Abstract
The gut is connected to the CNS by immunological mediators, lymphocytes, neurotransmitters, microbes and microbial metabolites. A mounting body of evidence indicates that the microbiome exerts significant effects on immune cells and CNS cells. These effects frequently result in the suppression or exacerbation of inflammatory responses, the latter of which can lead to severe tissue damage, altered synapse formation and disrupted maintenance of the CNS. Herein, we review recent progress in research on the microbial regulation of CNS diseases with a focus on major gut microbial metabolites, such as short-chain fatty acids, tryptophan metabolites, and secondary bile acids. Pathological changes in the CNS are associated with dysbiosis and altered levels of microbial metabolites, which can further exacerbate various neurological disorders. The cellular and molecular mechanisms by which these gut microbial metabolites regulate inflammatory diseases in the CNS are discussed. We highlight the similarities and differences in the impact on four major CNS diseases, i.e., multiple sclerosis, Parkinson’s disease, Alzheimer’s disease, and autism spectrum disorder, to identify common cellular and molecular networks governing the regulation of cellular constituents and pathogenesis in the CNS by microbial metabolites.
Subject terms: Autoimmune diseases, Chronic inflammation
Neurological disorders: bugs that benefit the brain
Disturbances of the microbial ecosystem in our gut create physiological disturbances that can accelerate the onset of neurological disease. The intestinal microbiome generates a diverse array of metabolites that can boost metabolic health, immune function, and other essential biological processes in the human host. Jeongho Park of Kangwon National University, Chuncheon, South Korea, and Chang Kim at the University of Michigan, Ann Arbor, USA, have reviewed research into how disruption of the healthy microbiome may contribute to Alzheimer’s, Parkinson’s, multiple sclerosis, and autism spectrum disorders. These studies indicate that such a “dysbiotic” state is generally associated with increased levels of inflammation. These disturbances are not sufficient to trigger neurological disorders on their own, but the resulting inflammatory state can act in concert with other biological risk factors to accelerate disease pathology.
Introduction
The central nervous system (CNS) is known as an immune-privileged tissue system but remains susceptible to inflammatory responses. While well protected by the blood–brain barrier (BBB), nevertheless the CNS hosts many types of inflammatory responses. A mounting body of evidence indicates the presence of close interactions between the neural system and gut microbes. These interactions involve endocrine, neurological, metabolic and immunological communication. The CNS and enteric nervous system (ENS) communicate via neurotransmitters, hormones, and metabolites1,2. Gut microbial metabolites are transported to the CNS, wherein they regulate CNS cells and immune cells. This review will examine our current understanding of how gut microbial metabolites impact four major CNS diseases and highlight the common disease-modifying functions of major microbial metabolites.
Multiple sclerosis (MS) is the most common form of inflammatory disease in the CNS and is associated with autoimmune-mediated demyelination3,4. Other less frequent inflammatory diseases include Rasmussen’s encephalitis5 and neuromyelitis optica (NMO)6. These diseases involve immune cell infiltrates in the brain and spinal cord, resulting in neuritis and myelitis7,8. Antigen-specific T and B lymphocytes are frequent immune cell infiltrates in CNS lesions. Inflammatory responses in the CNS are induced or suppressed by different types of T lymphocytes, such as Th1 and Th17 cells and Tregs9,10. Antibodies and B cells can augment or alter the pathogenesis of CNS inflammation11. Other cell types, such as microglia, macrophages and astrocytes, actively participate in immune cell regulation and neuroinflammation12. An increasing body of evidence indicates that other neurological diseases, such as Parkinson’s disease (PD), Alzheimer’s disease, and autism spectrum disorder (ASD), also involve significant inflammatory responses13–15. Therefore, we focus this review on these four CNS diseases, which have distinct tissue involvement, inflammatory responses and pathogenesis.
In recent years, we have witnessed a plethora of research on the functions of gut microbiota and their metabolites in regulating inflammatory responses in the CNS16–19. The intestine is rich in dietary materials and host secretions such as bile acids and mucins, which are subsequently metabolized by the cooperative activities of host enzymes and then by the gut microbiome. Various dietary fibers and complex carbohydrates can reach the terminal ileum and colon, which host the majority of the gut microbiome in the body. Microbial fermentation of sugars derived from carbohydrates in the colon produces short-chain fatty acids (SCFAs) such as acetate (C2), propionate (C3) and butyrate (C4)20. Another major group of metabolites includes tryptophan (Trp) metabolites. Dietary tryptophan is converted into indole derivatives, including indole-3-acetic acid (IAA), indole-3-propionic acid (IPA), indole-3-aldehyde (IAld), indole-3-acetaldehyde (IAAld), and indoleacrylic acid. Serotonin (5-HT), a neurotransmitter, is also produced in the gut lumen from Trp21–24. Primary bile acids are produced from cholesterol in the liver and are dehydroxylated by gut microbes to generate secondary bile acids25. These gut microbial metabolites function as agonists for various host receptors and intracellular molecules, such as G-protein-coupled receptors (GPCRs) and transcription factors, including GPR43, GPR41, TGR5, aryl hydrocarbon receptor (AhR), pregnane X receptor (PXR), farnesoid X receptor (FXR), vitamin D receptor (VDR), and others26. Moreover, many of these metabolites, particularly SCFAs, are metabolized in host cells to generate energy. Microbial metabolites regulate numerous host enzymes, such as histone deacetylases (HDACs). In general, microbial metabolites regulate host gene expression and metabolic activity in both immune cells and tissue cells. Importantly, these metabolites directly and indirectly affect the CNS to exert their regulatory functions19,27,28.
Basic functions of gut microbial metabolites
The most abundant metabolites in the colon are SCFAs, which have a combined luminal concentration of greater than 0.1 M in the human colon29. The three major SCFAs, C2, C3 and C4, function through four G-protein-coupled receptors (GPCRs), GPR41, GPR43, GPR109A and Olfr7830–32. In addition, SCFAs are natural HDAC inhibitors, and therefore, they can increase the acetylation and phosphorylation of host proteins such as histones, leading to increased gene expression and cell signaling33,34. SCFAs directly inhibit Type I and II HDACs and can also indirectly affect certain Type III HDACs, such as Sirtuin 135. Moreover, SCFAs are metabolized in host cells for energy production and thus have significant regulatory effects on host cell metabolism, leading to increased ATP levels, mTOR activity and fatty acid synthesis36. SCFAs are known to boost the differentiation of Th1 and Th17 cells and Tregs from naïve CD4 T cells, depending on the immunological milieu36. SCFAs increase IL-10 expression by lymphocytes (T and B cells) and macrophages37,38 in a manner largely mediated by their HDAC inhibitory activity. SCFAs regulate the activity of innate lymphocytes; they increase the number and activity of Group 3 innate lymphoid cells (ILC3s) but suppress Group 2 innate lymphoid cells (ILC2s) through GPCR activation and HDAC inhibition, respectively39–41. SCFAs also induce tolerogenic macrophages and dendritic cells by both GPCR activation and HDAC inhibition42.
Certain amino acid metabolites produced by microbes are also important regulators of host cells. Phenylalanine and tyrosine are precursors of dopamine, a key neurotransmitter, in the host, and some commensal bacteria produce amines related to these amino acids that activate host GPR5643. Trp is metabolized by both host cells and certain gut bacteria to serotonin (5-hydroxytryptamine), kynurenine (Kyn) and indole derivatives, which function as neurotransmitters and metabolic regulators24. Certain indole derivatives, such as IAA, IPA, and IAld, function as ligands for AhR, a member of the basic helix-loop-helix transcription factor family. These metabolites can also regulate host cells through EIF5a, IR76B, and PXR44. Certain microbial species, such as Peptostreptococcus russellii45 and Lactobacillus species46,47, can effectively produce indole derivatives45–47. Dietary indole derivatives, such as indole-3-carbinol (I3C) and diindolylmethane (DIM), can increase the activity of Tregs but suppress the generation of myelin oligodendrocyte glycoprotein (MOG)-specific Th17 cells48. Polyamines such as putrescine and spermidine are produced by intestinal bacteria and are present at 0.5 to 1 mM in the human colon49. Polyamines support the hypusine modification of eukaryotic translation initiation factor 5A (eIF5A) and promote polypeptide translation and cell proliferation50. Polyamines also have suppressive effects on Th17 cells51. Similarly, phytochemical metabolites, such as ginsenosides, resveratrol, and I3C, activate AhR to exert some of their regulatory functions52,53. Ginsenosides suppress Th1 and Th17 cells but increase Treg activity54. Resveratrol suppresses Th17 activity55.
Primary bile acid metabolites, such as cholic acid and chenodeoxycholic acid (CDCA), are made from cholesterol in the liver. Secondary bile acid metabolites, such as deoxycholate (DCA) and lithocholate (LCA), are formed by bacterial 7α-dehydroxylation of primary bile acids. Secondary bile acids activate multiple host receptors, such as PXR, VDR, LXR, FXR, TGR5, retinoid-related orphan receptor-γt (RORγt), and FoxP325,56. 3-OxoLCA inhibits Th17 polarization by binding to RORγt, whereas isoallo-LCA enhances Treg generation, in part by producing mitochondrial reactive oxygen species that induce FoxP3 expression57. Bile acid metabolites have both beneficial and pathogenic functions58–61. Primary and secondary bile acids activate the Nod-like receptor protein 3 (NLRP3) inflammasome and IL-1β production in macrophages61, which can increase innate immunity but cause chronic inflammation. However, activation of the bile acid receptors FXR and TGR5 is often associated with decreased inflammatory responses61,62.
Some microbial metabolites are pathogenic, causing inflammatory diseases and cancer. Pathogenic metabolites include trimethylamine N-oxide (TMAO)63, hydrogen sulfide64, phenol, p-cresol, N-nitrosamine, ammonia, 4-ethylphenylsulfate65, and uric acid66. These metabolites are risk factors for MS, AD, and other neurological disorders67–69. Certain environmental contaminants, such as the AhR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), are also pathogenic70.
Regulation of experimental neuroinflammation by microbial metabolites
The regulatory effects of microbiota on experimental neuroinflammation have been reported71–74. Rodent experimental autoimmune encephalitis (EAE) models are most frequently used75 because they largely mimic human MS. Antigen-induced EAE can be induced by immunization with several neuronal cell antigens, such as MOG, myelin basic protein (MBP), and proteolipid protein (PLP), along with potent adjuvants, such as complete Freund’s adjuvant and pertussis toxin75. Demyelination similar to that in MS is also induced by chemicals such as cuprizone (a copper-chelating agent) and lysolecithin76–78. Infection with Theiler’s murine encephalomyelitis virus induces demyelinating encephalitis79,80. Moreover, the engineered expression of T and B cell receptors specific for CNS antigens causes spontaneous EAE responses in mice81,82.
