bootstrap values are indicated at the nodes.
Historically, the classification and identification of rickettsiae have been based on differences in their epidemiology, serology, and intracellular growth characteristics. Because different species may have common ecological features (e.g., geographic distribution, arthropod vectors, etc.) and due to serological cross-reactivity, it is difficult to distinguish between rickettsiae. Moreover, DNA-DNA hybridization, a criterion used for the definition of species in the family Enterobacteriaceae, is not applicable to SFG rickettsiae (8). Other recognized tools for the taxonomic classification of bacteria, such as the study of protein profiles by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, monoclonal antibodies, Western blotting, and hybridization of labeled cloned DNA probes (1, 3, 17, 21, 31, 32, 34), are not widely applicable and lack interlaboratory reproducibility. Therefore, for over 10 years, the reference method for the identification of members of SFG rickettsiae has been the indirect microimmunofluorescence serologic typing test with mouse sera (18). Recently, phylogenetic analyses and the determination of taxonomic relationships based on comparisons of sequences of a combination of various genes have greatly facilitated the study of rickettsial taxonomy. The 16S rDNA (23, 26, 28), gltA (24), and ompA (11, 22) genes appear to be the most useful sequences. In our study, the 054 and HL-93 strains demonstrated ompA sequence homologies of 98.1% with each other and 96.6 and 95.6% with R. japonica, respectively, which are higher than those observed for strain HL-93 with R. rickettsii or R. conorii (94 and 90%, respectively). Although closely related, the data from our study show that strains 054 and HL-93 are distinct species (Fig. 2). Strains 054 and HL-93 and R. japonica were grouped into a well-supported cluster in the phylogenetic trees developed by the three analysis methods (Fig. 2). These results are the first showing isolates clustering with R. japonica and are in concordance with previous phenotypic and genotypic studies (5, 6, 15, 29, 30, 35–41; L. Chen, J. Z. Zhang, and D. Z. Bi, Program Abstr. EUWOG-ASR Joint Meeting, p. 12, 1999). Tick-associated rickettsiae are characterized by limited, specific geographical distribution areas (11). R. japonica was first isolated on Shikoku Island (27), the smallest of Japan's four main islands. The island is located between 132° to 135° East and 33° to 35° North. To the north is the Seto Inland Sea, while to the south is the Pacific Ocean. Heilongjiang Province of China, where the 054 and HL-93 strains were isolated, is located between about 130° to 140° East and 45° to 50° North. This suggests that an SFG rickettsia may have evolved separately in this area of the world and diverged more recently into Japan and in China. The current rickettsial taxonomy is based on serotyping (11), and, to date, there is no consensus on alternative genomic tools. However, based on data from previous genotypic and antigenic studies and on the phylogenetic analysis presented here, we believe that the 054 and HL-93 strains should be strongly considered as members of new species, and we formally propose that these be named “Rickettsia heilongjiangii” and “Rickettsia hulinii,” respectively.
Nucleotide sequence accession numbers.
The 16S rDNA GenBank accession numbers are AF172942 and AF178037 for 054 and HL-93, respectively. The gltA accession numbers are AF178034 and AF172943 for 054 and HL-93, respectively. The ompA accession numbers are AF179362 and AF179364 for the 5′ fragments of 054 and HL-93, respectively, and AF179363 and AF179366 for the 5′ fragments of 054 and HL-93, respectively).