While dysbiosis exacerbates EAE responses83,84, studies with germ-free (GF) animals or broad-spectrum antibiotics revealed that the microbiota is required for optimal EAE pathogenesis85,86, perhaps because of the general adjuvant effect of the microbiota in inducing immune responses. When specific microbial species or their products are suppressed, EAE activity may either increase or decrease, depending on the function of the affected microbial species87,88. For example, segmented filamentous bacteria (SFB), S. aureus and P. heparinolytica promote the generation of Th17 cells and exacerbate EAE responses89–91. Elimination of these Th17-inducing microbial species ameliorated EAE inflammation85,86. In contrast, treatment with SCFA-producing bacteria such as Clostridium tyrobutyricum ameliorated EAE immune responses92. Thus, the impact of the microbiota on CNS inflammation depends on the function of the altered microbial species in the gut84.
It is thought that the microbiota affects neuroinflammation by producing immune regulatory products and metabolites. For example, polysaccharide A (PSA), a capsular polysaccharide produced by Bacteroidetes fragilis, has significant immune regulatory function93. Oral administration of PSA suppressed demyelination and delayed the onset of EAE activity94. In this previous study, PSA increased the number of CD103+ dendritic cells (DCs) in draining lymph nodes and induced IL-10-producing T cells. PSA promoted Toll-like receptor 2 (TLR2) activation, which induced CD39+ regulatory T cells, in part by modulating DCs. In addition, poly-γ-glutamic acid (γ-PGA) has suppressive effects on EAE activity95. This finding is in line with decreased Th17 cell but increased Treg activity in the CNS following the administration of γ-PGA, which activates TLR4-MyD88-dependent and TLR4-MyD88-independent pathways to regulate T cell activity95. In contrast, some bacterial products can exacerbate CNS inflammation. Lipopolysaccharide (LPS) treatment accelerated inflammation-mediated demyelination in rats96. It is well established that peptidoglycans, the major cell wall component of gram-positive bacteria, promote DC maturation through TLR2 activation, which increases the secretion of proinflammatory cytokines and promotes effector T cell generation during EAE development89. Thus, microbial TLR ligands can have both inflammatory and suppressive roles in neuroinflammation.
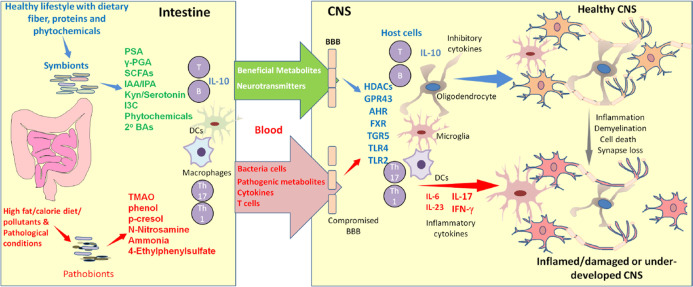
BBB disruption is observed in EAE and MS patients, and this can cause harmful exposure of CNS tissues to microbial products and microbial invasion following gut dysbiosis, leading to amplified inflammatory responses97,98. Under normal conditions, the microbiota and SCFAs support the integrity of the BBB99; it is important to protect the CNS from the harmful effects of inflammatory responses and toxicants. The BBB integrity is weakened in GF mice, and this defect is normalized by SCFA treatment in drinking water99 (Fig. 1). SCFAs, such as C2, C3, or C4, administered in drinking water ameliorated EAE activity. Interestingly, SCFAs appear to support CNS development beyond their immunoregulatory roles100,101. Microglial cells in GF mice had defective microglia with altered cell proportions and an immature phenotype, which were corrected by SCFA feeding in a GPR43-dependent manner100. In addition, C4 promoted oligodendrocyte maturation in the brain101. Thus, SCFAs support the functional maturation of CNS cells (Fig. 1). Increased levels of secondary bile acids, such as DCA, can weaken BBB function, perhaps because these compounds are amphiphilic steroids that loosen the cell membrane102,103. Thus, BBB function and associated CNS pathogenesis can be altered by changes in microbial metabolites.
Fig. 1. Microbial metabolites regulate CNS development, integrity, and inflammation.
Microbial metabolites positively and negatively influence CNS development and inflammatory responses. In the best case, beneficial metabolites are produced in symbiosis with a balanced population of diverse microbes in the gut. Together, these microbes produce myriad metabolites that are beneficial for the host. In dysbiosis, the production of harmful metabolites is increased while that of beneficial metabolites is decreased. In general, beneficial metabolites, such as SCFAs and Trp metabolites, reinforce the integrity of the gut barrier and BBB and support the functional maturation of CNS cells such as microglia, oligodendrocytes and astrocytes. Thus, these metabolites support CNS formation, neurological function, and the development of regulatory immune cells for immune tolerance. Moreover, these metabolites suppress harmful immune responses, such as the generation of pathogenic Th17 cells. These functions are mediated in part by various host receptors, such as GPCRs, transcription factors, nuclear ligand receptors (PXR and FXR), and TLRs. Conversely, harmful metabolites weaken the gut barrier and BBB and cause systemic inflammatory responses, neuronal cell death and tissue injury (e.g., demyelination), leading to inflammatory conditions that exacerbate CNS diseases. Not only harmful microbial metabolites but also pathogenic bacterial cells and T cells travel from the gut to the CNS to increase inflammation under pathological conditions.
Multiple groups have reported that SCFAs have EAE-suppressing activities (Table 1). Mice with elevated levels of SCFAs after feeding with SCFA water or dietary fiber had increased numbers of regulatory T cells in the intestine and CNS upon MOG immunization104,105. In addition, when mice were fed C4, cuprizone-mediated demyelination was ameliorated.18,74,101,105. The protective effect of C4 was further validated in a lysolecithin-induced demyelination model101. In addition to the major SCFAs (i.e., C2, C3, and C4), valerate (C5) can also regulate EAE activity27. C5 increased both regulatory B cells (Bregs) and T cells (Tregs). C5-treated B cells showed increased expression of IL-10, and adoptive transfer of these B cells ameliorated EAE activity. The protective effect was accompanied by decreased Th17 cell activity in the small intestine. C5, well known for its potent HDAC inhibitory activity, increased mTOR activation and glycolysis in lymphocytes27. Thus, both epigenetic (HDAC inhibition) and metabolic (mTOR) regulation appears to be involved in the protective effects of SCFAs in EAE pathogenesis.
Table 1.
Regulation of CNS diseases in animal models by gut microbial metabolites.
| Diseases in animal models | Metabolites | Effects of metabolites on disease | Effects of metabolites on cells and molecules | Ref |
|---|---|---|---|---|
| EAE and related demyelinating diseases | SCFAs | Exacerbation |
GPCR-mediated immune cell activation; Increased Th17 polarization |
18 |
| Suppression |
IL-10 production; Induction of regulatory T and B cells; Decreased MAPK activation; Th1 suppression; HDAC inhibition; Increased glycolysis and AKT/mTOR; Oligodendrocyte maturation |
18,27,102,104,105 | ||
| LCFA | Exacerbation |
MAPK activation; Increased Th17 and Th1 activity; Decreased Treg activity |
104,216 | |
| Trp metabolites (3.4-DAA I3S, I3C, DIM, IPA, IAld) | Suppression |
STAT1-mediated suppression of antigen presenting cells; Activation of microglial AhR; Decreased NF-κB activity; Th17 suppression; Treg expansion; Increased SOCS2 activity |
116,150,217 | |
| PSA | Suppression |
Increased activity of CD103+ DCs; Induction of IL-10+ T cells; TLR2-dependent increase in CD39+CD4+ T cell activity |
94,217 | |
| 2ND BA (TUDCA) | Suppression | Suppression of inflammatory responses in astrocytes and microglia cells in a GPBAR-dependent manner | 143 | |
| PD models | SCFAs | Exacerbation |
Microglia activation; Increased αSyn-mediated motor dysfunction; Activation of microglial and astrocytes; Higher expression of TLR4, Increased activity of TBK1, NF-κB, and TNF-α. |
71,218 |
| Suppression |
GPR41 activation; Suppression of dopaminergic neuronal loss; Enrichment of C4-producing bacteria; Increased gut occludin expression |
163,164 | ||
| 2ND BA (UDCA, and TUDCA) | Suppression |
Increased intracellular ATP levels; Enhanced contrast response function; Suppressed JNK activity; Suppressed ROS production |
179,183 | |
| AD models | SCFAs | Exacerbation |
Microglial activation; Increased amyloid β plaque deposition via apoE-TREM2 |
199 |
| Suppression |
Increased neuronal activity with hippocampal c-Fos expression; Decreased polymerization of amyloid β; Suppression of NF-κB and COX-2 in microglia |
73,188,195 | ||
| ASD models | SCFAs | Exacerbation |
Astrocyte activation; Increased TNF-α production; Altered hippocampus structure |
213 |
| Suppression | Increased excitatory/inhibitory balance in the prefrontal cortex | 215 |
AhR aryl hydrocarbon receptor, BDNF brain-derived neurotrophic factor, COX-2 cyclooxygenase-2, CREB cyclic AMP response element binding protein, GPBAR G-protein-coupled bile acid receptor, GPCR G-protein-coupled receptor, DAA digestible amino acid, DI diindolylmethane, DIM 3,3’-diindolylmethane, HDAC histone deacetylases, IAld indole aldehyde, IPA indole-3-propionic acid, I3C indole-3-carbinol, I3S 3-indoxyl sulfate, JNK c-Jun N-terminal kinase, MAPK mitogen-activated protein kinase, mTOR mechanistic target of rapamycin, NF-κB nuclear factor kappa-light-chain-enhancer of activated B cells, PSA polysaccharide A, ROS reactive oxygen species, SOCS suppressor of cytokine signaling, STAT signal transducer and activator of transcription, TBK1 TRIF-TANK binding kinase, TREM-2 triggering receptor expressed on myeloid cells-2, Trp tryptophan, TUDCA tauroursodeoxycholic acid, UDCA ursodeoxycholic acid.
The effects of SCFA-producing fiber-rich diets on EAE are mixed, and this appears to be due to considerable variation in the dietary fiber composition of high- and low-fiber diets used by different groups. Some groups used a diet containing no fiber (soluble or insoluble) as the control diet106–108. In general, soluble fibers such as pectin and inulin produce high levels of SCFAs, whereas insoluble fibers such as cellulose are not efficient sources of SCFAs. Other groups, including ours, compared the effects of high- and low-fiber diets containing the same amount of cellulose but different levels (0, 5, and 20%) of soluble fiber. When a high-fiber diet was compared with a zero-fiber diet, the protective effect was significant105. However, when the animal group fed a diet high in soluble fiber (pectin and inulin) was compared with that fed a diet containing zero soluble dietary fiber but the same level of cellulose, the difference was not significant18. This finding suggests the involvement of mechanisms other than SCFA production in the beneficial effects of dietary fiber. Despite its low bioavailability and SCFA-producing ability, cellulose seems to have a suppressive effect on EAE activity84. Cellulose decreased the number of neutrophils and Th2 cells in a spontaneous opticospinal encephalomyelitis mouse model expressing MOG-specific T and B cell antigen receptors84. While cellulose did not significantly change SCFA levels, it changed the microbiome composition in the gut, indicating that the protective effect of certain dietary fibers, such as cellulose, appears to be more dependent on microbial changes than on SCFA production108.
SCFAs appear to regulate EAE responses via several cellular and molecular mechanisms. SCFAs may function through astrocytes and microglial cells, which produce both suppressive and inflammatory cytokines and influence EAE responses. SCFAs can also act through immune cells such as Th17 cells and regulatory T cells, which differentially regulate inflammatory responses. Several cell types can produce IL-10 in response to SCFAs. Major cell types that produce IL-10 are T cells and macrophages, and this production can be further increased by SCFAs38,109–111 (Fig. 1). In our own study, SCFAs increased IL-10 production by microglial cells, and IL-10 production was required for the protective effect of SCFAs on EAE activity18.
While SCFAs have suppressive effects on EAE activity, their G-protein-coupled receptors, such as GPR43, can function unexpectedly to exacerbate EAE activity18. The mechanism is not entirely clear at this point, but the data imply that the protective function of SCFAs is perhaps mediated mainly through non-GPCR-dependent mechanisms. In this regard, the HDAC inhibitory activity of SCFAs may be important. GPCR activation by SCFAs can increase immune activity, including invigorating epithelial responses and activating inflammasomes in the gut112,113. We speculate that GPCR activation by SCFAs in the CNS could promote similar inflammatory responses. In this regard, the function of SCFAs in regulating EAE activity appears to be complex. When transferred into host mice, SCFA-treated MOG-specific effector Th17 cells have greater EAE activity than similarly prepared control Th17 cells18, indicating that SCFAs can affect both the inflammatory and regulatory arms of the CNS immune system. More work is required to understand the effects of SCFAs on key immune and CNS cell types, such as T and B cells, ILCs, glial cells, neurons, macrophages, and dendritic cells.
Indole derivatives and certain environmental contaminants, such as the AhR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), have been studied for their regulatory effects on EAE activity. AhR activation by FICZ and 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) (Trp-derived indole metabolites) suppressed EAE responses114,115, whereas AhR activation by TCDD exacerbated EAE pathogenesis70. Similarly, FICZ and TCDD differentially affected Th17 cells, Tregs, DCs, and astrocytes. The administration of dietary indoles was effective at suppressing EAE development48. Moreover, dietary indoles significantly ameliorated existing EAE48,116; this protection is thought to be mediated by the AhR-mediated expansion of regulatory T cells. These Trp-derived metabolites appear to suppress CNS inflammation by decreasing proinflammatory cytokine expression and inflammatory monocyte infiltration47,48,116, which may involve activation of the type-I IFN pathway in astrocytes to limit CNS inflammation in an AhR-dependent manner. Similarly, cruciferous plant-derived AhR ligands such as glucosinolates suppressed EAE pathogenesis117.
Significant associations between microbial metabolites and MS in humans
In humans, MS manifests in several ways with different patterns of disease progression and relapse; the common categorizations of MS include clinically isolated syndrome (CIS), relapsing-remitting MS (RRMS), primary progressive MS (PPMS), and secondary progressive MS (SPMS)118,119. RRMS, with repeated relapses and remissions, is most common (~85%). Most people with RRMS progress to SPMS with steadily worsening symptoms with or without remissions and relapses. PPMS is rarer (~10%), with slowly worsening symptoms from disease onset without remissions and relapses.
Changes in gut luminal microbial composition have been observed along with the abnormal invasion of microbes in the brains of RRMS patients120. RRMS patients have increased levels of Actinobacteria but decreased levels of Firmicutes and Bacteroidetes in feces. Within the Firmicutes family, fourteen species within Clostridium clusters XIVa and IV were found to be decreased in MS patients121. A cohort study found decreased Prevotella_9 but increased Streptococcus species, along with reduced levels of fecal SCFAs, in MS patients. In general, Tregs and Th17 cells in the blood indicate tolerogenic and inflammatory activity, respectively122. A positive correlation between Streptococcus species and Tregs was reported, and an inverse correlation between Prevotella_9 and Th17 cells was observed. Increased numbers of microbial peptidoglycan-containing macrophages and dendritic cells were detected in the brains of MS patients. Thus, MS patients have both dysbiosis and microbial invasion of the brain120,123. CCR9 is the small intestine-homing receptor for lymphocytes124,125 and is important for thymocyte localization in the thymic medulla126,127. Interestingly, CCR9+CD4+ T cells were found in the cerebrospinal fluid (CSF) of MS patients. T cell migration into inflamed CNS tissues involves another chemokine receptor, CCR6, that is highly expressed by Th17 cells128. CCR9+CD4+ T cells express RORγ and secrete IL-17 and IFN-γ in MS patients. Antibiotic treatment decreased these Th17 cells, suggesting that inflammatory CCR9+CD4+ T cells are induced by microbes129. While further study is required to determine the origin of these T cells, they appear to be generated in the intestine following dysbiosis and neural inflammation. These data imply the active movement of microbes and immune cells from the gut to CNS tissues under inflammatory conditions.
Normally, antigen-induced EAE induction in animals requires the use of potent adjuvants to break immune tolerance to self-antigens75. Fecal microbial transfer (FMT) from MS patients to animals induced EAE development following immunization without any adjuvant. FMT decreased certain microbial species, such as those in the Sutterella genus, and decreased IL-10 expression by T cells16,130. The effect of SCFA-generating dietary fibers on MS activity has been studied. A diet rich in plant fibers increased the fecal presence of the Lachnospiraceae phylum in MS patients and increased the numbers of Tregs and IL-10-producing PD-L1+ monocytes in the blood. While a fiber-rich diet failed to change MS clinical activity, patients on a Western diet had increased MS clinical activity131.
Negative correlations have been found between SCFA levels and T cell activity in MS patients. Higher fecal and blood SCFA levels were associated with increased Treg and suppressed Th1 cell activity122,131,132. Compared with healthy individuals, MS patients have decreased levels of blood C4 and SCFA-producing microbial species118. In line with the dysbiosis and reduced levels of SCFAs in MS patients118,133, decreased levels of all major SCFAs, such as C2, C3, and C4, were found in the blood of SPMS patients compared with that of healthy controls18. In addition, gut SCFA levels were decreased in RRMS patients122. In this regard, C3 increased Treg activity and decreased episodes of MS relapse134. In another study, fecal C3 levels were decreased in both RRMS and SPMS patients132. The therapeutic effect of C3 in MS patients was recently reported134. Regardless of MS subtype, C3 supplementation alleviated the clinical symptoms of MS, and MS patients with increased levels of SCFA-producing gut bacteria showed limited inflammatory T cell activity. C3 also increased regulatory T cells that produce IL-10, altered the expression of many genes in T cells, and increased oxygen consumption in T cell mitochondria. However, increased levels of plasma C2 and increased numbers of circulating Th17 cells were observed in MS patients with severe disability135,136. Because C2 is also produced by host cells under inflammatory and infection conditions137, the increased levels of C2 could originate from the host rather than microbial cells. These data, while contradictory to other results, seem to be in line with the effect of C2 on Th17 cell generation109. Therefore, more studies are required to establish the possible associations between SCFA levels and MS subtypes. More data may enable the utilization of SCFAs as biomarkers for MS activity and gut microbial status in MS patients.
Oral Trp administration boosted serotonin but decreased plasma cortisol levels in patients with neurological diseases. Acute MS and other neurologic symptoms were alleviated138,139. Dietary Trp can be metabolized by gut microbes or host cells after enteric absorption. While gut microbes produce various indole metabolites, host cells produce Kyn, kynurenic acid (KA) and quinolinic acid (QA) via the kynurenine pathway. The levels of urinary Kyn were decreased in RRMS patients140. KA levels were increased in RRMS patients but decreased in PPMS patients, while the level of QA was increased in most MS patients141. Trp administration improved the cognition and memory function of MS patients140,142–144. Pediatric MS is less frequent than adult MS, but relapses are more frequent and possibly more inflammatory in affected pediatric patients145. Increased Actinobacteria but decreased Clostridiales were observed in pediatric MS patients146. Decreased levels of Trp and indole lactate were also observed in pediatric MS patients. Interestingly, higher Kyn levels were associated with an increased relapse rate142. Many Trp metabolites, such as I3S, tryptamine (TA), IAA, Kyn and KA, function as AhR ligands147,148. KA also activates another host receptor, GPR35149. Trp metabolites increase AhR expression on microglia and astrocytes and control the inflammatory activities of these cells116,150. Secondary bile acids also regulate astrocytes by upregulating GPBAR1. Tauroursodeoxycholic acid (TUDCA), a taurine conjugate of ursodeoxycholic acid (UDCA), suppressed microglial and astrocytic polarization (Table 2)143. Overall, multiple gut metabolites can modulate microglial cells and astrocytes to suppress MS pathogenesis (Fig. 1).
Table 2.
Association of CNS diseases and gut microbial metabolites in humans.
| Diseases | Metabolites or their precursors | Effects of metabolites on disease | Effects of metabolites on cells and molecules | Ref |
|---|---|---|---|---|
| MS | SCFAs | Exacerbation |
Induction of IL-17+CD8 T cells; Increased C2 levels in MS patient serum |
135,136 |
| SCFAs | Suppression |
Increased Tregs but decreased Th1 cells; Increased mitochondrial oxidation |
118,122,132,134 | |
| Trp and Trp metabolites (indole, I3S, I3P, I3A) | Suppression |
Limited the pathogenic activity of microglia and astrocytes; AhR activation |
116,150 | |
| Secondary bile acids | Suppression |
Activation of GPBAR1 in astrocytes; TUDCA-mediated suppression of microglial and astrocytic inflammatory polarization |
143 | |
| PD | SCFAs | Exacerbation | Dysbiosis-mediated gut leakage | 219 |
| Suppression |
Decreased Prevotellaceae and Enterobacteriaceae; Increased blood levels of CXCL8 and IL-1β; Increased gut permeability |
160,167,174 | ||
| Secondary bile acids | Exacerbation |
Decreased lipid metabolism; Dysbiosis and elevated BA levels |
165,166 | |
| TMAO | Exacerbation | Increased α-syn aggregation and inflammation | 220 | |
| AD | SCFAs | Exacerbation |
Increased production of inflammatory cytokines; Endothelial dysfunction |
187 |
| Suppression | Increased IL-10 levels | 187 | ||
| Tryptophan | Suppression | Decreased tryptophan but increased blood Kyn/Trp ratio in AD patients | 201 | |
| Secondary Bile acid | Exacerbation |
Increased DCA but decreased primary BAs (cholic acid); Increased levels of TCA, 3-DCA and UDCA in the brain of AD patients |
202,203 | |
| ASD | SCFAs | Exacerbation |
Increased levels of butyrogenic bacteria; Increased levels of fecal SCFAs in ASD patients |
221,222 |
| Suppression | Decreased fecal levels of SCFAs in ASD patients | 211,212 |
AhR aryl hydrocarbon receptor, α-syn α-synuclein, BA bile acid, DCA deoxycholic acid, I3A GPBAR G-protein-coupled bile acid receptor, I3A indole-3-carboxaldehyde, I3P indole-3-propionic acid, I3S indoxyl 3-sulfate, TCA trichloroacetic acid, TGF transforming growth factor, TMAO trimethylamine N-oxide, Trp tryptophan, TUDCA tauroursodeoxycholic acid, UDCA ursodeoxycholic acid, VEGF vascular endothelial growth factor.