REFERENCES
- 1.Anacker R L, Philip R N, Williams J C, List R H, Mann R E. Biochemical and immunochemical analysis of Rickettsia rickettsii strains of various degrees of virulence. Infect Immun. 1984;44:559–564. doi: 10.1128/iai.44.3.559-564.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson B E, MacDonald G A, Jones D C, Regnery R L. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect Immun. 1990;58:2760–2769. doi: 10.1128/iai.58.9.2760-2769.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson B E, Tzianabos T. Comparative sequence analysis of a genus-common rickettsial antigen gene. J Bacteriol. 1989;171:5199–5201. doi: 10.1128/jb.171.9.5199-5201.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J K M. Bootstrap hypothesis tests for evolutionary trees and other dendrograms. Proc Natl Acad Sci USA. 1994;91:12293–12297. doi: 10.1073/pnas.91.25.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M, Fan M Y, Bi D Z, Zhang J Z. Sequence analysis of a fragment of rOmpA gene of several isolates of spotted fever group rickettsiae from China. Acta Virol. 1998;42:91–93. [PubMed] [Google Scholar]
- 6.Chen X R, Xu Z P, Chen W R, Li S L, Zhang Y G. Studies on the features of the strains of spotted fever group rickettsiae. Microbiology. 1997;24:31–31. [Google Scholar]
- 7.Dessen P, Fondrat C, Valencien C, Munier G. BISANCE: a French service for access to biomolecular databases. CABIOS. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- 8.Drancourt M, Raoult D. Taxonomic position of the rickettsiae: current knowledge. FEMS Microbiol Rev. 1994;13:13–24. doi: 10.1111/j.1574-6976.1994.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 9.Fan M Y, Zhang J Z, Chen M, Yu X J. Spotted fever group rickettsioses in China. In: Raoult D, Brouqui P, editors. Rickettsiae and rickettsial diseases at the turn of the third millenium. Paris, France: Elsevier; 1999. pp. 247–257. [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Fournier P E, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 12.Hanson B A, Wisseman C L, Jr, Waddell A, Silverman D J. Some characteristics of heavy and light bands of Rickettsia prowazekii on Renografin gradients. Infect Immun. 1981;34:596–604. doi: 10.1128/iai.34.2.596-604.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 14.Lou D, Wu Y M, Wang B, Wang W, Zhang Z, Zhang X W. Confirmation of patients with tick-borne spotted fever caused by Rickettsia heilongjiangi. Chin J Epidemiol. 1989;10:128–132. [Google Scholar]
- 15.Lou D, Wu Y, Wang B, Lui G, Li J, Wang W, Han Y F. A new member of the spotted fever group of rickettsiae-rickettsia. Chin J Microbiol Immunol. 1985;5:250–253. [Google Scholar]
- 16.Marrero M, Raoult D. Centrifugation-shell vial technique for rapid detection of Mediterranean spotted fever rickettsia in blood culture. Am J Trop Med Hyg. 1989;40:197–199. doi: 10.4269/ajtmh.1989.40.197. [DOI] [PubMed] [Google Scholar]
- 17.McDade J E, Black C M, Roumillat L F, Redus M A, Spruill C L. Addition of monoclonal antibodies specific for Rickettsia akari to the rickettsial diagnostic panel. J Clin Microbiol. 1988;26:2221–2223. doi: 10.1128/jcm.26.10.2221-2223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philip R N, Casper E A, Burgdorfer W, Gerloff R K, Hugues L E, Bell E J. Serologic typing of rickettsiae of the spotted fever group by micro immunofluorescence. J Immunol. 1978;121:1961–1968. [PubMed] [Google Scholar]
- 19.Raoult D, Brouqui P, Roux V. A new spotted-fever-group rickettsiosis. Lancet. 1996;348:412. doi: 10.1016/s0140-6736(05)65037-4. [DOI] [PubMed] [Google Scholar]
- 20.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694–719. doi: 10.1128/cmr.10.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regnery R L, Tzianabos T, Esposito J J, McDade J E. Strain differentiation of epidemic typhus rickettsiae (Rickettsia prowazekii) by DNA restriction endonuclease analysis. Curr Microbiol. 1983;8:355–358. [Google Scholar]