Microbial metabolites and Parkinson’s disease
Parkinson’s disease (PD) is caused by the loss of dopaminergic neurons in the substantia nigra (SN), leading to dopamine deficiency in the midbrain; in this region, dopamine is important for the regulation of body movement151. A number of genes and their polymorphisms have been implicated in PD pathogenesis152. The presence of neurotoxic protein inclusions called Lewy bodies, which are composed of α-synuclein oligomers, in the midbrain is a pathological feature of PD153,154. Th1 and Th17 cell-driven inflammation, potentially induced by Lewy bodies, plays significant roles in PD pathogenesis155,156. Because of the inflammatory nature of PD and the potential roles of microbiota and gut metabolites in regulating PD, we also include a discussion of PD in this review (Tables 1, 2). Both genetics and environmental factors, such as diet and exposure to neurotoxic chemicals, play significant roles in PD pathogenesis. Genetic risk factors include autosomal dominant mutations in the alpha-synuclein (SNCA), leucine-rich repeat kinase 2 (LRRK2) and vacuolar protein sorting ortholog 35 (VPS35) genes as well as autosomal recessive mutations in the PTEN-induced kinase 1 (PINK1), DJ-1 (also known as PARK7) and PARKIN genes157. Interestingly, polymorphisms in the LRRK2 gene are commonly found in PD and inflammatory bowel disease patients158,159.
Gut dysbiosis has been observed in PD patients71,160,161. Gut microbiota can regulate PD pathogenesis in animal models71,162–164. Mice colonized with fecal microbiota from PD patients developed PD-like pathological features, including α-synuclein aggregation in the brain71. Metabolite profiling of PD patients revealed decreased metabolism of unsaturated fatty acids and increased levels of secondary bile acid metabolites such as deoxycholic acid (DCA) and lithocholic acid (LCA). Importantly, significant decreases in SCFA-producing bacteria and decreased microbial carbohydrate processing activity were detected in a group of PD patients with reduced mobility165–167.
Among the LRRK2 gene mutations associated with PD, the G2019S mutation that affects the kinase domain is the most frequent168. The immunological characteristics of transgenic rats expressing human LRRK2 G2019S were examined169. Defective myelopoiesis and a decreased Th17 cell frequency were observed in these rats when they were challenged with colitis-inducing agents, such as 2,4,6-trinitrobenzene sulfonic acid (TNBS) and dextran sulfate sodium (DSS). Interestingly, we observed increased levels of Bacteroidetes in this animal model169. The MitoPark (MP) mouse model was created by deleting the mitochondrial transcription factor A (TFAM) gene in dopaminergic neurons170. Decreased gastrointestinal tract motility and gut dysbiosis were observed in this model. Moreover, increased levels of Prevotella were observed in this model and in PD patients. Young rats fed permethrin, a pesticide, developed PD-like symptoms and increased gut permeability. These animals also developed dysbiosis and showed decreased SCFA production.
The protective effects of valproic acid (a SCFA-related HDAC inhibitor) and C4 were observed in a model of PD induced with the chemicals manganese and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine160,163,171–174. Interestingly, a synthetic FFAR3 (GPR41) agonist (AR420626) corrected the movement disorder and neuronal loss in a 6-hydroxydopamine (OHDA, a neurotoxin)-induced PD mouse model163. In contrast, the microbiota and SCFAs were reported to promote PD pathogenesis in α-synuclein-transgenic mice71. Other researchers have also reported that SCFA administration exacerbates PD pathogenesis in another animal model175. The pathogenic role of SCFAs is unexpected but corresponds with the role of these metabolites in promoting the generation of inflammatory Th1 and Th17 cells, as reported previously by our group109,176. While these animal models provide insight into PD pathogenesis, the experimental conditions and pathogenesis in these models differ greatly from each other and from PD patients. Thus, the effects of SCFAs on PD pathogenesis are unclear, and more studies are required.
A previous study reported potential changes in bile acid metabolism with increased levels of secondary bile acids, such as LCA and DCA165. In a latent PD model in which α-syn fibrils were injected into the olfactory bulb to induce PD-like pathogenesis, the levels of bile acids such as ω-muricholic acid, TUDCA and UDCA were decreased in the blood177. Beneficial roles of TUDCA in acute, progressive and nigral transplant animal models of PD have been reported178–180. TUDCA ameliorated the decrease in dopaminergic fibers, mitochondrial dysfunction and neuroinflammation in an MPTP-induced animal model179,181,182. Moreover, TUDCA suppressed α-synuclein-induced oxidative stress via the activation of Nrf2, JNK and AKT179,182. In addition, UDCA decreased mitochondrial dysfunction in skin fibroblasts from patients harboring the LRRK2 G2019S mutation183. Thus, secondary bile acids have protective effects against PD pathogenesis.
Microbial metabolites and Alzheimer’s disease
AD is a progressive neural disease induced by the accumulation of toxic misfolded amyloid β-protein (Aβ) plaques in the brain. In this disease, extracellular β-amyloid plaques form in the basal, temporal, and orbitofrontal neocortex. In severe AD cases, the plaques spread to the hippocampus184. Dysbiosis is a risk factor for AD development. Mice humanized with stool from AD patients showed defects in cognitive function185. In addition to the gut bacterial composition, the diversity of gut fungal species (mycobiome) in patients with mild cognitive disorder was lower than that in control subjects. Dietary intervention with a Mediterranean ketogenic diet enhanced fungal microbiome diversity and decreased the levels of AD biomarkers in cerebral spinal fluid186.
Similar to MS and PD patients, AD patients have decreased levels of SCFAs186–189. In particular, AD patients with brain amyloid accumulation and endothelial dysfunction had decreased blood levels of C4187. AD pathology is associated with increased blood LPS and inflammatory cytokines. Interestingly, blood C4 and IL-10, but not acetate and valerate, were negatively associated with AD pathology, suggesting a role for SCFAs in AD pathogenesis. Aging and oxidative stress have been suggested to alter the gut microbial community and thus reduce the levels of SCFAs. Caloric restriction and dietary antioxidant supplementation can diminish AD development, in part by modulating microbial composition190–192. Dietary fiber and SCFAs have been reported to have beneficial effects on AD pathogenesis193–195. SCFA treatment decreased the polymerization of Aβ1-40 or Aβ1-42 peptides into neurotoxic multimeric Aβ forms188. Probiotic supplementation increased the hippocampal concentration of SCFAs and reduced anxiety-like behavior in a humanized AD mouse model73. The microbiota has been shown to regulate AD pathogenesis in several animal models. For example, amyloidosis with defective long-term synaptic potentiation develops in the APP/PS1 mouse model, created by overexpressing amyloid precursor protein (APP) and a mutant form of presellin 1 (PS1)196–198. Fecal microbiota transplantation into APP/PS1 mice increased C4 levels in the colon and increased the expression of synaptic plasticity-related proteins (PSD-95 and synapsin I) in the brain, indicating a decrease in synaptic disorder. The administration of C4-producing bacteria suppressed β-amyloid deposition and effector cytokine release from microglial cells73,195,199,200. In AD patients, the blood Kyn/Trp ratio was found to be increased due to decreased Trp levels, indicating an abnormality in Trp metabolism201. The levels of secondary bile acids such as DCA, TCA, 3-DCA, and UDCA were increased in AD patients202,203. Thus, AD patients have an imbalance in the levels of metabolites that regulate AD pathogenesis. SCFAs have both protective and pathogenic effects, Trp metabolites have protective effects, and bile acid metabolites have overall pathogenic effects on AD (Table 2).
Impacts on autism spectrum disorder
Autism spectrum disorder (ASD) is a group of developmental disabilities that involve social, behavioral and communication challenges. Both genetic and environmental factors have been identified as risk factors for the development of ASD, which is heritable. For example, chromosome 16p11.2 deletion and 15q11-q13 duplication are risk factors for ASD development. In addition, environmental factors such as maternal age, health status, infection/inflammation, perinatal hypoxia, medication, nutrition, and toxic exposure during fetal or early development are associated with ASD204,205. Pathogenic maternal immune activation of the IL-17A pathway was shown to promote ASD development in offspring206. The injection of synthetic dsRNA into pregnant dams, mimicking viral infection during pregnancy, induced placental IL-17A expression and fetal brain IL-17 receptor expression in animal models206. Th17 inflammation is associated with abnormal cortical development in animal models.
While ASD is different from MS, PD and AD in that it primarily involves CNS developmental issues rather than an ongoing inflammatory disease, a significant body of literature indicates an active role of dysbiosis in ASD. Some ASD patients report GI problems, such as abdominal pain, diarrhea, and food sensitivities207,208. Decreased levels of Firmicutes, Fusobacteria and Verrucomicrobia but increased levels of Bacteroidetes have been reported in ASD patients209. Elevated levels of Th17-inducing gut microbial species, such as segmented filamentous bacteria (SFB), Bifidobacterium adolescentis, and certain E. coli isolates, during pregnancy were shown to increase ASD development in offspring210. Intestinal CD11c+ DCs, which recognize microbial TLR3 ligands and produce IL-1β, IL-23, and IL-6, can induce pathogenic Th17 responses206,210. The significance of the microbial community in the onset of ASD has been validated in an FMT model72. When germ-free mice were humanized with microbes from ASD patient stool, their offspring showed behavioral disorders and decreased locomotion and interactions with other animals. The abundance of Bacteroides spp. and P. merdae was correlated with increased social behavior. Increased E. tayi, however, was associated with decreased social interaction in offspring72. Thus, dysbiosis can affect ASD at both the developmental and effector phases.
An altered microbial composition can impact metabolite production. It appears that microbial metabolites can bidirectionally regulate ASD activity. Decreased levels of fecal SCFAs (C2, C3, and C5) were observed in ASD patients211,212. Moreover, C4-producing bacteria and fecal SCFA levels were reported to be increased in ASD patients compared with unaffected individuals (Table 2). In a rat ASD model, C3 administration altered the patterning of human neural stem cells, leading to gliosis, disturbed neurocircuitry, inflammatory responses in the hippocampus and increased ASD-like behavior213,214. On the other hand, C4 administration improved social behavior and regulated autism-related genes related to the excitatory/inhibitory balance in the prefrontal cortex of mice215 (Table 1). Thus, the effect of SCFAs on ASD is currently equivocal. It is interesting that different SCFAs exert different effects on ASD development. Offspring from female animals that received an FMT from human ASD patients had lower levels of 5-aminovaleric acid (5AV) and taurine in the colon; these metabolites are gamma-aminobutyric acid (GABA) receptor agonists. When 5AV and taurine were administered to a mouse model of ASD (i.e., BTBR T+ tf/J mice), ASD-related behavior was suppressed72. Thus, microbial metabolites have significant regulatory effects on ASD activity.
Implications and concluding remarks
The gut is functionally linked to the CNS by immunological mediators, lymphocytes, neurotransmitters, microbes and microbial metabolites. Indeed, it is apparent that most organs and tissues in the body are regulated by gut microbiota and their metabolites, and the CNS is no exception. Importantly, because of the unique (i.e., neurological) functions of the CNS, the impact of gut microbes and their metabolites is manifested quite uniquely in this system. However, the effects of the microbiota and metabolites on different CNS diseases appear to be heterogeneous. Overall, the microbiota and their metabolites influence and strengthen CNS function in the healthy state; for example, SCFAs strengthen the BBB to maintain CNS integrity and protect the CNS from toxic and immunological damage. Various microbial metabolites regulate both inflammatory responses and neuronal activity in the CNS. Gut microbial metabolites promote the maturation of key CNS cells, such as oligodendrocytes, microglial cells and astrocytes, and regulate synapse formation. In addition, the gut microbial metabolites serotonin, dopamine, 5AV and taurine regulate neurotransmission. The production of these metabolites by microbes partly explains the changes in neurological activity following FMT from patients to experimental animals. Certain microbial metabolites, such as SCFAs, secondary bile acids and Trp metabolites, promote immune tolerance by increasing the expression of IL-10 and IL-22 to prevent inflammatory diseases. Under the highly variable pathological conditions of different diseases and hosts, SCFAs and other metabolites induce the generation of effector cells that produce neuroinflammatory cytokines. In this regard, microbial metabolites have important functions in regulating T cells, antigen-presenting cells, microglial cells and astrocytes.
We discussed the effects of microbiota and their metabolites on the pathogenesis of four different CNS diseases or disorders. A common pattern of regulation emerging from the discussed information is that disease pathogenesis is accelerated under conditions of dysbiosis (Fig. 2). While dysbiosis alone would not cause overt inflammatory responses in the CNS in normal hosts, it appears to increase pathological processes in hosts with underlying disease activity or risk factors. Changes in microbial metabolites are significant modifiers of pathogenesis in the CNS; these changes either positively or negatively affect disease onset and pathogenesis. Moreover, the altered production of microbe-derived neurotransmitters in dysbiosis appears to significantly affect neurological activity.
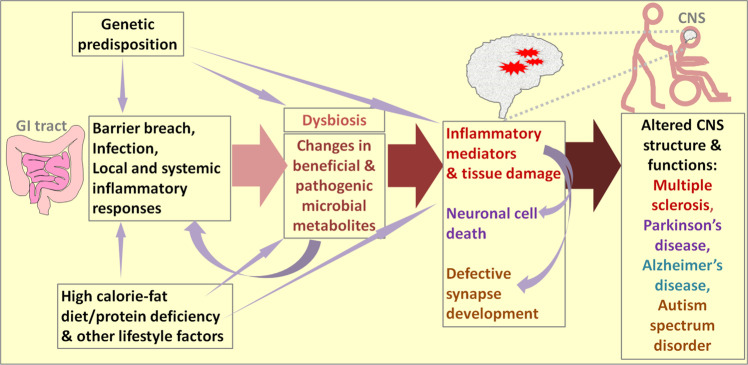
Fig. 2. The common regulatory network of microbial metabolites, inflammatory diseases and CNS disorders.
The common initiating factors for the four neurological diseases are genetic predisposition and environmental factors, which include diet and lifestyle. Under pathogenic conditions, the intestinal barrier can be breached, and systemic inflammatory responses can occur. These changes can be followed by dysbiosis (i.e., decreased gut microbial diversity leading to decreased levels of beneficial microbes). For example, consumption of a diet high in calories and fat but low in dietary fiber can accelerate pathogenic dysbiosis. As a result, decreased levels of beneficial gut microbial metabolites such as SCFAs, Trp metabolites and phytochemicals are produced, and simultaneously, the production of harmful metabolites such as long-chain fatty acids (LCFAs), certain bile acid metabolites, and toxic microbial metabolites increases, thereby affecting immune and tissue cells in both the intestine and CNS. These changes can decrease immune tolerance, which is important for preventing autoimmune diseases, and exacerbate pathogenic immune responses mediated by inflammatory cells such as Th17 and Th1 cells. These pathogenic inflammatory responses can contribute to tissue damage (demyelination in MS), neuronal cell death (PD and AD), and neuronal synapse development (ASD). Moreover, certain microbial metabolites regulate neurotransmission and, therefore, can directly affect neurological activity.
Despite research progress in this field, significant controversies remain regarding the functions of microbiota and their metabolites in regulating various CNS diseases. We still lack an understanding of the detailed functions of microbial metabolites. Importantly, microbes and microbial metabolites have both pro- and anti-inflammatory roles. What underlies the apparent bidirectional activities of gut microbial metabolites? SCFAs, which are largely anti-inflammatory and disease-suppressing, are sometimes associated with increased CNS disease activity. While more information is needed to elucidate the mechanisms underlying the apparent bidirectional regulation, it is clear that gut microbiota and their metabolites are significant modifiers rather than primary inducers of CNS diseases. Their effects on CNS pathogenesis can be altered depending on host factors, such as the type of inflammatory responses and stage of pathogenesis. Thus, an improved understanding of CNS regulation by microbial metabolites is required. The precise mechanisms by which these metabolites regulate immune responses individually or in combination to suppress or increase pathogenesis in the CNS have yet to be elucidated, particularly for relatively understudied diseases such as PD, AD, and ASD.
Acknowledgements
CHK is the Judy and Kenneth Betz Endowed Professor in the Mary H Weiser Food Allergy Center.
Author contributions
CHK and JP collected information and wrote the article.
Funding
This study was supported, in part, by the NIH (R01AI121302, R21AI14889801, R01AI074745, and R01AI080769 to CHK) and the National Research Foundation of Korea (2020R1G1A1099715 to JP).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassmann H, Brück W, Lucchinetti CF. The immunopathology of multiple sclerosis: an overview. Brain Pathol. 2007;17:210–218. doi: 10.1111/j.1750-3639.2007.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucchinetti C, et al. Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann. Neurol. 2000;47:707–717. doi: 10.1002/1531-8249(200006)47:6<707::aid-ana3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 5.Bien CG, et al. Destruction of neurons by cytotoxic T cells: a new pathogenic mechanism in Rasmussen’s encephalitis. Ann. Neurol. 2002;51:311–318. doi: 10.1002/ana.10100. [DOI] [PubMed] [Google Scholar]
- 6.Misu T, et al. Presence of six different lesion types suggests diverse mechanisms of tissue injury in neuromyelitis optica. Acta Neuropathol. 2013;125:815–827. doi: 10.1007/s00401-013-1116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlmann T, Lassmann H, Brück W. Diagnosis of inflammatory demyelination in biopsy specimens: a practical approach. Acta Neuropathol. 2008;115:275–287. doi: 10.1007/s00401-007-0320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höftberger R, Lassmann H. Inflammatory demyelinating diseases of the central nervous system. Handb. Clin. Neurol. 2017;145:263–283. doi: 10.1016/B978-0-12-802395-2.00019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradl M, et al. Neuromyelitis optica: pathogenicity of patient immunoglobulin in vivo. Ann. Neurol. 2009;66:630–643. doi: 10.1002/ana.21837. [DOI] [PubMed] [Google Scholar]
- 10.Linington C, Bradl M, Lassmann H, Brunner C, Vass K. Augmentation of demyelination in rat acute allergic encephalomyelitis by circulating mouse monoclonal antibodies directed against a myelin/oligodendrocyte glycoprotein. Am. J. Pathol. 1988;130:443. [PMC free article] [PubMed] [Google Scholar]
- 11.Cross AH, Trotter JL, Lyons J-A. B cells and antibodies in CNS demyelinating disease. J. Neuroimmunol. 2001;112:1–14. doi: 10.1016/s0165-5728(00)00409-4. [DOI] [PubMed] [Google Scholar]
- 12.Carson MJ, Thrash JC, Walter B. The cellular response in neuroinflammation: the role of leukocytes, microglia and astrocytes in neuronal death and survival. Clin. Neurosci. Res. 2006;6:237–245. doi: 10.1016/j.cnr.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heneka MT, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch EC, Hunot S. Neuroinflammation in Parkinson’s disease: a target for neuroprotection? Lancet Neurol. 2009;8:382–397. doi: 10.1016/S1474-4422(09)70062-6. [DOI] [PubMed] [Google Scholar]
- 15.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 16.Berer K, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl Acad. Sci. U. S. A. 2017;114:10719–10724. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mestre L, et al. Manipulation of gut microbiota influences immune responses, axon preservation, and motor disability in a model of progressive multiple sclerosis. Front. Immunol. 2019;10:1374. doi: 10.3389/fimmu.2019.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J, Wang Q, Wu Q, Mao-Draayer Y, Kim CH. Bidirectional regulatory potentials of short-chain fatty acids and their G-protein-coupled receptors in autoimmune neuroinflammation. Sci. Rep. 2019;9:8837. doi: 10.1038/s41598-019-45311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haase S, et al. The role of the gut microbiota and microbial metabolites in neuroinflammation. Eur. J. Immunol. 2020;50:1863–1870. doi: 10.1002/eji.201847807. [DOI] [PubMed] [Google Scholar]
- 20.Wong JM, De Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 21.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexeev EE, et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am. J. Pathol. 2018;188:1183–1194. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taleb S. Tryptophan dietary impacts gut barrier and metabolic diseases. Front. Immunol. 2019;10:2113. doi: 10.3389/fimmu.2019.02113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Sayin SamaI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR Antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Kim CH. Immune regulation by microbiome metabolites. Immunology. 2018;154:220–229. doi: 10.1111/imm.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luu M, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 2019;10:760. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker A, Fonseca S, Carding SR. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes. 2020;11:135–157. doi: 10.1080/19490976.2019.1638722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 30.Nøhr MK, et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154:3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 31.Pluznick JL. Microbial short-chain fatty acids and blood pressure regulation. Curr. Hypertens. Rep. 2017;19:25. doi: 10.1007/s11906-017-0722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thangaraju M, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14:115–121. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi M, et al. Short-chain fatty acids, GPR41 and GPR43 ligands, inhibit TNF-α-induced MCP-1 expression by modulating p38 and JNK signaling pathways in human renal cortical epithelial cells. Biochem. Biophys. Res. Commun. 2017;486:499–505. doi: 10.1016/j.bbrc.2017.03.071. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, et al. Short-chain fatty acids from periodontal pathogens suppress histone deacetylases, EZH2, and SUV39H1 to promote Kaposi’s sarcoma-associated herpesvirus replication. J. Virol. 2014;88:4466–4479. doi: 10.1128/JVI.03326-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim CH, Park J, Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw. 2014;14:277–288. doi: 10.4110/in.2014.14.6.