- 22.Roux V, Fournier P-E, Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. 1996;34:2058–2065. doi: 10.1128/jcm.34.9.2058-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux V, Raoult D. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res Microbiol. 1995;146:385–396. doi: 10.1016/0923-2508(96)80284-1. [DOI] [PubMed] [Google Scholar]
- 24.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for sequences. J Mol Biol. 1987;16:111–120. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Stothard D R, Fuerst P A. Evolutionary analysis of the spotted fever and typhus groups of Rickettsia using 16S rRNA gene sequences. Syst Appl Microbiol. 1995;18:52–61. [Google Scholar]
- 27.Uchida T, Tashiro F, Funato T, Kitamura Y. Isolation of a spotted fever group Rickettsia from a patient with febrile exanthematous illness in Shikoku, Japan. Microbiol Immunol. 1986;30:1323–1326. doi: 10.1111/j.1348-0421.1986.tb03053.x. [DOI] [PubMed] [Google Scholar]
- 28.Weisburg W G, Dobson M E, Samuel J E, Dasch G A, Mallavia L P, Baca O, Mandelco L, Sechrest J E, Weiss E, Woese C R. Phylogenetic diversity of the rickettsiae. J Bacteriol. 1989;171:4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y M, Liu X X, Wei A M, Hu L M, Yang Q. Genotypic analysis of spotted fever group rickettsia isolated in North part of China by PCR/RFLP. Chin Acta Public Health. 1998;17:71–72. [Google Scholar]
- 30.Wu Y M, Yu S R, Lou D. Western-blot analysis of Rickettsia heilongjiangi. J Prev Med PLA. 1994;12:28–30. [Google Scholar]
- 31.Xu W, Raoult D. Distribution of immunogenic epitopes on the two major immunodominant proteins (rOmpA and rOmpB) of Rickettsia conorii among the other rickettsiae of the spotted fever group. Clin Diagn Lab Immunol. 1997;4:753–763. doi: 10.1128/cdli.4.6.753-763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W, Raoult D. Production of monoclonal antibodies against Rickettsia massiliae and their use in antigenic and epidemiological studies. J Clin Microbiol. 1997;35:1715–1721. doi: 10.1128/jcm.35.7.1715-1721.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Jin Y, Fan M, Xu G, Liu Q, Raoult D. Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from China. J Clin Microbiol. 1993;31:83–88. doi: 10.1128/jcm.31.1.83-88.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X J, Fan M Y, Bi D Z, Xu G M. Antigenic analysis of Chinese strains of spotted fever group rickettsiae with monoclonal antibodies by immunoblotting. Chin J Microbiol Immunol. 1990;11:67–73. [Google Scholar]
- 35.Zhang J Z, Chen X R. Properties of monoclonal antibodies to Rickettsia heilongjiang (HLJ-054) strain and application it to identify SFGR Chinese isolates. Proceedings of 95 Shanghai International Epidemiological Association Congress (Regional); 1995. p. 64. [Google Scholar]
- 36.Zhang J Z, Chen X R, Chen W R, Xu Z P. Primary study on production and property of monoclonal antibody to Rickettsia heilongjiang (HLJ-054) strain. Chin J Zoonoses. 1994;10:119–122. [Google Scholar]
- 37.Zhang J Z, Fan M Y, Bi D Z. Cloning and sequence analysis of genomic fragment encoding 190KD protein antigen of HL-93 spotted fever group rickettsiae. Chin J Microbiol Immunol. 1996;3:227–230. [Google Scholar]
- 38.Zhang J Z, Fan M Y, Bi D Z. Isolation and identification of a new species of spotted fever group rickettsiae. Chin J Zoonoses. 1996;5:2–8. [Google Scholar]
- 39.Zhang J Z, Fan M Y, Bi D Z. Sequence analysis and comparison of 190 K of surface antigen gene of fragment of a new species of spotted fever group rickettsiae. Acta Virol. 1997;41:41–43. [PubMed] [Google Scholar]
- 40.Zhang J Z, Fan M Y, Bi D Z, Cui W F, Han Y F. Genotypic identification of three new strains of spotted fever group rickettsiae isolated in China. Acta Virol. 1996;40:215–219. [PubMed] [Google Scholar]
- 41.Zhang J Z, Fan M Y, Bi D Z, Song X P, Fang X. Analysis on structural polypeptide of Chinese new isolates of spotted fever group rickettsiae. J Shihezi Med College. 1996;2:71–74. [Google Scholar]
- 42.Zhang, J. Z., M. Y. Fan, X. J. Yu, and D. Raoult. Assessment of phylogenetic position of the Chinese isolate BJ-90. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]