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Säemann MD, et al. Anti‐inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL‐12 and up‐regulation of IL‐10 production. FASEB J. 2000;14:2380–2382. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 38.Sun M, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018;9:3555. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim CH. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 2021;18:1161–1171. doi: 10.1038/s41423-020-00625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sepahi A, Liu Q, Friesen L, Kim CH. Dietary fiber metabolites regulate innate lymphoid cell responses. Mucosal Immunol. 2021;14:317–330. doi: 10.1038/s41385-020-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chun E, et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut. Immun. Immun. 2019;51:871–884 e876. doi: 10.1016/j.immuni.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goverse G, et al. Diet-derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J. Immunol. 2017;198:2172–2181. doi: 10.4049/jimmunol.1600165. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. 2019;177:1217–1231 e1218. doi: 10.1016/j.cell.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proietti E, Rossini S, Grohmann U, Mondanelli G. Polyamines and kynurenines at the intersection of immune modulation. Trends Immunol. 2020;41:1037–1050. doi: 10.1016/j.it.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Wlodarska M, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37 e26. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Rouse M, Singh NP, Nagarkatti PS, Nagarkatti M. Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br. J. Pharmacol. 2013;169:1305–1321. doi: 10.1111/bph.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsumoto M, Benno Y. The relationship between microbiota and polyamine concentration in the human intestine: a pilot study. Microbiol. Immunol. 2007;51:25–35. doi: 10.1111/j.1348-0421.2007.tb03887.x. [DOI] [PubMed] [Google Scholar]
- 50.Landau G, Bercovich Z, Park MH, Kahana C. The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J. Biol. Chem. 2010;285:12474–12481. doi: 10.1074/jbc.M110.106419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tersey SA, Colvin SC, Maier B, Mirmira RG. Protective effects of polyamine depletion in mouse models of type 1 diabetes: implications for therapy. Amino Acids. 2014;46:633–642. doi: 10.1007/s00726-013-1560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chitrala KN, Yang X, Nagarkatti P, Nagarkatti M. Comparative analysis of interactions between aryl hydrocarbon receptor ligand binding domain with its ligands: a computational study. BMC Struct. Biol. 2018;18:15. doi: 10.1186/s12900-018-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Q, et al. Ginsenosides are novel naturally-occurring aryl hydrocarbon receptor ligands. PLoS One. 2013;8:e66258. doi: 10.1371/journal.pone.0066258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee MJ, et al. Korean red ginseng and ginsenoside-Rb1/-Rg1 alleviate experimental autoimmune encephalomyelitis by suppressing Th1 and Th17 cells and upregulating regulatory T Cells. Mol. Neurobiol. 2016;53:1977–2002. doi: 10.1007/s12035-015-9131-4. [DOI] [PubMed] [Google Scholar]
- 55.Xuzhu G, et al. Resveratrol modulates murine collagen-induced arthritis by inhibiting Th17 and B-cell function. Ann. Rheum. Dis. 2012;71:129–135. doi: 10.1136/ard.2011.149831. [DOI] [PubMed] [Google Scholar]
- 56.Campbell C, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581:475–479. doi: 10.1038/s41586-020-2193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hang S, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy BS, Watanabe K, Weisburger JH, Wynder EL. Promoting effect of bile acids in colon carcinogenesis in germ-free and conventional F344 rats. Cancer Res. 1977;37:3238–3242. [PubMed] [Google Scholar]
- 59.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakanaka T, et al. The effects of a TGR5 agonist and a dipeptidyl peptidase IV inhibitor on dextran sulfate sodium-induced colitis in mice. J. Gastroenterol. Hepatol. 2015;30(Suppl 1):60–65. doi: 10.1111/jgh.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hao H, et al. Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab. 2017;25:856–867 e855. doi: 10.1016/j.cmet.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo C, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:944. doi: 10.1016/j.immuni.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yazici C, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66:1983–1994. doi: 10.1136/gutjnl-2016-313321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo Z, et al. Intestinal microbiota distinguish gout patients from healthy humans. Sci. Rep. 2016;6:20602. doi: 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogt NM, et al. The gut microbiota-derived metabolite trimethylamine N-oxide is elevated in Alzheimer’s disease. Alzheimers Res. Ther. 2018;10:124. doi: 10.1186/s13195-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao X, et al. Uric acid induces cognitive dysfunction through hippocampal inflammation in rodents and humans. J. Neurosci. 2016;36:10990–11005. doi: 10.1523/JNEUROSCI.1480-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Hou Y, Wang G, Zheng X, Hao H. Gut microbial metabolites of aromatic amino acids as signals in host–microbe interplay. Trends Endocrinol. Metab. 2020;31:818–834. doi: 10.1016/j.tem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 70.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 71.Sampson TR, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s Disease. Cell. 2016;167:1469–1480 e1412. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharon G, et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell. 2019;177:1600–1618 e1617. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaur H, et al. Effects of probiotic supplementation on short chain fatty acids in the AppNL-G-F mouse model of Alzheimer’s Disease. J. Alzheimers Dis. 2020;76:1083–1102. doi: 10.3233/JAD-200436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haghikia A, et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity. 2015;43:817–829. doi: 10.1016/j.immuni.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 75.Constantinescu CS, Farooqi N, O’Brien K, Gran B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS) Br. J. Pharmacol. 2011;164:1079–1106. doi: 10.1111/j.1476-5381.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun X, Liu Y, Liu B, Xiao Z, Zhang L. Rolipram promotes remyelination possibly via MEK-ERK signal pathway in cuprizone-induced demyelination mouse. Exp. Neurol. 2012;237:304–311. doi: 10.1016/j.expneurol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 77.Blakemore WF. Demyelination of the superior cerebellar peduncle in the mouse induced by cuprizone. J. Neurol. Sci. 1973;20:63–72. doi: 10.1016/0022-510x(73)90118-4. [DOI] [PubMed] [Google Scholar]
- 78.Pavelko KD, Van Engelen BG, Rodriguez M. Acceleration in the rate of CNS remyelination in lysolecithin-induced demyelination. J. Neurosci. 1998;18:2498–2505. doi: 10.1523/JNEUROSCI.18-07-02498.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lipton HL. Theiler’s virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 1975;11:1147–1155. doi: 10.1128/iai.11.5.1147-1155.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu C, Collins J, Sharp E. The pathogenesis of Theiler’s GD VII encephalomyelitis virus infection in mice as studied by immunofluorescent technique and infectivity titrations. J. Immunol. 1967;98:46–55. [PubMed] [Google Scholar]
- 81.Krishnamoorthy G, Lassmann H, Wekerle H, Holz A. Spontaneous opticospinal encephalomyelitis in a double-transgenic mouse model of autoimmune T cell/B cell cooperation. J. Clin. Invest. 2006;116:2385–2392. doi: 10.1172/JCI28330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pöllinger B, et al. Spontaneous relapsing-remitting EAE in the SJL/J mouse: MOG-reactive transgenic T cells recruit endogenous MOG-specific B cells. J. Exp. Med. 2009;206:1303–1316. doi: 10.1084/jem.20090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johanson DM, et al. Experimental autoimmune encephalomyelitis is associated with changes of the microbiota composition in the gastrointestinal tract. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-72197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berer K, et al. Dietary non-fermentable fiber prevents autoimmune neurological disease by changing gut metabolic and immune status. Sci. Rep. 2018;8:10431. doi: 10.1038/s41598-018-28839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. U. S. A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ochoa-Reparaz J, et al. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 87.Benedek G, et al. Estrogen protection against EAE modulates the microbiota and mucosal-associated regulatory cells. J. Neuroimmunol. 2017;310:51–59. doi: 10.1016/j.jneuroim.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cekanaviciute E, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl Acad. Sci. U. S. A. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Visser L, et al. Proinflammatory bacterial peptidoglycan as a cofactor for the development of central nervous system autoimmune disease. J. Immunol. 2005;174:808–816. doi: 10.4049/jimmunol.174.2.808. [DOI] [PubMed] [Google Scholar]
- 90.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Calcinotto A, et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018;9:4832. doi: 10.1038/s41467-018-07305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen H, et al. Gut microbiota interventions with clostridium butyricum and norfloxacin modulate immune response in experimental autoimmune encephalomyelitis mice. Front. Immunol. 2019;10:1662. doi: 10.3389/fimmu.2019.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramakrishna C, et al. Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat. Commun. 2019;10:2153. doi: 10.1038/s41467-019-09884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ochoa-Reparaz J, et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–495. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 95.Lee K, et al. Bacillus-derived poly-gamma-glutamic acid reciprocally regulates the differentiation of T helper 17 and regulatory T cells and attenuates experimental autoimmune encephalomyelitis. Clin. Exp. Immunol. 2012;170:66–76. doi: 10.1111/j.1365-2249.2012.04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Felts PA, et al. Inflammation and primary demyelination induced by the intraspinal injection of lipopolysaccharide. Brain. 2005;128:1649–1666. doi: 10.1093/brain/awh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minagar A, Alexander JS. Blood-brain barrier disruption in multiple sclerosis. Mult. Scler. 2003;9:540–549. doi: 10.1191/1352458503ms965oa. [DOI] [PubMed] [Google Scholar]
- 98.Dopkins N, Nagarkatti PS, Nagarkatti M. The role of gut microbiome and associated metabolome in the regulation of neuroinflammation in multiple sclerosis and its implications in attenuating chronic inflammation in other inflammatory and autoimmune disorders. Immunology. 2018;154:178–185. doi: 10.1111/imm.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Braniste V, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014;6:263ra158–263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Erny D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen T, Noto D, Hoshino Y, Mizuno M, Miyake S. Butyrate suppresses demyelination and enhances remyelination. J. Neuroinflamm. 2019;16:165. doi: 10.1186/s12974-019-1552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mikov M, Kevresan S, Kuhajda K, Jakovljevic V, Vasovic V. 3Alpha,7alpha-dihydroxy-12-oxo-5beta-cholanate as blood-brain barrier permeator. Pol. J. Pharmacol. 2004;56:367–371. [PubMed] [Google Scholar]
- 103.Greenwood J, Adu J, Davey AJ, Abbott NJ, Bradbury MW. The effect of bile salts on the permeability and ultrastructure of the perfused, energy-depleted, rat blood-brain barrier. J. Cereb. Blood Flow. Metab. 1991;11:644–654. doi: 10.1038/jcbfm.1991.116. [DOI] [PubMed] [Google Scholar]
- 104.Haghikia A, et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity. 2016;44:951–953. doi: 10.1016/j.immuni.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 105.Mizuno M, Noto D, Kaga N, Chiba A, Miyake S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS One. 2017;12:e0173032. doi: 10.1371/journal.pone.0173032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schneeman BO, Gallaher D. Changes in small intestinal digestive enzyme activity and bile acids with dietary cellulose in rats. J. Nutr. 1980;110:584–590. doi: 10.1093/jn/110.3.584. [DOI] [PubMed] [Google Scholar]
- 107.Levrat M-A, Behr SR, Rémésy C, Demigné C. Effects of soybean fiber on cecal digestion in rats previously adapted to a fiber-free diet. J. Nutr. 1991;121:672–678. doi: 10.1093/jn/121.5.672. [DOI] [PubMed] [Google Scholar]
- 108.Fischer F, et al. Dietary cellulose induces anti-inflammatory immunity and transcriptional programs via maturation of the intestinal microbiota. Gut Microbes. 2020;12:1–17. doi: 10.1080/19490976.2020.1829962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cox MA, et al. Short-chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E(2) and cytokines. World J. Gastrol. 2009;15:5549–5557. doi: 10.3748/wjg.15.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 113.Macia L, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 114.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 115.Quintana FJ, et al. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. U. S. A. 2010;107:20768–20773. doi: 10.1073/pnas.1009201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Giacoppo S, et al. Protective role of (RS)-glucoraphanin bioactivated with myrosinase in an experimental model of multiple sclerosis. CNS Neurosci. Ther. 2013;19:577–584. doi: 10.1111/cns.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saresella M, et al. Alterations in circulating fatty acid are associated with gut microbiota dysbiosis and inflammation in multiple sclerosis. Front. Immunol. 2020;11:1390. doi: 10.3389/fimmu.2020.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Correale J, Gaitan MI, Ysrraelit MC, Fiol MP. Progressive multiple sclerosis: from pathogenic mechanisms to treatment. Brain. 2017;140:527–546. doi: 10.1093/brain/aww258. [DOI] [PubMed] [Google Scholar]
- 120.Branton WG, et al. Brain microbiota disruption within inflammatory demyelinating lesions in multiple sclerosis. Sci. Rep. 2016;6:37344. doi: 10.1038/srep37344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miyake S, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeng Q, et al. Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem. Int. 2019;129:104468. doi: 10.1016/j.neuint.2019.104468. [DOI] [PubMed] [Google Scholar]
- 123.Schrijver IA, et al. Bacterial peptidoglycan and immune reactivity in the central nervous system in multiple sclerosis. Brain. 2001;124:1544–1554. doi: 10.1093/brain/124.8.1544. [DOI] [PubMed] [Google Scholar]
- 124.Campbell DJ, Butcher EC. Intestinal attraction: CCL25 functions in effector lymphocyte recruitment to the small intestine. J. Clin. Invest. 2002;110:1079–1081. doi: 10.1172/JCI16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kang SG, Wang C, Matsumoto S, Kim CH. High and low vitamin A therapies induce distinct FoxP3+ T-cell subsets and effectively control intestinal inflammation. Gastroenterology. 2009;137:1391–1402. doi: 10.1053/j.gastro.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu C, et al. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 127.Uehara S, Song K, Farber JM, Love PE. Characterization of CCR9 expression and CCL25/thymus-expressed chemokine responsiveness during T cell development: CD3(high)CD69+ thymocytes and gammadeltaTCR+ thymocytes preferentially respond to CCL25. J. Immunol. 2002;168:134–142. doi: 10.4049/jimmunol.168.1.134. [DOI] [PubMed] [Google Scholar]
- 128.Wang C, Kang SG, Lee J, Sun Z, Kim CH. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2009;2:173–183. doi: 10.1038/mi.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kadowaki A, Saga R, Lin Y, Sato W, Yamamura T. Gut microbiota-dependent CCR9+CD4+ T cells are altered in secondary progressive multiple sclerosis. Brain. 2019;142:916–931. doi: 10.1093/brain/awz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miller PG, Bonn MB, Franklin CL, Ericsson AC, McKarns SC. TNFR2 deficiency acts in concert with gut microbiota to precipitate spontaneous sex-biased central nervous system demyelinating autoimmune disease. J. Immunol. 2015;195:4668–4684. doi: 10.4049/jimmunol.1501664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saresella M, et al. Immunological and clinical effect of diet modulation of the gut microbiome in multiple sclerosis patients: a pilot study. Front. Immunol. 2017;8:1391. doi: 10.3389/fimmu.2017.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takewaki D, et al. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc. Natl Acad. Sci. U. S. A. 2020;117:22402–22412. doi: 10.1073/pnas.2011703117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen J, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Duscha A, et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell. 2020;180:1067–1080. doi: 10.1016/j.cell.2020.02.035. [DOI] [PubMed] [Google Scholar]
- 135.Perez-Perez S, et al. Acetate correlates with disability and immune response in multiple sclerosis. PeerJ. 2020;8:e10220. doi: 10.7717/peerj.10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moussallieh FM, et al. Serum analysis by 1H nuclear magnetic resonance spectroscopy: a new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult. Scler. 2014;20:558–565. doi: 10.1177/1352458513504638. [DOI] [PubMed] [Google Scholar]
- 137.Crabtree B, Souter MJ, Anderson SE. Evidence that the production of acetate in rat hepatocytes is a predominantly cytoplasmic process. Biochem. J. 1989;257:673–678. doi: 10.1042/bj2570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hyyppä MT, Jolma T, Riekkinen P, Rinne UK. Effects of L-tryptophan treatment on central indoleamine metabolism and short-lasting neurologic disturbances in multiple sclerosis. J. Neural Transm. 1975;37:297–304. doi: 10.1007/BF01258656. [DOI] [PubMed] [Google Scholar]
- 139.Hyyppä MT, Jolma T, Långvik V-A, Kytömäki O, Syvälahti E. l-Tryptophan and neuroendocrine regulation in neurologic patients: Hormone responses to l-tryptophan loading in patients with hypothalamic lesions. Psychoneuroendocrinology. 1977;2:349–357. doi: 10.1016/0306-4530(77)90004-x. [DOI] [PubMed] [Google Scholar]
- 140.Gaetani L, et al. Host and microbial tryptophan metabolic profiling in multiple sclerosis. Front . Immunol. 2020;11:157. doi: 10.3389/fimmu.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lim CK, et al. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017;7:41473. doi: 10.1038/srep41473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nourbakhsh B, et al. Altered tryptophan metabolism is associated with pediatric multiple sclerosis risk and course. Ann. Clin. Transl. Neurol. 2018;5:1211–1221. doi: 10.1002/acn3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bhargava P, et al. Bile acid metabolism is altered in multiple sclerosis and supplementation ameliorates neuroinflammation. J. Clin. Invest. 2020;130:3467–3482. doi: 10.1172/JCI129401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lieben CK, et al. Intake of tryptophan-enriched whey protein acutely enhances recall of positive loaded words in patients with multiple sclerosis. Clin. Nutr. 2018;37:321–328. doi: 10.1016/j.clnu.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 145.Gorman MP, Healy BC, Polgar-Turcsanyi M, Chitnis T. Increased relapse rate in pediatric-onset compared with adult-onset multiple sclerosis. Arch. Neurol. 2009;66:54–59. doi: 10.1001/archneurol.2008.505. [DOI] [PubMed] [Google Scholar]
- 146.Tremlett H, Waubant E. The gut microbiota and pediatric multiple sclerosis: recent findings. Neurotherapeutics. 2018;15:102–108. doi: 10.1007/s13311-017-0574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heath-Pagliuso S, et al. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37:11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- 148.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 149.Wang J, et al. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006;281:22021–22028. doi: 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 150.Rothhammer V, et al. Microglial control of astrocytes in response to microbial metabolites. Nature. 2018;557:724–728. doi: 10.1038/s41586-018-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Maetzler W, Berg D. Parkinson disease in 2017: changing views after 200 years of Parkinson disease. Nat. Rev. Neurol. 2018;14:70–72. doi: 10.1038/nrneurol.2017.183. [DOI] [PubMed] [Google Scholar]
- 152.Singh M, et al. Polymorphism in environment responsive genes and association with Parkinson disease. Mol. Cell. Biochem. 2008;312:131–138. doi: 10.1007/s11010-008-9728-2. [DOI] [PubMed] [Google Scholar]
- 153.Spillantini MG, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 154.Winner B, et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl Acad. Sci. U. S. A. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kustrimovic N, et al. Parkinson’s disease patients have a complex phenotypic and functional Th1 bias: cross-sectional studies of CD4+ Th1/Th2/T17 and Treg in drug-naive and drug-treated patients. J. Neuroinflamm. 2018;15:205. doi: 10.1186/s12974-018-1248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen S, et al. Increased abundance of myeloid-derived suppressor cells and Th17 cells in peripheral blood of newly-diagnosed Parkinson’s disease patients. Neurosci. Lett. 2017;648:21–25. doi: 10.1016/j.neulet.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 157.Hernandez DG, Reed X, Singleton AB. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016;139(Suppl 1):59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fujioka S, et al. Occurrence of Crohn’s disease with Parkinson’s disease. Parkinsonism Relat. Disord. 2017;37:116–117. doi: 10.1016/j.parkreldis.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Unger MM, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 161.Keshavarzian A, et al. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015;30:1351–1360. doi: 10.1002/mds.26307. [DOI] [PubMed] [Google Scholar]
- 162.Perez-Pardo P, et al. Role of TLR4 in the gut-brain axis in Parkinson’s disease: a translational study from men to mice. Gut. 2019;68:829–843. doi: 10.1136/gutjnl-2018-316844. [DOI] [PubMed] [Google Scholar]
- 163.Hou YF, et al. Gut microbiota-derived propionate mediates the neuroprotective effect of osteocalcin in a mouse model of Parkinson’s disease. Microbiome. 2021;9:34. doi: 10.1186/s40168-020-00988-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bordoni L, et al. Positive effect of an electrolyzed reduced water on gut permeability, fecal microbiota and liver in an animal model of Parkinson’s disease. PLoS One. 2019;14:e0223238. doi: 10.1371/journal.pone.0223238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li P, et al. Gut microbiota dysbiosis is associated with elevated bile acids in Parkinson’s Disease. Metabolites. 2021;11:29. doi: 10.3390/metabo11010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shao Y, et al. Comprehensive metabolic profiling of Parkinson’s disease by liquid chromatography-mass spectrometry. Mol. Neurodegener. 2021;16:4. doi: 10.1186/s13024-021-00425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cirstea MS, et al. Microbiota composition and metabolism are associated with gut function in Parkinson’s Disease. Mov. Disord. 2020;35:1208–1217. doi: 10.1002/mds.28052. [DOI] [PubMed] [Google Scholar]
- 168.Funayama M, et al. An LRRK2 mutation as a cause for the parkinsonism in the original PARK8 family. Ann. Neurol. 2005;57:918–921. doi: 10.1002/ana.20484. [DOI] [PubMed] [Google Scholar]
- 169.Park J, et al. Parkinson disease-associated LRRK2 G2019S transgene disrupts marrow myelopoiesis and peripheral Th17 response. J. Leukoc. Biol. 2017;102:1093–1102. doi: 10.1189/jlb.1A0417-147RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ekstrand MI, et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl Acad. Sci. U. S. A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu J, et al. Sodium butyrate exerts protective effect against Parkinson’s disease in mice via stimulation of glucagon like peptide-1. J. Neurol. Sci. 2017;381:176–181. doi: 10.1016/j.jns.2017.08.3235. [DOI] [PubMed] [Google Scholar]
- 172.Johnson J, Jr, et al. Valproate and sodium butyrate attenuate manganese-decreased locomotor activity and astrocytic glutamate transporters expression in mice. Neurotoxicology. 2018;64:230–239. doi: 10.1016/j.neuro.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kidd SK, Schneider JS. Protective effects of valproic acid on the nigrostriatal dopamine system in a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. Neuroscience. 2011;194:189–194. doi: 10.1016/j.neuroscience.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Aho VTE, et al. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease. Mol. Neurodegener. 2021;16:6. doi: 10.1186/s13024-021-00427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Qiao CM, et al. Sodium butyrate exacerbates Parkinson’s disease by aggravating neuroinflammation and colonic inflammation in MPTP-Induced Mice Model. Neurochem. Res. 2020;45:2128–2142. doi: 10.1007/s11064-020-03074-3. [DOI] [PubMed] [Google Scholar]
- 176.Park J, Goergen CJ, HogenEsch H, Kim CH. Chronically elevated levels of short-chain fatty acids induce T cell-mediated ureteritis and hydronephrosis. J. Immunol. 2016;196:2388–2400. doi: 10.4049/jimmunol.1502046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Graham SF, et al. Metabolomic profiling of bile acids in an experimental model of prodromal Parkinson’s Disease. Metabolites. 2018;8:71. doi: 10.3390/metabo8040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cuevas, E. et al. Tauroursodeoxycholic acid (TUDCA) is neuroprotective in a chronic mouse model of Parkinson’s disease. Nutr. Neurosci. 1–18 (2020). [DOI] [PubMed]
- 179.Castro-Caldas M, et al. Tauroursodeoxycholic acid prevents MPTP-induced dopaminergic cell death in a mouse model of Parkinson’s disease. Mol. Neurobiol. 2012;46:475–486. doi: 10.1007/s12035-012-8295-4. [DOI] [PubMed] [Google Scholar]
- 180.Duan WM, Rodrigues CM, Zhao LR, Steer CJ, Low WC. Tauroursodeoxycholic acid improves the survival and function of nigral transplants in a rat model of Parkinson’s disease. Cell Transpl. 2002;11:195–205. [PubMed] [Google Scholar]
- 181.Rosa AI, et al. Tauroursodeoxycholic acid improves motor symptoms in a mouse model of Parkinson’s Disease. Mol. Neurobiol. 2018;55:9139–9155. doi: 10.1007/s12035-018-1062-4. [DOI] [PubMed] [Google Scholar]
- 182.Moreira S, et al. Nrf2 activation by tauroursodeoxycholic acid in experimental models of Parkinson’s disease. Exp. Neurol. 2017;295:77–87. doi: 10.1016/j.expneurol.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 183.Mortiboys H, et al. UDCA exerts beneficial effect on mitochondrial dysfunction in LRRK2(G2019S) carriers and in vivo. Neurology. 2015;85:846–852. doi: 10.1212/WNL.0000000000001905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tiwari S, Atluri V, Kaushik A, Yndart A, Nair M. Alzheimer’s disease: pathogenesis, diagnostics, and therapeutics. Int. J. Nanomed. 2019;14:5541–5554. doi: 10.2147/IJN.S200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fujii Y, et al. Fecal metabolite of a gnotobiotic mouse transplanted with gut microbiota from a patient with Alzheimer’s disease. Biosci. Biotechnol. Biochem. 2019;83:2144–2152. doi: 10.1080/09168451.2019.1644149. [DOI] [PubMed] [Google Scholar]
- 186.Nagpal R, et al. Gut mycobiome and its interaction with diet, gut bacteria and alzheimer’s disease markers in subjects with mild cognitive impairment: a pilot study. EBioMedicine. 2020;59:102950. doi: 10.1016/j.ebiom.2020.102950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marizzoni M, et al. Short-chain fatty acids and lipopolysaccharide as mediators between gut dysbiosis and amyloid pathology in Alzheimer’s Disease. J. Alzheimers Dis. 2020;78:683–697. doi: 10.3233/JAD-200306. [DOI] [PubMed] [Google Scholar]
- 188.Ho L, et al. Protective roles of intestinal microbiota derived short chain fatty acids in Alzheimer’s disease-type beta-amyloid neuropathological mechanisms. Expert. Rev. Neurother. 2018;18:83–90. doi: 10.1080/14737175.2018.1400909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu S, Gao J, Zhu M, Liu K, Zhang HL. Gut microbiota and dysbiosis in Alzheimer’s Disease: implications for pathogenesis and treatment. Mol. Neurobiol. 2020;57:5026–5043. doi: 10.1007/s12035-020-02073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Luchsinger JA, Tang M-X, Shea S, Mayeux R. Caloric intake and the risk of Alzheimer Disease. Arch. Neurol. 2002;59:1258–1263. doi: 10.1001/archneur.59.8.1258. [DOI] [PubMed] [Google Scholar]
- 191.Engelhart MJ, et al. Dietary intake of antioxidants and risk of Alzheimer disease. JAMA. 2002;287:3223–3229. doi: 10.1001/jama.287.24.3223. [DOI] [PubMed] [Google Scholar]
- 192.Spychala MS, et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann. Neurol. 2018;84:23–36. doi: 10.1002/ana.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J. Alzheimers Dis. 2011;26:187–197. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- 194.Martins IJ, Fernando WMADB. High fibre diets and Alzheimer’s Disease. Food Nutr. Sci. 2014;05:410–424. [Google Scholar]
- 195.Sun J, et al. Effect of Clostridium butyricum against microglia-mediated neuroinflammation in Alzheimer’s Disease via regulating gut microbiota and metabolites Butyrate. Mol. Nutr. Food Res. 2020;64:e1900636. doi: 10.1002/mnfr.201900636. [DOI] [PubMed] [Google Scholar]
- 196.Da Silva SV, et al. Early synaptic deficits in the APP/PS1 mouse model of Alzheimer’s disease involve neuronal adenosine A 2A receptors. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Spires TL, Hyman BT. Transgenic models of Alzheimer’s disease: learning from animals. NeuroRx. 2005;2:423–437. doi: 10.1602/neurorx.2.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Trinchese F, et al. Progressive age-related development of Alzheimer-like pathology in APP/PS1 mice. Ann. Neurol. 2004;55:801–814. doi: 10.1002/ana.20101. [DOI] [PubMed] [Google Scholar]
- 199.Colombo AV, et al. Microbiota-derived short chain fatty acids modulate microglia and promote Aβ plaque deposition. eLife. 2021;10:e59826. doi: 10.7554/eLife.59826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sun J, et al. Fecal microbiota transplantation alleviated Alzheimer’s disease-like pathogenesis in APP/PS1 transgenic mice. Transl. Psychiatr. 2019;9:189. doi: 10.1038/s41398-019-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Widner B, et al. Tryptophan degradation and immune activation in Alzheimer’s disease. J. Neural Transm. (Vienna) 2000;107:343–353. doi: 10.1007/s007020050029. [DOI] [PubMed] [Google Scholar]
- 202.MahmoudianDehkordi S, et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimers Dement. 2019;15:76–92. doi: 10.1016/j.jalz.2018.07.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Baloni P, et al. Metabolic network analysis reveals altered bile acid synthesis and metabolism in Alzheimer’s Disease. Cell Rep. Med. 2020;1:100138. doi: 10.1016/j.xcrm.2020.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lord C, Elsabbagh M, Baird G, Veenstra-Vanderweele J. Autism spectrum disorder. Lancet. 2018;392:508–520. doi: 10.1016/S0140-6736(18)31129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bölte S, Girdler S, Marschik PB. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell. Mol. Life Sci. 2019;76:1275–1297. doi: 10.1007/s00018-018-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Choi GB, et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J. Autism Dev. Disord. 2014;44:1117–1127. doi: 10.1007/s10803-013-1973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Madra M, Ringel R, Margolis KG. Gastrointestinal issues and autism spectrum disorder. Child Adolesc. Psychiatr. N. Am. 2020;29:501–513. doi: 10.1016/j.chc.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.De Angelis M, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kim S, et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism – comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu S, et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-018-36430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Choi J, et al. Pathophysiological and neurobehavioral characteristics of a propionic acid-mediated autism-like rat model. PLoS One. 2018;13:e0192925. doi: 10.1371/journal.pone.0192925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Abdelli LS, Samsam A, Naser SA. Propionic acid induces gliosis and neuro-inflammation through modulation of PTEN/AKT pathway in Autism Spectrum Disorder. Sci. Rep. 2019;9:8824. doi: 10.1038/s41598-019-45348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kratsman N, Getselter D, Elliott E. Sodium butyrate attenuates social behavior deficits and modifies the transcription of inhibitory/excitatory genes in the frontal cortex of an autism model. Neuropharmacology. 2016;102:136–145. doi: 10.1016/j.neuropharm.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 216.Hammer A, et al. Impact of combined sodium chloride and saturated long-chain fatty acid challenge on the differentiation of T helper cells in neuroinflammation. J. Neuroinflamm. 2017;14:184. doi: 10.1186/s12974-017-0954-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Platten M, et al. Treatment of autoimmune neuroinflammation with a synthetic tryptophan metabolite. Science. 2005;310:850–855. doi: 10.1126/science.1117634. [DOI] [PubMed] [Google Scholar]
- 218.Sun MF, et al. Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson’s disease mice: Gut microbiota, glial reaction and TLR4/TNF-alpha signaling pathway. Brain Behav. Immun. 2018;70:48–60. doi: 10.1016/j.bbi.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 219.Shin C, Lim Y, Lim H, Ahn TB. Plasma short-chain fatty acids in patients With Parkinson’s Disease. Mov. Disord. 2020;35:1021–1027. doi: 10.1002/mds.28016. [DOI] [PubMed] [Google Scholar]
- 220.Chen SJ, et al. The gut metabolite trimethylamine N-oxide is associated with Parkinson’s Disease severity and progression. Mov. Disord. 2020;35:2115–2116. doi: 10.1002/mds.28246. [DOI] [PubMed] [Google Scholar]
- 221.Coretti L, et al. Gut microbiota features in young children with Autism Spectrum Disorders. Front. Microbiol. 2018;9:3146. doi: 10.3389/fmicb.2018.03146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang L, et al. Elevated fecal short chain fatty acid and ammonia concentrations in children with Autism Spectrum Disorder. Dig. Dis. Sci. 2012;57:2096–2102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]